Abstract
The key elements of the initiation of Helicobacter pylori chromosome replication, DnaA protein and putative oriC region, have been characterized. The gene arrangement in the H.pylori dnaA region differs from that found in many other eubacterial dnaA regions (rnpA-rmpH-dnaA-dnaN-recF-gyrB). Helicobacter pylori dnaA is flanked by two open reading frames with unknown function, while dnaN-gyrB and rnpA-rmpH loci are separated from the dnaA gene by 600 and 90 kb, respectively. We show that the dnaA gene encoding initiator protein DnaA is expressed in H.pylori cells. The H.pylori DnaA protein, like other DnaA proteins, can be divided into four domains. Here we demonstrate that the C-terminal domain of H.pylori DnaA protein is responsible for DNA binding. Using in silico and in vitro studies, the putative oriC region containing five DnaA boxes has been located upstream of the dnaA gene. DNase I and gel retardation analyses show that the C-terminal domain of H.pylori DnaA protein specifically binds each of five DnaA boxes.
INTRODUCTION
The events that occur at the replication origin (oriC) are central to the processes regulating DNA replication (1). In bacteria, replication of a circular chromosome starts from a replication origin oriC and proceeds bi-directionally until the replication forks reach the termination site (ter). The structure of the oriC region has been analyzed in Gram-negative and Gram-positive bacteria. The sequences of oriC regions are conserved only among closely related organisms. Sequence analysis revealed that the origins of various bacteria contain short, conserved motifs that are essential for oriC function: AT-rich regions and non-palindromic 9 bp sequences named DnaA boxes (2). Spacer regions, which vary in nucleotide composition and length, separate these conserved sequences. The initiator protein DnaA plays an important role in the initiation and regulation of chromosomal replication. It binds to the origin of replication, specifically to the DnaA boxes. Among bacteria, the initiation of replication is best understood in Escherichia coli (2,3). Within the E.coli oriC region five DnaA boxes are present. Binding of 10–20 DnaA protein molecules promotes a local unwinding within the AT-rich region of the oriC. The unwound region provides the entry site for the helicase complex (DnaB6–DnaC6), followed by other proteins required to form replication forks (2–4).
Helicobacter pylori is a Gram-negative, spiral-shaped pathogenic bacterium that was first isolated and cultured from biopsy specimens by Marshall and Warren (5). Since that time extensive studies of H.pylori biology have been carried out. This organism is a human gastric pathogen associated with peptic ulcer disease as well as chronic gastritis. Recent epidemiological studies demonstrated that H.pylori is a primary risk factor for the development of intestinal type gastric adenocarcinoma.
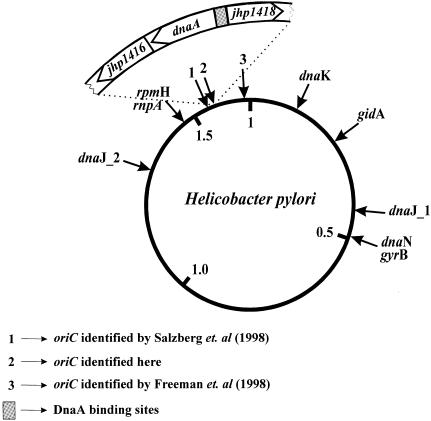
Recently the genome sequences of two unrelated isolates of H.pylori, 26695 and J99, have been determined. The 26695 and J99 circular chromosomes are 1 667 867 bp (6) and 1 643 831 bp (7) in size, respectively. The gene content of two sequenced H.pylori genomes suggests that the basic mechanism of chromosomal replication is similar to that of other eubacteria (6,7). However, experimental data concerning the replication of H.pylori are scarce. The genomic analysis revealed few surprising features, in particular in the initiation of replication. The typical eubacterial block of replication genes, dnaA-dnaN-recF-gyrB (2,8,9) does not exist; the dnaA gene is located ∼600 kb away from the dnaN-gyrB genes (see Fig. 8), while the recF gene is missing. The dnaC gene encoding DnaC protein, which delivers the DnaB helicase to the prepriming complex, is absent. Moreover, an origin of DNA replication has not been identified and is not evident from the genome sequence. Thus, it is interesting to better understand the initiation of chromosomal replication in this extensively studied pathogen.
Figure 8.
Locations of the typical eubacterial origin genes in the H.pylori chromosome (according to the H.pylori databases: http://www. tigr.org and http://scriabin.astra zeneca-boston.com/hpylori ).
Here we characterize the key elements of chromosomal replication initiation from H.pylori, DnaA protein and the putative oriC region. We demonstrate that the dnaA gene encoding DnaA protein is expressed in H.pylori and that the binding domain of the H.pylori DnaA protein specifically recognizes the DnaA boxes from the E.coli oriC region. Using in silico and in vitro methods, the putative H.pylori oriC region containing five DnaA boxes has been located upstream of the dnaA gene.
MATERIALS AND METHODS
Bacterial strains, media and culture conditions
The E.coli and H.pylori strains used and their origins are listed in Table 1. Escherichia coli DH5α served as host for all plasmid constructs and AG115 strain was utilized as host for the overproduction of the fusion protein glutathione S-transferase (GST)-DnaA(IV). Escherichia coli strains were grown in Luria–Bertani medium at 37°C. The H.pylori J99 strain was cultivated on Columbia agar medium supplemented with horse blood (10%) under micro-aerobic atmosphere (5% O2, 15% CO2, 80% N2) at 37°C for 48–72 h.
Table 1. Bacterial strains and plasmids used in this study.
| Strain/plasmid |
Genotype/relevant characteristics |
Reference |
| Escherichia coli AG115 | lacX74, galU, galK, araD139, strA, hsdR17/F’, lacIq lacZ::Tn10 | (39) |
| Escherichia coli DH5α | supE44, hsdR17, recA1, endA1, gyrA96, thi-1, relA1 | (11) |
| H.pylori J99 | cagA+ vacA+ isolated from a patient with duodenal ulcer | (7) |
| pGEX-KG | Ptac, gst, oripBR322 | (40) |
| GHPAQ41 | ATCCa Molecular Biology, pUC18 derivative containing the entire H.pylori dnaA gene (1741 bp fragment) | (6) |
| pGEXHpdnaA(IV) | pGEX-KG derivative containing the 343 bp EcoRI–BamHI fragment encoding domain IV of H.pylori DnaA protein | This study |
| pOC170 | pBR322 derivative containing the E.coli oriC region (five DnaA boxes) | (41) |
aATCC, American Type Culture Collection.
DNA manipulations
To prepare chromosomal DNA from H.pylori, cells were scraped from an agar plate and suspended in 200 µl of ice-cold STE buffer (150 mM NaCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA) containing lysozyme (100 µg/ml), and incubated at 37°C for 10 min. Sodium dodecyl sulfate (SDS) was then added to final concentrations of 1% and the cell lysate was incubated at 65°C for 10 min. After addition of proteinase K (final concentration 25 µg/ml), the samples were incubated at 50°C for 2 h (10). Chromosomal DNA was extracted using phenol–chloroform and precipitated with ethanol.
Purification of plasmids and DNA fragments was done using kits according to the manufacturers’ protocols (Qiagen). DNA fragments for footprinting experiments and gel retardation were PCR amplified using primers, one of which was 5′-end-labeled using [γ-32P]ATP and T4 polynucleotide kinase (11). Enzymes were supplied by Roche, Fermentas MBI and Gibco BRL. Isotopes were obtained from Amersham. The oligonucleotides used for PCR or for sequencing were chemically synthesized (Sigma-ARK Scientific, Darmstadt, Germany). DNA sequencing was performed using a Thermo Sequenase cycle sequencing kit (Amersham). For both strands, the nucleotide sequences were determined.
RNA isolation and RT–PCR analysis
Colonies of H.pylori were harvested from blood agar plate and resuspended in phosphate-buffered saline (0.8% NaCl, 0.02% KCl, 0.144% Na2HPO4, 0.024% KH2PO4, pH 7.4). Total RNA was extracted with TRI REAGENT (Molecular Research Center, Inc.). Subsequent steps of RNA isolation were carried out according to Chomczynski and Sacchi (12).
RT–PCR reactions were carried out using a Gibco BRL RT–PCR kit. After digestion of total RNA (3.6 µg) with DNase I, the samples were incubated at 65°C for 10 min and placed on ice. An oligonucleotide (20 pmol), PHPGR (Table 2) complementary to the 3′ region of the dnaA gene, was added to the RNA samples and then a reverse transcription (RT) reaction was performed at 37°C for 90 min. An aliquot of 5 µl out of 20 µl RT solution was subjected to subsequent PCR. The amplification reaction was carried out in 50 µl using primers PHPGR and PHPGF (Table 2) for 40 cycles, with each cycle consisting of denaturation, 15 s at 96°C; annealing, 30 s at 47°C for 1–20 cycles and 66°C for 21–40 cycles; and elongation, 30 s at 72°C. Products of the RT–PCR reaction were analyzed by agrose gel electrophoresis as well as by Southern hybridization. The DNA fragments were transferred onto nylon membrane and hybridized with the digoxygenin-labeled DNA probe. Immunodetection of the hybrids was performed according to the manufacturer’s protocol (Roche).
Table 2. Oligonucleotides used in this study.
| Oligonucleotide |
Sequencea |
| PHPGR | 5′-CGGAATTCTCATTCACTTGAATTGAA-3′ |
| PHPGF | 5′-CGGGATCCGATCATGCTGAAGGTTCA-3′ |
| HpS1a | 5′-CGCAAAGCAGCATGAAAATC-3′ |
| HpS1b | 5′-CAATATTGTTGTTGGTATCC-3′ |
| EcboxR | 5′-ACTCAAATAAGTATACAGATC-3′ |
| EcboxF | 5′-TGTGATCTCTTATTAGGATC-3′ |
| FHpbox1 | 5′-AAAGCAAGCATTATAGACAAACCCTTAAA-3′ |
| RHpbox1 | 5′-TTTAAGGGTTTGTCTATAATGCTTGCTTT-3′ |
| FHpbox2 | 5′-TTTTAAGGCTTCATTCACATTTCATTCAC-3′ |
| RHpbox2 | 5′-GTGAATGAAATGTGAATGAAGCCTTAAAA-3′ |
| FHpbox4 | 5′-CATTCACCACTTATTCACGCTATAATAAC-3′ |
| RHpbox4 | 5′-GTTATTATAGCGTGAATAAGTGGTGAATG-3′ |
| FHpbox5 | 5′-TATTCCTTTTTCATTCACCAACCCTTAAA-3′ |
| RHpbox5 | 5′-TTTAAGGGTTGGTGAATGAAAAAGGAATA-3′ |
aBold letters indicate restriction sites.
DnaA purification
The C-terminal domain (IV) of the H.pylori DnaA protein was fused to the C-terminus of GST. Part of the H.pylori dnaA gene encoding the domain IV of the DnaA protein was amplified using the primers PHPGF and PHPGR (Table 2). The amplified fragment digested with EcoRI and BamHI was cloned into the EcoRI and BamHI sites of the pGEX-KG expression vector. The fusion protein GST-HpDnaA(IV) was overexpressed in E.coli AG115 and purified using glutathione–Sepharose 4B beads (Pharmacia) as described previously (13,14). The purified proteins were analysed by SDS–polyacrylamide gel electrophoresis (PAGE). The E.coli DnaA protein was isolated as described earlier (15).
Preparation of antisera
Antisera were obtained from rabbits by immunization with the purified GST-HpDnaA(IV) fusion protein and mixed with Freund’s complete adjuvant. Serum samples were taken 10 days after the second booster injection. Cellular particles were removed by centrifugation, and antisera were stored at –20°C.
SDS–PAGE and western blotting
SDS–PAGE was performed according to the method established by Laemmli (16). Proteins were separated by 10 or 12% SDS–PAGE and transferred to a nitrocellulose membrane. The membrane was blocked for 1 h at room temperature with TBST (10 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.05% Tween-20) containing 3% bovine serum albumin (BSA), and subsequently incubated with a polyclonal anti-GST-HpDnaA(IV) antibody. Afterwards the DnaA protein was detected using a goat anti-rabbit secondary antibody conjugated with alkaline phosphatase. The membrane was stained with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium.
Electrophoretic mobility shift assay
For binding assays, DNA (50 ng) was incubated with the GST-HpDnaA(IV) or E.coli DnaA protein in the presence of a competitor ΦX 174 RF DNA (100 ng) at 20°C for 20 min in a binding buffer (20 mM HEPES–KOH pH 8.0, 5 mM Mg-acetate, 1 mM Na2-EDTA, 4 mM DTT, 0.2% Triton X-100, 5 mg/ml BSA and 1 mM ATP). The bound complexes were analyzed by electrophoresis on 1% agarose gels (0.25× TBE, at 4 V/cm, 4°C). Gels were stained with ethidium bromide. For radioactive shifts, 32P-labeled DNA (8 ng) was incubated with the GST-HpDnaA(IV) or E.coli DnaA in the presence of competitor poly(dI·dC) (100 ng) under conditions described above. The bound complexes were separated by electrophoresis in 8% polyacrylamide gels (0.25× TBE, at 4 V/cm, 4°C). Gels were dried and analyzed by autoradiography.
DNase I footprinting
For footprinting experiments, the promoter region of the H.pylori dnaA gene was amplified by PCR using HpS1a and HpS1b primers (Table 2). The 5′-radiolabeled DNA fragments (∼10 fmol) were incubated with different amounts of the GST-HpDnaA(IV) protein in a binding buffer (20 mM HEPES pH 7.6, 5 mM Mg-acetate, 4 nM DTT, 1 mM EDTA, 3 mM ATP, 0.2% Triton X-100, 5% glycerol, 100 mM K-acetate, 5 mM Ca-acetate) at room temperature for 30 min. Then DNase I digestion was carried out according to the described procedure (17). The DNase I cleavage products were separated in 8% polyacrylamide–urea sequencing gels. Gels were dried and analyzed by autoradiography.
Computer analysis
The sequences were analyzed with the Wisconsin Package v9.0 Genetics Computer Group (GCG, Madison, WI). Sequence alignments were created with the Bestfit, while the pI value was predicted with the IsoElectric program modules. Secondary structure predictions were obtained from the PHD server: http ://www.embl-heidelberg. de /predictprot ein/predictprotein.html, which includes the algorithms developed by Rost and Sander (18). Sequences of dnaA genes were from the National Center for Biotechnology: Bacillus subtilis (P05648), Campylobacter jejuni (CAB72494), E.coli (BAA16384), H.pylori J99 (AAD06996).
DNA walks
The computational analysis was performed on the sequence of the H.pylori J99 genome downloaded from: http: //www.ncbi.nlm.nih.gov. To find the point where the DNA asymmetry changes its sign, we performed DNA walks on the complete chromosome sequence. The data obtained have been presented in the form of a diagram of detrended DNA walks as described previously (19,20). In this walk, the shift of the walker in the two-dimensional space is [1,1] for guanine, [1,0] for adenine and thymine and [1,(G/C)] for cytosine, where G and C are total numbers of guanine and cytosine in the analyzed sequence. In such a walk, the walker starts and completes its walk at position zero on the y-axis. Coordinates on the x-axis correspond to the position on the chromosome. After a rough estimation of the region of the origin of replication, we performed detailed DNA walks in that region, analyzing differences between G and C content in the DNA sequence. Methods are also described in detail at web site http://smORFland.microb.uni.wroc.pl.
RESULTS
Characterization of the H.pylori J99 dnaA gene and its product, initiator protein DnaA
The predicted molecular mass of the H.pylori DnaA protein is 52.7 kDa. Based on a phylogentic tree of eubacterial DnaA proteins (not shown), H.pylori DnaA is most similar to DnaA of C.jejuni (39.9% identity). DnaA protein from distantly related E.coli still reveals 32.9% identity to H.pylori DnaA.
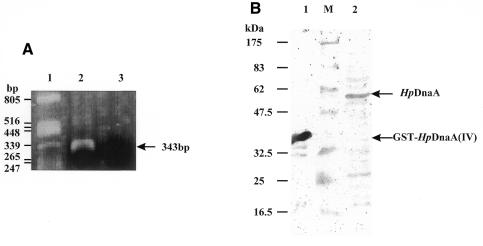
Expression of the dnaA gene in H.pylori cells was proved by RT–PCR (Fig. 1A) and western blotting (Fig. 1B). The antibodies against the H.pylori DnaA protein (see Materials and Methods) detected a 53 kDa H.pylori protein corresponding in size to the deduced dnaA gene product (Fig. 1B).
Figure 1.
Expression of the dnaA in H.pylori J99 cells. (A) RT–PCR amplification of dnaA mRNA. The RT–PCR reactions were performed as described in Materials and Methods. Molecular size markers are indicated on the left and the product of RT–PCR is indicated on the right. Lane 1, marker; lane 2, RNA (3.6 µg) digested with DNase I; lane 3, control: RNA (3.6 µg) digested with DNase I, PCR reaction performed without reverse transcriptase. (B) Western analysis of total H.pylori cell proteins using a rabbit polyclonal antiserum directed against domain IV of the H.pylori DnaA protein. Molecular mass markers (M) are indicated on the left and the positions of reactive antigens are indicated on the right. Lane 1, purified fusion protein GST-HpDnaA(IV); lane 2, total H.pylori cell proteins.
Based on the homology pattern, bacterial DnaA proteins have been divided into four domains, to which different functions could subsequently be assigned (2,21; Fig. 2). The DNA binding domain of E.coli DnaA has been localized in the C-terminus. In order to check whether the C-terminus of the H.pylori DnaA protein is responsible for DNA binding, its interaction with DNA was analyzed by gel retardation assay. The PCR-amplified DNA fragment of the dnaA gene encoding domain IV of H.pylori DnaA protein fused to the gst gene (Materials and Methods and Table 1) was overexpressed in E.coli AG115. The resulting fusion protein, GST-HpDnaA(IV) (38 kDa) was purified by affinity chromatography on glutathione–Sepharose 4B as described earlier (13,14). Since the H.pylori oriC region had not been known, the E.coli oriC region containing five DnaA boxes was chosen to study the GST-HpDnaA(IV) protein interaction with DNA (Fig. 3). The GST-HpDnaA(IV) protein was incubated with the E.coli oriC fragment and then the DNA–protein complexes were analyzed by a gel retardation assay (Fig. 3). The interaction of the GST-HpDnaA(IV) protein with the E.coli oriC region led to formation of discrete nucleoprotein complexes (Fig. 3). The non-specific competitor (ΦX 174 RF DNA) was not bound at all by the fusion protein. Binding of the GST protein alone to the E.coli oriC fragment was not observed (data not shown). Nucleoprotein complex formation between GST-HpDnaA(IV) protein and a single DnaA box from the E.coli oriC region occurred already at the lowest protein concentration (at a GST-HpDnaA(IV)/DnaA box molar ratio of 0.6:1). At higher protein concentration, sequential binding of GST-HpDnaA(IV) molecules to the five DnaA boxes caused the formation of high molecular weight complexes (Fig. 3). Thus, the data specify the C-terminal domain of the H.pylori DnaA protein as the DNA binding domain.
Figure 2.
Protein sequence alignment of homologous DnaA proteins. The borders of different domains have been adjusted according to Messer et al. (21). Identical amino acids are shown with a black background, similar amino acids are shaded. Secondary structure prediction for the domain IV of E.coli and H.pylori DnaA proteins was obtained from PHD server as described in Materials and Methods.
Figure 3.
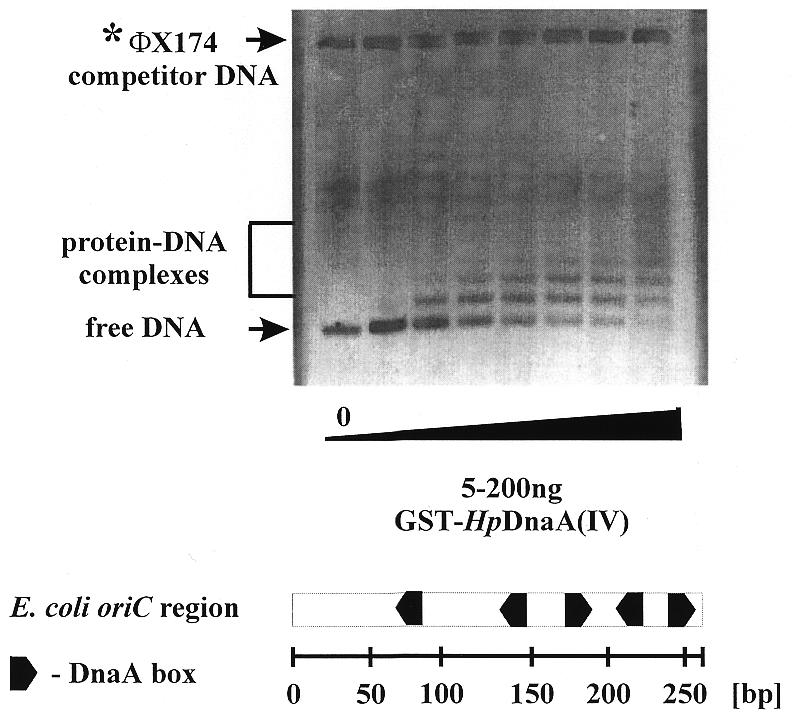
Interaction of GST-HpDnaA(IV) with the replication origin of E.coli. Gel retardation assay was carried out as described in Materials and Methods using the 460 bp XhoI–SmaI fragment of pOC170 containing the E.coli oriC region (Table 1) and varying concentrations of protein. The GST-HpDnaA(IV)–E.coli oriC complexes formed were separated in a 1% agarose gel. The bottom part shows the structure of the E.coli oriC region.
In silico identification of the putative H.pylori origin of replication
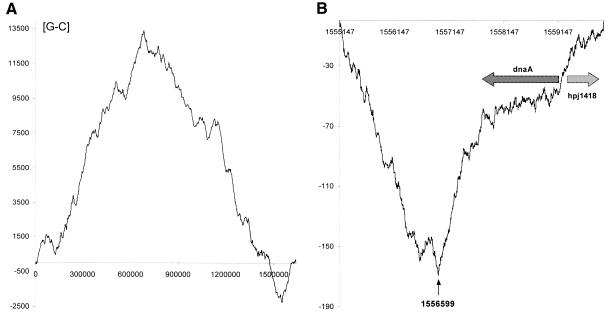
It is well documented that all eubacterial chromosomes are divided by origin (oriC) and terminus (terC) of replication (or termini in linear chromosomes) into two replichores. It has been noted that in many bacterial genomes sequenced so far, the leading strand contains more G than C. Thus, the oriC and terC regions of chromosome replication can be detected by plotting this GC skew along the genome (22). The best graphical method indicating the point where the bias in nucleotide composition of the DNA molecule changes its sign is the detrended DNA walk (19,20). This demonstrates the local deviations from the average composition (i.e. G and C content) in the analyzed consecutive DNA sequences (Fig. 4). A diagram showing the result of [G-C] DNA walks on the whole H.pylori chromosome is presented in Figure 4A. There are two evident global switch points of DNA asymmetry. To obtain more precise results, we performed a [G-C] DNA walk nucleotide by nucleotide in the region close to the putative origin of replication (Fig. 4B). Also in Figure 4B, the positions of the dnaA gene and the open reading frame (ORF) jhp1418 are indicated. According to the computational analysis the switch of DNA asymmetry is located downstream of the dnaA gene at the position 1 556 599 bp.
Figure 4.
In silico identification of the putative H.pylori oriC region. Detrended [G-C] DNA walks on the H.pylori J99 chromosome performed for (A) the whole chromosome and (B) the region close to the putative origin of replication. In such walks, the walker goes up when it finds G and down when it finds C in its position. Steps up and down are corrected in such a way that the walker completes its walk at position zero on the y-axis. Coordinates on the x-axis correspond to the position on the chromosome. After a rough estimation of the region of the origin of replication, we performed detailed DNA walks in that region, analyzing the difference between G and C content on the DNA sequence. The positions of the dnaA gene and of the ORF jhp1418 are shown. The switch of DNA asymmetry is located downstream of the dnaA gene at position 1 556 599 bp. The scale on the x-axis corresponds to coordinates of the chromosome sequence in base pairs as annotated in the database http://www.ncbi.nlm.nih.gov. Numbers on the y-axis indicate the relative cumulative abundance of the nucleotides in the analyzed region.
In vitro identification of the replication origin from H.pylori
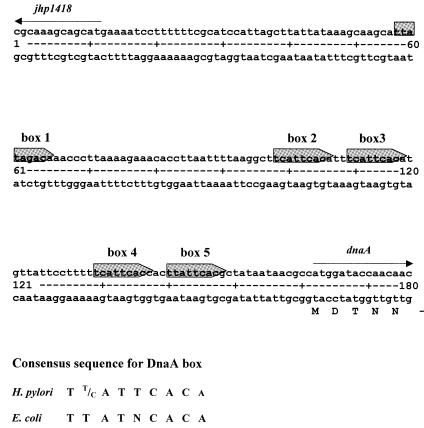
In many eubacterial chromosomes including B.subtilis (23), Mycobacterium tuberculosis (24) and Streptomyces lividans (13,25), the dnaA gene is located close to the functional replication origin. The close vicinity of the global switch point of DNA asymmetry (the minimum, Fig. 4B) and the dnaA gene on the H.pylori chromosome prompted us to look for putative origin sequences (DnaA boxes). A search for DnaA box motifs whose sequence differs up to 2 nt from the most stringent consensus sequence for the E.coli DnaA box (5′-TTATNCACA-3′) allowed the identification of five DnaA boxes upstream of the dnaA gene (Fig. 5). All five DnaA boxes have the same orientation; the pairs of DnaA boxes 2, 3 and 4, 5 are closely spaced (2 bp in between) while the distance between DnaA boxes 1, 2 and 3, 4 is 33 and 13 bp, respectively (Fig. 5).
Figure 5.
Structure of the putative H.pylori oriC region. The DnaA boxes are shaded in gray. The bottom part shows the consensus sequence for the H.pylori and the E.coli DnaA box.
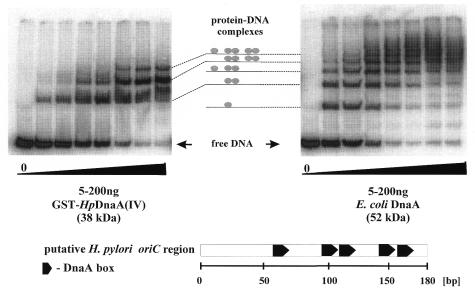
To determine whether the GST-HpDnaA(IV) protein interacts with the DNA fragment containing five H.pylori DnaA boxes, a gel retardation assay was performed. In addition, we also evaluated whether the E.coli DnaA protein specifically binds these boxes. The putative H.pylori oriC region was amplified using a pair of primers: HpS1a and 32P-labeled HpS1b (Table 2). After incubation of the labeled 187 bp DNA fragment with different amounts of the GST-HpDnaA(IV) fusion protein or the E.coli DnaA protein, the resulting nucleoprotein complexes were analyzed in an 8% native polyacrylamide gel (Fig. 6). At higher concentrations of the E.coli DnaA protein, five nucleoprotein complexes were observed. Binding of the GST-HpDnaA(IV) protein resulted in the formation of three nucleoprotein complexes. Probably, the GST-HpDnaA(IV) binds two closely spaced DnaA boxes (2, 3 and 4, 5) as a dimer due to interaction via its N-terminus (i.e. GST) (Fig. 6). The affinity of the GST-HpDnaA(IV) protein to the single DnaA box 1 seems to be low since the band corresponding to the nucleoprotein complex with the lowest mobility (Fig. 6) was weak.
Figure 6.
Interaction of the GST-HpDnaA(IV) protein (left) and the E.coli DnaA protein (right) with the putative H.pylori oriC region. Gel retardation assay was carried out as described in Materials and Methods using 32P-labeled 187 bp H.pylori oriC fragment (amplified by HpS1a and 32P-labeled HpS1b primers, Table 2) and varying concentrations of protein. The DNA–protein complexes were separated in an 8% polyacrylamide gel. The bottom part shows the structure of the putative H.pylori oriC region.
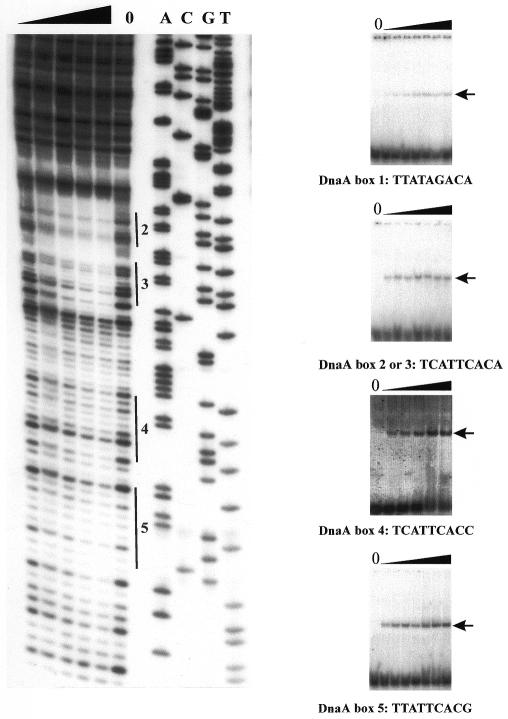
To confirm the location of the DnaA binding sites within the H.pylori oriC region, DNaseI footprinting analysis was performed. The 187 bp PCR-amplified DNA fragment, labeled at one 5′-end (lower strand, see above) was incubated with various amounts of the GST-HpDnaA(IV) protein and then subjected to limited DNase I cleavage (Fig. 7). The protected sites correspond to the locations of four DnaA boxes 2, 3, 4, 5 within the analyzed oriC region. At higher protein concentrations the entire DNA fragment (53 bp) containing four DnaA boxes and a short spacer (13 bp) between DnaA boxes 3 and 4 was protected. Using the upper strand (32P-labeled primer HpS1a), identical protection sites were determined (data not shown). However, protection of the DnaA box 1 was not observed for both strands.
Figure 7.
Interaction of the GST-HpDnaA(IV) protein with individual DnaA boxes from the putative H.pylori oriC region. Left, DNase I footprinting analysis (lower strand). DNase I footprinting was performed using the 187 bp H.pylori oriC fragment that was amplified by HpS1a and 32P-labeled HpS1b primers. DNA fragments were incubated with increasing amounts of the GST-HpDnaA(IV) as described in Materials and Methods. Only protected DnaA boxes (2, 3, 4, 5) are marked. Lanes A, C, G and T are sequencing reactions prepared with the 32P-labeled HpS1b primer. Right, gel retardation assay. The assay was performed using labeled double-stranded oligonucleotides containing a single DnaA box 1, 2, 3, 4 or 5. DNA fragments were incubated with increasing amounts of the GST-HpDnaA(IV) as described in Materials and Methods. The DNA–protein complexes were separated in an 8% polyacrylamide gel. The arrow indicates the nucleoprotein complex.
In addition, we evaluated in detail the interaction of the GST-HpDnaA(IV) protein with individual DnaA boxes from the putative H.pylori oriC, using a gel retardation assay. Double-stranded, 32P-labeled oligonucleotides containing a single DnaA box 1, 2 (box 3 is identical to box 2), 4 and 5 (Table 2) were incubated with various amounts of the GST-HpDnaA(IV) protein and then nucleoprotein complexes were separated in an 8% native polyacrylamide gel (Fig. 7). For each analyzed oligonucleotide a single nucleoprotein complex was already visible at a low protein concentration (at a GST-HpDnaA(IV)/DnaA box ratio of 1:1 or 1:2, respectively). However, binding of GST-HpDnaA(IV) protein to the DnaA box 1 resulted in a faint nucleoprotein complex suggesting that this complex is quite unstable. This is in agreement with the observation from the DNase I footprinting analysis where DnaA box 1 was not protected.
DISCUSSION
Until recently, nothing was known about the initiation of chromosome replication in H.pylori, one of the most extensively studied pathogens. Despite the fact that the complete genome sequence of two different strains of H.pylori, 26695 (6) and J99 (7), has been determined, a typical eubacterial origin of replication was not identified. Here we characterize the key elements of the initiation of H.pylori chromosome replication: DnaA protein and the putative oriC region. In addition, we analyzed the interaction between these key elements using gel retardation and DNase I footprinting.
Comparison of numerous eubacterial genetic maps around the chromosome replication initiator gene, dnaA, showed conservation of the gene cluster rnpA-rmpH-dnaA-dnaN-recF-gyrB (2,8,9). However, the gene arrangement in the H.pylori dnaA region differs from that found in other eubacterial dnaA regions. Helicobacter pylori dnaA is flanked by two ORFs with unknown function (Fig. 8). The expression of the dnaA gene in H.pylori cells was confirmed by RT–PCR and western blotting analysis using antibodies against H.pylori DnaA protein (Fig. 1).
Amino acid sequence comparison of DnaA proteins and secondary structure prediction (data not shown) have shown that the DnaA protein of H.pylori, like other DnaA proteins, can be divided into four domains (Fig. 2). Mutational analysis of several DnaA proteins (26–30) has shown that the C-terminus of the DnaA protein (domain IV) is necessary and sufficient for specific DNA binding. The DNA binding motif of bacterial DnaA proteins consists presumably of two α-helices (A and B) with a basic loop between them, and a third long α-helix (C1-2) (21,30,31). The predicted secondary structure of the H.pylori DnaA protein reveals the presence of three potential α-helices within the C-terminus (domain IV) (Fig. 2). Four basic amino acid residues are located between two α-helices of the H.pylori domain IV (Fig. 2, KSSEIKVSSRQR). Gel-retardation assay clearly demonstrated that the H.pylori domain IV is responsible for specific DNA binding (Figs 3, 6 and 7).
In many eubacterial chromosomes the dnaA gene is located close to the origin of replication. In the case of H.pylori J99 we have found that the dnaA is also located close to the oriC region as predicted in silico analysis, 2.5 kb away from the putative oriC region (Fig. 4). On the basis of the DNA walks almost the same topology was obtained for the other H.pylori strain, 26695; the switch in DNA asymmetry is located 1.4 kb from the dnaA gene (at the position 1 606 201 bp; data not shown). In the chromosome of the related organism C.jejuni, the bias in [G-C] content also indicates co-localization of the oriC region and the dnaA gene (32). The region of the potential replication origin of H.pylori 26695 has been suggested by Freeman et al. (33) to be ∼60 kb distant from the dnaA gene using strand asymmetry in a sliding windows analysis (Fig. 8). Salzberg et al. (34) found the switch of the asymmetry in oligomer frequency about 18 kb away from the dnaA gene (Fig. 8). Recently, Grigoriev (35), analyzing cumulative skew diagrams for short windows, found the switch in asymmetry close to the dnaA gene for both sequenced H.pylori genomes. In the case of H.pylori J99, the global minimum determined here is located only ∼500 bp away from the centre of the region predicted by Grigoriev (35). Thus, it is not a very significant difference. It may result from different methods of DNA walks. Grigoriev (35) used an adjacent windows analysis for the cumulative skew diagrams construction while in our detailed analysis we performed a walk nucleotide by nucleotide. In numerous genomes the in silico predicted origin is in close proximity to a classical progenitor origin region containing at least a few DnaA binding motifs (DnaA boxes) (36). In these organisms, the functional oriC region is located within the cluster of genes rnpA-rmpH-dnaA-dnaN-recF-gyrB rnpA, usually next to the dnaA gene. Moreover, for B.subtilis (23) and S.lividans (25), it was experimentally determined that the region containing numerous DnaA boxes in close vicinity of the dnaA gene can replicate as an autonomous circular minichromosome.
The correspondence between the location of the dnaA gene and oriC as predicted in silico prompted us to look for typical repeated origin sequences: DnaA boxes and AT-rich regions in the vicinity of the H.pylori J99 dnaA gene. Five DnaA boxes have been found next to the start of the dnaA gene (Fig. 5); the sequences of four differ by 1 nt from the most stringent consensus sequence of E.coli DnaA box (TTATNCACA), whereas the sequence of the fifth DnaA box differs by 2 nt. It has to be noted that except for the putative oriC region we did not find a region that contains five DnaA boxes within ∼200 bp. Comparison of five DnaA boxes from the putative H.pylori oriC region allowed us to propose the consensus sequence: TT/CATTCACA (Fig. 5). As in other bacteria, the bases in the positions 4, 5, 6, 7 and 8 of the H.pylori DnaA boxes were found to be highly conserved (37), and at least three of the four most important T residues (2, 4, 7′, 9′) are conserved in each box (38). There are many examples in which a cluster of four or more DnaA boxes indicates a functional chromosomal initiation region. Moreover, domain IV of H.pylori DnaA protein specifically binds each of the five DnaA boxes from the identified origin (Fig. 7).
The G+C content of the putative H.pylori oriC region is almost 10% lower than the overall G+C content of H.pylori DNA (39%). Since the entire oriC region is AT-rich (70%) it is difficult to predict where unwinding occurs. Typical AT-rich 13mers direct repeats have not been found. Currently, we are trying to determine whether the opening of DNA strands takes place within DnaA boxes or in a region adjacent to the DnaA boxes cluster.
Altogether, our results suggest that we have identified the putative H.pylori oriC region that is located upstream of the dnaA gene. The putative oriC region contains five DnaA boxes. DNase I and gel retardation assays clearly demonstrated that the C-terminal domain of the H.pylori initiator DnaA protein specifically binds each of the five DnaA boxes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jurek Majka for help in isolation of the GST-HpDnaA(IV) protein. We are grateful to Tomasz Cierpicki for helping in secondary structure prediction of H.pylori DnaA. A.Z. and D.J gratefully acknowledge financial support received from the Committee of Scientific Studies (KBN grant 6P04A 033 18) and from the Foundation for Polish Science, respectively. S.C. and P.M were supported by KBN grant 6PO4A 025 18. This work was in part supported by grant 436 POL 113/82/0 of the Deutsche Forschungsgemeinschaft.
References
- 1.Baker T.A.and Bell,S.P. (1988) Polymerases and the replisome: machines within machines. Cell, 92, 295–305. [DOI] [PubMed] [Google Scholar]
- 2.Messer W. and Weigel,C. (1996) Initiation of chromosome replication. In Neidhardt,F.C., Curtis,R.,III, Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella. Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1579–1601.
- 3.Kornberg A. and Baker,T. (1992) The DNA Replication, 2nd Edn. Freeman,W.H. & Co., New York, NY.
- 4.Fang L.H., Davey,M.J. and O’Donnell,M. (1999) Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell, 4, 541–553. [DOI] [PubMed] [Google Scholar]
- 5.Marshall B.J. and Warren,J.R (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet, 16, 1311–1315. [DOI] [PubMed] [Google Scholar]
- 6.Tomb J.-F., White,O., Kerlavage,A.-R., Clayton,R.A., Sutton,G.G., Fleischmann,R.D., Ketchum,K.A., Klenk,H.P., Gill,S., Dougherty,B.A. et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature, 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 7.Alm R.A., Ling,L.-S.L., Moir,D.T., King,B.L., Brown,E.D., Doig,P.C., Smith,D.R., Noonan,B., Guild,B.C., deJonge,B.L. et al. (1999) Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature, 397, 176–180. [DOI] [PubMed] [Google Scholar]
- 8.Ogasawara N. and Yoshikawa,H. (1992) Genes and their organization in the replication origin region of the bacterial chromosome. Mol. Microbiol., 6, 629–634. [DOI] [PubMed] [Google Scholar]
- 9.Yoshikawa H. and Ogasawara,N. (1991) Structure and function of DnaA and the DnaA-box in eubacteria: evolutionary relationships of bacterial replication origins. Mol. Microbiol., 5, 2589–2597. [DOI] [PubMed] [Google Scholar]
- 10.Covacci A. and Rappuolo,R. (1996). PCR amplification of gene sequences from Helicobacter pylori. In Lee,A. and Megraud,F. (eds), Helicobacter pylori: Techniques for Clinical Diagnosis and Basis Research. WB Saunders Company Ltd, London, UK, pp. 94–111.
- 11.Sambrook J.F., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- 12.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 13.Jakimowicz D., Majka,J., Messer,W., Speck,C., Fernandez,M., Martin,M.C., Sanchez,J., Schauwecker,F., Keller,U., Schrempf,H. and Zakrzewska-Czerwinska,J. (1998) Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology, 144, 1281–1290. [DOI] [PubMed] [Google Scholar]
- 14.Majka J., Jakimowicz,D., Zarko-Postawka,M. and Zakrzewska-Czerwinska,J. (1997) Glutathione S-transferase fusions proteins as an affinity reagent for rapid isolation of specific sequence directly from genomic DNA. Nucleic Acids Res., 25, 2537–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaper S. and Messer,W. (1995) Prediction of the structure of the replication initiator protein DnaA. J. Biol. Chem., 270, 17622–17626. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U.K. (1970) Cleavage of structural proteins during assembly of the head of the bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 17.Roth A., Urmoneit,B. and Messer,W. (1994) Functions of histone-like proteins in the initiation of DNA replication at oriC of Escherichia coli. Biochimie, 76, 917–923. [DOI] [PubMed] [Google Scholar]
- 18.Rost B. and Sander,C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins, 19, 55–74. [DOI] [PubMed] [Google Scholar]
- 19.Cebrat S. and Dudek,M. (1998) The effect of DNA phase structure on DNA walks. Eur. Phys. J., B3, 271–276. [Google Scholar]
- 20.Mackiewicz P., Gierlik,A., Kowalczuk,M., Dudek,M.R. and Cebrat,S. (1999) Asymmetry of nucleotide composition of prokaryotic chromosomes. J. Appl. Genet., 40, 1–14. [Google Scholar]
- 21.Messer W., Blaesing,F., Majka,J., Nardmann,J., Schaper,S., Schmidt,A., Seitz,H., Speck,C., Tungler,D., Wegrzyn,G., Weigel,C., Welzeck,M. and Zakrzewska-Czerwi_ska,J. (1999) Functional domains of DnaA proteins. Biochimie, 81, 819–825. [DOI] [PubMed] [Google Scholar]
- 22.Lobry J.R. (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol., 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 23.Moriya S., Atlung,T., Hansen,F.G., Yoshikawa,H. and Ogasawara,N. (1992) Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol. Microbiol., 6, 309–315. [DOI] [PubMed] [Google Scholar]
- 24.Salazar L., Fsihi,H., De Rossi,E., Riccardi,G., Rios,C., Cole,S.T. and Takiff,H.E. (1996) Organization of the origins of replication of the chromosomes of Mycobacterium tuberculosis and isolation of a functional origin from M. smegmatis. Mol. Microbiol., 20, 283–293. [DOI] [PubMed] [Google Scholar]
- 25.Zakrzewska-Czerwinska J. and Schrempf,H. (1992) Characterization of an autonomously replicating region from the Streptomyces lividans chromosome. J. Bacteriol ., 174, 2688–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth A. and Messer,W. (1995) The DNA binding domain of the initiator protein DnaA. EMBO J., 14, 2106–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richter S. and Messer,W. (1995) Genetic structure of the dnaA region of the cyanobacterium Synechocystis sp. strain PCC6803. J. Bacteriol., 177, 4245–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutton M.D. and Kaguni,J.M. (1997) Novel alleles of the Escherichia coli dnaA gene. J. Mol. Biol., 271, 693–703. [DOI] [PubMed] [Google Scholar]
- 29.Majka J., Jakimowicz,D., Messer,W., Schrempf,H., Lisowski,M. and Zakrzewska-Czerwinska,J. (1999) Interactions of the Streptomyces lividans initiator protein DnaA with its target. Eur. J. Biochem., 260, 325–335. [DOI] [PubMed] [Google Scholar]
- 30.Blaesing F., Weigel,C., Welzeck,M. and Messer,W. (2000) Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol. Microbiol., 36, 557–569. [DOI] [PubMed] [Google Scholar]
- 31.Schaper S. and Messer,W. (1997) Prediction of the structure of the replication initiator protein DnaA. Proteins, 28, 1–9. [PubMed] [Google Scholar]
- 32.Parkhill J., Wren,B.W., Mungal,K., Ketley,J.M., Churcher,C., Basham,D., Chillingworth,T., Davies. R.M., Feltwell,T., Holroyd,S., Jagels,K., Karlyshev,A.V., Moule,S., Pallen,M.J., Penn,C.W., Quail,M.A., Rajandream,M.-A., Rutherford,K.M., van Vliet,A.H.M., Whitehead,S. and Barrell,B.G. (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature, 403, 665–668. [DOI] [PubMed] [Google Scholar]
- 33.Freeman J.M., Plasterer,T.N., Smith,T.F. and Mohr,S.C. (1998) Patterns of genome organization in bacteria. Science, 279, 1827a. [Google Scholar]
- 34.Salzberg S.L., Salzberg,A.J., Kerlavage,A.R. and Tomb,J.F. (1998) Skewed oligomers and origins of replication Gene, 217, 57–67. [DOI] [PubMed] [Google Scholar]
- 35.Grigoriev A. (2000) Graphical genome comparison: rearrangements and replication origin of Helicobacter pylori. Trends Genet., 16, 376–378. [DOI] [PubMed] [Google Scholar]
- 36.Grigoriev A. (1998) Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res., 26, 2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaefer C. and Messer,W. (1991) DnaA protein/DNA interaction. Modulation of the recognition sequence. Mol. Gen. Genet., 226, 34–40. [DOI] [PubMed] [Google Scholar]
- 38.Speck C., Weigel,C. and Messer,W. (1997) From footprint to toeprint: a close-up of the DnaA box, the binding site for the bacterial initiator protein DnaA. Nucleic Acids Res., 25, 3242–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattern S. (1992) Regulation of the galactose operon of Streptomyces lividans. Thesis. Universität Osnabrück, Osnabrück, Germany.
- 40.Guan K.L. and Dixon,J.E. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem., 192, 262–267. [DOI] [PubMed] [Google Scholar]
- 41.Messer W., Hartmann-Kuhlein,H., Langer,U., Mahlow,E., Roth,A., Schaper,S., Urmoneit,B. and Woelker,B. (1992) The complex for replication initiation of Escherichia coli. Chromosoma, 102, S1–S6. [DOI] [PubMed] [Google Scholar]