Abstract
Background and Aims Wild olive (Olea europaea subsp. europaea var. sylvestris) is important from an economic and ecological point of view. The effects of anthropogenic activities may lead to the genetic erosion of its genetic patrimony, which has high value for breeding programmes. In particular, the consequences of the introgression from cultivated stands are strongly dependent on the extent of gene flow and therefore this work aims at quantitatively describing contemporary gene flow patterns in wild olive natural populations.
Methods The studied wild population is located in an undisturbed forest, in southern Spain, considered one of the few extant hotspots of true oleaster diversity. A total of 225 potential father trees and seeds issued from five mother trees were genotyped by eight microsatellite markers. Levels of contemporary pollen flow, in terms of both pollen immigration rates and within-population dynamics, were measured through paternity analyses. Moreover, the extent of fine-scale spatial genetic structure (SGS) was studied to assess the relative importance of seed and pollen dispersal in shaping the spatial distribution of genetic variation.
Key Results The results showed that the population under study is characterized by a high genetic diversity, a relatively high pollen immigration rate (0·57), an average within-population pollen dispersal of about 107 m and weak but significant SGS up to 40 m. The population is a mosaic of several intermingled genetic clusters that is likely to be generated by spatially restricted seed dispersal. Moreover, wild oleasters were found to be self-incompatible and preferential mating between some genotypes was revealed.
Conclusions Knowledge of the within-population genetic structure and gene flow dynamics will lead to identifying possible strategies aimed at limiting the effect of anthropogenic activities and improving breeding programmes for the conservation of olive tree forest genetic resources.
Keywords: SSRs, gene flow, paternity assignment, SGS, oleaster, wild olive, pollen dispersal, seed dispersal, conservation genetics, Olea europaea
INTRODUCTION
Gene flow promotes the diffusion of genes, producing a change in their spatial distribution. In sessile organisms such as plants, it occurs mainly through pollen and seed dispersal (Slatkin, 1985; Robledo-Arnuncio et al., 2014). Gene flow, together with the main characteristics of the mating system (e.g. outcrossing and selfing rates, biparental inbreeding), is considered one of the most important determinants of the genetic structure of plant populations (Robledo-Arnuncio et al., 2014). The extent of gene flow can be assessed at two spatial scales by studying short-distance dispersal (SDD) and long-distance dispersal (LDD): while the former shapes within-population dynamics, the latter determines the connectivity between populations, spreading advantageous mutations and counteracting the effect of genetic drift (Oddou-Muratorio et al., 2005; Sork and Smouse, 2006; Marchelli et al., 2012). For wind-pollinated trees, pollen-mediated gene flow (hereafter ‘pollen flow’) can be considered the most important mechanism to maintain genetic connectivity over long distances (Buschbom et al., 2011; Robledo-Arnuncio, 2011; Kremer et al., 2012; Bertolasi et al., 2015). For these reasons, the assessment of pollen flow and its effects at different spatial scales is important to predict the impact of landscape alterations (e.g. fragmentation) on reproduction and other evolutionary processes, besides being crucial for designing proper conservation strategies (Sork et al., 1999; Craft and Ashley, 2010, and references therein).
Levels of gene flow have traditionally been assessed through indirect methods from the extent of subpopulation differentiation (Neigel, 1997; Sork et al., 1999; Godoy and Jordano, 2001). However, these methods only measure so-called ‘historical gene flow’, i.e. gene flow over several generations that generated the current genetic structure. Generally, they do not provide any information on ‘contemporary’ gene flow, i.e. gamete movements determined by recent reproductive events (Craft and Ashley, 2010; Piotti et al., 2012). It is instead the study of contemporary gene flow, by means of direct methods, that can provide an accurate description of the distribution of pollen and seed dispersal distances (Sork et al., 1999). The ease in developing hypervariable molecular markers, such as microsatellites (simple sequence repeats, SSRs), and the availability of refined statistical models have allowed an accurate measurement of contemporary SDD and LDD (Ashley, 2010; Jones et al., 2010; Leonarduzzi et al., 2012; Kremer et al., 2012; Robledo-Arnuncio et al., 2014).
Wild olive (Olea europaea subsp. europaea var. sylvestris, also known as oleaster olive) represents a botanical variety of Olea europaea subsp. europaea (Brot.) and, as the cultivated olive (O. europaea subsp. europaea var. europaea), it is mainly distributed along the Mediterranean Basin (Green, 2002). It is a monoecious, predominantly allogamous, self-incompatible tree. In both cultivated and wild olive, pollination is predominantly anemophilous and seed dispersal is mainly mediated by birds (Villemur et al., 1984; Jordano, 1987; Rey and Alcántara, 2000). In ‘wild olive’, two distinct forms have been recognized: true oleaster, the wild form present in undisturbed areas, and feral forms occurring in secondary habitats, such as disturbed areas or abandoned fields. The latter originates from seedlings of cultivated clones or from hybridization between true oleasters and cultivars (Zohary and Hopf, 1994; Angiolillo et al., 1999; Lumaret et al., 2004; Belaj et al., 2007). Wild and cultivated olives have the same chromosome number (2n = 46) and are interfertile (Zohary and Spiegel-Roy, 1975; Besnard and Bervillé, 2000). In the literature, there are several studies that have characterized the genetic diversity of wild olive populations, assessing their relationships with cultivated stands (Belaj et al., 2010, and references therein). In particular, some studies showed that isolated populations of ancient indigenous true oleaster are still present, in both the western and the eastern Mediterranean. These undisturbed forests of true oleaster can be considered as real ‘hotspots’ of var. sylvestris genetic diversity (Lumaret and Ouazzani, 2001; Erre et al., 2010; Besnard et al., 2013).
From an ecological point of view, wild olive is considered a valuable natural resource because its great resistance to wind, bushfire, frost and drought might be crucial in preventing desertification (Mulas et al., 2004; Belaj et al., 2007). In recent years, fruit breeding programmes have increased interest in wild germplasm as a source of useful traits (e.g. resistance to diseases, stress) to be introduced into the genome of cultivated plants (Fischer and Fischer, 2004; Hajjar and Hodgkin, 2007; Sorkheh et al., 2009; De la Rosa et al., 2014). Indeed, Klepo et al. (2013, 2014) and Arias-Calderon et al. (2015) demonstrated the usefulness of wild olive for generating new genotypes with relevant characteristics. Despite their acknowledged importance, wild olive populations are being affected by several threats due to human activities, climatic pressure and gametic introgression from plantations. Wild olive populations are likely to be restricted to very few isolated forest areas (Zohary and Spiegel-Roy, 1975; Lumaret et al., 2004; Yoruk and Taskin, 2014) and the need to implement conservation programmes to maintain their genetic peculiarities has been stressed in several studies (Lumaret and Ouazzani, 2001; Médail et al., 2001; Belaj et al., 2007; Erre et al., 2010; Muñoz-Diez et al., 2011). Estimating the extent of pollen flow at the population level is an important contribution in delineating conservation strategies for viable, natural populations, and few studies have focused on this topic so far in wild olive subspecies (Breton et al., 2006; Besnard et al., 2009a, b, 2013; Garcia-Verdugo et al., 2010). However, only in the work by Besnard et al. (2009a) was the effectiveness of pollen flow within and among O. europaea subsp. laperrinei patches studied to define a conservation strategy. And to our knowledge, pollen flow has never been quantified by direct methods in wild olive populations. In this context, by using highly variable SSRs and applying paternity analysis, we investigated pollen flow patterns and fine-scale spatial genetic structure (SGS) in a large, endemic wild olive stand forest in Andalusia (southern Spain). The specific objectives of the study were: (1) to directly measure the levels of contemporary pollen flow quantifying the pollen immigration rate and within-population pollen dispersal distances, and (2) to assess the presence and extent of SGS by spatial autocorrelation and Bayesian clustering techniques to understand the relative importance of seed and pollen dispersal in shaping the spatial distribution of genetic variation in Olea europaea subsp. europaea var. sylvestris.
MATERIALS AND METHODS
Study area and data sampling
The study area is located in the province of Cádiz (Andalusia region, southern Spain, 36°30′29″N, 5°44′32″W), within a large forest patch (∼350 ha) on a plateau at about 250 m. a.s.l. This area is characterized by a large undisturbed wild olive forest mixed with Quercus coccifera and Quercus ilex, and a very low presence of olive cultivation (Belaj et al., 2007, 2010, 2011).
All 225 adult trees (putative fathers) present in an area of ∼6 ha were sampled (Fig. 1). Among them, five trees were selected as ‘mother trees’ for seed collection based on their regular and abundant fructification. They were individually labelled as M1–M5 and 20 seeds per mother tree were collected. All seeds (hereafter ‘offspring’) were later germinated in a greenhouse at IFAPA Centro ‘Alameda del Obispo’ Córdoba, Spain, according to standard procedures (Santos-Antunes et al., 2005). At the end of the germination procedure, 88 offspring were obtained for genetic characterization.
Fig. 1.
Map of the study plot. Open squares indicate maternal trees and filled circles indicate other adult trees.
DNA extraction and SSR analysis
Total genomic DNA (225 adults and 88 offspring) was extracted from fresh leaves after germination of seeds, as described by De la Rosa et al. (2004). The samples were characterized by eight SSRs: ssrOeUA-DCA-(03, 09, 11, 16, 18) (Sefc et al., 2000), ssrOe-UDO99-(024, 043) (Cipriani et al., 2002) and ssrOe-GAPU-103A (Carriero et al., 2002). The selected loci have been extensively used, showing a high discrimination power (e.g. Belaj et al., 2007, 2011, 2012; Erre et al., 2010; Trujillo et al., 2014; Noormohammadi et al., 2014). In particular, six among them had been listed by Baldoni et al. (2009) among the best SSRs for fingerprinting and for evolutionary and population genetic analyses. Loci are hereafter referred to as DCA-(03, 09, 11, 16, 18), UDO-(024, 043) and GAPU-103A.
Amplifications were performed according to the PCR conditions described by De la Rosa et al. (2004), and the detection of amplification products was carried out with an automated sequencer (ABI PRISM 3100, sequencer, Applied Biosystems, Foster City, CA, USA). Sizing was performed using the program GENESCAN 3.7 and GENOTYPER 3.7 from Applied Biosystems. Three reference samples were used in all runs.
Data analysis
Population genetic analysis.
GENEPOP 4.0 (Raymond and Rousset, 1995) was used to calculate the number of alleles per locus (Na), the observed (HO) and expected heterozygosity (HE) and the inbreeding coefficient (FIS), as well as to test genotypic frequencies at each locus for conformance to Hardy–Weinberg (HW) expectations. The same software was used to text for possible linkage disequilibrium among SSRs in the population. Each locus was evaluated for the possible presence of null alleles using MICRO-CHECKER 2.2.3 (Van Oosterhout et al., 2004).
Spatial genetic structure.
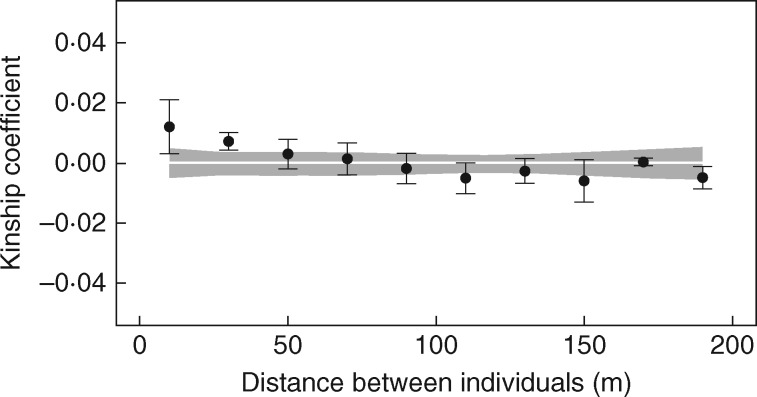
Fine-scale spatial genetic structure was investigated by classical spatial autocorrelation analysis performed using SPAGeDi 1.4 (Hardy and Vekemans, 2002) and estimating the pairwise kinship coefficient (Fij) of Loiselle et al. (1995). Tests for statistical significance of mean kinship coefficient values in each distance class were conducted by (1) random shuffling (5000 times) of individual geographical locations to define the upper and lower bounds of the 95 % confidence interval around the null hypothesis that Fij and pairwise distances were uncorrelated, and (ii) estimating 95 % confidence intervals around mean Fij values by bootstrapping loci (5000 repeats). Analyses were performed using both the even distance classes option (using 20-m-wide distance classes) and the even sample size option (distributing all possible pairs in ten distance classes with similar numbers of pairs per class). The intensity of SGS was also measured by the Sp statistic (Vekemans and Hardy, 2004), computed as Sp = bF/(F1–1), where bF is the regression slope of the kinship estimator Fij computed among all pairs of individuals against their geographical distances, and F1 is the average kinship coefficient between individuals of the first distance class (0–20 m). Sp has the desirable characteristic of being comparable among studies. The statistical significance of F1 and bF was tested based on 5000 permutations of spatial coordinates among individuals.
To explore the possible presence of subtle spatial signals in the genetic structure of the population, the spatially explicit Bayesian clustering algorithm implemented in the R package GENELAND v. 4.0.2 aimed at assigning individuals to putative sub-populations was used (Guillot et al., 2005, 2008). The program was run exploiting both genetic and spatial information. We used the spatial model with correlated allele frequencies, setting the maximum number of sub-populations to 20, and running the analyses for 2 × 106 iterations, with a thinning value of 1000. The possible presence of null alleles was explicitly taken into account by using the filter.null.alleles option. For each number of sub-populations tested, the analysis was repeated ten times and the highest median number of clusters was chosen as the most representative one.
Paternity analysis.
The final dataset for paternity analysis was decided after checking for the possible presence of null alleles. Loci with a non-negligible frequency of possible null alleles were treated following the substitution procedure proposed by Bacles and Ennos (2008). Briefly, for these loci homozygous genotypes were systematically changed to heterozygotes with null allele, and non-amplifying genotypes were transformed into null allele homozygotes. All paternity data analysis was also run on the original dataset for comparison. Paternity analysis was conducted using the maximum-likelihood procedure implemented in the software FaMoz (Gerber et al., 2003). For each offspring, paternity was categorically assigned to the individual with the highest LOD score above an estimated threshold. The threshold for statistical significance of LOD scores was estimated following the procedure described by Gerber et al. (2000), based on the simulation of 5 × 104 locally and externally pollinated seeds, to minimize both type I error (i.e. when a seed pollinated by local pollen is not assigned to local fathers), and type II error (i.e. when a paternity is attributed to a local father whereas the true father is outside the sampling area) (Gerber et al., 2000). Thresholds were estimated for a genotyping error value 0·02 and accounting for the heterozygote deficit estimated from adult genotypes. Sampled seeds were considered as locally pollinated if at least one compatible father with an LOD score above the threshold was found, otherwise they were classified as pollinated by external trees. Therefore, in the following, ‘pollen immigration rate’ refers to the proportion of sample seeds pollinated from outside the study plot.
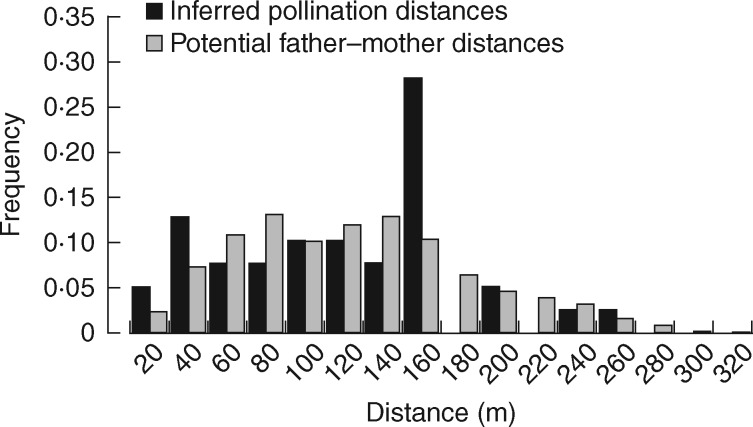
Once paternity was assessed, pollination distances were calculated as Euclidean distances between the assigned fathers and the mother trees. To determine whether the distribution of pollen dispersal resembled that of distances between all potential fathers and mother trees, we compared the two distributions using the Kolmogorov–Smirnov test (Sokal and Rohlf, 1995).
RESULTS
Genetic diversity in the wild population under study
In total, 225 adult trees were genotyped with eight SSRs, which yielded a total of 133 alleles, with average Na and HE equal to 16·92 and 0·771, respectively (Table 1). Significant departures from HW equilibrium and the possible presence of null alleles according to the Micro-Checker test were observed at three out of eight loci (DCA-03, GAPU-103A, UDO-43). The null allele frequencies, as estimated using Brookfield’s formula (Brookfield, 1996), were 0·077 (DCA-03), 0·086 (GAPU-103A) and 0·100 (UDO-43) (Table 1). All SSRs were unlinked.
Table 1.
Genetic diversity of eight SSRs in the wild olive tree population (n = 225)
| Marker | Na | HO | HE | FIS | fnull |
|---|---|---|---|---|---|
| DCA-03 | 13 | 0·482 | 0·607 | 0·206*** | 0·077 |
| DCA-09 | 18 | 0·897 | 0·892 | −0·006ns | – |
| DCA-11 | 8 | 0·717 | 0·663 | −0·082ns | – |
| DCA-16 | 26 | 0·897 | 0·915 | 0·020** | – |
| DCA-18 | 15 | 0·890 | 0·891 | 0·001ns | – |
| GAPU-103A | 18 | 0·512 | 0·656 | 0·220*** | 0·086 |
| UDO-24 | 13 | 0·712 | 0·711 | −0·002ns | – |
| UDO-43 | 22 | 0·647 | 0·832 | 0·224*** | 0·100 |
Na, number of alleles; HO, observed heterozygosity; HE, expected heterozygosity; FIS, inbreeding coefficient. Significance of the departures from Hardy–Weinberg equilibrium: ***, significance at the 0·1 % nominal level; **, significance at the 1 % nominal level; *, significance at the 5 % nominal level and ‘ns’ depicts non-significant values; fnull, null-allele frequency as estimated by Brookfield’s formula (Brookfield, 1996) for the loci for which the presence of null alleles was detected using MICRO-CHECKER (Van Oosterhout et al., 2004).
Spatial genetic structure
Spatial autocorrelation analyses detected a weak but significant spatial genetic structure in the first two investigated distance classes, until 40 m (Fig. 2), and the estimated Sp statistic was 0·0049 (±0·0046 s.e.). GENELAND spatially explicit clustering analysis, which was intended to investigate even subtler characteristics of SGS, indicated that the population was composed of a mosaic of intermingled clusters (best grouping at K=16, Supplementary Data Fig. S1). The distribution of intra-cluster pairwise distances is L-shaped with a median value of 43·08 m.
Fig. 2.
Correlograms from spatial autocorrelation analysis using the correlation coefficient Fij of Loiselle et al. (1995) and even distance classes. The grey area represents the 95 % confidence intervals around the null hypothesis of absence of spatial genetic structuring, and black lines around mean Fij values represent their 95 % confidence intervals generated by jackknifing loci.
Paternity analysis
We took into account the possible presence of genotyping errors by accommodating the three loci for which the presence of null alleles was suspected with a transformation of the original dataset. Both the original and the transformed datasets provided high resolution in terms of cumulative exclusion probability (>0·999). Paternity results of the 88 offspring presented hereafter are those obtained from paternity analysis carried out on the transformed dataset, although the estimates of gene flow parameters from the two datasets were almost identical (results not shown).
The pollen immigration rate, which indicates the proportion of seed pollinated by external trees, was 0·57. On the other hand, a total of 39 (43 %) offspring were assigned to local fathers. Twenty-eight of the 225 local adult trees (∼12 %) were implicated in the pollination process. While in most of the cases local adults were identified as fathers of a single offspring, five of them (nos. 47, 89, 135, 142 and 243) had more than one assigned seed. These five trees were the fathers of 16 offspring out of the 39 (41 %) locally assigned. The proportion of offspring assigned to local fathers varied considerably among half-sib families, ranging from 16·6 % (M4) to 76·4 % (M3). In the half-sib family M3, seven offspring were sired by tree no. 142 and three offspring by tree no. 135. No cases of self-pollination were observed (Table 2).
Table 2.
Half-sib family, number of offspring (Noffspring), number of offspring with local paternities assigned (Nloc), number of identified local fathers (Nfathers), number of offspring not assigned to local fathers (Next), number of self-pollinated offspring (Nself), mean and maximum pollination distances between each mother tree and the identified fathers
| Half-sib family | Noffspring | Nloc | Nfathers | Next | Nself | Pollination distance (m) |
|
|---|---|---|---|---|---|---|---|
| Mean | Max | ||||||
| M1 | 18 | 10 | 9 | 8 | 0 | 132·8±65·4 | 256·8 |
| M2 | 17 | 4 | 4 | 13 | 0 | 87·1±51·0 | 137·0 |
| M3 | 17 | 13 | 5 | 4 | 0 | 147·9±17·8 | 185·8 |
| M4 | 18 | 3 | 3 | 15 | 0 | 83·5±7·2 | 91·7 |
| M5 | 18 | 9 | 7 | 9 | 0 | 35·1±20·7 | 82·5 |
The mean pollen dispersal distance between mother trees and the identified fathers was 106·8 ± 58·9 and the maximum dispersal distance was 256·8 m (Fig. 3). The difference between the distribution of inferred pollen dispersal distances and the distribution of all distances between potential fathers and maternal trees was statistically significant (Kolmogorov–Smirnov test: D = 0·8, P < 0·001). This result could be due to the presence of a larger number of pollinations in the distance class 150–160 m (Fig. 3). In particular, tree no. 142 sired six offspring from M3, with the two trees located at a distance of 143·5 m. For this reason, the Kolmogorov–Smirnov test was repeated after removing the distance class 150–160 m. However, the differences observed between the distributions were confirmed (D = 0·9, P < 0·001), probably due to an excess of detected pollination events <40 m.
Fig. 3.
Distribution of inferred pollen dispersal distances (black bars), and distribution of all distances between potential fathers and maternal trees (grey bars).
DISCUSSION
Wild olive populations are experiencing an increasing fragmentation and size reduction, which it particularly urgent to implement conservation programmes for maintaining their genetic peculiarities. The investigation of within-population neutral genetic dynamics and, in particular, gene flow patterns will greatly help in identifying correct approaches to preserve olive tree genetic resources. For this reason, we studied pollen flow and fine-scale SGS in an area which is considered one of the few extant hotspots of wild olive genetic diversity. The results of what, to the best of our knowledge, is the first investigation of contemporary gene flow patterns in Olea spp. showed the relative importance of seed and pollen movements in shaping the spatial distribution of genetic variation at the local scale. Besides providing greatly improved knowledge of the spatial extent of dispersal patterns in Olea spp., the assessment of mating patterns and pollen immigration will represent an important contribution to identify possible conservation and utilization strategies for olive tree.
Population genetic diversity
A high genetic diversity was observed in the investigated population. The mean values of HO and HE (0·719 and 0·771, respectively) are similar to those observed for this subspecies in other populations from the Western Mediterranean Basin (Belaj et al., 2007, 2010) and higher than those reported by Yoruk and Taskin (2014) for wild olive populations in the Eastern Mediterranean Basin. This trend is in agreement with several previous studies that showed a high genetic variability in the Western Mediterranean Basin, in particular in the Iberian Peninsula and on both sides of the Maghreb (Breton et al., 2006; Rubio De Casas et al., 2006; Besnard et al., 2013; Muñoz Diez et al., 2015). It should be noted, however, that comparing the data in the literature is not simple as the differences found in the number of alleles per locus and the heterozygosity values may be due to several factors, such as the different loci and the number of genotypes studied (Lopes et al., 2004). In a previous work on wild populations from different Andalusian provinces, the Cádiz population was the one with the highest genome proportion assigned to the true oleaster gene pool (Belaj et al., 2010). This result is in agreement with several studies (Lumaret et al., 2004; Belaj et al., 2007; Besnard et al., 2013) that considered the Cádiz population as one of the world’s most important ‘hotspots’. Therefore, the wild olive population of Cádiz might represent an unexploited reservoir of genetic variability for Olea europaea subsp. europaea var. sylvestris ideal for in situ conservation programmes. Wild genotypes from the forest stand investigated in the present study, as well as progenies obtained from crosses with cultivar ‘Picual’, have been recently studied with the aim of exploiting the characteristics of this subspecies to improve the Spanish olive breeding programme from a qualitative and an agronomic point of view (Klepo et al., 2013; Arias-Calderon et al., 2015). The results stressed that the gene pool of this wild population could actually represent a future source of genetic diversity linked to interesting agronomical and ecophysiological characteristics, and suggested the high conservation value of this forest genetic resource.
Contemporary pollen flow
By means of paternity analysis, we estimated the pollen immigration rate and quantitatively described within-population pollen movements. Based on highly a polymorphic marker set and taking into account the possible influence of genotyping errors, the paternity analysis showed that 57 % of sampled seeds were sired by foreign fathers. The estimated level of pollen immigration is similar to what has been estimated in populations characterized by relatively low levels of geographical isolation in wind-pollinated tree species (an average value of ∼54 % according to Bittencourt and Sebbenn, 2007). The population under study is actually within a large, continuous forest patch, but nevertheless the identified level of pollen immigration is similar to that reported for a fragmented population of Laperrine's olive (Besnard et al., 2009a) and in general to that of other wind-pollinated species of the same family (Oleaceae) in fragmented landscapes. For example, in fragmented populations of Fraxinus excelsior pollen immigration was between 43 and 68 % (Bacles and Ennos, 2008). The possibility of LDD by pollen in wild and cultivated olive has been observed in different studies (Alba et al., 2006; Pinillos and Cuevas, 2009; Fernández-Rodríguez et al., 2014; Rojo et al., 2016). In particular, studying the potential sources of airborne Olea pollen in the south-west Iberian Peninsula, Fernández-Rodríguez et al. (2014) showed that olive pollen can spread even at the regional scale (up to 200–300 km). There is evidence that reproductive isolation in olive populations is unlikely and that pollen exchanges are frequent even among isolated populations. In particular, such pollen LDD ability was demonstrated by investigating the existence of independent lineages in the wild olive across the Mediterranean (Rubio de Casas et al., 2006) and analysing the genetic diversity and population structure of wild olive from the north-western Mediterranean Basin (Belaj et al., 2007). However, our study was mainly focused on local pollination dynamics and only the pollen immigration rate provided an indication of gene flow coming from outside the study plot without any reference to the distance travelled from potential pollen sources. To obtain such information, future studies on olive gene flow should involve the characterization of surrounding populations with a different experimental approach (see, for example, Robledo-Arnuncio, 2011).
The within-population pollen dispersal distribution is determined by several factors, such as landscape topography, wind direction, local density and other intrinsic characteristics of the population, for example the spatial distribution of individuals (Gaino et al., 2010; Piotti et al., 2012; Fernández-Rodrígues et al., 2014; Bertolasi et al., 2015). Paternity analysis showed that 43 % of pollinations were between local individuals, with an average distance of ∼107 m and a slight excess of pollination events at a distance <40 m. Such a distribution of pollination distances is similar to what has been reported for other wind-pollinated forest species within continuous populations [e.g. 140 m in Pinus flexilis, Schuster and Mitton (2000); 135·5 m in Pinus sylvestris, Robledo-Arnuncio and Gil (2005); 111·9 m in Pinus pinaster, De Lucas et al. (2008); 83 m in Araucaria angustifolia, Bittencourt and Sebbenn (2007); 100 m in Ulmus, Nielsen and Kjær (2010)]. In Olea europaea L. var. europaea, the distribution of within-population pollen dispersal distances during an artificial pollination experiment was studied by Pinillos and Cuevas (2009). These authors found that a maximum pollen dispersal of 100 m from the source could be reached although the concentration of pollen grains fell rapidly with distance from the application point. A similar dispersal pattern and an effective pollination distance of 90 m were recorded by Griggs et al. (1975) in an olive seed orchard. In subsp. laperrinei, within-population pollen dispersal distances were highly variable depending on the stand studied, with a maximum recorded distance of ∼2000 m (Besnard et al., 2009a).
The within-population pollen dispersal distribution described may be influenced by olive density, which is not uniform in the stand under study (Fig 1), and by other factors such as an uneven distribution of reproductive success and the presence of several cases of multiple paternity; for example, trees 142 and 135 are fathers of 7/13 and 3/13 seeds, respectively, of mother M3. As reported by Besnard et al. (2009a) such findings are probably due to gametophytic compatibilities between some genets, but they could also be due to synchronized floral phenology, to inter-individual variation in pollen production and to preferential wind direction. In agreement with several previous studies on O. europaea, no cases of self-pollination were observed. In fact, paternity tests revealed that most olive cultivars are self-incompatible [De la Rosa et al., 2004; Díaz et al., 2006; but see Mookerjee et al. (2005) for some exceptions] and that subsp. laperrinei presents a low percentage of auto pollination [<2 % of selfings; Besnard et al. (2009a)].
Spatial genetic structure
To highlight the outcome of neutral processes in a natural olive tree population, the possible existence and extent of SGS were investigated through spatial autocorrelation analysis. Before our study, the only available information on the spatial distribution of genetic variation in Olea spp. was from the investigation by Besnard et al. (2007), aimed at exploring SGS at the regional scale by analysing fragmented populations of subsp. laperrinei. At the population level, the intensity of SGS is mainly determined by effective population density and dispersal, with seedling spatial distribution being particularly dependent on seed dispersal distances (Sagnard et al., 2011). A weak SGS (Sp = 0·0049) with clusters of related individuals up to 40 m wide was found in the investigated population. The low intensity of SGS characterizing the population is typical for wind-pollinated and animal-dispersed trees (Vekemans and Hardy, 2004). Moreover, it is known that self-incompatible or outcrossing species, such as the olive, tend to show a lower SGS than selfing species (Doligez et al., 1998; Vekemans and Hardy, 2004), presumably due to high gene flow through pollen.
Although our study was not aimed at providing a detailed description of seed dispersal patterns, SGS provides an indication about the extent of genetically homogeneous groups of individuals possibly generated by spatial limitations to seed dispersal. Several studies have investigated the loss of fruits and the consequent dispersal of seeds in wild and cultivated olive. Spenneman and Allen (2000) reported several studies that document how wild olives and cultivated olives are an important source of nourishment for many species of birds and small animals, thus becoming dispersal vectors for their seeds. However, fruit consumption and resulting zoochorous seed dispersal can vary according to several factors (Alcántara et al., 1997; Nathan and Muller-Landau et al., 2000) such as the size of drupes, which is highly variable among different olive genotypes. In a study in which the agro-morphological traits of 47 wild olive trees from southern Spain were characterized, Belaj et al. (2011) reported that fruit and seed weight ranged from 150 to 1260 mg and from 50 to 360 mg, respectively. Similar results were found by Alcántara et al. (2000) (seed size, 22·8–325·6 mg). Besides, fruit size can shape the diversity of potential dispersers, as previously shown in California (Aslan and Rejmánek, 2011). Consequently, the distance travelled by seeds from the mother tree after primary and secondary dispersal can greatly vary (Rey et al., 2004) due to different seed weight and dispersal vectors (small mammals, rodents or several species of birds) (Spenneman and Allen, 1998). In an experiment on germination and early traits of olive seedlings, Rey et al. (2004) found that only 19 % of seeds germinated after two years. Growth performance at the seedling stage was correlated with larger fruit size and influenced by the microhabitat regardless of seedling density. The behaviour of dispersal agents and differential survival of the largest seeds can contribute to seed dispersal being restricted to short distances, generating, along with local pollen flow, the weak genetic structure identified in our population.
Implications for conservation and use of O. sylvestris genetic resources
Wild olive is a distinctive element of the Mediterranean flora of the Iberian Peninsula (Rubio de Casas et al., 2006; Belaj et al., 2010). Wild olive populations are not considered as to be directly threatened by demographic fluctuations in the near future (Belaj et al., 2007). However, the monitoring of wild olive gene pools has been judged essential to prevent genetic erosion brought about by the introgression from cultivated to wild germplasm or other anthropogenic effects (e.g. habitat fragmentation) (Belaj et al., 2007; Perea and Gutiérrez-Galán, 2016).
Our study, besides showing the high genetic diversity of the investigated stand, has allowed us to obtain a first overview of pollen dispersal patterns and the fine-scale spatial genetic structure of wild olive populations in a particular ecological context for which no previous information was available. Parentage and spatial analyses based on genetic data are powerful tools for detailed study of the scale of dispersal patterns within forest tree populations. The short distance component of dispersal patterns has a strong influence on the characteristics of fine-scale genetic structure that, in turn, may determine the rate and direction of microevolutionary changes at the population level in forest trees (Pluess et al., 2009; Oddou-Muratorio et al., 2011; Brousseau et al., 2015). The results of our study have therefore provided important information which can be used to plan conservation and utilization of wild olive genetic resources based on improved knowledge of the spatial extent of dispersal in wild olive. Wild olive has become increasingly of interest for breeding programmes of cultivated olive due to its resistance to biotic stresses and earliness of bearing (Klepo et al., 2013, 2014; Arias-Calderon et al., 2015). The presence of weak but significant SGS suggests that, for future ex situ conservation and breeding strategies, seeds need to be collected from selected parents belonging to different genetic clusters at a distance >40 m to avoid collecting seeds from genetically related individuals. Another criterion which might be applied is that, given that the average distance of within-population pollen dispersal was 107 m, seeds could be collected at about this distance to increase the probability of maximizing the genetic variation embedded in populations selected for seed collection.
Besides the implications of our results to ex-situ conservation programmes, the quantification of pollen immigration and within-population pollen dispersal distances can also help in predicting introgression from cultivated to natural stands when they are in close contact, as in the case of several wild olive populations. Crop-to-wild gene flow has been considered a threat to natural genetic resources and has received particular attention in recent decades. In fruit trees, introgression from cultivars to wild populations has been investigated and shown to be widespread (Cornille et al., 2015, and references therein). For example, crop-to-wild gene flow was shown to be responsible for a decrease in fitness in apple wild populations, leading researchers to consider non-introgressed populations with high levels of genetic diversity as a priority for conservation programmes (Bleeker et al., 2007; Cornille et al., 2015). In olive, recent studies have demonstrated frequent introgression from seed orchards and cultivated stands to wild populations (Besnard et al., 2011, 2013; Perea and Gutiérrez-Galán, 2016) showing the importance of assessing gene flow patterns when natural and cultivated stands are in close contact.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: Bayesian clustering results. (a) Distribution of results from the ten repetitions of GENELAND analyses. The highest median number of clusters (k=16) was chosen as the most representative one. (b) Assignment of adult individuals to the 16 clusters. Each cluster is represented as a different colour. (c) Distribution of within-cluster pairwise distances.
Supplementary Material
ACKNOWLEDGEMENTS
The present study was partly financed by the INIA projects RF 2009-0005-00-00 and INIA: ERA-NET: 219262 FP7-ERANET ARIMNET of the National Institute of Agricultural Research (INIA), Ministry of Education and Culture, Spain, partially funded by the European Regional Development Fund (ERDF). Part of this study was carried out within a postdoctoral INIA contract for A. Belaj (Subprograma DOC-INIA), National Institute of Agricultural Research (INIA), Ministry of Education and Culture, Spain. We thank all the people involved with the sampling and monitoring of the genotypes at the first stages of the study.
LITERATURE CITED
- Alba F, Nieto-Lugilde D, Comtois P, de la Guardia C, De Linares C, Ruiz L.. 2006. Airborne-pollen map for Olea europaea L. in eastern Andalusia (Spain) using GIS: estimation models. Aerobiologia 22: 107–116. [Google Scholar]
- Alcántara JM, Rey PJ, Valera F, Sánchez-Lafuente AM, Gutiérrez JE.. 1997. Habitat alteration and plant intra-specific competition for seed dispersers. An example with Olea europaea var. sylvestris. Oikos 79: 291–300. [Google Scholar]
- Alcántara JM, Rey PJ, Valera F, Sánchez-Lafuente AM.. 2000. Factors shaping the seed fall pattern of a bird-dispersed plant. Ecology 81: 1937–1950. [Google Scholar]
- Angiolillo A, Mencuccini M, Baldoni L.. 1999. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theoretical and Applied Genetics 98: 411–421. [Google Scholar]
- Arias-Calderon R, León L, Bejarano-Alcázar J, Belaj A, De la Rosa R, Rodríguez-Jurado D.. 2015. Resistance to Verticillium wilt in olive progenies from open-pollination. Scientia Horticulturae 185: 34–42. [Google Scholar]
- Ashley MV. 2010. Plant parentage, pollination, and dispersal: how DNA microsatellites have altered the landscape. Critical Reviews in Plant Sciences 29: 148–161. [Google Scholar]
- Aslan CE, Rejmánek M.. 2011. Smaller Olea europaea fruits have more potential dispersers: implication for olive invasiveness in California. Madroño 58: 86–91. [Google Scholar]
- Bacles CFE, Ennos RA.. 2008. Paternity analysis of pollen-mediated gene flow for Fraxinus excelsior L. in a chronically fragmented landscape. Heredity 101: 368–380. [DOI] [PubMed] [Google Scholar]
- Baldoni L, Cultrera NG, Mariotti R, et al. 2009. A consensus list of microsatellite markers for olive genotyping. Molecular Breeding 24: 213–231. [Google Scholar]
- Belaj A, Muñoz-Diez C, Baldoni L, Porceddu A, Barranco D, Satovic Z.. 2007. Genetic diversity and population structure of wild olives from North-Western Mediterranean assessed by SSR markers. Annals of Botany 100: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaj A, Muñoz-Diez C, Baldoni L, Satovic Z, Barranco D.. 2010. Genetic diversity and relationships of wild and cultivated olives at regional level in Spain. Scientia Horticolturae 124: 323–330. [Google Scholar]
- Belaj A, León L, Satovic Z, De la Rosa R.. 2011. Variability of wild olives (Olea europaea subsp. europaea var. sylvestris) analyzed by agro-morphological traits and SSR markers. Scientia Horticulturae 129: 561–569. [Google Scholar]
- Belaj A, del Carmen, Dominguez-García M, et al. 2012. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genetics & Genomes 8: 365–378. [Google Scholar]
- Bertolasi B, Leonarduzzi C, Piotti A, et al. 2015. A last stand in the Po valley: genetic structure and gene flow patterns in Ulmus minor and U. pumila. Annals of Botany 115: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Bervillé A.. 2000. Multiple origins for Mediterranean olive (Olea europaea L. ssp. europaea) based upon mitochondrial DNA polymorphisms. Comptes Rendus de l’Académie des Sciences-Series III-Sciences de la Vie 323: 173–181. [DOI] [PubMed] [Google Scholar]
- Besnard G, Christin PA, Baali-Cherif D, Bouguedoura N, Anthelme F.. 2007. Spatial genetic structure in the Laperrine’s olive (Olea europaea subsp. laperrinei), a long-living tree from the central Saharan mountains. Heredity 99: 649–657. [DOI] [PubMed] [Google Scholar]
- Besnard G, Baali-Cherif D, Bettinelli-Riccardi S, Parietti D, Bouguedoura N.. 2009a. Pollen-mediated gene flow in a highly fragmented landscape: consequences for defining a conservation strategy of the relict Laperrine’s olive. Comptes Rendum Biologies 332: 662–672. [DOI] [PubMed] [Google Scholar]
- Besnard G, Rubio de Casas R, Christin PA, Vargas P.. 2009b. Phylogenetics of Olea (Oleaceae) based on plastid and nuclear ribosomal DNA sequences: tertiary climatic shifts and lineage differentiation times. Annals of Botany 104: 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Hernández P, Khadari B, Dorado G, Savolainen V.. 2011. Genomic profiling of plastid DNA variation in the Mediterranean olive tree BMC Plant Biology 11: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, El Bakkali A, Haouane H, Baali-Cherif D, Moukhli A, Khadari B.. 2013. Population genetics of Mediterranean and Saharan olives: geographic patterns of differentiation and evidence for early generations of admixture. Annals of Botany 112: 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker W, Schmitz U, Ristow M.. 2007. Interspecific hybridisation between alien and native plant species in Germany and its consequences for native biodiversity. Biological Conservation 137: 248–253. [Google Scholar]
- Breton C, Tersac M, Bervillé A.. 2006. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. Journal of Biogeography 34: 1916–1928. [Google Scholar]
- Bittencourt JVM, Sebbenn AM.. 2007. Patterns of pollen and seed dispersal in a small, fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 99: 580–591. [DOI] [PubMed] [Google Scholar]
- Brookfield JFY. 1996. A simple new method for estimating null allele frequency from heterozygote deficiency. Molecular Ecology 5: 453–455. [DOI] [PubMed] [Google Scholar]
- Brousseau L, Foll M, Scotti-Saintagne C, Scotti I.. 2015. Neutral and adaptive drivers of microgeographic genetic divergence within continuous populations: the case of the neotropical tree Eperua falcata (Aubl.). PLoS ONE 10: e0121394. doi:10.1371/journal.pone.0121394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbom J, Yanbaev Y, Degen B.. 2011. Efficient long-distance gene flow into an isolated relict oak stand. Journal of Heredity 100: 106–113. [DOI] [PubMed] [Google Scholar]
- Carriero F, Fontanazza G, Cellini F, Giorgio G.. 2002. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theoretical and Applied Genetics 104: 301–307. [DOI] [PubMed] [Google Scholar]
- Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R.. 2002. Microsatellite markers isolated in olive are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars (Olea europaea L.). Theoretical and Applied Genetics 104: 223–228. [DOI] [PubMed] [Google Scholar]
- Cornille A, Feurtey A, Gélin U, et al. 2015. Anthropogenic and natural drivers of gene flow in a temperate wild fruit tree: a basis for conservation and breeding programs in apples. Evolutionary Applications 8: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft KJ, Ashley MV.. 2010. Pollen-mediated gene flow in isolated and continuous stands of bur oak, Quercus macrocarpa (Fagaceae). American Journal Botany 97: 1999–2006. [DOI] [PubMed] [Google Scholar]
- De Lucas AI, Robledo-Arnuncio JJ, Hidalgo E, González-Martínez SC.. 2008. Mating system and pollen gene flow in Mediterranean maritime pine. Heredity 100: 390–399. [DOI] [PubMed] [Google Scholar]
- De la Rosa R, James CM, Tobutt KR.. 2004. Using microsatellites for paternity testing in olive progenies. HortScience 39: 351–354. [Google Scholar]
- De la Rosa R, Klepo T, Arias-Calderón R, et al. 2014. Corrent status of conservation, evaluation and usefulness of wild olive germplasm. Acta Horticolturae 1057: 515–519. [Google Scholar]
- Díaz A, Martín A, Rallo P, Barranco D, De la Rosa R.. 2006. Self-incompatibility of ‘Arbequina’ and ‘Picual’ Olive Assessed by SSR Markers. Journal of the American Society for Horticultural Science 131: 250–255. [Google Scholar]
- Doligez A, Baril C, Joly HI.. 1998. Fine-scale spatial genetic structure with non-uniform distribution of individuals. Genetics 148: 905–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erre P, Chessa I, Muñoz-Diez C, Belaj A, Rallo L, Trujillo I.. 2010. Genetic diversity and relationships between wild and cultivated olives (Olea europaea L.) in Sardinia as assessed by SSR markers. Genetic Resources and Crop Evolution 57: 41–54. [Google Scholar]
- Fernández-Rodríguez S, Skjøth CA, et al. 2014. Identification of potential sources of airborne Olea pollen in the Southwest Iberian Peninsula. International Journal of Biometeorology 58: 337–348. [DOI] [PubMed] [Google Scholar]
- Fischer M, Fischer C.. 2004. Genetic resources as basis for new resistant apple cultivars. Journal of Fruit and Ornamental Plant Research 64: 63–76. [Google Scholar]
- Gaino AP, Silva AM, Moraes MA, et al. 2010. Understanding the effects of isolation on seed and pollen flow, spatial genetic structure and effective population size of the dioecious tropical tree species Myracrodruon urundeuva. Conservation Genetics 11: 1631–1643. [Google Scholar]
- García-Verdugo C, Forrest AD, Fay MF, Vargas P.. 2010. The relevance of gene flow in metapopulation dynamics of an oceanic island endemic, Olea europaea subsp. guanchica. Evolution 64: 3525–3536. [DOI] [PubMed] [Google Scholar]
- Gerber S, Mariette S, Streiff R, Bodénès C, Kremer A.. 2000. Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Molecular Ecology 9: 1037–1048. [DOI] [PubMed] [Google Scholar]
- Gerber S, Chabrier P, Kremer A.. 2003. FAMOZ: a software for parentage analysis using dominant, codominant and uniparentally inherited markers. Molecular Ecology Notes 3: 479–481. [Google Scholar]
- Godoy JA, Jordano P.. 2001. Seed dispersal by animals: exact identification of source trees with endocarp DNA microsatellites. Molecular Ecology 10: 2275–2283. [DOI] [PubMed] [Google Scholar]
- Green PS. 2002. A version of Olea L. (Oleaceae). Kew Bulletin 57: 91–140. [Google Scholar]
- Griggs WH, Hartmann HT, Bradley BT, Ivakiri BT, Whisler JE.. 1975. Olive pollination in California. California Agricultural Experimental Station (Bulletin) 869: 1–50. [Google Scholar]
- Guillot G, Mortier F, Estoup A.. 2005. GENELAND: a computer package for landscape genetics. Molecular Ecology Notes 5: 712–715. [Google Scholar]
- Guillot G, Santos F, Estoup A.. 2008. Analysing georeferenced population genetics data with GENELAND: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics 24: 1406–1407. [DOI] [PubMed] [Google Scholar]
- Hajjar R, Hodgkin T.. 2007. The use of wild relatives for crop improvement: a survey of developments over the past 20 years. Euphytica 156: 1–13. [Google Scholar]
- Hardy OJ, Vekemans X.. 2002. SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 618–620. [Google Scholar]
- Jones AG, Small CM, Paczolt KA, Ratterman NL.. 2010. A practical guide to methods of parentage analysis. Molecular Ecology Res 10: 6–30. [DOI] [PubMed] [Google Scholar]
- Jordano P. 1987. Patterns of mutualistic interactions in pollination and seed dispersal: connectance, dependence asymmetries, and coevolution. American Naturalist 129: 657–677. [Google Scholar]
- Yoruk B, Taskin V.. 2014. Genetic diversity and relationships of wild and cultivated olives in Turkey. Plant Systematics and Evolution 300: 1247–1258. [Google Scholar]
- Klepo T, De la Rosa R, Satovic Z, León L, Belaj A.. 2013. Utility of wild germplasm in olive breeding. Scientia Horticulturae 152: 92–101. [Google Scholar]
- Klepo T, Toumi A, De la Rosa R, León L, Belaj A.. 2014. Agronomic evaluation of seedlings from crosses between the main Spanish olive cultivar ‘Picual’ and two wild olive trees. Journal Horticultural Scientie & Biotechnology 89: 508–512. [Google Scholar]
- Kremer A, Ronce O, Robledo-Arnuncio JJ, et al. 2012. Long‐distance gene flow and adaptation of forest trees to rapid climate change. Ecology Letters 15: 378–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonarduzzi C, Leonardi S, Menozzi P, Piotti A.. 2012. Towards an optimal sampling effort for paternity analysis in forest trees: what do the raw numbers tell us? iForest-Biogeosciences and Forestry 5: 18–25. [Google Scholar]
- Loiselle BA, Sork VL, Nason J, Graham C.. 1995. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). American Journal of Botany 82: 1420–1425. [Google Scholar]
- Lopes MS, Mendoca D, Sefc KM, Sabino Gil F, Da Camara MA.. 2004. Genetic evidence intra-cultivar variability within Iberian olive cultivars. Hortscience 39: 1562–1565. [Google Scholar]
- Lumaret R, Ouazzani N.. 2001. Plant genetics: ancient wild olives in Mediterranean forests. Nature 413: 700–700. [DOI] [PubMed] [Google Scholar]
- Lumaret R, Ouazzani N, Michaud H, Vivier G, Deguilloux MF, Di Giusto F.. 2004. Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin. Heredity 92: 343–351. [DOI] [PubMed] [Google Scholar]
- Marchelli P, Smouse PE, Gallo LA.. 2012. Short-distance pollen dispersal for an outcrossed, wind-pollinated southern beech (Nothofagus nervosa (Phil.) Dim. et Mil.). Tree Genetics & Genomes 8: 1123–1134. [Google Scholar]
- Médail F, Quézel P, Besnard G.. 2001. Systematics, ecology and phylogeographic significance of Olea europaea L. ssp. maroccana (Greuter & Burdet) P.Vargas et al. a relictual olive tree in south-west Morocco. Botanical Journal of the Linnean Society 137: 249–266. [Google Scholar]
- Mookerjee S, Guerin J, Collins G, Ford C, Sedgley M.. 2005. Paternity analysis using microsatellite markers to identify pollen donors in an olive grove. Theoretical and Applied Genetics 111: 1174–1182. [DOI] [PubMed] [Google Scholar]
- Mulas M, Fadda A, Cauli E.. 2004. Prime osservazioni su cloni di oleastro (Olea europaea var. sylvestris Hoff. E Link) selezionati per l’utilizzo forestale. Italus Hortus 4: 214–217. [Google Scholar]
- Muñoz-Diez CM, Trujillo I, Barrio E, Belaj A, Barranco D, Rallo L.. 2011. Centennial olive trees as a reservoir of genetic diversity. Annals of Botany 108: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz Diez CM, Trujillo I, Martinez-Urdiroz N, et al. 2015. Olive domestication and diversification in the Mediterranean Basin. New Phytologist 206: 436–447. [DOI] [PubMed] [Google Scholar]
- Nathan R, Muller-Landau HC.. 2000. Spatial patterns of seed dispersal, their determinants and consequences for recruitment. Tree 15: 278–285. [DOI] [PubMed] [Google Scholar]
- Neigel JE. 1997. A comparison of alternative strategies for estimating gene flow from genetic markers. Annual Review of Ecology and Systematics 28: 105–128. [Google Scholar]
- Nielsen LR, Kjær ED.. 2010. Gene flow and mating patterns in individuals of wych elm (Ulmus glabra) in forest and open land after the influence of Dutch elm disease. Conservation Genetics 11: 257–268. [Google Scholar]
- Noormohammadi Z, Trujillo I, Belaj A, Ataei S, Hosseini-Mazinan M.. 2014. Genetic structure of Iranian olive cultivars and their relationship with Mediterranean’s cultivars revealed by SSR markers. Scientia Horticulturae 178: 175–183. [Google Scholar]
- Oddou-Muratorio S, Klein EK, Austerlitz F.. 2005. Pollen flow in the wild service tree, Sorbus torminalis (L.) Crantz: II. Pollen dispersal and heterogeneity in mating success inferred from parent offspring analysis. Molecular Ecology 14: 4441–4452. [DOI] [PubMed] [Google Scholar]
- Oddou-Muratorio S, Klein EK, Vendramin G, Fady B.. 2011. Spatial vs. temporal effects on demographic and genetic structures: the roles of dispersal, masting and differential mortality on patterns of recruitment in Fagus sylvatica. Molecular Ecology 20: 1997–2010. [DOI] [PubMed] [Google Scholar]
- Perea R, Gutiérrez-Galán A.. 2016. Introducing cultivated trees into the wild: Wood pigeons as dispersers of domestic olive seeds. Acta Oecologica 71: 73–79. [Google Scholar]
- Pinillos V, Cuevas J.. 2009. Open-pollination provides sufficient levels of cross-pollen in spanish monovarietal olive orchards. HortScience 44: 499–502. [Google Scholar]
- Piotti A, Leonardi S, Buiteveld J, et al. 2012. Comparison of pollen gene flow among four European beech (Fagus sylvatica L.) populations characterized by different management regimes. Heredity 108: 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluess AR, Sork VL, Dolan B, et al. 2009. Short distance pollen movement in a wind-pollinated tree, Quercus lobata (Fagaceae). Forest Ecology and Management 258: 735–744. [Google Scholar]
- Raymond M, Rousset F.. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal Heredity 86: 248–249. [Google Scholar]
- Rey PJ, Alcántara JM.. 2000. Recruitment dynamics of a fleshy‐fruited plant (Olea europaea): connecting patterns of seed dispersal to seedling establishment. Journal of Ecology 88: 622–633. [Google Scholar]
- Rey PJ, Alcántara JM, Valera F, et al. 2004. Seedling establishment in Olea europaea: seed size and microhabitat affect growth and survival. Ecoscience 11: 310–320. [Google Scholar]
- Robledo-Arnuncio JJ. 2011. Wind pollination over mesoscale distances: an investigation with Scots pine. New Phytologist 190: 222–233. [DOI] [PubMed] [Google Scholar]
- Robledo-Arnuncio JJ, Gil L.. 2005. Patterns of pollen dispersal in a small population of Pinus sylvestris L. revealed by total-exclusion paternity analysis. Heredity 94:13–22. [DOI] [PubMed] [Google Scholar]
- Robledo-Arnucio JJ, Klein EK, Muller-Landau HC, Santamaria L.. 2014. Space, time and complexity in plant dispersal ecology. Movement Ecology 2:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo J, Orlandi F, Pérez-Badia R, et al. 2016. Modeling olive pollen intensity in the Mediterranean region through analysis of emission sources. Science of the Total Environment 551: 73–82. [DOI] [PubMed] [Google Scholar]
- Rubio De Casas R, Besnard G, Schönswetter P, Balaguer L, Vargas P.. 2006. Extensive gene flow blurs phylogeographic but not phylogenetic signal in Olea europaea L. Theoretical and Applied Genetics 113: 575–583. [DOI] [PubMed] [Google Scholar]
- Sagnard F, Oddou-Muratorio S, Pichot C, Vendramin GG, Fady B.. 2011. Effects of seed dispersal, adult tree and seedling density on the spatial genetic structure of regeneration at fine temporal and spatial scales. Tree Genetics & Genomes 7: 37–48. [Google Scholar]
- Santos-Antunes AR, Léon L, De la Rosa R, et al. 2005. The length of the juvenile period in olive as influenced by vigor of the seedling and the precocity of the parents. HortScience40:1213–1215. [Google Scholar]
- Schuster WS, Mitton JB.. 2000. Paternity and gene dispersal in limber pine (Pinus flexilis James). Heredity 84: 348–361. [DOI] [PubMed] [Google Scholar]
- Sefc KM, Lopes MS, Mendonça D, et al. 2000. Identification of microsatellite loci in olive (Olea europaea L.) and their characterization in Italian and Iberian olive trees. Molecular Ecology 9:1171–1173. [DOI] [PubMed] [Google Scholar]
- Slatkin M. 1985. Gene flow in natural populations. Annual Review of Ecology and Systematics 16: 393–430. [Google Scholar]
- Sokal RR, Rohlf FJ.. 1995. Biometry: principles and practices of statistics in biological research, 3rd edn.New York: W. H. Freeman. [Google Scholar]
- Sork VL, Smouse PE.. 2006. Genetic analysis of landscape connectivity in tree populations. Landscape Ecology 21:821–836. [Google Scholar]
- Sork VL, Nason J, Campbell DR, Fernandez JF.. 1999. Landscape approaches to historical and contemporary gene flow in plants. Trends in Ecology & Evolution 14: 219–224. [DOI] [PubMed] [Google Scholar]
- Sorkheh K, Shiran B, Rouhi V, et al. 2009. Phenotypic diversity within native Iranian almond (Prunus spp.) species and their breeding potential. Genetic Resources and Crop Evolution 56: 947–961. [Google Scholar]
- Spenneman DHR, Allen LR.. 1998. Nathan Cobb’s laboratory conservation and interpretation project. The spread of olives (Olea sp.) on Wagga Wagga Campus. II. Distance, rate and vectors of seed dispersal. The Jonestone Centre. No. 101. Charles Sturt University, Albury, NSW, Australia.
- Spenneman DHR, Allen LR.. 2000. Feral olives (Olea europaea) as future woody weeds in Australia: a review. Australian Journal of Experimental Agriculture 40: 889–901. [Google Scholar]
- Trujillo I, Ojeda MA, Urdiroz NM, et al. 2014. Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) using SSR and morphological markers. Tree Genetic & Genomes 10: 141–155. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P.. 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538. [Google Scholar]
- Vekemans X, Hardy OJ.. 2004. New insights from fine-scale spatial genetic structure analyses in plant populations. Molecular Ecology 13: 921–935. [DOI] [PubMed] [Google Scholar]
- Villemur P, Musho US, Delmas JM, Maamar M, Ouksili A.. 1984. Contribution à l’étude de la biologie florale de l’olivier (Olea europaea L.): stérilité mâle, flux pollinique et période effective de pollinisation. Fruits 39: 467–473. [Google Scholar]
- Zohary D, Hopf M.. 1994. Domestication of plants in the Old World. Oxford: Clarendon Press. [Google Scholar]
- Zohary D, Spiegel-Roy P.. 1975. Beginnings of fruit growing in the Old World. Science 187: 319–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.