ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) is a multidrug-resistant pathogen that poses a significant risk to global health today. We have developed a promising new FtsZ-targeting agent (TXA707) with potent activity against MRSA isolates resistant to current standard-of-care antibiotics. We present here results that demonstrate differing extents of synergy between TXA707 and a broad range of β-lactam antibiotics (including six cephalosporins, two penicillins, and two carbapenems) against MRSA. To explore whether there is a correlation between the extent of synergy and the preferential antibacterial target of each β-lactam, we determined the binding affinities of the β-lactam antibiotics for each of the four native penicillin-binding proteins (PBPs) of S. aureus using a fluorescence anisotropy competition assay. A comparison of the resulting PBP binding affinities with our corresponding synergy results reveals that β-lactams with a high affinity for PBP2 afford the greatest degree of synergy with TXA707 against MRSA. In addition, we present fluorescence and electron microscopy studies that suggest a potential mechanism underlying the synergy between TXA707 and the β-lactam antibiotics. In this connection, our microscopy results show a disruption of septum formation in TXA707-treated MRSA cells, with a concomitant mislocalization of the PBPs from midcell to nonproductive peripheral sites. Viewed as a whole, our results indicate that PBP2-targeting β-lactam antibiotics are optimal synergistic partners with FtsZ-targeting agents for use in combination therapy of MRSA infections.
KEYWORDS: combination therapy, FtsZ-targeting agents, MRSA, PBP binding affinity, PBP mislocalization, PBP2-targeting β-lactam antibiotics, synergy, TXA707
INTRODUCTION
The emergence of multidrug-resistant (MDR) bacterial pathogens has become a global threat to public health (1, 2). Methicillin-resistant Staphylococcus aureus (MRSA) is an MDR bacterial pathogen associated with a high mortality rate in both hospital and community settings (3–5). In particular, MRSA is resistant to most β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems (6). The antibacterial targets of β-lactam antibiotics are the penicillin-binding proteins (PBPs), with S. aureus expressing four native PBPs (PBP1, PBP2, PBP3, and PBP4) (7, 8). In addition to these four PBPs, MRSA also expresses a fifth PBP (PBP2A) which, unlike the native S. aureus PBPs, is a poor target for the β-lactams (9, 10).
The PBPs are a family of proteins that are involved in the final steps of bacterial cell wall assembly, particularly at the septa of dividing cells (11, 12). In S. aureus, all four native PBPs are known to localize to the septa of dividing cells, with each possessing a catalytic transpeptidase (TPase) activity that is important for cross-linking peptidoglycan strands in the newly forming cell wall (13–17). PBP2 is unique among the PBPs in that it has two distinct catalytic domains, making it the only bifunctional PBP in S. aureus (7, 18, 19). One catalytic domain of PBP2 is responsible for its TPase activity, while the other catalytic domain has transglycosylation (TGase) activity, which catalyzes the cross-linking of glycan moieties to form peptidoglycan strands (19). Pinho and coworkers have demonstrated that the ability of MRSA to survive in the presence of β-lactam antibiotics requires cooperative functioning between PBP2 and PBP2A (18, 20, 21).
The function of the PBPs in S. aureus are controlled both spatially and temporally throughout the cell cycle (22). A key protein involved in this spatial and temporal regulation is FtsZ, which ensures the proper localization of the PBPs to the septum during division (23–26). We have previously shown that the FtsZ-targeting agent TXA707 (see structure in Fig. S1 in the supplemental material) acts synergistically with the third-generation cephalosporin cefdinir against MRSA (27). We sought to determine the extent to which other β-lactam antibiotics synergize with TXA707 against MRSA and how the synergy is impacted by the PBP binding affinity and selectivity of the β-lactams. Although the PBPs have long been established as the targets of β-lactam antibiotics, the binding affinities and selectivities of these drugs for the individual PBPs of S. aureus have not been well characterized. Here, we determined the binding affinities of 10 representative β-lactam antibiotics (including six cephalosporins, two penicillins, and two carbapenems) for the four native PBPs of S. aureus. We then determined how the PBP binding selectivities of the β-lactams correlate with the extent to which the drugs synergize with TXA707 against MRSA. Our results indicate that β-lactam antibiotics with a high affinity for PBP2 afford the greatest degree of synergy with TXA707.
RESULTS
Intrinsic activities of TXA707 and the β-lactam antibiotics against MRSA COL.
We investigated 10 different β-lactam antibiotics, including 6 cephalosporins (cefradine, cephalexin, cefdinir, cefotaxime, ceftriaxone, and cefepime), 2 penicillins (oxacillin and ticarcillin), and 2 carbapenems (ertapenem and imipenem), for synergy with TXA707 against MRSA COL. As an initial step in these investigations, we determined the intrinsic activities of the 10 β-lactams and TXA707 against this bacterial strain, with the resulting MIC values being listed in Table 1. TXA707 exhibits potent activity against MRSA COL (MIC = 2 μg/ml). In contrast, all the β-lactams studied are associated with poor activity, with MICs ranging from 32 to 2,048 μg/ml.
TABLE 1.
Intrinsic activities of TXA707 and the β-lactam antibiotics against MRSA COL
| Agent | MIC (μg/ml) |
|---|---|
| TXA707 | 2 |
| Cefradine | 128 |
| Cephalexin | 128 |
| Cefdinir | 256 |
| Cefotaxime | 1024 |
| Ceftriaxone | 2048 |
| Cefepime | 256 |
| Oxacillin | 512 |
| Ticarcillin | 512 |
| Ertapenem | 256 |
| Imipenem | 32 |
Binding affinities of the β-lactam antibiotics for the PBPs of S. aureus.
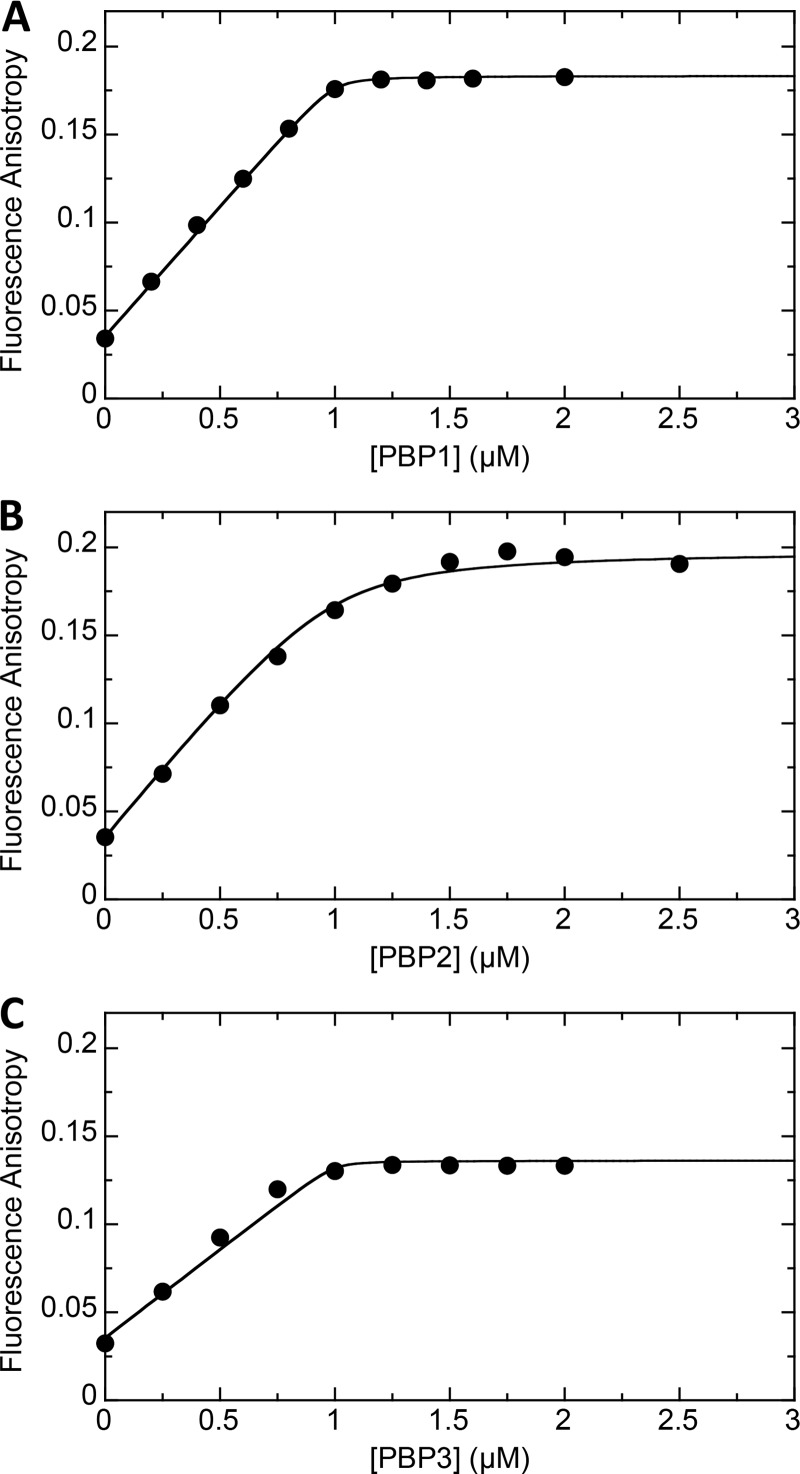
We sought to determine the binding affinities of the 10 β-lactam antibiotics for PBP1, PBP2, PBP3, and PBP4 of S. aureus using a competition fluorescence anisotropy assay and Bocillin (Thermo Fisher Scientific, Waltham, MA) as the fluorescent ligand. Before conducting these competition experiments, we first characterized the binding of Bocillin itself to each of the PBPs. To this end, we monitored the impact of increasing concentrations of PBP1, PBP2, or PBP3 on the fluorescence anisotropy of 1 μM Bocillin, with the resulting anisotropy titration curves being shown in Fig. 1. Note that the addition of each protein increases the fluorescence anisotropy of Bocillin, indicative of ligand binding to each host protein. Furthermore, each anisotropy titration curve is associated with an inflection point at approximately 1 μM protein, consistent with Bocillin binding to each target protein with a 1:1 stoichiometry. The anisotropy titration results also reveal that 1 μM Bocillin is fully saturated by a 2 μM concentration of each PBP. These results suggest that Bocillin targets S. aureus PBP1, PBP2, and PBP3 with a similar affinity.
FIG 1.
Fluorescence anisotropy of 1 μM Bocillin as a function of increasing concentrations of PBP1 (A), PBP2 (B), or PBP3 (C). All experiments were conducted at 25°C in solution containing 100 mM Tris-HCl (pH 7.4), 250 mM NaCl, and 5% (vol/vol) glycerol.
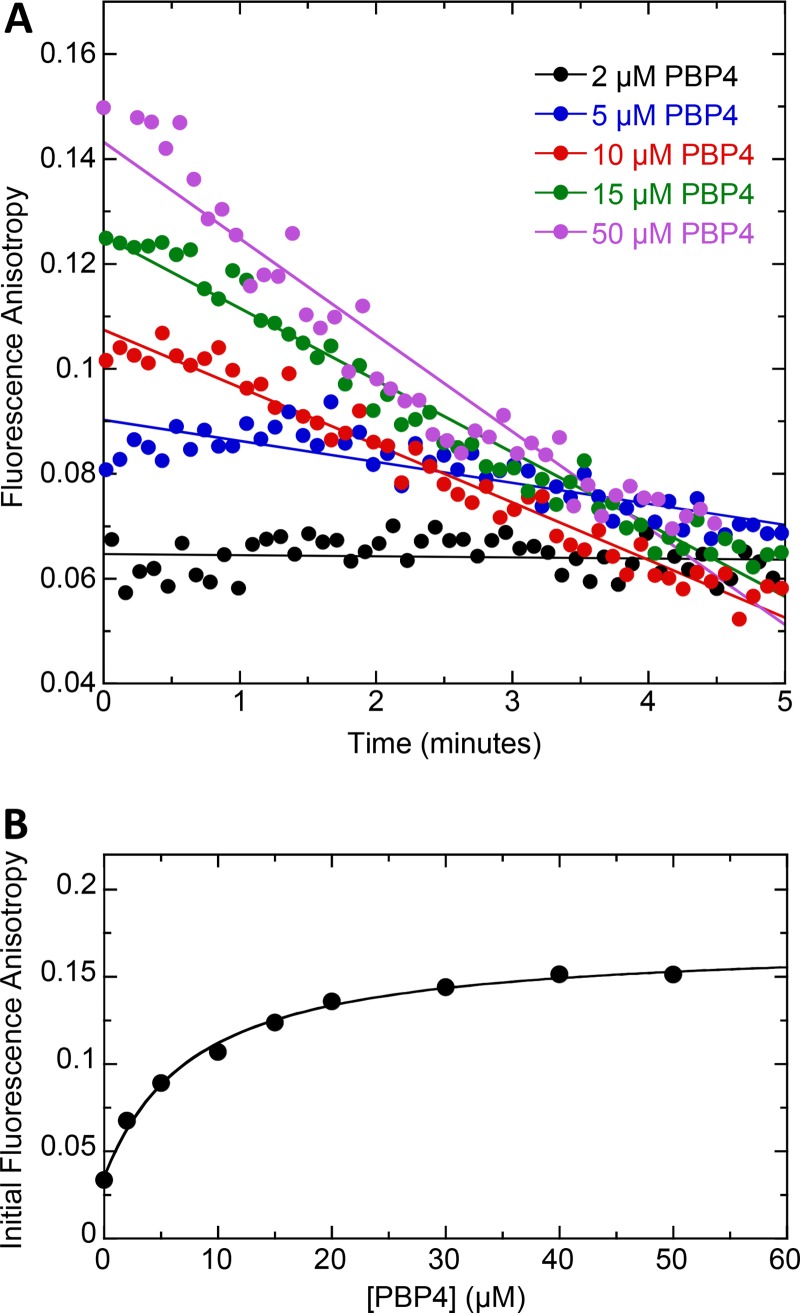
S. aureus PBP4 has been previously reported to exhibit β-lactamase activity (28). Our fluorescence anisotropy studies with PBP4 and Bocillin are consistent with this observation. Specifically, we find that the fluorescence anisotropy of Bocillin decreases as a function of time in the presence of PBP4 (Fig. 2A). This observation suggests that Bocillin is being hydrolyzed by PBP4, with the hydrolysis product being subsequently released from the protein binding pocket. We therefore monitored the time-dependent fluorescence anisotropy of 1 μM Bocillin as a function of PBP4 concentration, with representative time-dependent profiles at PBP4 concentrations of 2, 5, 10, 15, and 50 μM being shown in Fig. 2A. A plot of initial fluorescence anisotropy versus PBP4 concentration reveals that 50 μM PBP4 is required to fully complex with 1 μM Bocillin (Fig. 2B), a protein concentration 25-fold higher than that indicated above for the full saturation of 1 μM Bocillin by PBP1, PBP2, or PBP3. These results suggest that Bocillin binds PBP4 with a lower affinity than the other three PBPs.
FIG 2.
(A) Time dependence of the fluorescence anisotropy of 1 μM Bocillin in the presence of the indicated concentrations of PBP4. (B) Initial fluorescence anisotropy of 1 μM Bocillin as a function of increasing concentrations of PBP4. All experiments were conducted at 25°C in solution containing 20 mM Tris-HCl (pH 7.4), 500 mM NaCl, and 5% (vol/vol) glycerol.
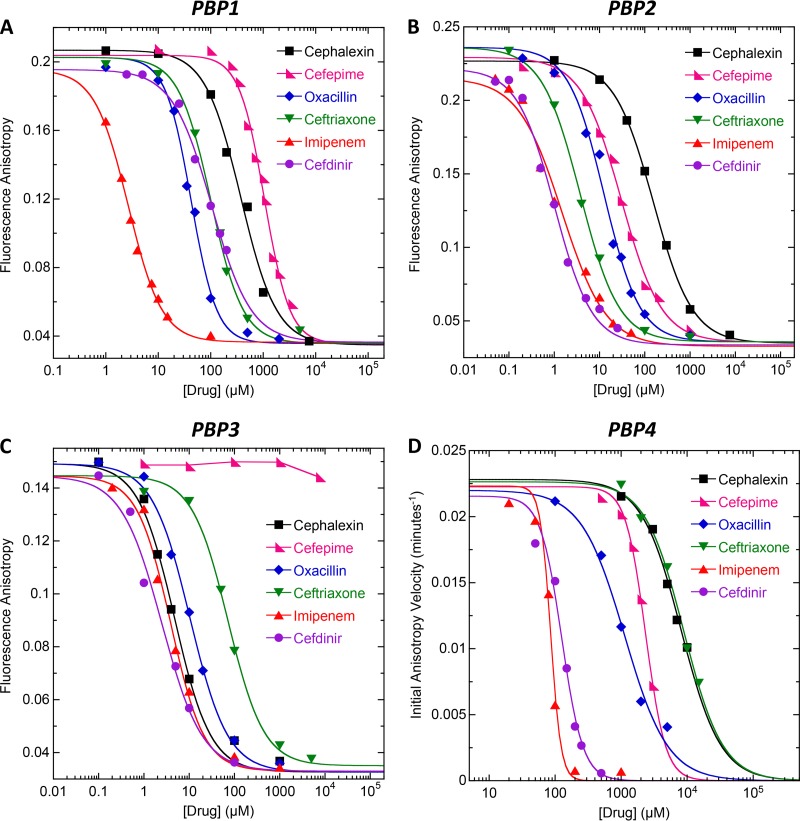
Armed with the PBP concentrations required to fully complex 1 μM Bocillin, we then proceeded to investigate the binding of the 10 β-lactam antibiotics noted above to PBP1, PBP2, PBP3, and PBP4 by monitoring the fluorescence anisotropy of 1 μM Bocillin in the presence of a saturative PBP concentration and increasing concentrations of β-lactam. In this competition anisotropy assay, β-lactam binding to PBP1, PBP2, or PBP3 is correlated with a proportional decrease in the observed anisotropy of Bocillin. With regard to PBP4, β-lactam binding is correlated with a proportional decrease in the initial velocity of Bocillin hydrolysis, as determined from time-dependent anisotropy profiles like those shown in Fig. 2A. Representative competition anisotropy curves for each of the four native PBPs are shown in Fig. 3. These representative anisotropy curves reflect the competition results for 6 of the 10 β-lactams examined, with the corresponding competition results for the other 4 β-lactams being shown in Fig. S2 in the supplemental material.
FIG 3.
(A to C) Fluorescence anisotropy of 1 μM Bocillin in the presence of the indicated β-lactam antibiotic and 2 μM of either PBP1 (A), PBP2 (B), or PBP3 (C). (D) Initial anisotropy velocity of 1 μM Bocillin in the presence of the indicated β-lactam and 50 μM PBP4. The solid lines reflect the nonlinear least-squares fits of the data with equation 1, with the exception of the curve for cefepime and PBP3, which could not be fit due to the weak binding of the drug. Experimental conditions were as described in the legends to Fig. 1 (for PBP1, PBP2, and PBP3) and Fig. 2 (for PBP4).
The binding of β-lactams to the PBPs is associated with an irreversible (covalent) acylation interaction that precludes determination of binding affinity in the form of an equilibrium Kd value. Instead, we assessed binding affinity in the form of an inhibitory concentration reflecting the concentration of β-lactam required to reduce the observed anisotropy of Bocillin by 50% (IC50). To this end, we analyzed the competition anisotropy curves with the following relationship:
| (1) |
In this relationship, C is the concentration of β-lactam, robs is the observed fluorescence anisotropy of Bocillin, nH is the Hill slope, and rf and rb are the fluorescence anisotropies of free and PBP-bound Bocillin, respectively.
The IC50 values resulting from our analyses described above are summarized in Table 2, with lower IC50s being indicative of higher PBP binding affinities. Inspection of these data provides information with regard to the PBP binding selectivities of the β-lactams, with a difference in IC50 of ≥3-fold being viewed as reflecting selective binding. In this connection, cefradine and cephalexin are selective for PBP3, whereas ceftriaxone, cefotaxime, ertapenem, and cefepime are selective for PBP2. Cefdinir, oxacillin, and ticarcillin selectively target both PBP2 and PBP3, while imipenem selectively targets PBP1, PBP2, and PBP3. None of the β-lactams examined target PBP4 with a high degree of affinity or selectivity.
TABLE 2.
IC50 values for the binding of the β-lactam antibiotics to the four native PBPs of S. aureus
| β-Lactam | IC50 (μM)a |
|||
|---|---|---|---|---|
| PBP1 | PBP2 | PBP3 | PBP4 | |
| Cefradine | 516 ± 69 | 180 ± 7 | 3.0 ± 0.2 | 8,250 ± 280 |
| Cephalexin | 374 ± 28 | 154 ± 4 | 4.7 ± 0.4 | 8,108 ± 304 |
| Cefepime | 1,059 ± 38 | 30.3 ± 1.5 | >7,500 | 2,346 ± 39 |
| Ticarcillin | 147 ± 15 | 22.4 ± 1.5 | 9.2 ± 0.3 | 392 ± 26 |
| Oxacillin | 42.2 ± 1.7 | 12.6 ± 0.9 | 10.4 ± 0.6 | 1,128 ± 90 |
| Ertapenem | 89.7 ± 1.8 | 7.8 ± 0.5 | 82.8 ± 6.6 | 233 ± 7 |
| Cefotaxime | 88 ± 3.9 | 4.2 ± 0.1 | 531 ± 50 | 2,567 ± 102 |
| Ceftriaxone | 100 ± 6 | 3.8 ± 0.1 | 71.9 ± 5.3 | 8,906 ± 321 |
| Imipenem | 2.7 ± 0.1 | 1.3 ± 0.1 | 3.9 ± 0.3 | 85.4 ± 2.4 |
| Cefdinir | 104 ± 4 | 1.0 ± 0.1 | 2.5 ± 0.3 | 126 ± 6 |
IC50 reflects the concentration of β-lactam antibiotic that reduces the anisotropy (in the case of PBP1, PBP2, and PBP3) or initial anisotropy velocity (in the case of PBP4) of 1 μM Bocillin by 50%. The indicated uncertainties reflect the standard deviation of the experimental anisotropy data points in Figures 3 and S3 from the corresponding fitted curves.
Synergistic activities of the β-lactam antibiotics with TXA707 against MRSA COL.
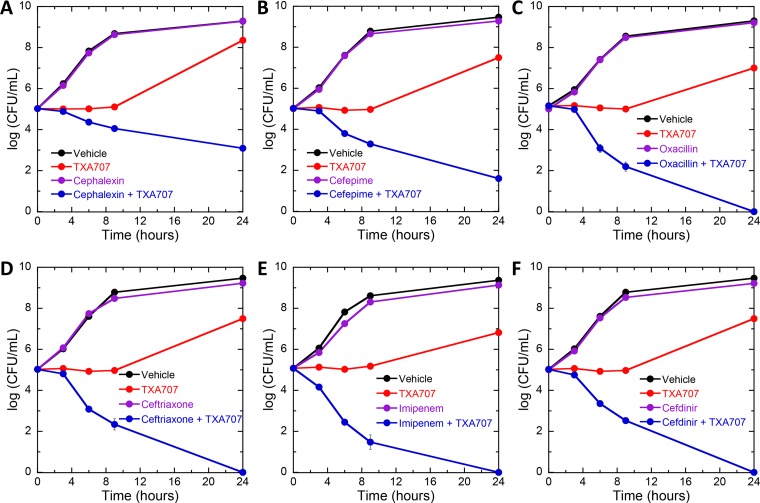
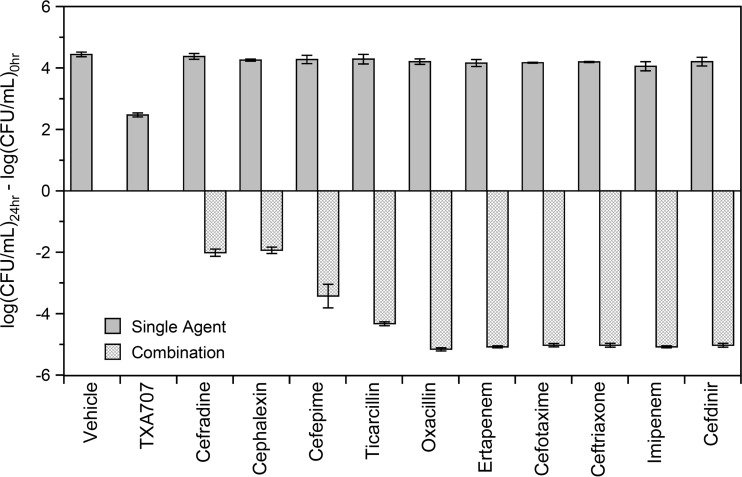
We used a time-kill approach to assess the relative potential of the 10 β-lactams to act synergistically with TXA707 against MRSA COL. To this end, we monitored bactericidal activity in the presence of a sub-MIC concentration of TXA707 alone (at 0.5× MIC), β-lactam alone (at 0.008× MIC), or a combination of both. Figure 4 shows the resulting kill curves for the same six β-lactam antibiotics represented in the competition anisotropy curves depicted in Fig. 3, with the corresponding kill curves for the other four β-lactams examined being shown in Fig. S3 in the supplemental material. No killing is evident in the presence of any of the β-lactams alone, with growth comparable to that associated with vehicle being observed instead. Similarly, growth is also observed in the presence of TXA707 alone, albeit less than that associated with vehicle or β-lactam alone. In striking contrast to TXA707 or β-lactam alone, the combination of TXA707 with each of the β-lactams is bactericidal. Thus, all 10 β-lactams exhibit bactericidal synergy with TXA707. However, the magnitude of kill, and therefore synergy, varies depending on the β-lactam. Figure 5 highlights this variability by plotting the calculated difference between the log(CFU/ml) at 24 relative to 0 h [log(CFU/ml)24 − log(CFU/ml)0] for each agent alone and in combination. Inspection of these data reveals that cefdinir, imipenem, ceftriaxone, cefotaxime, ertapenem, ticarcillin, and oxacillin result in >4 logs of kill when combined with TXA707. The combination of cefepime and TXA707 results in a less robust degree of kill (∼3 logs), with the combination of cefradine or cephalexin and TXA707 yielding the lowest degree of kill (∼2 logs).
FIG 4.
(A to F) Time-kill curves for MRSA COL showing synergy between TXA707 and cephalexin (A), cefepime (B), oxacillin (C), ceftriaxone (D), imipenem (E), or cefdinir (F). Bacteria were treated with DMSO vehicle (black), β-lactam alone at 0.008× MIC (violet), TXA707 alone at 0.5× MIC (red), or a combination of β-lactam at 0.008× MIC and TXA707 at 0.5× MIC (blue).
FIG 5.
Change in the log(CFU/ml) at 24 h {log(CFU/ml)24 − log(CFU/ml)0} of MRSA COL treated with either a single agent (DMSO vehicle, the indicated β-lactam at 0.008× MIC, or TXA707 at 0.5× MIC) or with a combination of β-lactam at 0.008× MIC and TXA707 at 0.5× MIC.
Impact of TXA707 alone or in combination with select β-lactam antibiotics on septum formation in MRSA COL.
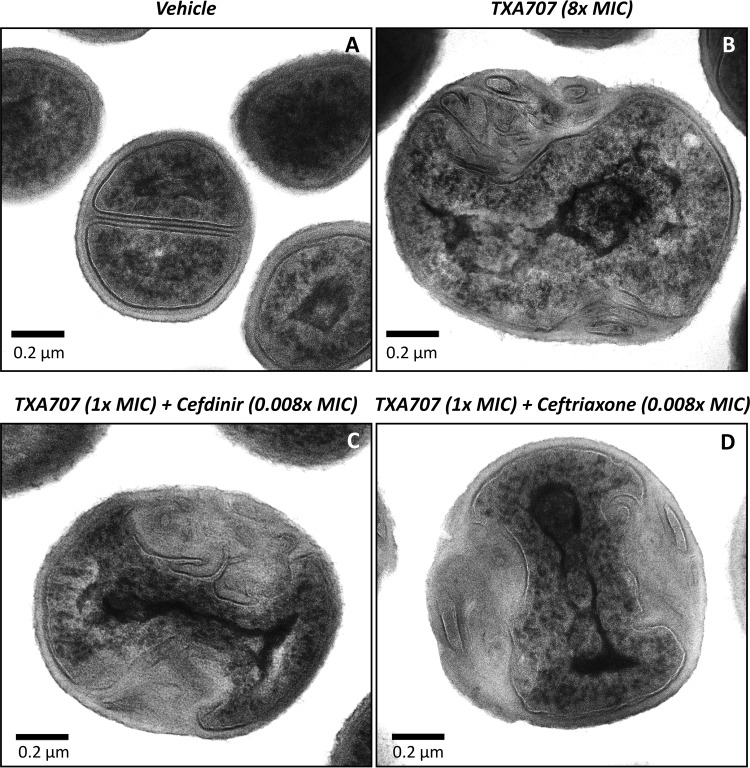
We used transmission electron microscopy (TEM) to compare the morphology of MRSA COL cells treated with vehicle relative to those treated with TXA707 at 8× MIC for 3 h. As expected, vehicle-treated cells are able to form septa at midcell and undergo division (Fig. 6A). In contrast, TXA707-treated cells become enlarged, increasing in average diameter from 0.8 to 1.6 μm. In addition, no midcell septa are evident in TXA707-treated cells, with pronounced invaginations of the cell wall being induced at distinct sites on the cell periphery (Fig. 6B). MRSA COL cells treated for 3 h with a combination of TXA707 at only 1× MIC (an 8-fold lower concentration than that used for treatment with the compound alone) and either cefdinir or ceftriaxone at 0.008× MIC exhibit a similar morphological behavior, with the induced cell wall invaginations being even larger and more pronounced (Fig. 6C and D).
FIG 6.
Impact of TXA707 alone or in combination with either cefdinir or ceftriaxone on septal formation in MRSA COL. (A to D) TEM micrographs of MRSA COL cells treated for 3 h with either vehicle (A), TXA707 alone at 8× MIC (B), TXA707 at 1× MIC in combination with cefdinir at 0.008× MIC (C), or TXA707 at 1× MIC in combination with ceftriaxone at 0.008× MIC (D).
Impact of TXA707 on the septal localization of the PBPs in MRSA COL.
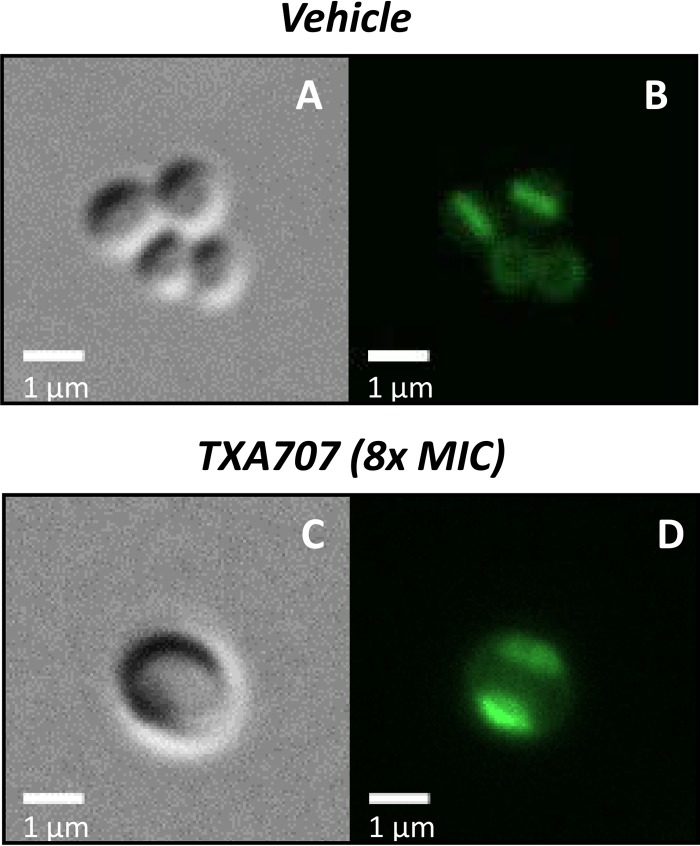
We next used fluorescence microscopy to monitor the impact of TXA707 treatment (at 8× MIC for 3 h) on localization of the PBPs in MRSA COL. To this end, we labeled MRSA COL cells with 2 μM Bocillin and compared the fluorescence pattern associated with vehicle- versus TXA707-treated cells. Recall that at 1 μM, Bocillin binds stoichiometrically to PBP1, PBP2, and PBP3, with its affinity for these PBPs being significantly higher than its affinity for PBP4. Thus, it is likely that Bocillin is labeling PBP1, PBP2, and PBP3 in the MRSA cells. In cells treated with vehicle alone, the majority of the Bocillin fluorescence is localized to the septum that is able to form at midcell (Fig. 7A and B). This observation is consistent with the localization of PBP1, PBP2, and PBP3 to the septa of dividing cells. In contrast, in TXA707-treated cells, no septal Bocillin fluorescence is evident at midcell. Instead, the Bocillin fluorescence is localized to distinct regions of the cell periphery (Fig. 7C and D), a behavior suggesting that the TXA707-induced invaginations of the cell wall observed in the TEM characterizations may reflect mislocalized PBPs and possibly other septal components (compare Fig. 6B and 7D). MRSA COL cells treated with a combination of TXA707 and β-lactam could not be sufficiently labeled with Bocillin, as the nonfluorescent β-lactam competes with the Bocillin fluorophore, thereby reducing or eliminating the observed fluorescence. In the aggregate, our microscopy results suggest that TXA707 treatment disrupts septum formation by causing the mislocalization of key septal components, including PBP1, PBP2, and PBP3.
FIG 7.
Impact of TXA707 on PBP localization in MRSA COL. (A to D) Differential interference contrast (A and C) and fluorescence (B and D) micrographs of MRSA COL cells treated for 3 h with either vehicle (A and B) or TXA707 at 8× MIC (C and D). In these micrographs, the PBPs in the cells were labeled by treatment with 2 μM Bocillin for 10 min just prior to visualization.
DISCUSSION
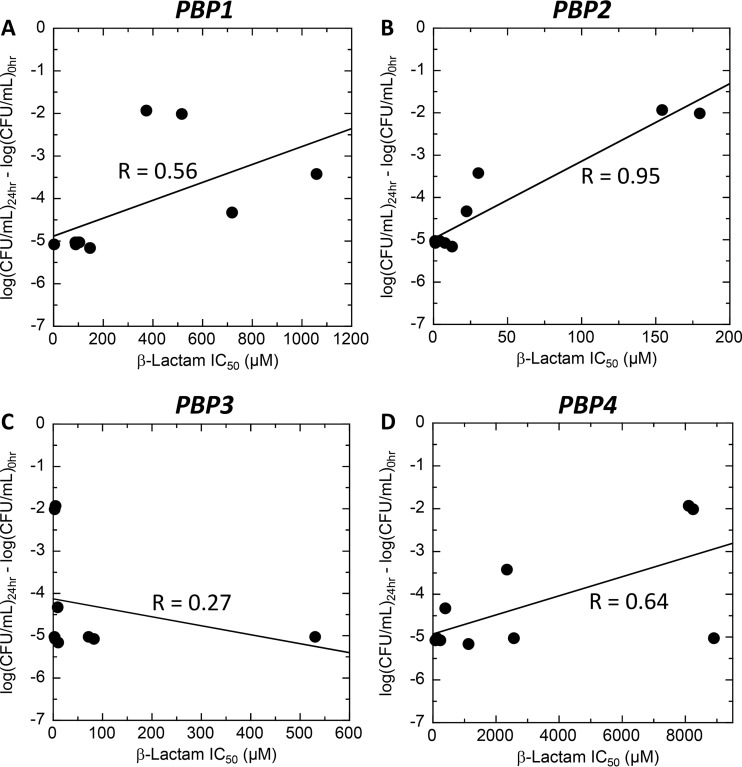
In this study, we show that a broad range of β-lactam antibiotics from three different chemical classes act synergistically with the FtsZ-targeting agent TXA707 against MRSA. Our time-kill synergy results demonstrate that the synergistic actions of the β-lactams in combination with TXA707 are bactericidal in nature. However, the degree of synergistic kill afforded by the β-lactams depends on the specific agent. The question thus arises as to the nature of the correlation, if any, between the magnitude of synergistic kill and the PBP binding affinities and selectivities of the β-lactams. Correlation plots of the PBP binding affinities of the β-lactams (as reflected by the IC50 values in Table 2) versus their corresponding extents of synergistic kill when combined with TXA707 reveals a clear correlation (R = 0.95) between the degree of bactericidal synergy and affinity for PBP2 (Fig. 8B). β-Lactams with a high affinity for PBP2 (low IC50) synergize the best with TXA707. No such correlation is evident for the other three PBPs, with R values of 0.56, 0.27, and 0.64 for PBP1, PBP3, and PBP4, respectively (Fig. 8A, C, and D). In this connection, PBP2-selective β-lactams (e.g., ceftriaxone, cefotaxime, and ertapenem) are better synergizers with TXA707 than PBP3-selective agents (e.g., cephalexin and cefradine) (Table 2 and Fig. 8). That said, β-lactams need not be purely PBP2-selective (e.g., cefdinir, oxacillin, and imipenem) in order to synergize well with TXA707, as long as their PBP binding behavior includes high-affinity targeting of PBP2.
FIG 8.
(A to D) Correlation plots of the extents to which the β-lactams synergize with TXA707 against MRSA COL versus their affinities (as reflected by the IC50 values from Table 2) for PBP1 (A), PBP2 (B), PBP3 (C), and PBP4 (D). In these plots, each datum point corresponds to a different β-lactam antibiotic, with the solid lines reflecting linear regression fits of the data. The correlation constant (R value) derived from the linear regression analysis of each data set is indicated.
We have previously shown that TXA707 inhibits bacterial cell division by altering the dynamics of FtsZ polymerization and disrupting FtsZ Z-ring formation at midcell (29). The formation of the Z-ring is a critical first step toward formation of the septum in dividing cells (25). Consistent with this critical linkage between Z-ring and septum formation, our TEM results indicate that the FtsZ-targeting actions of TXA707 interfere with the formation of septa in MRSA cells (Fig. 6). The Z-ring serves as a scaffold for the recruitment of essential cell division components to the newly forming septum (30). Among these essential cell division components are the PBPs. Our fluorescence microscopy results reveal that the disruptive actions of TXA707 on septum formation in MRSA cause a mislocalization of key PBPs (likely to be PBP1, PBP2, and PBP3) away from the septum to nonproductive peripheral sites (Fig. 7). This observation suggests a potential mechanism underlying the synergy between β-lactams and TXA707. Inhibition of FtsZ function by TXA707 interferes with the septal localization of the PBPs. In so doing, the number of PBPs that are appropriately localized becomes greatly reduced, thereby sensitizing the bacteria to the β-lactams. It is also possible that the actions of the β-lactams on the peripherally localized PBPs adversely impact the integrity of the bacterial cell wall at those peripheral sites, resulting in β-lactam-induced cell lysis and death.
Why then do PBP2-targeting β-lactams synergize the best with TXA707 against MRSA? Pinho and coworkers have demonstrated the cooperative localization and functioning of PBP2 and PBP2A (18, 20). It is likely that mislocalization of PBP2 by the actions of TXA707 results in the concomitant mislocalization of PBP2A. This effect would significantly diminish the molecular basis for the MRSA phenotype, and render MRSA cells particularly susceptible to the dual actions of FtsZ inhibitors and PBP2-targeting β-lactams. Consistent with this notion, previous studies have indicated that FtsZ inhibitors sensitize MRSA to β-lactams to a much greater extent than they do methicillin-susceptible S. aureus (27).
The results of our studies suggest that FtsZ-targeting agents offer an opportunity to repurpose β-lactam antibiotics for use against β-lactam-resistant MRSA strains through combination therapy. Combination therapy with synergistic drug partners has numerous advantages, including a reduced dosage of each drug required for efficacy, a reduced potential for toxicity, and a reduced potential for the emergence of resistance (31–33). Viewed as a whole, the results of our studies indicate that PBP2-targeting β-lactam antibiotics are optimal synergistic partners with FtsZ-targeting agents for combination therapy of MRSA infections.
MATERIALS AND METHODS
Compounds, antibiotics, and MRSA COL.
TXA707 was synthesized as previously described (29). Cefdinir, ceftriaxone (sodium salt), cefotaxime (sodium salt), cefepime HCl, and cefradine were obtained from TOKU-E (Bellingham, WA). Oxacillin (sodium salt), ticarcillin (disodium salt), and cephalexin were obtained from Sigma-Aldrich (St. Louis, MO). Imipenem and ertapenem were obtained from LKT Laboratories (St. Paul, MN) and Ontario Chemicals (Guelph, Ontario, Canada), respectively. Bocillin was obtained from Thermo Fisher Scientific. MRSA COL was provided by Alexander Tomasz (Rockefeller University, New York, NY).
Cloning, expression, and purification of the S. aureus PBPs.
The pbpA, pbpB, pbpC, and pbpD genes that encode S. aureus PBP1, PBP2, PBP3, and PBP4 were amplified by PCR using genomic DNA extracted from MRSA COL. The primers used for these amplifications are listed in Table S1 in the supplemental material. For PBP1, PBP2, and PBP3, the primers were designed to remove the N-terminal transmembrane domain. For PBP4, the primers were designed to remove both the N-terminal signal peptide sequence and the C-terminal transmembrane domain. Each amplified gene was cloned into the pET-22b(+) plasmid (Novagen-EMD Chemicals, Inc.) using the NEBuilder HiFi DNA assembly cloning kit (New England BioLabs, Inc.), such that a 6× His tag was introduced at the C terminus of each recombinant gene product. The sequences of all recombinant plasmids were verified by sequence analyses, and the plasmids were then transformed into Escherichia coli BL21(DE3) cells.
Transformed E. coli were grown on Luria-Bertani (LB) agar plates containing 100 μg of ampicillin/ml. Single colonies were isolated and grown in 20 ml of ampicillin-containing LB broth overnight at 37°C. The overnight cultures were diluted into 4 liters of autoinduction terrific broth (34), followed by incubation at 37°C for 6 h. The cultures were then incubated for an additional 24 to 32 h at 30°C. The cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C. The cell pellets were then resuspended in 50 ml of 10 mM sodium phosphate (pH 7.6), 250 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10% (vol/vol) glycerol and stored at −70°C.
Cells were lysed by ultrasonication for 15 min at 0°C, with an on/off cycle of 10 s at 60 W. The lysates centrifuged at 10,000 × g for 30 min at 4°C. The resulting supernatants were added to 5 ml of Talon metal affinity resin (Clontech Laboratories, Inc.) and shaken for 20 min at 4°C. The resin was then washed with 50 ml of buffer containing 10 mM sodium phosphate (pH 7.6), 250 mM NaCl, and 10% (vol/vol) glycerol (buffer A) and packed into a gravity flow column. The column was washed with 10 ml of buffer A, followed by 25 ml of buffer containing 10 mM sodium phosphate (pH 7.6), 10 mM imidazole, and 250 mM NaCl (buffer B). The protein was then eluted by using 15 ml of elution buffer containing 10 mM sodium phosphate (pH 7.6), 150 mM imidazole, and 250 mM NaCl (buffer C), and 500 μl fractions were collected.
Each fraction was analyzed by SDS-PAGE, and fractions containing protein were combined. For, PBP1, PBP2, and PBP3, the combined fractions were dialyzed overnight against 4 liters of buffer containing 100 mM Tris-HCl (pH 7.4), 250 mM NaCl, and 10% (vol/vol) glycerol. The resulting dialysates were concentrated to 1 ml of volume using Amicon Ultra 10K filters (EMD Millipore, Inc.). The combined elution fractions of PBP4 were dialyzed overnight against 4 liters of buffer containing 20 mM Tris-HCl (pH 7.4), 500 mM NaCl, and 5% (vol/vol) glycerol and then concentrated to 2 ml. Protein concentrations were quantified by using a bicinchoninic acid assay, with final protein concentrations ranging from 13 to 26 mg/ml.
Fluorescence anisotropy assays for Bocillin binding to the PBPs.
Fluorescence anisotropy experiments were performed using an Aviv model ATF105 spectrofluorometer (Aviv Biomedical, Lakewood, NJ). In these experiments, bandwidths were set to 4 nm in both the excitation and the emission directions, with the excitation and emission wavelengths being set at 488 and 510 nm, respectively. For PBP1, PBP2, and PBP3, Bocillin (1 μM) was titrated with increasing concentrations (ranging from 0 to 2.5 μM) of protein in 150 μl of buffer containing 100 mM Tris-HCl (pH 7.4), 250 mM NaCl, and 5% (vol/vol) glycerol. After each protein addition, the samples were equilibrated for 5 min at 25°C, whereupon the fluorescence anisotropy was measured at 25°C. For PBP4, individual samples containing 1 μM Bocillin and a protein concentration ranging from 2 to 50 μM were prepared in 150 μl of buffer containing 20 mM Tris-HCl (pH 7.4), 500 mM NaCl, and 5% (vol/vol) of glycerol. The fluorescence anisotropy of each sample was then measured at 25°C every 6 s for 15 min. A quartz ultra-micro cell (Hellma, Inc.) with a 2- by 5-mm aperture and a 15-mm center height was used for all measurements. The path lengths in the excitation and emissions directions were 1 and 0.2 cm, respectively. All steady-state anisotropy experiments were conducted in triplicate, with the reported anisotropies reflecting the average values.
Competition fluorescence anisotropy assays.
For PBP1, PBP2, and PBP3, individual samples containing 1 μM Bocillin and β-lactam antibiotic at a concentration ranging from 0 to 7.5 mM were prepared in 150 μl of buffer containing 100 mM Tris-HCl (pH 7.4), 250 mM NaCl, and 5% (vol/vol) of glycerol. The reactions were then initiated by addition of 2 μM protein. After 5 min of equilibration at 25°C, the fluorescence anisotropy was then recorded as described above. For PBP4, individual samples containing 1 μM Bocillin and β-lactam antibiotic at a concentration ranging from 0 to 15 mM were prepared in 150 μl of buffer containing 20 mM Tris-HCl (pH 7.4), 500 mM NaCl, and 5% (vol/vol) of glycerol. The reaction was then initiated by the addition of 50 μM protein. Time-dependent fluorescence anisotropy was then acquired as described above.
MIC assay.
MIC assay was conducted in accordance with Clinical and Laboratory Standards Institute guidelines for broth microdilution (35). Briefly, log-phase MRSA COL was added to 96-well microtiter plates (at 5 × 105 CFU/ml) containing 2-fold serial dilutions of compound or β-lactam antibiotic in cation-adjusted Mueller-Hinton (CAMH) broth, with each compound concentration being present in duplicate. The final volume in each well was 0.1 ml, and the microtiter plates were incubated aerobically for 24 h at 37°C. Bacterial growth was monitored by measuring the optical density at 600 nm using a VersaMax plate reader (Molecular Devices, Inc.), with the MIC being defined as the lowest compound concentration at which growth was ≥90% inhibited.
Time-kill assay for synergy.
Exponentially growing MRSA COL was diluted in CAMH broth to a final count of 105 CFU/ml. The colony count at time zero was verified by plating serial dilutions of the culture in duplicate on tryptic soy agar (TSA) plates. The initial culture was aliquoted into tubes containing either 0.5× MIC of TXA707, 0.008× MIC of the β-lactam antibiotic, or both agents combined. An equivalent volume of dimethyl sulfoxide (DMSO) was added to the vehicle control tube. The cultures were then incubated at 37°C with shaking. The CFU/ml in each culture was determined over time by withdrawing samples at time points ranging from 3 to 24 h and plating appropriate serial dilutions on to TSA plates. All TSA plates were incubated at 37°C, and the CFU/ml at each time point determined by counting colonies after 24 h.
Transmission electron microscopy.
Log-phase MRSA COL cells were diluted to an optical density at 600 nm of 0.1 and then cultured at 37°C for 3 h in the presence of DMSO vehicle, TXA707 alone at a concentration of 16 μg/ml (8× MIC), or a combination of TXA707 at a concentration of 2 μg/ml (1× MIC) and either cefdinir at a concentration of 2 μg/ml (0.008× MIC) or ceftriaxone at a concentration of 16 μg/ml (0.008× MIC). The cultures were then centrifuged at 16,000 × g for 3 min at room temperature. The resulting bacterial pellets were washed with 1 ml of phosphate-buffered saline (PBS) and resuspended in 500 μl of 100 mM cacodylate buffer (pH 7.2) containing 2.5% glutaraldehyde and 4% paraformaldehyde. The fixed bacterial cells were then postfixed in buffered osmium tetroxide (1%), subsequently dehydrated in a graded series of ethanol, and embedded in epon resin. Thin sections (90 nm) were cut on a Leica EM UC6 ultramicrotome. Sectioned grids were then stained with a saturated solution of uranyl acetate and lead citrate. Images were captured with an AMT XR111 digital camera at 80 kV on a Philips CM12 transmission electron microscope.
Fluorescence microscopy.
Log-phase MRSA COL cells were diluted to an optical density at 600 nm of 0.1 and then cultured at 37°C for 3 h in the presence of DMSO vehicle or TXA707 at a concentration of 16 μg/ml (8× MIC). The cultures were then centrifuged at 16,000 × g for 3 min at room temperature. The resulting bacterial pellets were washed with 1 ml of PBS, resuspended in 100 μl of PBS containing 2 μM Bocillin, and incubated in the dark for 10 min at room temperature. The cells were then washed with 1 ml of PBS and resuspended in 500 μl of PBS. Each cell suspension (2.5 μl) was placed on a thin layer of 1.5% agarose in PBS mounted on a microscope slide. Differential interference contrast and fluorescence visualization of the cells were performed using an Olympus BX50 microscope equipped with an X-cite Exacte 200W mercury lamp and a 100× Olympus UPLSAPO oil immersion objective (1.40 aperture). Images were captured using a QImaging Retiga R3 charge-coupled device camera and processed with the Ocular version 1 software package (QImaging).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grant R56/R01 AI118874.
We are indebted to Alexander Tomasz (Rockefeller University, New York, NY) for providing the MRSA COL.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00863-17.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States 2013. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 3.Bassetti M, Peghin M, Trecarichi EM, Carnelutti A, Righi E, Del Giacomo P, Ansaldi F, Trucchi C, Alicino C, Cauda R, Sartor A, Spanu T, Scarparo C, Tumbarello M. 2017. Characteristics of Staphylococcus aureus bacteremia and predictors of early and late mortality. PLoS One 12:e0170236. doi: 10.1371/journal.pone.0170236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastagia M, Kleinman LC, Lacerda de la Cruz EG, Jenkins SG. 2012. Predicting risk for death from MRSA bacteremia. Emerg Infect Dis 18:1072–1080. doi: 10.3201/eid1807.101371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis 13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers HF, Deck DH. 2009. Beta-lactam and other cell wall- and membrane-active antibiotics, p 773–793. In Katzung BG, Masters SB, Trevor AJ (ed), Basic and clinical pharmacology, 11th ed McGraw-Hill, New York, NY. [Google Scholar]
- 7.Pinho MG, Kjos M, Veening J-W. 2013. How to get (a) round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 8.Reed P, Atilano ML, Alves R, Hoiczyk E, Sher X, Reichmann NT, Pereira PM, Roemer T, Filipe SR, Pereira-Leal JB, Ligoxygakis P, Pinho MG. 2015. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog 11:e1004891. doi: 10.1371/journal.ppat.1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartman BJ, Tomasz A. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol 158:513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fishovitz J, Hermoso JA, Chang M, Mobashery S. 2014. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 66:572–577. doi: 10.1002/iub.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 12.Egan AJ, Cleverley RM, Peters K, Lewis RJ, Vollmer W. 2017. Regulation of bacterial cell wall growth. FEBS J 284:851–867. doi: 10.1111/febs.13959. [DOI] [PubMed] [Google Scholar]
- 13.Pinho MG, de Lencastre H, Tomasz A. 2000. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J Bacteriol 182:1074–1079. doi: 10.1128/JB.182.4.1074-1079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira SFF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. 2009. Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol Microbiol 72:895–904. doi: 10.1111/j.1365-2958.2009.06687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira SFF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J Bacteriol 189:3525–3531. doi: 10.1128/JB.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monteiro JM, Fernandes PB, Vaz F, Pereira AR, Tavares AC, Ferreira MT, Pereira PM, Veiga H, Kuru E, Van Nieuwenhze MS, Brun YV, Filipe SR, Pinho MG. 2015. Cell shape dynamics during the staphylococcal cell cycle. Nat Commun 6:8055. doi: 10.1038/ncomms9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 187:1815–1824. doi: 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinho MG, Errington J. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol Microbiol 55:799–807. doi: 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- 19.Lovering AL, de Castro LH, Lim D, Strynadka NC. 2007. Structural insight into the transglycosylation step of bacterial cell wall biosynthesis. Science 315:1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 20.Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci U S A 98:10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinho MG, Filipe SR, de Lencastre H, Tomasz A. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J Bacteriol 183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roemer T, Schneider T, Pinho MG. 2013. Auxiliary factors: a chink in the armor of MRSA resistance to beta-lactam antibiotics. Curr Opin Microbiol 16:538–548. doi: 10.1016/j.mib.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Kirkpatrick CL, Viollier PH. 2011. New(s) to the (Z-)ring. Curr Opin Microbiol 14:691–697. doi: 10.1016/j.mib.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance, and disassembly of the Z ring. Nat Rev Microbiol 7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 26.Scheffers DJ, Pinho MG. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaul M, Mark L, Parhi AK, LaVoie EJ, Pilch DS. 2016. Combining the FtsZ-targeting prodrug TXA709 and the cephalosporin cefdinir confers synergy and reduces the frequency of resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 60:4290–4296. doi: 10.1128/AAC.00613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navratna V, Nadig S, Sood V, Prasad K, Arakere G, Gopal B. 2010. Molecular basis for the role of Staphylococcus aureus penicillin binding protein 4 in antimicrobial resistance. J Bacteriol 192:134–144. doi: 10.1128/JB.00822-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaul M, Mark L, Zhang Y, Parhi AK, Lyu YL, Pawlak J, Saravolatz S, Saravolatz LD, Weinstein MP, LaVoie EJ, Pilch DS. 2015. TXA709, an FtsZ-targeting benzamide prodrug with improved pharmacokinetics and enhanced in vivo efficacy against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 59:4845–4855. doi: 10.1128/AAC.00708-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutkenhaus J, Pichoff S, Du S. 2012. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton 69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roemer T, Boone C. 2013. Systems-level antimicrobial drug and drug synergy discovery. Nat Chem Biol 9:222–231. doi: 10.1038/nchembio.1205. [DOI] [PubMed] [Google Scholar]
- 32.Fischbach MA. 2011. Combination therapies for combating antimicrobial resistance. Curr Opin Microbiol 14:519–523. doi: 10.1016/j.mib.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worthington RJ, Melander C. 2013. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabski A, Mehler M, Drott D. 2005. The overnight express autoinduction system: high-density cell growth and protein expression while you sleep. Nat Methods 2:233–235. doi: 10.1038/nmeth0305-233. [DOI] [Google Scholar]
- 35.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. CLSI document M07-A8, 8th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.