ABSTRACT
Streptococcus pneumoniae isolates of serotype 3 were collected from cases of invasive pneumococcal disease (n = 124) throughout Japan between April 2010 and March 2013. A penicillin-resistant S. pneumoniae (PRSP) isolate from an adult patient, strain KK0981 of serotype 3, was identified among these strains. Whole-genome analysis characterized this PRSP as a recombinant strain derived from PRSP of serotype 23F with the cps locus (20.3 kb) replaced by that of a penicillin-susceptible strain of serotype 3.
KEYWORDS: Streptococcus pneumoniae, capsular switching, gPRSP, serotype 3
TEXT
Streptococcus pneumoniae is the most important pathogen in community-acquired respiratory and invasive infections in all age groups.
In Japan, 7-valent pneumococcal conjugate vaccine (PCV7) was given to children under 5 years old since November 2010 until its replacement by PCV13 in November 2013. A direct vaccine effect on the incidence of invasive pneumococcal disease (IPD) in young children was obtained immediately (1, 2), followed shortly by an indirect effect on the incidence of IPD in adults (3). Similar effects were reported in the United States (4) and the European Union (5). However, serotype 3, included in PCV13, remains the leading cause of IPD in adults (5–7).
Penicillin-resistant S. pneumoniae (PRSP), which mostly represents serotypes 6B, 14, 19F, 23F, and 6A of the vaccine type (VT), has amino acid substitutions near or in conserved amino acid motifs in 3 penicillin-binding proteins (PBPs), PBP1A, PBP2X, and PBP2B, encoded by pbp1a, pbp2x, and pbp2b, respectively (2, 3). These resistances, identified by pbp gene analysis, are expressed as the genotype (g) (8). Clinical isolates of serotype 3 usually are penicillin-susceptible or penicillin-intermediate S. pneumoniae having amino acid substitutions in PBP2X (gPISP [pbp2x]), with susceptibility to penicillin G (PEN) ranging from 0.031 to 0.125 μg/ml (3, 8).
Of pneumococcal strains (n = 1,317) collected throughout Japan from April 2010 to March 2013, 124 strains were identified as serotype 3 of the mucoid type in children (n = 10) and adults (n = 114) (2, 3). All but one of the strains were PEN susceptible, having an MIC of ≤0.125 μg/ml, and belonged to the sequence type 180 (ST180) group. The remaining strain, non-ST180 strain KK0981, was identified as a new gPRSP belonging to ST242, which has not been reported in any other country (3).
In the present study, we used a comparative genomic approach to determine how evolution to gPRSP in that serotype 3 isolate occurred through recombination of the capsular (cps) locus region flanked by the pbp2x and pbp1a genes.
Table 1 shows characteristics, sequence types (STs), genotypic resistance, and β-lactam susceptibilities determined by bacteriologic methods in the 3 strains analyzed in this study; KK0981 was a new gPRSP (serotype 3, ST242), KK0381 a gPISP (serotype 3, ST180), and KK1157 a gPRSP (serotype 23F, ST242) derived from the Taiwan23F-15 clone. KK0381 was selected as a candidate donor of the cps locus among serotype 3 strains mostly identified as ST180 (see Fig. S1 in the supplemental material). KK1157 was selected as a candidate recipient from among serotype 23F ST242 strains identified as gPRSP (n = 30).
TABLE 1.
Characteristics, sequence type, genotypic penicillin and macrolide resistances, and MICs of pneumococcal strains analyzed
| Strain | Serotype | ST | Penicillin resistance genotypea | Macrolide resistance gene(s) | MICs (μg/ml) forb: |
Disease (source) | Year of isolation | City isolated | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | AMP | CTX | MEM | ||||||||
| KK0381 (candidate donor) | 3 | 180 | gPISP (pbp2x) | erm(B) | 0.063 | 0.125 | 0.25 | 0.063 | Pneumonia (blood) | 2010 | Niigata |
| KK0981 (putative recombinant) | 3 | 242 | gPRSP (pbp1a + pbp2x + pbp2b) | mef(A) + erm(B) | 1 | 2 | 1 | 1 | Bacteremia (blood) | 2011 | Amagasaki |
| KK1157 (candidate recipient) | 23F | 242 | gPRSP (pbp1a + pbp2x + pbp2b) | mef(A) + erm(B) | 2 | 2 | 1 | 1 | Septic arthritis (blood) | 2011 | Sapporo |
pbp gene alterations identified by real-time PCR are given in parentheses.
PEN, penicillin G; AMP, ampicillin; CTX, cefotaxime; MEM, meropenem.
Whole-genome sequencing for these strains was performed by PacBio single-molecule real-time (SMRT) technology. The resulting sequence data were analyzed using the MiGAP pipeline (see http://www.migap.org/index.php/en) (9). The cps locus and its neighbors were annotated manually with the OXC141 and ATCC 700669 reference strains with GenomeMatcher (10). Gene names for KK0981 were correlated with those for OXC141 (11). Among the genomes of these strains, homology and single-nucleotide variant (SNV) comparisons were performed using BLAST Ring Image Generator (BRIG) software (12) and the MUMmer program, respectively.
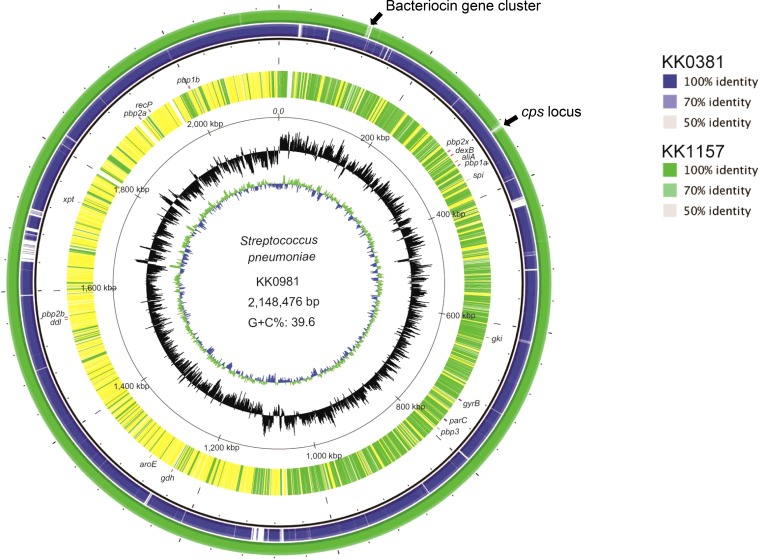
The KK0981 (gPRSP/serotype 3) genome was found to have uniformly greater similarity and closer identity of SNVs with the KK1157 (gPRSP/serotype 23F) genome than with that the KK0381 (gPISP/serotype 3) genome (Fig. 1). Amino acid sequences of pbp1a, pbp2x, and pbp2b genes of KK0981 were identical with those of KK1157, which showed the same ST242 but a different serotype (see Fig. S2 in the supplemental material). These data indicated that the KK0981 and KK1157 strains share the same genetic background. Specifically, these original data suggest that recombination of the cps locus region flanked by pbp2x and pbp1a genes occurred between the genomes of KK0381 and KK1157.
FIG 1.
Circular representation of the genome of S. pneumoniae strain KK0981 (gPRSP/serotype 3, ST242) compared with the genomes of the KK0381 (gPISP/serotype 3, ST180) and KK1157 (gPRSP/serotype 23F, ST242) strains. KK0981 presented a chromosome 2,148,476 bp long, which contained 2164 coding sequences (CDSs), 12 rRNAs, and 58 tRNAs, showing an overall 39.6% GC content (accession number AP017971). Three contiguous sequences were obtained for KK0381, with a total length of 2,158,744 bp (i, 2,074,518 bp; ii, 43,298 bp; iii; 40,928 bp), and for KK1157, with a total length of 2,267,745 bp (i; 2,200,203 bp, ii, 36,583 bp, iii, 30,959 bp). The concentric circles represent the following information (from the outside in): circle 1, BLAST comparisons of the KK1157 draft genome against KK0981 (green); circle 2, BLAST comparisons of the KK0381 against KK0981 (blue); circle 3, annotated CDSs encoded on forward (green) and reverse (yellow) chromosomal strands, respectively; circle 4, distance from the putative origin of replication; circle 5, GC skew (black); circle 6, GC content (green/blue). Two arrows indicate the region with no homology between KK0981 and KK1157. The circular map was constructed using BRIG software and ArcWithColor software (12) with modification.
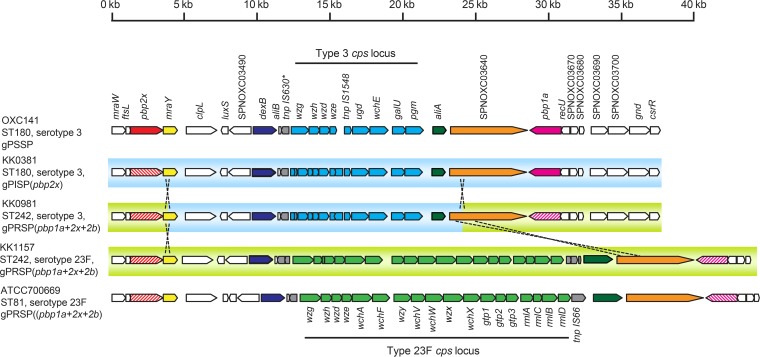
To identify the recombinant position in the KK0981 (gPRSP/serotype 3) genome, nucleotide sequence alignments from pbp2x, the cps locus, and pbp1a (total length, 29,629 bp) were compared with those of KK1157 (gPRSP/serotype 23F) and KK0381 (gPISP/serotype 3) (Fig. 2). The 20,291-bp length from bp 312701 located in the mraY gene to bp 332991 located in the SPNOXC03640 gene (encoding endo-alpha-N-acetylgalactosaminidase) of KK0981 was identical to that of KK0381 (see Fig. S3 and S4 in the supplemental material). Sequences of the cps loci and their flanking regions in the 3 strains were aligned with those of the reference strains OXC141 (NC_017592.1; PEN susceptible, serotype 3, ST180) and ATCC 700669 (NC_011900.1; PEN resistant, serotype 23F, ST81). No traces of phage or insertion sequences (IS) were clearly identified in the near vicinity of the recombination position.
FIG 2.
Comparison of the cps locus and its flanking region. Genomic organization of cps loci and their flanking regions in KK0381, KK0981, and KK1157 were aligned. OXC141 (NC_017592.1) and ATCC 700669 (NC_011900.1) were reference strains for the locus tag or gene names of type 3 and type 23F cps loci, respectively. Arrows represent CDSs and their functions in color: pbp2x (red), mraY (yellow), dexB (blue), transposase (gray), aliA (green), SPNOXC03640 (orange), pbp1a (pink), serotype 3 cps genes (cyan), serotype 23F cps genes (light green). Hatched arrows represent genotypic (g) resistant pbp genes. KK0981 of gPRSP of serotype 3 and ST242 was identified definitely as a transformant caused by recombination in regions from mraY to the SPOXC03640 gene of the serotype 3 pneumococcal strain of ST180 as a donor type and KK1157 of gPRSP of serotype 23F and ST242 derived from the Taiwan23F-15 clone as a recipient type. The cyan or light green bar behind the CDSs represents the genetic background of KK0381 or KK1157, respectively, based upon SNV analysis. The alignment was generated with GenomeMatcher software (10); tnpIS630 is a pseudogene.
From the results described above, KK0981 (gPRSP/serotype 3 and ST242) was concluded to occur by homologous recombination of the 20.3-kb region from the mraY gene to the SPOXC03640 gene, with a pneumococcal strain of serotype 3 and ST180 as a donor type and a PRSP strain of serotype 23F and ST242 derived from a Taiwan23F-15 clone as a recipient type.
Details of polysaccharide structures and capsule biosynthetic genes have been elucidated (13). Homologous recombination is known to occur in the cps locus and its flanking regions, changing serotypes from 4 to 19A (14), from 6A to 6C, or from 7B to 9N (15). Characteristically, the pbp2x and pbp1a genes involved in β-lactam resistance adjoin the 2 ends of the cps locus. Recombination of a large DNA fragment including the cps locus and pbp genes with lengths from 19.0 kb to over 58.2 kb have been noted in 11 events occurring in 36 independent cases of capsular switching (15). Previously, a pneumococcal strain of serotype 3 found to be PEN nonsusceptible (MIC ≥ 1 μg/ml), which belonged to the PMEN clone Spain23F-1 (CC81 and ST81), was isolated from a patient with IPD in the United Kingdom (16). While a recombination event was reported to occur at the cps locus region involving pbpX (pbp2x), variations in pbp1a and pbp2b genes affecting β-lactam resistance are unexplained.
In our study, KK1157 (gPRSP/serotype 23F, ST242), the recipient type, belonged to ST242 of the Taiwan23F-15 clone registered in the Pneumococcal Molecular Epidemiology Network (PMEN). Most ST242 pneumococcal strains registered in multilocus sequence type (MLST) databases (see http://pubmlst.org/spneumoniae/) were from Asia; approximately half of them were not susceptible to PEN (MIC ≥ 1 μg/ml). The present study is the first report of a PRSP derived from a Taiwan23F-15 clone prevalent in Asia that acquired a cps locus region encoding a type 3 capsule by homologous recombination, resulting in occurrence of PRSP of serotype 3. Additionally, this recombinant event most likely occurred in recent years because the SNVs in the recombination region of KK0981 were completely consistent with those of KK0381 of serotype 3.
Evolution to penicillin resistance by capsular switching will continue to occur in S. pneumoniae strains not included in the present vaccine type because of selection pressures involving both the spread of pneumococcal vaccination and excessive use of chemotherapeutic agents. Ongoing high-quality molecular epidemiologic surveillance is essential.
Accession number(s).
The whole-genome sequences of the 3 strains have been registered in the DNA Data Bank of Japan (DDBJ) under the accession numbers AP017971 (KK0981), AP018043 (KK0381), and AP018044 (KK1157).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by two grants, one from JSPS KAKENHI (26870568) and the other from the Japanese Ministry of Health, Labor and Welfare (Research on Emerging and Re-emerging Infectious Diseases H22-013) (to K.U.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00478-17.
REFERENCES
- 1.Chiba N, Morozumi M, Shouji M, Wajima T, Iwata S, Sunakawa K, Ubukata K; Invasive Pneumococcal Diseases Surveillance Study Group. 2013. Invasive Pneumococcal Diseases Surveillance Study Group. Rapid decrease of 7-valent conjugate vaccine coverage for invasive pneumococcal diseases in pediatric patients in Japan. Microb Drug Resist 19:308–315. doi: 10.1089/mdr.2012.0180. [DOI] [PubMed] [Google Scholar]
- 2.Chiba N, Morozumi M, Shouji M, Wajima T, Iwata S, Ubukata K; Invasive Pneumococcal Diseases Surveillance Study Group. 2014. Changes in capsule and drug resistance of peumococci after introduction of PCV7, Japan, 2010-2013. Emerg Infect Dis 20:1132–1139. doi: 10.3201/eid2007.131485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ubukata K, Chiba N, Hanada S, Morozumi M, Wajima T, Shouji M, Iwata S; Invasive Pneumococcal Diseases Surveillance Study Group. 2015. Serotype changes and drug resistance in invasive pneumococcal diseases in adults after vaccinations in children, Japan, 2010-2013. Emerg Infect Dis 21:1956–1965. doi: 10.3201/eid2111.142029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis 201:32–41. doi: 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 5.Harboe ZB, Dalby T, Weinberger DM, Benfield T, Mølbak K, Slotved HC, Suppli CH, Konradsen HB, Valentiner-Branth P. 2014. Impact of 13-valent pneumococcal conjugate vaccination in invasive pneumococcal disease incidence and mortality. Clin Infect Dis 59:1066–1073. doi: 10.1093/cid/ciu524. [DOI] [PubMed] [Google Scholar]
- 6.Horácio AN, Silva-Costa C, Lopes JP, Ramirez M, Melo-Cristino J; Portuguese Group for the Study of Streptococcal Infections. 2016. Serotype 3 remains the leading cause of invasive pneumococcal disease in adults in Portugal (2012-2014) despite continued reductions in other 13-valent conjugate vaccine serotypes. Front Microbiol 7:1616. doi: 10.3389/fmicb.2016.01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanada S, Iwata S, Kishi K, Morozumi M, Chiba N, Wajima T, Takata M, Ubukata K; Invasive Pneumococcal Diseases Surveillance Study Group. 2016. Host factors and biomarkers associated with poor outcomes in adults with invasive pneumococcal disease. PLoS One 11:e0147877. doi: 10.1371/journal.pone.0147877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiba N, Morozumi M, Ubukata K. 2012. Application of the real-time PCR method for genotypic identification of β-lactam resistance in isolates from invasive pneumococcal diseases. Microb Drug Resist 18:149–156. doi: 10.1089/mdr.2011.0102. [DOI] [PubMed] [Google Scholar]
- 9.Noguchi H, Taniguchi T, Itoh T. 2008. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res 15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. 2008. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croucher NJ, Mitchell AM, Gould KA, Inverarity D, Barquist L, Feltwell T, Fookes MC, Harris SR, Dordel J, Salter SJ, Browall S, Zemlickova H, Parkhill J, Normark S, Henriques-Normark B, Hinds J, Mitchell TJ, Bentley SD. 2013. Dominant role of nucleotide substitution in the diversification of serotype 3 pneumococci over decades and during a single infection. PLoS Genet 9:e1003868. doi: 10.1371/journal.pgen.1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golubchik T, Brueggemann AB, Street T, Gertz RE Jr, Spencer CCA, Ho T, Giannoulatou E, Link-Gelles R, Harding RM, Beall B, Peto TE, Moore MR, Donnelly P, Crook DW, Bowden R. 2012. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multifragment recombinant event. Nat Genet 44:352–355. doi: 10.1038/ng.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyres KL, Lambertsen LM, Croucher NJ, McGee L, von Gottberg A, Liñares J, Jacobs MR, Kristinsson KG, Beall BW, Klugman KP, Parkhill J, Hakenbeck R, Bentley SD, Brueggemann AB. 2013. Pneumococcal capsular switching: a historical perspective. J Infect Dis 207:439–449. doi: 10.1093/infdis/jis703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.