Abstract
This study sought to evaluate the potential of circulating long non‐coding RNAs (lncRNAs) as biomarkers for heart failure (HF). We measured the circulating levels of 13 individual lncRNAs which are known to be relevant to cardiovascular disease in the plasma samples from 72 HF patients and 60 non‐HF control participants using real‐time reverse transcription‐polymerase chain reaction (real‐time RT‐PCR) methods. We found that out of the 13 lncRNAs tested, non‐coding repressor of NFAT (NRON) and myosin heavy‐chain‐associated RNA transcripts (MHRT) had significantly higher plasma levels in HF than in non‐HF subjects: 3.17 ± 0.30 versus 1.0 ± 0.07 for NRON (P < 0.0001) and 1.66 ± 0.14 versus 1.0 ± 0.12 for MHRT (P < 0.0001). The area under the ROC curve was 0.865 for NRON and 0.702 for MHRT. Univariate and multivariate analyses identified NRON and MHRT as independent predictors for HF. Spearman's rank correlation analysis showed that NRON was negatively correlated with HDL and positively correlated with LDH, whereas MHRT was positively correlated with AST and LDH. Hence, elevation of circulating NRON and MHRT predicts HF and may be considered as novel biomarkers of HF.
Keywords: heart failure, LncRNA, NRON, MHRT, plasma
Introduction
HF is a major public health problem afflicting a large population (>25 million patients) in the world 1 and an intricate pathophysiological syndrome consequent to feeble cardiac contraction and inadequate blood ejection 2. The clinical manifestations of HF mainly arise from myocardial infarction (MI), hypertension, myocarditis and inherited cardiomyopathy 3, 4. Without successful intervention within a certain timeframe, HF can cause sudden cardiac death or severe disability, being the most devastating cardiovascular disease in terms of mortality, morbidity and the quality of life. One of the difficulties for timely treatment of HF is our current dearth of sensitive and specific biomarkers for early diagnosis of the malady. A number of clinically validated biomarkers such as cardiac troponin, natriuretic peptide, B‐type natriuretic peptide (BNP) and N‐terminal proBNP (NT‐proBNP) have been used in the diagnosis of HF 5, 6, 7, 8, 9, 10. Nonetheless, these traditional biomarkers have some limitations in defining the aetiology or prognosis of HF 5, 6, 7. For example, none of these markers are specific to HF, but their serum/plasma levels can rise in a number of other diseases such as cardiopulmonary disease, kidney failure and hepatic cirrhosis. Quest for more reliable biomarkers is therefore highly desirable. It is known that aberrant changes in the expression of multiple genes in myocardium are a major cause, as well as useful predictors of the pathologic remodelling in failing heart. Identification of such genes, particularly those that are highly sensitive and specific to HF, may be the key step towards reliable early prediction of HF.
Non‐coding RNAs (ncRNAs), including microRNAs (miRNAs) and lncRNAs, have recently been found to play important regulatory roles in the development and progression of cardiovascular diseases 11, 12, 13, 14. These RNAs have also been implicated in the diagnosis of cardiovascular diseases owing to the characteristic alterations of their circulating levels with different categories and grades of pathological processes. LncRNAs belong to a newly discovered class of functional mRNA‐like transcripts that lack significant open reading frames or protein‐coding capacity 14 and have emerged as an important player in cardiovascular diseases, including a number of cardiac‐specific or cardiac‐related lncRNAs such as SRA, DIO3OS, SAF, NESPAS, MIAT, NRON, CARL, HCG22, FENDRR, MHRT, aHIF, ZFAS1 and CDR1AS 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26 (http://cmbi.bjmu.edu.cn/lncrnadisease). Recent research data have also suggested the roles of lncRNAs in HF 15, 16, 17, 18. Most prominently, circulating lncRNAs are exceptionally stable in the bloodstream and readily detectable in human subjects, such as in patients with cancers or acute kidney injury, implying that circulating lncRNAs might be a non‐invasive and rapid diagnostic tool for disease diagnosis and prognosis 15. However, studies on circulating lncRNAs for the prediction of HF have been sparse. A comprehensive study using microarray analysis compared expression alterations of lncRNAs in the heart, whole blood and plasma in a mouse model of acute HF 17. Their results revealed 32 differentially expressed lncRNAs with changes greater than twofold. Another study conducted with serum samples from HF patients suggested the potential of LIPCAR (the mitochondrial lncNA uc022bqs.1) to predict survival in patients with HF. Yet, none of these deregulated lncRNAs belong to the cardiac‐specific or cardiac‐related ones mentioned above.
This study was therefore designed to explore the possibility of the known cardiac‐specific and cardiac‐related lncRNAs in plasma samples from patients with HF as circulating biomarkers for HF. Quantitative RT‐PCR was employed to determine the plasma levels of the test lncRNAs. Our results identified NRON and MHRT as possible novel biomarkers for predicting HF.
Materials and Methods
Participants
Between February 2014 and January 2015, 104 HF patients and 109 non‐HF control participants presented to the First Affiliated Hospital, the Second Affiliated Hospital, the Third Affiliated Hospital and the Fourth Affiliated Hospital of the Harbin Medical University (Harbin, China). Diagnosis of HF and the criteria for inclusion of patients were as previously described in detail 27, 28 (see Supplementary Methods). The clinical characteristics of the study population are summarized in Table 1 and Tables S1 and S2.
Table 1.
The demographic characteristics and HF‐relevant indicators in HF patients and non‐HF control participants
| Characteristics | Non‐HF | HF | P value |
|---|---|---|---|
| Age | |||
| N (missing) | 60 (0) | 72 (0) | 0.8710 |
| Mean (Std) | 60.08 (11.97) | 59.31 (11.19) | |
| Min, max | 36, 88 | 28, 83 | |
| Median | 58 | 60.50 | |
| Range | 52~67.50 | 51~67 | |
| Gender | |||
| Male | 37 | 47 | 0.6676 |
| Female | 23 | 25 | |
| Total (missing) | 60 (0) | 72 (0) | |
| Hypertension | |||
| Yes | 17 | 39 | 0.2858 |
| No | 19 | 28 | |
| Total (missing) | 36 (24) | 67 (5) | |
| Diabetes | |||
| Yes | 7 | 17 | 0.4480 |
| No | 29 | 48 | |
| Total (missing) | 36 (24) | 65 (7) | |
| CHOL | |||
| N (missing) | 57 (3) | 63 (9) | 0.0344 |
| Mean (Std) | 4.72 (0.70) | 4.40 (1.11) | |
| Min, max | 3.24, 6.33 | 1.98, 8.84 | |
| Median | 4.69 | 4.31 | |
| Range | 4.33~5.21 | 3.71~5.03 | |
| TG | |||
| N (missing) | 57 (3) | 63 (9) | 0.3930 |
| Mean (Std) | 1.32 (0.52) | 1.49 (0.74) | |
| Min, max | 0.66, 2.43 | 0.49, 3.65 | |
| Median | 1.16 | 1.26 | |
| Range | 0.91, 1.51 | 0.92~1.89 | |
| HDL | |||
| N (missing) | 57 (3) | 63 (9) | 0.00739 |
| Mean (std) | 1.20 (0.23) | 1.12 (0.30) | |
| Min, max | 0.79, 1.88 | 0.79, 2.85 | |
| Median | 1.20 | 1.07 | |
| Range | 1.06~1.31 | 0.95~1.21 | |
| LDL | |||
| N (missing) | 57 (3) | 63 (9) | 0.6399 |
| Mean (Std) | 2.84 (0.56) | 2.95 (0.92) | |
| Min, max | 1.70, 4.31 | 0.93, 6.36 | |
| Median | 2.86 | 2.75 | |
| Range | 2.46~3.23 | 2.28~3.44 | |
| Glycemia | |||
| N (missing) | 57 (3) | 67 (5) | 0.13911 |
| Mean (Std) | 5.99 (1.90) | 6.93 (4.25) | |
| Min, max | 3.98, 14.20 | 2.89, 36.6 | |
| Median | 5.46 | 5.86 | |
| Range | 5.11~6.04 | 5.03~7.34 | |
| ALT | |||
| N (missing) | 26 (34) | 65 (7) | 0.0737 |
| Mean (Std) | 23.27 (12.23) | 30.49 (20.31) | |
| Min, max | 11.00, 59.00 | 0.26, 108.00 | |
| Median | 21.50 | 25 | |
| Range | 15.00~27.00 | 17.00~35.00 | |
| AST | |||
| N (missing) | 26 (34) | 65 (7) | 0.02176 |
| Mean (Std) | 20.73 (3.81) | 28.14 (15.27) | |
| Min, max | 14.00, 28.00 | 7.00, 92.00 | |
| Median | 20.50 | 24.00 | |
| Range | 18.00~23.00 | 19.00~32.00 | |
| AST/ALT | |||
| N (missing) | 23 (37) | 65 (7) | 0.2975 |
| Mean (Std) | 1.06 (0.36) | 1.39 (3.02) | |
| Min, max | 0.50, 1.80 | 0.40, 25.00 | |
| Median | 1.00 | 0.85 | |
| Range | 0.80~1.30 | 0.70~1.20 | |
| BUN | |||
| N (missing) | 55 (5) | 67 (5) | <0.0001 |
| Mean (Std) | 5.76 (1.46) | 7.36 (2.65) | |
| Min, max | 3.30, 10.34 | 3.07, 18.04 | |
| Median | 5.50 | 6.94 | |
| Range | 4.53~6.85 | 5.64~8.77 | |
| Cr | |||
| N (missing) | 57 (3) | 68 (4) | 0.00215 |
| Mean (Std) | 72.96 (16.64) | 90.11 (34.60) | |
| Min, max | 8.20, 103.60 | 6.31, 239.30 | |
| Median | 75.00 | 81.55 | |
| Range | 62.20~85.00 | 67.70~100.80 | |
| Bun/Cr | |||
| N (missing) | 23 (37) | 68 (4) | <0.0001 |
| Mean (Std) | 84.25 (24.31) | 48.90 (40.44) | |
| Min, max | 45.92, 159.08 | 0.05, 124.58 | |
| Median | 83.69 | 58.73 | |
| Range | 69.00~93.00 | 0.12~78.58 | |
| UA | |||
| N (missing) | 56 (4) | 68 (4) | <0.0001 |
| Mean (Std) | 316.21 (79.21) | 422.88 (144.25) | |
| Min, max | 156.40, 516.00 | 88.75, 799.70 | |
| Median | 304.30 | 387.15 | |
| Range | 257.10~374.30 | 320.45~532.95 | |
| Co2CP | |||
| N (missing) | 52 (8) | 48 (24) | 0.4233 |
| Mean (Std) | 36.48 (47.05) | 37.44 (67.67) | |
| Min, max | 23.20, 281.00 | 10.00, 495.70 | |
| Median | 27.35 | 27.75 | |
| Range | 26.10~28.50 | 26.00~30.00 | |
| NT‐proBNP | |||
| N (missing) | 0 (60) | 65 (7) | NA |
| Mean (Std) | 3786.62 (6091.64) | ||
| Min, max | 104.00, 35,000.00 | ||
| Median | 2144.00 | ||
| Range | 635.00, 3788.00 | ||
CHOL, total cholesterol; TG, triglyceride; HDL, high‐density cholesterol; LDL, low‐density cholesterol; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; Cr, creatinine; UA, uric acid; Co2CP, carbon dioxide combining power; NT‐proBNP, amino‐terminal pro‐brain natriuretic peptide.
Ethical approval of studies and informed consent
The study protocols and the procedures for handling human samples were approved by the Institutional Research Board of the Harbin Medical University (No.HMUIRB‐20140027). The written informed consents were obtained from all subjects recruited to our study.
Collection and handling of human blood samples
Whole blood samples (1 ml per patient) were drawn from the study subjects via a direct venous puncture into the tubes containing sodium citrate. The human whole blood samples in sodium citrate vacuum tubes were kept at 4°C and then centrifuged at 2000 × g/min. at 4°C for 10 min. to obtain plasma samples.
RNA extraction and quantitative real‐time reverse transcription (RT)‐polymerase chain reaction (qPCR)
Total RNA was extracted from the prepared plasma samples using Trizol LS reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer's instructions. In brief, each plasma sample (0.25 ml) was mixed well with 1 ml Trizol reagents in a tube. Chloroform (0.2 ml) was added into the sample and shaken vigorously by hand for 15 sec. The sample was incubated at room temperature for 5 min. and then centrifuged at 12000 × g/min. at 4°C for 15 min. The supernatant was transferred to a new tube, and an equal volume of isopropanol was added to the aqueous phase. After mixing and incubation at room temperature for 10 min., the sample was again centrifuged at 12000 × g/min. at 4°C for 10 min. After removal of the supernatant, the pellet was washed with 1 ml of 75% ethanol for the initial homogenization. Then, the sample was centrifuged at 10,600 r.p.m./min. at 4°C for 5 min. The RNA pellet was dissolved in DEPC water. The quality of our RNA samples was first measured by NanoDrop ND‐8000 (Thermo Fisher Scientific, Waltham, MA, USA). To ensure the RNA/DNA ratio 1.8–2.0. Then, the integrity of the RNA samples was assessed by standard denaturing agarose gel electrophoresis and confirmed by discrete 28 s and 5 s bands without smear.
The SYBR Green PCR Master Mix Kit (cat#: 4367659, Life technology, USA) was used for qPCR for relative quantification of lncRNAs (see Supplementary Materials online for detail). The qPCR primer pairs used in our study are listed in Table S4 online.
Statistical analysis
Categorical data are presented as count and percentile. Continuous variables are described as mean ± S.E.M. (standard error of measurement), min, max, median or interquartile range, as specified in the data descriptions. The statistical analyses are described in detail in supplementary methods. All analyses were carried out with SAS 9.1 (Serial No. 989155) except that ROC was carried out with SPSS v17.0 software. The significant level was set at 0.05, and two‐tailed P values <0.05 were considered statistically significant.
Results
Clinical characteristics of the study population
Plasma samples were collected from a total of 104 HF patients and 109 control participants for measuring lncRNAs. To have more rational comparisons between HF and control participants, we filtered the plasma samples based upon the clinical or demographic characteristics of the patients recruited. We identified 32 HF patients and 49 control participants who did not have matched clinical or demographic characteristics between the two groups, and we therefore discarded these samples leaving 72 HF patients and 60 control participants for detailed statistical analysis. Of the 72 HF patients, 65 had an elevated NT‐proBNP at enrolment during the study period. Table 1 shows the clinical and demographic characteristics of the patients enrolled in this study (also see Tables S1 and S2 online for the complete data sets of all 104 HF patients and 109 control participants). There were no age and gender differences between the test patients and control participants, nor was any difference in blood pressure.
Reciprocal changes in NRON and MHRT blood levels in AMI patients
Our initial quantitative real‐time RT‐PCR (qPCR) analysis included 13 known cardiac‐specific or cardiac‐related lncRNAs: SRA, DIO3OS, SAF, NESPAS, MIAT, NRON, CARL, HCG22, FENDRR, MHRT, aHIF, ZFAS1 and CDR1AS. As illustrated in Figure 1A, of 13 lncRNAs tested, only two, NRON and myosin heavy–chain‐associated RNA transcripts (MHRT), demonstrated significant differences in plasma samples between HF and non‐HF. Specifically, the circulating level of NRON was significantly higher in HF than in non‐HF subjects (3.17 ± 0.30 versus 1.0 ± 0.07; P < 0.0001) (Fig. 1B and C; Table 2). Similarly, the plasma level of MHRT was also markedly elevated in HF (1.66 ± 0.14) relative to that in non‐HF subjects (1.00 ± 0.12; P < 0.0001). The median Ct value for NRON was 26.3 by 40 cycles of qPCR with standard deviation of 2.2, and the median Ct value for MHRT was 27.0 with standard deviation of 1.6, indicating that these two lncRNAs are fairly abundant in plasma.
Figure 1.
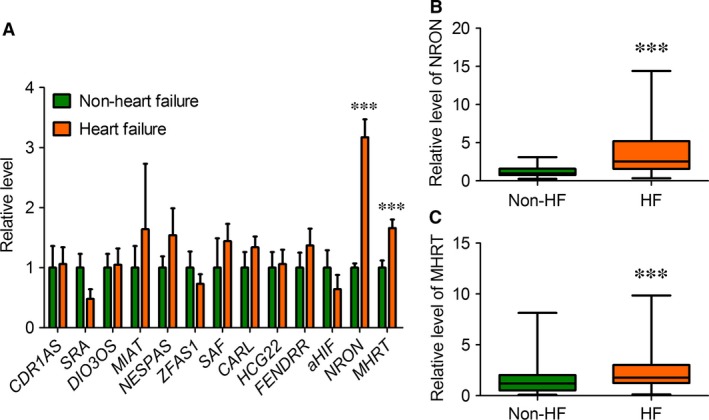
Changes in circulating lncRNA levels in patients with HF relative to non‐HF control participants. (A) Circulating levels of lncRNAs were determined by quantitative real‐time RT‐PCR (qPCR) with the plasma samples prepared from HF patients and non‐HF control participants. Note that only NRON and MHRT demonstrated significant differences between patients HF and non‐HF control participants. Data are presented as mean ± S.E.M. ***P < 0.0001, n = 72 for HF and n = 60 for non‐HF control participants. (B & C) Box plot of plasma NRON and MHRT levels, respectively, providing a non‐parametric illustration of numerical data displaying the degree of dispersion (spread), skewness in the data (asymmetry of distribution) and outliers, without making any assumptions of the underlying statistical distribution. ***P < 0.0001, n = 72 for HF and n = 60 for non‐HF control participants.
Table 2.
Statistical analysis of the circulating NRON and MHRT
| LncRNA | Non‐HF | HF | P value |
|---|---|---|---|
| MHRT | |||
| N (missing) | 60 (0) | 72 (0) | <0.0001* |
| Mean (Std) | 1.0 (0.89) | 1.66 (1.2) | |
| Min, max | 0.06, 5.85 | 0.10, 7.08 | |
| Median | 0.86 | 1.27 | |
| Range (Q1, Q3) | 0.38~1.43 | 0.90~2.15 | |
| NRON | |||
| N (missing) | 60 (0) | 72 (0) | <0.0001* |
| Mean (Std) | 1.0 (0.54) | 3.17 (2.58) | |
| Min, max | 0.09, 8.14 | 0.13, 9.86 | |
| Median | 1.20 | 1.77 | |
| Range (Q1, Q3) | 0.54~1.26 | 1.26~2.99 | |
P values are for comparisons between HF patients versus non‐HF control participants. * P < 0.001 vs. Non‐HF.
Similar elevations of the circulating levels of NRON and MHRT were consistently observed when all plasma samples (104 HF patients and 109 control participants) were included in our analysis (Table S3 online).
Evaluation of circulating NRON and MHRT as new biomarkers for HF
Having established that NRON and MHRT are present in the peripheral circulation and their plasma levels are anomaly altered in HF patients, we sought to determine the potential utility of circulating NRON and MHRT as diagnostic biomarkers of HF. To this end, ROC analysis was performed to evaluate the predictive power of circulating NRON and MHRT alone for HF. Our results showed that the area under ROC curve was 0.865 (95% CI = 0.805~0.926) for NRON alone (Fig. 2A), 0.702 (95% CI = 0.612~0.791) for MHRT alone (Fig. 2B).
Figure 2.
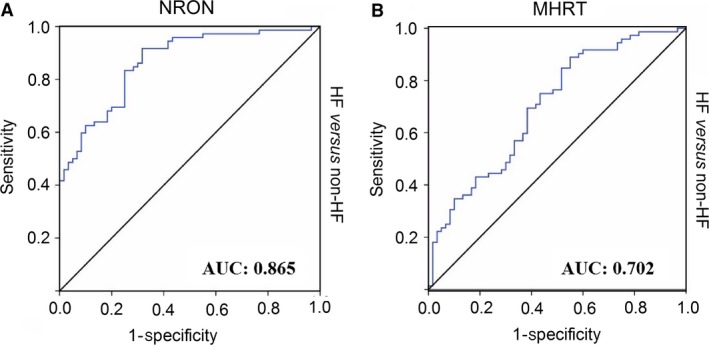
Receiver operator characteristic analysis of circulating NRON and MHRT for predicting HF. The area under ROC curve was determined to evaluate the predictive power of circulating NRON (A) and MHRT (B) levels for HF using non‐HF participants as control.
The univariate analysis with logistic regression showed that the odds ratios (OR) were 4.505 (95% CI: 2.393~8.478) for NRON (P < 0.0001), and 1.701 (95% CI: 1.225~2.363) for MHRT (P = 0.0015) between HF and non‐HF (Table 3).
Table 3.
Univariate regression analysis for the association of NRON and MHRT with demographic characteristics between HF patients and non‐HF control participants
| Variable | B | S.E. | λ2 | P | OR | 95% CI | |
|---|---|---|---|---|---|---|---|
| Low | High | ||||||
| NRON | 1.5051 | 0.3226 | 21.7608 | <0.0001 | 4.505 | 2.393 | 8.478 |
| MHRT | 0.5313 | 0.1676 | 10.0523 | 0.0015 | 1.701 | 1.225 | 2.363 |
| Age | −0.00592 | 0.0153 | 0.1504 | 0.6982 | 0.994 | 0.965 | 1.024 |
| Gender | 0.1558 | 0.3630 | 0.1843 | 0.6677 | 1.169 | 0.574 | 2.381 |
| HDL | −1.1380 | 0.7586 | 2.2509 | 0.1335 | 0.320 | 0.072 | 1.417 |
| LDL | 0.1928 | 0.2424 | 0.6326 | 0.4264 | 1.213 | 0.754 | 1.950 |
| TG | 0.4278 | 0.2985 | 2.0546 | 0.1518 | 1.534 | 0.855 | 2.754 |
| CHOL | −0.3756 | 0.2072 | 3.2842 | 0.0699 | 0.687 | 0.458 | 1.031 |
The multivariate logistic regression analysis further verified NRON and MHRT as independent predictors for HF (Table 4): The OR values were 3.377 (95% CI: 1.441~7.915) for NRON (P = 0.0051) and 1.679 (95% CI: 1.068~2.639 for MHRT (P = 0.0248) between HF and non‐HF (Table 4).
Table 4.
Multivariate regression analysis for the association of NRON and MHRT with demographic characteristics between HF patients and non‐HF control participants
| Variable | B | S.E. | λ2 | P | OR | 95% CI | |
|---|---|---|---|---|---|---|---|
| Low | High | ||||||
| NRON | 1.2170 | 0.4346 | 7.8412 | 0.0051 | 3.377 | 1.441 | 7.915 |
| MHRT | 0.5182 | 0.2308 | 5.0395 | 0.0248 | 1.679 | 1.068 | 2.639 |
| Age | 0.00199 | 0.0315 | 0.0040 | 0.9496 | 1.002 | 0.942 | 1.066 |
| Gender | −0.6181 | 0.7135 | 0.7505 | 0.3863 | 0.539 | 0.133 | 2.182 |
| HDL | 7.6350 | 2.0206 | 14.2775 | 0.0002 | >999.999 | 39.437 | >999.999 |
| LDL | 6.1932 | 1.4468 | 18.3247 | <0.0001 | 489.417 | 28.720 | >999.999 |
| TG | −6.3369 | 1.4537 | 19.0036 | <0.0001 | 0.002 | <0.001 | 0.031 |
| CHOL | 2.6408 | 0.7249 | 13.2708 | 0.0003 | 14.024 | 3.387 | 58.066 |
Relation of NRON and MHRT to conventional prognostic markers
To further evaluate the usefulness of circulating NRON and MHRT as HF biomarkers, we tested whether their levels were correlated with cardiac risk factors, conventional HF markers and cardiac function parameters. The data summarized in Table 5 show that NRON was negatively correlated with HDL and positively correlated with LDL, whereas MHRT was positively correlated with AST and LDH (Table 6). Neither NRON nor MHRT was correlated with age, gender, diabetes mellitus, hypertension, smoking history, total cholesterol, triglyceride (TG), cardiac troponin I (cTnI), aspartate aminotransferase 29, creatine kinase (CK), creatine kinase‐myocardial band (CKMB), NT‐proNBP or cardiac function parameters.
Table 5.
Spearman's rank correlation analysis for the association of NRON with cardiac risk factors, cardiac biomarkers and cardiac function parameters in HF patients
| NRON | ||
|---|---|---|
| Coefficient | P | |
| Cardiovascular risk factors | ||
| Age | 0.02786 | 0.7512 |
| Gender | 0.13804 | 0.1145 |
| Diabetes | 0.09415 | 0.3490 |
| Hypertension | −0.03081 | 0.7573 |
| Smoking | 0.15804 | 0.2319 |
| HDL | −0.22658 | 0.0128 |
| LDL | 0.11300 | 0.2191 |
| CHOL | −0.12143 | 0.1864 |
| TG | 0.04193 | 0.6493 |
| Cardiac biomarkers | ||
| cTnI | 0.06742 | 0.6088 |
| AST | 0.09553 | 0.4085 |
| LDH | 0.53876 | <0.0001 |
| CK | 0.14788 | 0.1905 |
| CKMB | 0.05194 | 0.7814 |
| NT‐proBNP | 0.10304 | 0.4141 |
| Cardiac function | ||
| E/A | −0.36332 | 0.1263 |
| EF | −0.22937 | 0.0952 |
| FS | 0.02309 | 0.9071 |
cTnI, cardiac troponin I; CK, creatine kinase; CKMB, creatine kinase‐myocardial band; E/A, E, peak velocity of the early diastolic filling wave, A, peak velocity of the late diastolic filling wave; EF, ejection fraction; FS, fractional shortening.
Table 6.
Spearman's rank correlation analysis for the association of MHRT with cardiac risk factors, cardiac biomarkers and cardiac function parameters in HF patients
| MHRT | ||
|---|---|---|
| Coefficient | P | |
| Cardiovascular risk factors | ||
| Age | 0.03310 | 0.7063 |
| Gender | 0.16987 | 0.0515 |
| Diabetes | 0.11410 | 0.2559 |
| Hypertension | −0.00328 | 0.9738 |
| Smoking | −0.00405 | 0.9757 |
| HDL | −0.14948 | 0.1032 |
| LDL | −0.07218 | 0.4334 |
| CHOL | −0.10753 | 0.2424 |
| TG | −0.01565 | 0.8653 |
| Cardiac biomarkers | ||
| cTnI | 0.08386 | 0.5241 |
| AST | 0.35285 | 0.0016 |
| LDH | 0.43344 | <0.0001 |
| CK | 0.02483 | 0.8270 |
| CKMB | 0.23464 | 0.2039 |
| NT‐proBNP | 0.07810 | 0.5363 |
| Cardiac function | ||
| E/A | 0.15182 | 0.5350 |
| EF | −0.14227 | 0.3048 |
| FS | 0.11656 | 0.5547 |
Discussion
In the present study, we analysed the levels of a selected set of lncRNAs in the plasma samples of HF patients for their potential as biomarkers for the diagnosis of HF. These lncRNAs were selected for our study because they have been documented to play important roles in shaping developmental process of the heart and in the pathogenesis and progression of cardiac diseases 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26. Our results identified two lncRNAs, NRON and MHRT out of 13 known cardiac‐relevant lncRNAs examined, as promising candidate biomarkers for HF in the light of the significant elevations of their circulating levels in HF patients relative to non‐HF control participants and the close correlation between the circulating levels of NRON and MHRT.
Published studies on circulating LncRNAs as HF biomarkers
NRON (Non‐coding RNA repressor of NFAT) is enriched in muscles (including cardiac muscle), placenta, spleen, thymus and lymph nodes and has been denoted as a repressor of the nuclear factor of activated T cells (NFAT) by influencing its nuclear trafficking 30, 31. NFAT is known to be a critical protein in the regulation of intracellular Ca2+ homoeostasis and of gene expression as a transcription factor in the heart, and its expression and activity are tremendously increased in HF 32, 33. It is conceivable that by regulating NFAT, NRON can participate in the genesis and development of HF. Yet, such a notion requires rigorous experimentation to verify. MHRT was initially identified as a cardiac‐specific and cardiac enriched protective lncRNA by Han et al. 21. It acts to protect the heart against pathological hypertrophy; yet, pathological stress such as hypertrophy and HF inhibits MHRT transcription in the heart 21. Recently, MHRT was found to suppress cardiomyocyte apoptosis induced by H2O2 to simulate the acute ischaemic condition 26, and under such context, MHRT expression in cardiomyocytes was activated by oxidative stress. In this same study, the authors found that the plasma MHRT level is markedly elevated in patients with acute MI. Nevertheless, the potential usefulness of NRON and MHRT as biomarkers of HF has not been previously evaluated.
Here, we revealed that NRON and MHRT were both elevated in their plasma levels in patients with HF relative to non‐HF control participants. We also found that these two lncRNAs are fairly abundant RNA species in human serum samples based on the relatively low Ct values of qPCR experiments (the median Ct value for NRON was 26.3, ranging from 22.3 to 31.4; and the median Ct value for MHRT was 27.0 with a range from 23.4 to 30.4 in patients with HF). These facts prompted us to propose that either of these two lncRNAs is a reasonable predictor of HF. For years, NT‐proBNP has been believed to be an established risk marker for HF. Our analysis indicated that the predictive power of NRON is comparable to that of NT‐proBNP: The reported value of AUC is 0.844 for NT‐proBNP 34, and the value for NRON was 0.865. By comparison, the AUC value for MHRT is lower (0.702); yet, it still falls into the ‘good’ category for clinical applications according to the guide for classifying the accuracy of a diagnostic test with the traditional academic point system (0.9–1.0 excellent; 0.8–0.9 very good; 0.7–0.8 good; 0.6–0.7 sufficient; 0.5–0.6 bad; < 0.5 test not useful) 35, 36.
In an earlier study, Kumarswamy et al 16. conducted global transcriptomic analyses in plasma RNA from patients with or without left ventricular remodelling after MI with three independent patient cohorts developing cardiac remodelling and HF. The authors found that LIPCAR is down‐regulated early after MI but up‐regulated during later stages. Plasma levels of LIPCAR can predict patients developing cardiac remodelling and future cardiovascular deaths. Li et al. 17 analysed the expression levels of lncRNAs in whole blood, tissue and plasma in a mouse model of acute HF. The study revealed that 518 lncRNAs are up‐regulated while 908 are down‐regulated in the heart with microarray‐based analyses with 32 differentially expressed lncRNAs with changes greater than twofold. Greco et al. 29 profiled and validated lncRNAs in left ventricle biopsies of 18 patients affected by non‐end‐stage dilated ischaemic cardiomyopathy and 17 matched controls. Fourteen lncRNAs were significantly modulated in non‐end‐stage HF patients, identifying a HF lncRNA signature. In particular, CDKN2B‐AS1/ANRIL (antisense non‐coding RNA in the INK4 locus), HOTAIR (HOX transcript antisense RNA) and LOC285194/TUSC7 (tumour suppressor candidate 7) showed similar modulation in peripheral blood mononuclear cells and heart tissue, suggesting a potential role as disease biomarkers. Yan et al. 37 identified an lncRNA UCA1 (urothelial carcinoma‐associated 1) as a biomarker for acute myocardial infarction (AMI) with its plasma level significantly decreased in AMI patients, compared with non‐AMI subjects.
In one of our previous studies, we reported two lncRNAs zinc finger antisense 1 (ZFAS1) and Cdr1 antisense (CDR1AS) as novel biomarkers of acute MI, with their reciprocal changes in the whole blood samples (ZFAS1 was down‐regulated, whereas CDR1AS was up‐regulated) independently predicting acute MI 38. Intriguingly, in the context of HF as in the present study, these two lncRNAs did not show significant alterations in their circulating levels, indicating that they may be specific for predicting acute MI. To the best of our knowledge, there have been no other published studies on the circulating lncRNAs in HF patients. Our study therefore represents the first of such efforts to identify biomarkers with the potential to predict HF in humans.
Significance of our findings
NRON and MHRT as biomarkers could offer a number of advantages. First, lncRNAs have been found to be overall more stable than protein markers in circulation and can be easily detected in blood samples (whole blood, plasma and serum); thus, it is possible that NRON and MHRT might also be more stable in the blood than the traditional protein markers 15, 39, 40. Second, NRON and MHRT can be detected in a quantitative manner by highly sensitive methods such as real‐time PCR. And finally, changes in NRON and MHRT in the bloodstream may reflect alterations of cardiac function and structure during the development of heart disease thereby helping us to infer the underlying molecular mechanisms. This is in resembling miRNAs as biomarkers of heart disease. For example, the early elevation of circulating miR‐1 during acute myocardial infarction can be interpreted as increased apoptotic cardiomyocyte death 41, 42, 43.
Limitations of our study
In the present study, we focused on only a subset of lncRNAs that are known (at the time we initiated our study) to be relevant to cardiac disease without dealing with the global transcriptome profiling. Thus, our findings do not provide a panorama for comprehensive understanding of all lncRNAs identified thus far, but might have missed out many other important lncRNAs that were not included in the present study for their potential as HF biomarkers. Nonetheless, the lncRNAs selected for our study are those that have been shown to be able to cause cardiac disorders or are abundantly expressed in heart cells. Another limitation of the study is the unknown sources of NRON and MHRT in the bloodstream: Are they released from dead cells in the failing heart or are they secreted by blood cells in response to the damaged heart?
Conflict of interest
None declared.
Author contributions
LNX, LHS, YZ, YJL and BFY designed, performed study and supervised all aspects of the research and analysis. LNX, YJL and BFY finalized the manuscript. LNX, LHS, YZ, YCH, YH, QQL, YG, BBF, LNC, XXW, CQX, MYZ and ZGW assisted in research, data analysis and interpretation. BY, YT, SW, ZMD and YJL were responsible for collect blood samples and for the final approval of the manuscript. YH was responsible for the statistical analysis involved in the study. BY, YT, SW and ZMD reviewed the clinical aspects and writing of manuscript.
Supporting information
Table S1 The demographic characteristics and HF‐relevant indicators in HF patients, non‐HF control participants for NRON
Table S2 The demographic characteristics and HF‐relevant indicators in HF patients, non‐HF control participants for MHRT
Table S3 The Statistical Analysis of Circulating NRON and MHRT
Table S4 Human gene‐specific primers for real‐time PCR
Acknowledgements
This work was supported in part by grants from the Funds for Creative Research Groups of the National Natural Science Foundation of China (81421063), the Key Project of National Science Foundation of China (81230081 & 81130088), the National Basic Research Program of China (973 program, 2013CB531104), Major Program of National Natural Science Foundation of China (81530010) and the Natural Science Foundation of China (81170219, 81371211 & 81570357).
Contributor Information
Yanjie Lu, Email: yjlu2008@163.com.
Bao Feng Yang, Email: yangbf@ems.hrbmu.edu.cn.
References
- 1. Ambrosy AP, Fonarow GC, Butler J, et al The global health and economic burden of hospitalizations for heart failure: lessons learned from hospitalized heart failure registries. J Am Coll Cardiol. 2014; 63: 1123–33. [DOI] [PubMed] [Google Scholar]
- 2. Mozaffarian D, Benjamin EJ, Go AS, et al Executive Summary: Heart Disease and Stroke Statistics‐2015 Update A Report From the American Heart Association. Circulation. 2015; 131: 434–41. [DOI] [PubMed] [Google Scholar]
- 3. Johnson FL. Pathophysiology and etiology of heart failure. Cardiol Clin. 2014; 32: 9–19. [DOI] [PubMed] [Google Scholar]
- 4. Kehat I, Molkentin JD. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation. 2010; 122: 2727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oremus M, Mckelvie R, Donwauchope A, et al A systematic review of BNP and NT‐proBNP in the management of heart failure: overview and methods. Heart Fail Rev. 2014; 19: 813–5. [DOI] [PubMed] [Google Scholar]
- 6. Sato Y, Fujiwara H, Takatsu Y. Cardiac troponin and heart failure in the era of high‐sensitivity assays. J Cardiol. 2012; 60: 160–7. [DOI] [PubMed] [Google Scholar]
- 7. Burke MA, Cotts WG. Interpretation of B‐type natriuretic peptide in cardiac disease and other comorbid conditions. Heart Fail Rev. 2007; 12: 23–36. [DOI] [PubMed] [Google Scholar]
- 8. Gaggin HK, Mohammed AA, Bhardwaj A, et al Heart failure outcomes and benefits of NT‐proBNP‐guided management in the elderly: results from the prospective, randomized ProBNP outpatient tailored chronic heart failure therapy (PROTECT) study. J Cardiac Fail. 2012; 18: 626–34. [DOI] [PubMed] [Google Scholar]
- 9. Bettencourt P. NT‐proBNP and BNP: biomarkers for heart failure management. Eur J Heart Fail. 2004; 6: 359–63. [DOI] [PubMed] [Google Scholar]
- 10. Gaggin HK, Truong QA, Rehman SU, et al Characterization and prediction of natriuretic peptide “nonresponse” during heart failure management: results from the ProBNP Outpatient Tailored Chronic Heart Failure (PROTECT) and the NT‐proBNP‐Assisted Treatment to Lessen Serial Cardiac Readmissions and Death (BATTLESCARRED) study. Congest Heart Fail. 2013; 19: 135–42. [DOI] [PubMed] [Google Scholar]
- 11. Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res. 2012; 111: 1349–62. [DOI] [PubMed] [Google Scholar]
- 12. Tijsen AJ, Pinto YM, Creemers EE. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am J Physiol Heart Circ Physiol. 2012; 303: H1085–95. [DOI] [PubMed] [Google Scholar]
- 13. Mishra PK, Tyagi N, Kumar M, et al MicroRNAs as a therapeutic target for cardiovascular diseases. J Cell Mol Med. 2009; 13: 778–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Archer K, Broskova Z, Bayoumi AS, et al Long non‐coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci. 2015; 16: 23651–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumarswamy R, Thum T. Non‐coding RNAs in cardiac remodeling and heart failure. Circ Res. 2013; 113: 676–89. [DOI] [PubMed] [Google Scholar]
- 16. Kumarswamy R, Bauters C, Volkmann I, et al Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014; 114: 1569–75. [DOI] [PubMed] [Google Scholar]
- 17. Li D, Chen G, Yang J, et al Transcriptome analysis reveals distinct patterns of long noncoding RNAs in heart and plasma of mice with heart failure. PLoS One. 1932; 8: e77938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JH, Gao C, Peng G, et al Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. 2011; 109: 1332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Congrains A, Kamide K, Oguro R, et al Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012; 220: 449–55. [DOI] [PubMed] [Google Scholar]
- 20. Grote P, Wittler L, Hendrix D, et al The tissue‐specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013; 24: 206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han P, Li W, Lin CH, et al A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014; 514: 102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishii N, Ozaki K, Sato H, et al Identification of a novel non‐coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006; 51: 1087–99. [DOI] [PubMed] [Google Scholar]
- 23. Matkovich SJ, Edwards JR, Grossenheider TC, et al Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci USA. 2014; 111: 12264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang K, Long B, Zhou LY, et al CARL lncRNA inhibits anoxia‐induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR‐539‐dependent PHB2 downregulation. Nat Commun. 2014; 5: 3596. [DOI] [PubMed] [Google Scholar]
- 25. Yan B, Yao J, Liu JY, et al lncRNA‐MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015; 116: 1143–56. [DOI] [PubMed] [Google Scholar]
- 26. Zhang J, Gao C, Meng M, et al Long noncoding RNA MHRT protects cardiomyocytes against H2O2‐induced apoptosis. Biomol Ther. 2016; 24: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alpert JS, Thygesen K, Antman E, et al Myocardial infarction redefined: a consensus document of the Joint European Society of Cardiology/American College of Cardiology Committee for the redefinition of myocardial infarction. J Am Coll Cardiol. 2000; 36: 959–69. [DOI] [PubMed] [Google Scholar]
- 28. Morrow DA, Cannon CP, Jesse RL, et al; National Academy of Clinical Biochemistry . National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines: clinical characteristics and utilization of biochemical markers in acute coronary syndromes. Circulation. 2007; 115: e356–75. [DOI] [PubMed] [Google Scholar]
- 29. Greco S, Zaccagnini G, Perfetti A, et al Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. 2016; 14: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharma S, Findlay GM, Bandukwala HS, et al Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA‐protein scaffold complex. Proc Natl Acad Sci. 2011; 108: 11381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Willingham AT, Orth AP, Batalov S, et al A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005; 309: 1570–3. [DOI] [PubMed] [Google Scholar]
- 32. Dirkx E, Gladka MM, Philippen LE, et al Nfat and miR‐25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat Cell Biol. 2013; 15: 1282–93. [DOI] [PubMed] [Google Scholar]
- 33. Wilkins BJ, Windt LJD, Bueno OF, et al Targeted disruption of NFATc3, but not NFATc4, reveals an intrinsic defect in calcineurin‐mediated cardiac hypertrophic growth. Mol Cell Biol. 2002; 22: 7603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Q, Cao X, Wang X, et al Correction: utility of NT‐proBNP for identifying LV failure in patients with acute exacerbation of chronic bronchitis. PLoS One. 2013; 8: e52553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bossuyt PM, Reitsma JB, Bruns DE, et al The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Clin Chem. 2003; 49: 7–18. [DOI] [PubMed] [Google Scholar]
- 36. Irwig L, Bossuyt P, Glasziou P, et al Evidence base of clinical diagnosis: designing studies to ensure that estimates of test accuracy are transferable. BMJ. 2002; 324: 669–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yan Y, Zhang B, Ning L, et al Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed Res Int. 2016; 2016: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ying Z, Sun L, Xuan L, et al Reciprocal changes of circulating long non‐coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016; 6: 22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jing Z, Shi C, Zhu L, et al Chronic cerebral hypoperfusion induces vascular plasticity and hemodynamics but also neuronal degeneration and cognitive impairment. J Cereb Blood Flow Metab. 2015; 35: 1249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Teng KY, Ghoshal K. Role of noncoding RNAs as biomarker and therapeutic targets for liver fibrosis. Gene Expr. 2015; 16: 155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ai J, Zhang R, Li Y, et al Circulating microRNA‐1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010; 391: 73–7. [DOI] [PubMed] [Google Scholar]
- 42. Kuwabara Y, Ono K, Horie T, et al Increased microRNA‐1 and microRNA‐133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011; 4: 446–54. [DOI] [PubMed] [Google Scholar]
- 43. Pan Z, Sun X, Ren J, et al miR‐1 exacerbates cardiac ischemia‐reperfusion injury in mouse models. PLoS One. 2012; 7: e50515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The demographic characteristics and HF‐relevant indicators in HF patients, non‐HF control participants for NRON
Table S2 The demographic characteristics and HF‐relevant indicators in HF patients, non‐HF control participants for MHRT
Table S3 The Statistical Analysis of Circulating NRON and MHRT
Table S4 Human gene‐specific primers for real‐time PCR


