Abstract
Recoverin is a neuronal calcium sensor involved in vision adaptation that reversibly associates with cellular membranes via its calcium-activated myristoyl switch. While experimental evidence shows that the myristoyl group significantly enhances membrane affinity of this protein, molecular details of the binding process are still under debate. Here, we present results of extensive molecular dynamics simulations of recoverin in the proximity of a phospholipid bilayer. We capture multiple events of spontaneous membrane insertion of the myristoyl moiety and confirm its critical role in the membrane binding. Moreover, we observe that the binding strongly depends on the conformation of the N-terminal domain. We propose that a suitable conformation of the N-terminal domain can be stabilized by the disordered C-terminal segment or by binding of the target enzyme, i.e., rhodopsin kinase. Finally, we find that the presence of negatively charged lipids in the bilayer stabilizes a physiologically functional orientation of the membrane-bound recoverin.
Short abstract
Molecular dynamics simulations provide atomistic insights into the reversible membrane binding of recoverin, demonstrating the key role of the myristoyl moiety for the membrane association.
Introduction
Myristoylation represents a lipidation modification occurring in numerous proteins involved in intracellular signaling.1 A hydrophobic myristoyl moiety, covalently attached to the N-terminus of a signaling protein, can enhance the affinity of the protein for cellular membranes and thus contribute to its membrane targeting.2 This type of targeting can be regulated in a reversible way by modulating the exposure of the myristoyl lipid chain at the protein surface. In proteins featuring a myristoyl switch, such modulation is accomplished by conformational changes triggered by ligand binding.2
Recoverin, which represents a member of the neuronal calcium sensor (NCS) family, is a 23 kDa protein possessing a calcium-activated myristoyl switch. Recoverin is primarily expressed in photoreceptor cells of the vertebrate retina, where it participates in light adaptation via calcium-dependent inhibition of the enzyme rhodopsin kinase (RK).3−6 Apart from an N-terminal myristoyl chain, the molecular structure of recoverin contains four evolutionarily conserved helix–loop–helix motifs called the EF hands, two of which can coordinate Ca2+ (Figure 1). Solution NMR structures of recoverin exist for both low and high concentrations of calcium ions.7,8 They reveal that at resting intracellular calcium concentrations, the myristoyl moiety is sequestered in a hydrophobic cavity inside the N-terminal domain (Figure 1, left). However, when the cytoplasmic concentration of calcium increases, the second and the third EF hand (EF2 and EF3) both bind a Ca2+ ion, and the protein undergoes a structural rearrangement of its two domains. The conformational transition exposes the myristoyl group and also creates a binding pocket for RK (Figure 1, right). This allows the calcium-activated recoverin to reversibly associate with rod outer segment (ROS) disk membranes and inhibit RK.
Figure 1.
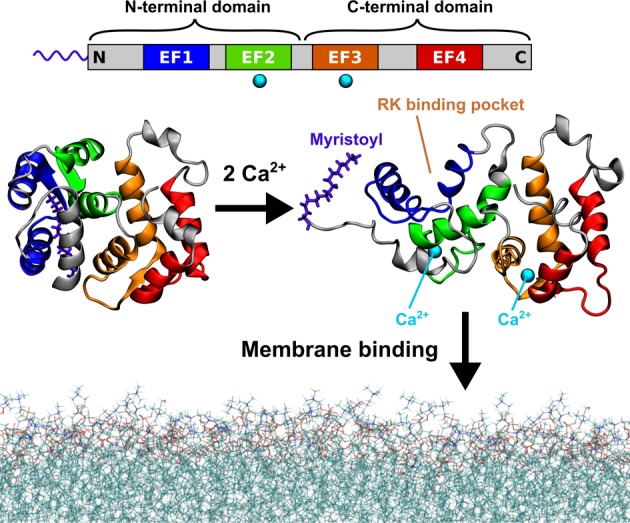
Calcium-activated myristoyl switch of recoverin. At low intracellular concentrations of calcium ions, the myristoyl group of recoverin is hidden inside the N-terminal domain of the protein (left). When the concentration of calcium rises, two of the four evolutionarily conserved EF hand motifs (top) each bind a calcium ion and the protein undergoes a conformational transition exposing the myristoyl group, as well as opening up a binding site for RK (right). The calcium-loaded recoverin is capable of reversible membrane binding to rod outer segment (ROS) disk membranes. The protein structures shown in this figure were determined by solution NMR7,8 (PDB IDs 1IKU and 1JSA). The last 13 C-terminal amino acid residues are missing from the structures as their geometry was not resolved in the NMR experiments.
While the myristoyl moiety was shown to promote the membrane binding of recoverin,9,10 its membrane insertion was inferred only indirectly from experimental data.11 Moreover, the contribution of other factors to the reversible membrane association of recoverin still remains to be clarified. In particular, the role of protein–membrane electrostatic interactions, which are known to participate in the membrane binding of other N-myristoylated proteins,12,13 has been discussed repeatedly in the literature.11,14,15 Recent experiments on monolayers suggest that the calcium-dependent membrane binding of recoverin is assisted by the presence of negatively charged lipids in the membrane.15 Apart from a patch of basic residues located near the N-terminus, the highly charged C-terminal segment of recoverin may also be involved in the electrostatic interaction with the lipid bilayer as proposed for S-modulin, which is a frog homologue of recoverin.16 This 13-residue C-terminal segment, containing six lysines and two glutamates, forms a short helix in the crystal structure of non-myristoylated recoverin,17 but its geometry has not been resolved in the solution NMR structures.7,8 Although more recent studies did not directly confirm the membrane interaction of the C-terminus,11,14,18 they showed that its removal affects the conformation of recoverin18 and the affinity of the sensor for calcium.18 The precise function of the charged C-terminus and whether it modulates the membrane association thus remain unclear.
In order to elucidate the mechanistic details of the process of membrane anchoring via the myristoyl group, we perform here extensive atomistic and coarse-grained molecular dynamics (MD) simulations of calcium-loaded recoverin in the proximity of phospholipid bilayers with varying lipid compositions. These simulations allow us to establish the key factors governing successful membrane insertion of the myristoyl moiety and decide about relative contributions of hydrophobic and electrostatic interactions to the reversible membrane binding of the protein. This work thus preconditions a detailed molecular understanding of the biological functionality of recoverin.
Results and Discussion
To gain atomistic insight into the membrane binding of recoverin, we performed all-atom MD simulations of a recoverin molecule in the proximity of a phospholipid bilayer. In most of our all-atom simulations, the lipid composition was chosen to be 80% dioleoylphosphatidylcholine (PC) and 20% dioleoylphosphatidylglycerol (PG) to allow for a direct comparison with previous solid-state NMR measurements.11 Moreover, the presence of 20% of negatively charged PG lipids mimics the charge distribution found in ROS disk membranes.19 In total, we performed 22 all-atom MD simulations, including several trajectories with a PC-only or a PG-only membrane (see Table S1). In addition, we conducted coarse-grained MD simulations of recoverin in the proximity of PC:PG (4:1), PC-only, and PG-only membranes, with at least three trajectories obtained for each of the membrane compositions (Table S2). All simulations were started with the N-terminal domain of recoverin facing initially the membrane surface, but without any direct contact of the amino acid residues with the lipids (i.e., with a minimum protein–membrane distance of 1 nm).
Mechanism of Myristoyl Insertion and Its Energetics
Our all-atom simulations captured multiple events of a spontaneous membrane insertion of the myristoyl moiety (see Table S1). In all these cases, after initial reorientation, recoverin approached the lipid bilayer with its N-terminal domain before myristoyl insertion. Once the methyl end of the myristoyl chain reached a suitable gap between lipid head groups, it penetrated the carbonyl region of the bilayer and became accommodated among the acyl chains of the surrounding phospholipids (Figure 2A). This process of membrane penetration took 2–9 ns (Figure S1) in the individual runs. During the process of myristoyl membrane embedding, the basic residues K5 and K37 interacted with the bilayer, while the positively charged C-terminus stayed away from the membrane (Figure S1). After recoverin became anchored to the membrane by the myristoyl chain, its orientation stabilized by the interaction of positively charged amino acid residues with negatively charged PG lipids, which were locally enriched around these residues (Figure S2). Some of the basic amino acid residues even penetrated into the lipid headgroup region (Figure 2B). Most importantly, once inserted, the myristoyl group remained membrane-embedded for the 1 μs duration of the simulations (see Figure 2B).
Figure 2.
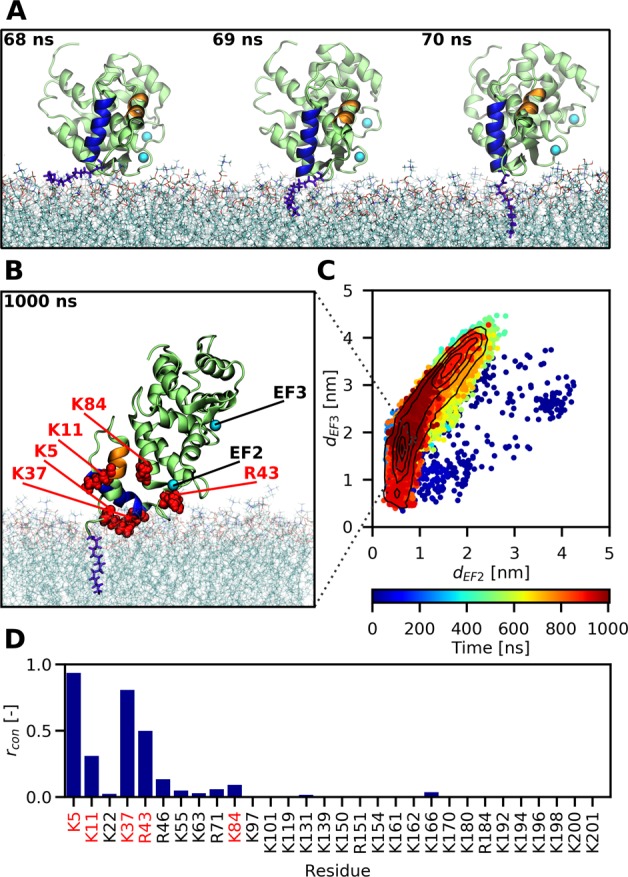
All-atom MD simulations reveal how the myristoyl group anchors recoverin to a PC:PG (4:1) membrane. (A) Snapshots capturing the fast process of myristoyl insertion. The myristoyl moiety is displayed in blue-violet, while the two N-terminal helices A and B of recoverin are highlighted in orange and blue, respectively. (B) Snapshot obtained at the end of a 1 μs trajectory, showing the membrane-embedded myristoyl group (violet) and five positively charged residues (red) reported by previous NMR measurements11 to interact with the membrane. (C) Membrane orientation of recoverin during the course of a 1 μs trajectory described in terms of the distances of the two calcium ions to the membrane. On average, the protein exhibits a tilted orientation toward the lipid bilayer, with one calcium closer to the membrane surface than the other one. Importantly for the biological function of recoverin, the binding pocket for RK remains accessible during the trajectory. (D) Relative proportions rcon of simulation time that each of the basic residues of recoverin spent in contact with the membrane, i.e., at a distance <0.6 nm (for details see Table S1).
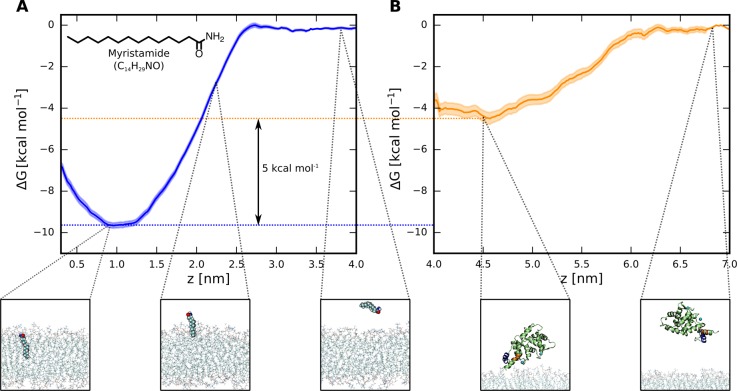
To assess the importance of the myristoyl anchor for the membrane binding of recoverin, we deleted the myristoyl group from the final snapshot of a trajectory featuring myristoyl insertion (for details see Table S1) and continued the simulation. We found that, without the myristoyl anchor, the orientation of recoverin relative to the membrane became destabilized, and ultimately, recoverin detached itself completely from the membrane surface (see Figure S3). This result demonstrates that, in agreement with experimental results,9,10 a myristoyl moiety is required for stable membrane binding. To further quantify the free energy gain resulting from myristoyl insertion, we employed the method of umbrella sampling to calculate the free energy profile of a myristamide molecule (as the closest proxy to the myristoyl moiety in recoverin) penetrating a PC:PG (4:1) bilayer. The resulting profile (Figure 3A) shows a free-energy preference of about 10 kcal mol–1 for the membrane over the aqueous phase, which is consistent with previous estimates based on experimental data.20,21 Importantly, our additional umbrella sampling simulations of the membrane detachment of a non-myristoylated recoverin indicate that the remaining binding free energy of the protein to the membrane is significantly smaller, equaling ∼4 kcal mol–1 (Figure 3B). Taken together, these results underscore the crucial importance of the myristoyl moiety for the membrane association of recoverin.
Figure 3.
(A) Free energy profile of myristamide insertion from water into a PC:PG (4:1) membrane, showing the dependence of the free energy on the distance of the center of mass of myristamide from the central plane of the bilayer. The robustness of this result was verified with respect to the choice of the force field and the set of initial geometries (Figure S4). (B) Free energy profile of the membrane detachment of a non-myristoylated recoverin, calculated along the distance between the center of mass of the protein and the central plane of the bilayer.
Exposure of Myristoyl Needed for Anchoring
We observed membrane insertion of the myristoyl moiety for several but not all of the trajectories. In several cases, recoverin approached the lipid bilayer with an unfavorable orientation, interacting with lipid head groups via its positively charged C-terminus (Figure S5). This orientation brought the myristoyl anchor in a position away from the membrane and also caused the binding site for RK to be blocked by the lipids. Our coarse-grained simulations (vide infra) indicate that such a geometry represents a shallow local minimum; nevertheless, it persisted on the time scales accessible to our all-atom MD simulations.
For yet other trajectories, membrane insertion of the myristoyl moiety did not occur even with a favorable orientation of recoverin, which approached the membrane with its N-terminal domain. We observed that this situation occurred both for the PC:PG (4:1) bilayer and for simulations with pure PC and pure PG bilayers (see Table S1 and Figure S6 for more details). A closer look at the protein structure in these trajectories reveals that the hydrophobic myristoyl moiety moved in between the amino acid side chains and became shielded from the polar water environment. Moreover, the move of the myristoyl group was accompanied by structural changes in the N-terminal domain, affecting the conformation of two N-terminal helices A and B (Figure S7). In particular, the length of the helix A changed as well as the angle between helices A and B, and helix B became shifted toward the center of the protein, narrowing the binding pocket for RK.
In contrast, in trajectories featuring myristoyl insertion, a close-to-perpendicular relative orientation of helices A and B kept the myristoyl moiety away from hydrophobic protein residues and maintained the exposure of the myristoyl anchor to the polar aqueous environment (Figures S7 and S8). The conformation of these two helices thus has to be stable enough to resist the tendency of the hydrophobic myristoyl anchor to return to the hydrophobic core of the protein. At the same time, the two helices, forming an important part of the myristoyl switch, have to be sufficiently flexible to allow for the motion of the myristoyl moiety out of the protein core during the calcium-driven conformational transition. This subtle balance between stability and flexibility may be a critical feature determining the efficacy of the calcium–myristoyl switch. In this respect, it is notable that our simulations suggest that a subset of the calcium-loaded structures of recoverin exists in a state structurally similar to the available set of NMR structures (PDB ID 1JSA), but with their myristoyl anchors sticking to hydrophobic residues of the protein and, therefore, being incapable of membrane insertion. Overall, our simulations show that myristoyl insertion occurs as long as two conditions are fulfilled: (1) a favorable orientation of recoverin with respect to the membrane and (2) exposure of the myristoyl group to the aqueous environment, maintained by the conformation of the N-terminal domain.
Rhodopsin Kinase Stabilizes the N-Terminus
The observation that the shielding of the myristoyl group from water was associated with conformational changes in the N-terminus motivated us to explore possible factors enhancing the stability of an N-terminal conformation that would favor myristoyl exposure and subsequent membrane insertion. Since a significant aspect of the conformational changes was the displacement of helix B in the direction of closing the RK binding pocket, we tested whether a ligand filling the binding pocket could prevent this motion and thus stabilize the geometry of the N-terminus. Inspired by ref (6), suggesting that recoverin may bind RK in the cytoplasm before associating with the membrane, we performed three simulations with a 16-residue fragment of RK present in the RK binding site (see Table S1). Indeed, the presence of the RK fragment increased the conformational stability of the N-terminal domain, with the Cα root-mean-square deviation of residues 2–97 (N-terminal domain) from the initial NMR structure equaling 0.31 nm, as contrasted to 0.48 nm in a run without RK (for more details see the Supporting Information). Consequently, the myristoyl group remained extruded to the aqueous environment, and as soon as it was brought to an immediate vicinity of the lipid bilayer by a favorable orientation of the protein, it spontaneously penetrated the membrane and stayed there (Figure S9). This result suggests that the cytoplasmic prebinding of RK accelerates the subsequent membrane association of the recoverin–RK complex.
Orientation of Membrane-Anchored Recoverin
The orientation of recoverin on a PC:PG (4:1) bilayer was inferred previously from the fitting of solid-state NMR spectra,11 with the conclusion that recoverin predominantly assumes a tilted orientation relative to the membrane. Such an orientation brings the EF2-bound calcium close to the bilayer and the EF3-bound calcium farther away. Moreover, the solid-state NMR measurements identified five basic residues of the N-terminal domain, K5, K11, K37, R43, and K84, as being in contact with the bilayer. In comparison, our all-atom MD simulations provided a more dynamic picture of the membrane-anchored protein, which adopts a relatively broad range of orientations toward the membrane (Figure 2C). We found that the tilted orientation of the protein predicted by NMR was indeed prominent (Figure 2B,C); nevertheless, we also observed more parallel orientations of the protein relative to the bilayer, with both calcium-loaded EF hands interacting with lipid head groups (Figure S10). Importantly, we confirmed that the positively charged residues K5, K11, K37, R43, and K84 identified by NMR were in frequent contact with the bilayer during the simulations (Figures 2D and S10) and that the binding site for RK was not blocked by the lipids.
To further examine how the negative charge in the membrane modulates recoverin–membrane interactions, we performed coarse-grained MD simulations of the anchoring of recoverin to membranes with three different lipid compositions (see Table S2). The use of the coarse-grained MARTINI model22 allowed us to reach simulation times needed for exploring all possible orientations of recoverin at the membrane, at the expense of sacrificing atomistic details and the possibility to explore changes in the protein structure. The membrane was formed by a pure PC bilayer, a PC:PG (4:1) bilayer, or a pure PG bilayer, ranging from electroneutral to weakly or highly negatively charged systems (Figure S11). Irrespective of the membrane composition, we always observed a spontaneous membrane insertion of the myristoyl moiety. This may have been facilitated by the fact that the structure of the protein was fixed in the coarse-grained model (see Methods) and by the relatively smooth energy landscape pertinent to the coarse-grained description.22 Nevertheless, the membrane binding occurred significantly more slowly for the pure PC bilayer than for the negatively charged ones (see Figure S12). Intriguingly, the successful anchoring to the PC-only membrane did not result in any stable orientation of recoverin relative to the bilayer (Figure 4). As a consequence, the binding site for RK was often blocked by the lipids. The presence of negatively charged PG molecules in the membrane significantly accelerated the anchoring of recoverin to the bilayer (Figure S12) since the interaction of its negative charge with the patch of positively charged amino acid residues near the N-terminus allowed recoverin to faster adopt orientations favorable for myristoyl insertion. In the case of the mixed membrane (PC:PG, 4:1), the membrane-anchored recoverin adopted predominantly a tilted orientation toward the membrane, with the binding site for RK remaining accessible (Figure 4). This orientation agreed with results obtained from atomistic simulations (Figure 2). Note that this orientational effect is somewhat attenuated, albeit still present when the electrostatic screening is enhanced via employing polarizable MARTINI force field (see Figure S15). Interestingly, in the case of the PG-only membrane, we also occasionally found an interaction of the C-terminal part with PG molecules (Figure 4), but this interaction had only a transient character. This finding supports our hypothesis that the geometry resulting from interactions mediated by the positively charged C-terminus represents only a shallow local minimum. Altogether, the results of our coarse-grained MD simulations point to an active role of negatively charged phospholipids in the fine-tuning of the orientation of recoverin toward the membrane. Lipid composition thus may play an important role in regulating the physiological action of recoverin, i.e., binding of RK. A similar active role of negatively charged phospholipids was found recently for other peripheral membrane proteins, such as the yeast small GTPases Rho1p, human focal adhesion kinase, and cytochrome P450.23−25
Figure 4.
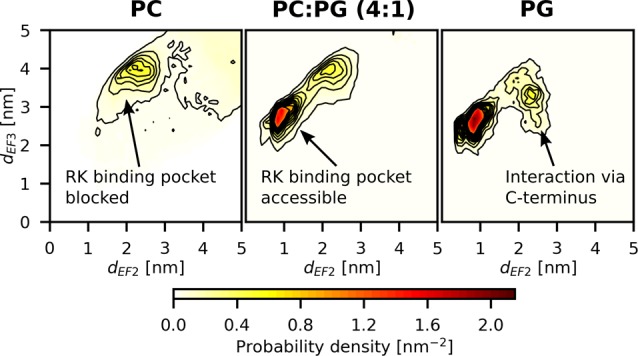
Probability density of membrane orientation of recoverin expressed in terms of the distances of the two calcium ions from the membrane. Each plot represents an average of three 5 μs coarse-grained MD trajectories (see Table S2). The successful anchoring to the PC-only membrane did not result in a fixed orientation of recoverin relative to the bilayer, and the binding site for RK frequently became blocked by the lipids. For the mixed membrane (PC:PG, 4:1), recoverin adopted a tilted orientation toward the membrane. In the PG-only membrane, the C-terminal part also interacted with PG molecules, but this interaction only occurred occasionally and had a transient character.
Role of the Charged C-Terminus
The structure of the highly charged 13-residue C-terminal segment has not been resolved by solution NMR.8 Therefore, we modeled the initial geometry of the C-terminus by adopting it from a crystal structure17 of a non-myristoylated recoverin (PDB ID 1OMR, see Methods), where it forms a short helix (see Figure S13). Throughout our atomistic simulations, the helical structure of the C-terminus was essentially preserved. As mentioned above, owing to the high density of positively charged residues, we frequently observed that recoverin interacted with the membrane via the C-terminus, resulting in a protein orientation parallel with the bilayer, which blocked the binding site for RK. Our coarse-grained simulations provide a strong support for the hypothesis that this mode of membrane interaction is a transient local minimum only. To find out how removing the C-terminus would affect the overall orientation of the protein we performed additional atomistic simulations (Table S1) with the last 13 C-terminal residues deleted. We found that the removal of the C-terminal segment eliminated the unfavorable orientation which moved the myristoyl anchor away from the membrane, but at the same time, it also prolonged the time recoverin spent in solution before landing at the membrane. Moreover, in some of the simulations (for details see the Supporting Information), removing the C-terminus altered the conformation of the loop connecting the two domains of recoverin, resulting in a mutual displacement of the two central helices and a change in the relative orientation of the two domains. This is in line with previous results for another member of the NCS protein family, the NCS-1 protein, for which the deletion of the C-terminal segment increased the structural flexibility of the protein and weakened the interdomain correlation.26
Importantly, the NMR data show that the C-terminus is disordered, i.e., it is likely to adopt a number of conformations different from that observed in crystals. We suggest that some of these more unstructured conformations may transiently fill the binding pocket for RK and thereby stabilize the N-terminal domain. A similar stabilizing effect was already predicted for the C-terminus of the related NCS-1 protein.27 In this way, the C-terminus may prolong the time that the myristoyl moiety spends exposed to the aqueous phase and thus increase the odds of successful membrane anchoring of recoverin.
Conclusion
In the present study, we elucidated with atomistic detail the process of membrane insertion of recoverin at varying conditions. Our simulations not only relate directly to experiments, whenever the latter are available, but also provide experimentally hardly accessible interpretation of the membrane embedding process of recoverin in terms of the underlying molecular interactions. The key results of this work are 3-fold:
Direct MD simulations of the protein with and without the myristoyl anchor demonstrate that it is the critical determinant for efficient membrane binding of recoverin. The physiological functionality of the myristoyl switch stems from a subtle balance between its stability and flexibility.
Approaching the membrane, recoverin samples a relatively broad distribution of geometries and orientations. Successful membrane insertion of myristoyl requires, however, a rather specific conformation of the N-terminus.
Simulations show that protein–membrane electrostatic interactions stabilize the biologically functional orientation (i.e., that allowing for efficient binding of RK) of recoverin after membrane anchoring.
In summary, this study presents carefully designed and benchmarked molecular simulations that allow us to elucidate the mechanism of membrane insertion of recoverin, ultimately leading us to understanding of the key factors for its biological functionality.
Methods
Atomistic MD simulations were performed using the GROMACS 5.1.2 package.28 The simulation box contained one calcium-loaded recoverin molecule, a bilayer comprising 190 phospholipid molecules, sufficiently large to avoid the interaction of recoverin with its periodic images, and a 150 mM KCl aqueous solution containing ∼20,000 water molecules, with extra K+ ions added to neutralize the system. The lipid species used in our simulations were dioleoylphosphatidylcholine (PC) and dioleoylphosphatidylglycerol (PG). The phospholipid bilayers were assembled and hydrated by employing the CHARMM-GUI membrane builder.29−31 The structure of calcium-loaded recoverin was obtained from an ensemble of 24 NMR structures of myristoylated bovine recoverin8 (PDB ID 1JSA, structures no. 1 and 8). Since the last 13 C-terminal residues were not resolved by the NMR measurements, we transferred the C-terminal segment (residues 185–202) from an X-ray crystal structure of non-myristoylated recoverin17 (PDB ID 1OMR). Recoverin was inserted approximately 1 nm above the surface of the bilayer so that its residues were not in contact with the lipid head groups. The initial orientation of the protein relative to the membrane was close to perpendicular, with the N-terminus facing the bilayer. Prior to inserting recoverin to the vicinity of the bilayer, we pre-equilibrated the structures of both the protein and the membrane separately in aqueous solutions. To ensure robustness of the results, we performed our atomistic MD simulations with two different sets of force field parameters: CHARMM and AMBER/Slipids. In the former case, we used the CHARMM36 force field32 to parametrize the lipids and CHARMM22/CMAP33,34 to describe the protein. In the latter case, the AMBER ff99SB-ILDN force field35 was used for the protein, while the lipids were described with the AMBER-compatible Slipids force field.36−38 In both CHARMM and AMBER simulations, water molecules were described with the TIP3P model.39 Free energy profiles characterizing membrane insertion of myristamide and membrane interaction of non-myristoylated recoverin were obtained for the CHARMM force field from umbrella sampling simulations by employing the weighted histogram analysis (WHAM) method.40 Coarse-grained simulations presented in this article utilized the MARTINI model41 and were performed in GROMACS 4.6.5.42 We used the VMD program43 to visualize the system and prepare figures. For both the atomistic and coarse-grained simulations, additional details on the simulation setup, force field parameters, system building, and structure equilibration can be found in the Supporting Information.
Acknowledgments
P.J. acknowledges support from the Czech Science Foundation (Grant No. P208/12/G016) and from the Academy of Finland via the FiDiPro award. This work was supported by The Ministry of Education, Youth and Sports from the Large Infrastructures for Research, Experimental Development and Innovations project “IT4Innovations National Supercomputing Center—LM2015070”. Access to computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum provided under the program “Projects of Large Research, Development, and Innovations Infrastructures” (CESNET LM2015042) is greatly appreciated.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00210.
Further details on computational methods and additional analysis of MD trajectories (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Farazi T. A.; Waksman G.; Gordon J. I. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 2001, 276 (43), 39501–39504. 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- Resh M. D. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2006, 2 (11), 584–590. 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- Burgoyne R. D. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 2007, 8 (3), 182–193. 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D.; O'Callaghan D. W.; Hasdemir B.; Haynes L. P.; Tepikin A. V. Neuronal Ca2+-sensor proteins: multitalented regulators of neuronal function. Trends Neurosci. 2004, 27 (4), 203–209. 10.1016/j.tins.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Burgoyne R. D. The neuronal calcium-sensor proteins. Biochim. Biophys. Acta, Mol. Cell Res. 2004, 1742, 59–68. 10.1016/j.bbamcr.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Ames J. B.; Lim S. Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim. Biophys. Acta, Gen. Subj. 2012, 1820 (8), 1205–1213. 10.1016/j.bbagen.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.; Ames J. B.; Harvey T. S.; Stryer L.; Ikura M. Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 1995, 376 (6539), 444–447. 10.1038/376444a0. [DOI] [PubMed] [Google Scholar]
- Ames J. B.; Ishima R.; Tanaka T.; Gordon J. I.; Stryer L.; Ikura M. Molecular mechanics of calcium–myristoyl switches. Nature 1997, 389 (6647), 198–202. 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- Desmeules P.; Grandbois M.; Bondarenko V. A.; Yamazaki A.; Salesse C. Measurement of Membrane Binding between Recoverin, a Calcium-Myristoyl Switch Protein, and Lipid Bilayers by AFM-Based Force Spectroscopy. Biophys. J. 2002, 82 (6), 3343–3350. 10.1016/S0006-3495(02)75674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmeules P.; Penney S.-É.; Desbat B.; Salesse C. Determination of the Contribution of the Myristoyl Group and Hydrophobic Amino Acids of Recoverin on its Dynamics of Binding to Lipid Monolayers. Biophys. J. 2007, 93 (6), 2069–2082. 10.1529/biophysj.106.103481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine K. G.; Mesleh M. F.; Opella S. J.; Ikura M.; Ames J. B. Structure, topology, and dynamics of myristoylated recoverin bound to phospholipid bilayers. Biochemistry 2003, 42 (21), 6333–6340. 10.1021/bi0206816. [DOI] [PubMed] [Google Scholar]
- McLaughlin S.; Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem. Sci. 1995, 20 (7), 272–276. 10.1016/S0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- Murray D.; Ben-Tal N.; Honig B.; McLaughlin S. Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure 1997, 5 (8), 985–989. 10.1016/S0969-2126(97)00251-7. [DOI] [PubMed] [Google Scholar]
- Senin I. I.; Churumova V. A.; Philippov P. P.; Koch K.-W. Membrane binding of the neuronal calcium sensor recoverin – modulatory role of the charged carboxy-terminus. BMC Biochem. 2007, 8, 24. 10.1186/1471-2091-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvez P.; Schmidt T. F.; Cantin L.; Klinker K.; Salesse C. Phosphatidylserine allows observation of the calcium-myristoyl switch of recoverin and its preferential binding. J. Am. Chem. Soc. 2016, 138 (41), 13533–13540. 10.1021/jacs.6b04218. [DOI] [PubMed] [Google Scholar]
- Matsuda S.; Hisatomi O.; Tokunaga F. Role of Carboxyl-Terminal Charges on S-Modulin Membrane Affinity and Inhibition of Rhodopsin Phosphorylation. Biochemistry 1999, 38 (4), 1310–1315. 10.1021/bi982117u. [DOI] [PubMed] [Google Scholar]
- Weiergraber O. H.; Senin I. I.; Philippov P. P.; Granzin J.; Koch K. W. Impact of N-terminal myristoylation on the Ca2+-dependent conformational transition in recoverin. J. Biol. Chem. 2003, 278 (25), 22972–22979. 10.1074/jbc.M300447200. [DOI] [PubMed] [Google Scholar]
- Weiergraber O. H.; Senin I. I.; Zernii E. Y.; Churumova V. A.; Kovaleva N. A.; Nazipova A. A.; Permyakov S. E.; Permyakov E. A.; Philippov P. P.; Granzin J.; Koch K. W. Tuning of a neuronal calcium sensor. J. Biol. Chem. 2006, 281 (49), 37594–37602. 10.1074/jbc.M603700200. [DOI] [PubMed] [Google Scholar]
- Wu G.; Hubbell W. L. Phospholipid asymmetry and transmembrane diffusion in photoreceptor disc membranes. Biochemistry 1993, 32 (3), 879–88. 10.1021/bi00054a020. [DOI] [PubMed] [Google Scholar]
- Pool C. T.; Thompson T. E. Chain length and temperature dependence of the reversible association of model acylated proteins with lipid bilayers. Biochemistry 1998, 37 (28), 10246–10255. 10.1021/bi980385m. [DOI] [PubMed] [Google Scholar]
- Peitzsch R. M.; McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry 1993, 32 (39), 10436–43. 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; Tieleman D. P. Perspective on the Martini model. Chem. Soc. Rev. 2013, 42 (16), 6801–6822. 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- Pleskot R.; Cwiklik L.; Jungwirth P.; Zarsky V.; Potocky M. Membrane targeting of the yeast exocyst complex. Biochim. Biophys. Acta, Biomembr. 2015, 1848 (7), 1481–1489. 10.1016/j.bbamem.2015.03.026. [DOI] [PubMed] [Google Scholar]
- Herzog F. A.; Braun L.; Schoen I.; Vogel V. Structural Insights How PIP2 Imposes Preferred Binding Orientations of FAK at Lipid Membranes. J. Phys. Chem. B 2017, 121 (15), 3523–3535. 10.1021/acs.jpcb.6b09349. [DOI] [PubMed] [Google Scholar]
- Navratilova V.; Paloncyova M.; Berka K.; Otyepka M. Effect of Lipid Charge on Membrane Immersion of Cytochrome P450 3A4. J. Phys. Chem. B 2016, 120 (43), 11205–11213. 10.1021/acs.jpcb.6b10108. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Yang S.; Qi R.; Zou Y.; Ma B.; Nussinov R.; Zhang Q. Effects of the C-Terminal Tail on the Conformational Dynamics of Human Neuronal Calcium Sensor-1 Protein. J. Phys. Chem. B 2015, 119, 14236–14244. 10.1021/acs.jpcb.5b07962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellucci L.; Corni S.; Felice R. D.; Paci E. The Structure of Neuronal Calcium Sensor-1 in Solution Revealed by Molecular Dynamics Simulations. PLoS One 2013, 8 (9), e74383. 10.1371/journal.pone.0074383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Jo S.; Kim T.; Iyer V. G.; Im W. Software news and updates - CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29 (11), 1859–1865. 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Jo S.; Lim J. B.; Klauda J. B.; Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009, 97 (1), 50–58. 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E. L.; Cheng X.; Jo S.; Rui H.; Song K. C.; Davila-Contreras E. M.; Qi Y. F.; Lee J. M.; Monje-Galvan V.; Venable R. M.; Klauda J. B.; Im W. CHARMM-GUI Membrane Builder Toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35 (27), 1997–2004. 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda J. B.; Venable R. M.; Freites J. A.; O’Connor J. W.; Tobias D. J.; Mondragon-Ramirez C.; Vorobyov I.; MacKerell A. D.; Pastor R. W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114 (23), 7830–7843. 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A. D.; Bashford D.; Bellott M.; Dunbrack R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; Joseph-McCarthy D.; Kuchnir L.; Kuczera K.; Lau F. T. K.; Mattos C.; Michnick S.; Ngo T.; Nguyen D. T.; Prodhom B.; Reiher W. E.; Roux B.; Schlenkrich M.; Smith J. C.; Stote R.; Straub J.; Watanabe M.; Wiorkiewicz-Kuczera J.; Yin D.; Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102 (18), 3586–3616. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- MacKerell A. D.; Feig M.; Brooks C. L. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004, 25 (11), 1400–1415. 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K.; Piana S.; Palmo K.; Maragakis P.; Klepeis J. L.; Dror R. O.; Shaw D. E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct., Funct., Genet. 2010, 78 (8), 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambeck J. P. M.; Lyubartsev A. P. Derivation and Systematic Validation of a Refined All-Atom Force Field for Phosphatidylcholine Lipids. J. Phys. Chem. B 2012, 116 (10), 3164–3179. 10.1021/jp212503e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambeck J. P. M.; Lyubartsev A. P. An Extension and Further Validation of an All-Atomistic Force Field for Biological Membranes. J. Chem. Theory Comput. 2012, 8 (8), 2938–2948. 10.1021/ct300342n. [DOI] [PubMed] [Google Scholar]
- Jambeck J. P. M.; Lyubartsev A. P. Another Piece of the Membrane Puzzle: Extending Slipids Further. J. Chem. Theory Comput. 2013, 9 (1), 774–784. 10.1021/ct300777p. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79 (2), 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Kumar S.; Bouzida D.; Swendsen R. H.; Kollman P. A.; Rosenberg J. M. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. 1. The Method. J. Comput. Chem. 1992, 13 (8), 1011–1021. 10.1002/jcc.540130812. [DOI] [Google Scholar]
- Monticelli L.; Kandasamy S. K.; Periole X.; Larson R. G.; Tieleman D. P.; Marrink S. J. The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theory Comput. 2008, 4 (5), 819–834. 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4 (3), 435–447. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics 1996, 14 (1), 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.