Abstract
Normal physiology undergoes 24-hour changes in function, that include daily rhythms in circulating/hormones, most notably melatonin and cortical steroids. This study focuses on N-acetyltryptamine, a little-studied melatonin receptor mixed agonist/antagonist and the likely evolutionary precursor of melatonin. The central issue addressed was whether N-acetyltryptamine is physiologically present in the circulation. N-Acetyltrypamine was detected by LC-MS/MS in daytime plasma of three different mammals in subnanomolar levels (mean ± SEM: rat, 0.29 ± 0.05 nM, N=5; rhesus macaque, 0.54 ± 0.24 nM, N=4; human, 0.03 ± 0.01 nM, N=32). Twenty four hour blood collections from rhesus macaques revealed a nocturnal increase in plasma N-acetyltryptamine (P < 0.001), which varied from 2- to 15- fold over daytime levels among the four animals studied. Related RNA sequencing studies indicated that the transcript encoding the tryptamine acetylating enzyme arylalkylamine N-acetyltransferase (AANAT) is expressed at similar levels in the rhesus pineal gland and retina, thereby indicating that either tissue could contribute to circulating N-acetyltryptamine. The evidence that N-acetyltryptamine is a physiological component of mammalian blood and exhibits a daily rhythm, together with known effects as a melatonin receptor ligand shifts the status of N-acetyltryptamine from pharmacological tool to that of a candidate for a physiological role. This provides a new opportunity to extend our understanding of 24-hour biology.
Keywords: N-Acetyltryptamine, Melatonin, Pineal, Retina, Human, Nonhuman primate, Daily rhythm, Transcriptome profiling
INTRODUCTION
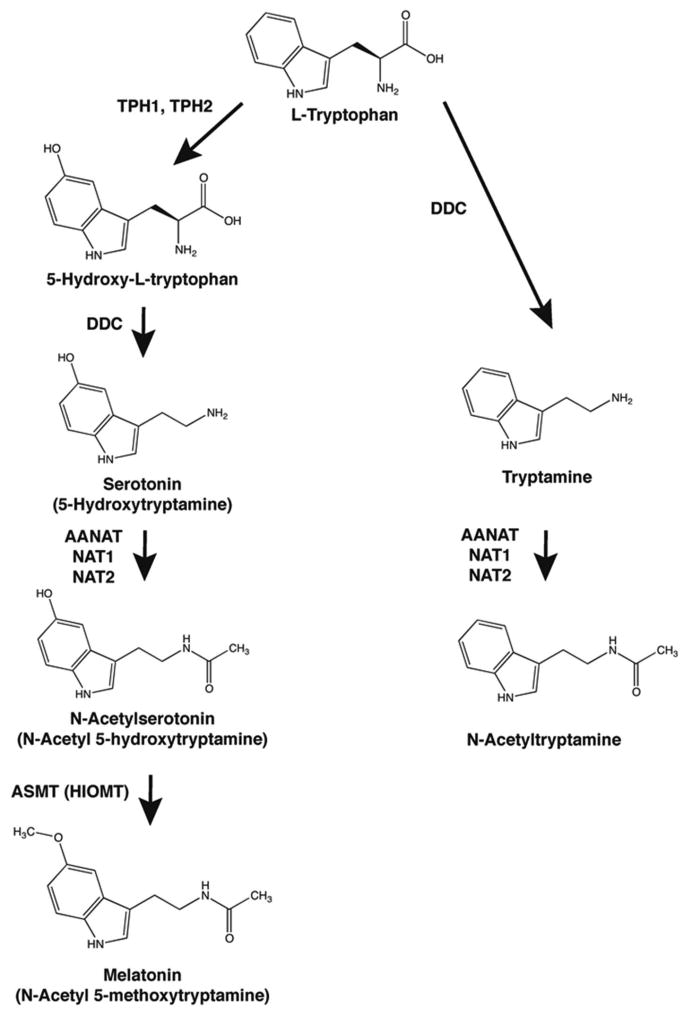
N-Acetyltryptamine is an analog of melatonin (5-methoxy N-acetyltryptamine) (Figure 1) and its likely evolutionary precursor (Klein 2004; Klein 2006). Whereas thousands of publications over the last half century focused on melatonin and the role it plays as the hormonal output signal of the circadian system (Wood and Loudon 2014), N-acetyltryptamine has received considerably less attention. This is surprising because it has been known for decades from pharmacological studies that N-acetyltryptamine is a mixed melatonin receptor agonist/antagonist (Dubocovich, Delagrange et al. 2010). These include investigations using amphibian skin by Heward and Hadley (Heward and Hadley 1975), mammalian retina by Dubocovich and colleagues (Dubocovich 1984; Dubocovich 1985), suprachiasmatic cells by Mason and Brooks (Mason and Brooks 1988) and amphibian melanocytes and chicken brain by Sugden and coworkers (Chong and Sugden 1994; Sugden, Chong et al. 1995).
Figure 1. Synthesis of N-acetyltryptamine and melatonin from tryptophan.
The biosynthesis of N-acetyltryptamine from tryptophan by a pathway sharing enzymes with the melatonin pathway. TPH1, tryptophan hydroxylase-1; DDC, dopa decarboxylase; AANAT, arylalkylamine -acetyltransferase; NAT1, arylamine N-acetyltransferase 1; NAT2, arylamine N-acetyltransferase 2; ASMT, acetylserotonin O-methyltransferase; HIOMT, hydroxyindole-O-methyltransferase.
Both N-acetyltryptamine and melatonin are derivatives of tryptophan (Figure 1). Synthesis of N-acetyltryptamine in involves decarboxylation of tryptophan to tryptamine by a broadly distributed aromatic amino acid decarboxylase (DDC); the next step in the conversion is N-acetylation. The latter can occur in vertebrates through the action of two enzyme families.
The first is the arylalkylamine N-acetyltransferase (AANAT) family, that selectively and efficiently acetylate arylalkylamines (Klein 2007; Pavlicek, Sauzet et al. 2010; Falcon, Coon et al. 2014); AANAT controls the daily rhythm in pineal melatonin synthesis (Klein 2007). In addition to being highly expressed in the pineal gland, it is also present in vertebrate retinas at variable levels and at distinctly lower levels in the pituitary gland (Coon, Roseboom et al. 1995; Fleming, Barrett et al. 1999; Coon, Del Olmo et al. 2002; Iuvone, Tosini et al. 2005; Tosini, Pozdeyev et al. 2008) and brain (Gaudet, Palkovits et al. 1991; Gaudet and Namboodiri 1993). A single AANAT gene is found in most vertebrate genomes; however, several are present fish genomes (Klein 2007).
The second family of enzymes that can acetylate arylalkylamines (Steinberg, Cohen et al. 1969) includes arylamine N-acetyltransferase 1 (NAT1) and 2 (NAT2) (Huang, Louie et al. 2013), which catalyze the acetylation of arylamines and hydrazines. The ability of these enzymes to acetylate arylalkylamines may explain the observation that N-acetylserotonin is a minor metabolite of serotonin by perfused rat liver (Tyce, Flock et al. 1968; Tyce, Flock et al. 1968). NAT1 is expressed widely across human tissues, whereas human NAT2 is expressed primarily in the liver and gastrointestinal tract. Human NAT2 has been of special interest because it exhibits a common polymorphism resulting in rapid, intermediate and slow acetylator phenotypes (Hein 2009).
The paucity of knowledge of the biology of N-acetyltryptamine may reflect its status as a pharmacological tool and that it has far lower affinity than melatonin at melatonin receptors (Dubocovich, Delagrange et al. 2010). In addition, and perhaps most importantly, the physiological presence of N-acetyltryptamine has not been reported. Here, this issue was addressed using mass spectrometry, a method that has been used previously for melatonin analysis (Lewy and Markey 1978; Carter, Calcutt et al. 2012). The results of this study establish that N-acetyltryptamine is physiologically present in plasma and exhibits a daily rhythm. This advance points to a possible physiological role of N-acetyltryptamine as a chronobiological signal.
MATERIALS AND METHODS
Biological materials
Rhesus macaques
The rhesus macaques (Macaca mulatta) used in this study came from two sources. Those used for 24-hour plasma collection were housed at the Oregon National Primate Center indoors under controlled lighting comprising 12 h of light and 12 h of darkness per day (12:12 light:dark; lights on at 7:00 h). Care was in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals at the Oregon National Primate Center. Monkey chow (LabDiet High Protein Monkey Chow, LabDiet, Inc., St. Louis, MO), supplemented with fresh fruit and vegetables was provided daily at 8:00 h and 15:00 h, and fresh drinking water was available ad libitum. Serial blood samples were collected across the day and night using a surgically implanted subclavian vein catheter and a remote blood sampling system (Urbanski 2011; Urbanski, Sorwell et al. 2014); plasma samples were prepared and stored frozen. The animals were part of unrelated previous studies.
The rhesus macaques used for RNA sequencing and LC-MS/MS analysis of tissues were obtained through the Veterinary Resources Program Primate Recycle Program of NIH over a period of 11 years (Protocols 99-012 and 11-055). These animals were 3 to 33 years of age, male and female, 4 to 14 kg body weight. All protocols and handling of the animals conformed to the Guidelines for Care and Use of Laboratory Animals (NIH Publication 80-23). Animals were housed under a 12-h light, 12-h dark lighting regimen (lights on at 06:00 h) in NIH facilities for at least 6 weeks. The light level in the cages was approximately 325 Lux. Animals (three per time point) were sacrificed between 11:00 and 13:00 h (day), 17:00 and 19:00 h (dusk), 23:00 and 01:00 h (night) and 05:00 and 07:00 h (dawn). Animals were anesthetized by intramuscular injection (0.1 ml/kg body weight) of Ketamine. HCl, 100 mg/ml; a catheter was subsequently placed in the saphenous vein prior to transport to the procedure room. Animals were sacrificed using Euthasol (Virbac, Fort Worth, TX) or Beuthanasia D (Schering Plough, Union, NJ). Those animals sampled at night or dawn were anesthesized under dim red light and blindfolded; euthanasia was done under dim white light and the blindfold was removed after decapitation. Removal of tissues, including the pineal gland, retina, and others (corpus callosum, frontal cortex, cerebellum, thalamus, hippocampus, caudate, medulla, pituitary, heart, liver, lung, kidney, skeletal muscle, small intestine, testis and ovary) was complete within 45 minutes following euthanasia. The interval between euthanasia and freezing for pineal glands and retinal tissues were 15 to 20 and 20 to 45 minutes, respectively. Following dissection, all tissues were immediately placed in containers on solid CO2 except in the case of the retina which was washed (phosphate buffered saline; 0.5 to 1 minutes) prior to freezing. Tissue was stored at −80°.
Rats
Rats (male Sprague Dawley, 200 to 250 grams) were bred and raised in facilities at the National Institutes of Health (NIH). The lighting cycle was 14:10 light:dark. Blood was collected at 13:00 h from CO2-anesthetized rats immediately following decapitation; animals were handled in accordance with NIH guidelines.
Human
Thirty-two human donors were recruited through the Research Donor Program of the NIH Blood Bank (Department of Transfusion Medicine, Research Support Services); donors were anonymized. Blood was drawn (7:30 to 14:00 h) into EDTA-treated tubes and stored at 4° C prior to plasma preparation. Samples were analyzed individually and also used to make a pool for methods development. A summary profile of the donors is in Table S1. The human plasma pool used for methods development was prepared from these anonymized samples.
Materials
The following were obtained (sources): melatonin (RBI Research Biochemicals, Natick, MA); d4-N-acetyl-5-methoxytryptamine, [α,α,β,β -d4, 98% d] (Medical Isotopes, Inc., Pelham, NH); Nacetylserotonin (Sigma-Aldrich, St. Louis, MO); N-acetyltryptamine (Tocris Bioscience, Bristol, UK). d7-N-acetylserotonin [α, α, β, β, acetyl-d7] (NIMH Chemical Synthesis and Drug Supply Program; https://nimh-repository.rti.org/index.cfm). Deuterium labeled N-acetyltryptamine (d3-N-acetyltryptamine [acetyl-d3]) was synthesized from tryptamine and acetic anhydride-d6 (MP Biomedicals, Solon, Ohio) on wet ice in 0.1 M phosphate buffer (pH 7.0), following a procedure based on the Schotten-Baumann reaction (Smith and March 2007). The product was extracted into chloroform and the extract was washed sequentially with five volumes of water (2 ×), 1M NaOH (2×), 1 M HCl (2×) and water (2×) to remove acetic acid, salts and unreacted tryptamine. The chloroform was dehydrated (MgCl2) and taken to dryness. The residue was re-dissolved and crystallized (Desilva and Snieckus 1978). The product (MP= 74–75° C, MW= 205.2) comigrated with N-acetyltryptamine and was >99% pure according to thin layer chromatography and high performance chromatography-mass spectrometry.
LC-MS/MS measurement of N-acetyltryptamine, N-acetylserotonin and melatonin
Sample preparation
Pineal glands and retinal samples were weighed and homogenized in 500 ml of phosphate buffered saline containing 1% RNaseOUT (Thermo Fisher, Waltham, MA). Samples of the homogenate were subsequently used for LC-MS/MS analysis of N-acetyltryptamine, melatonin and N-acetylserotonin and for analysis of total RNA, which was used for normalization.
To a volume (0.2 to 1 mL) of plasma or homogenate a 20 µL volume of methanol: water (1:1) containing 100 pg (0.42 pmol) of d4-N-acetyl-5-methoxytryptamine [α,α,β,β -d4], 100 pg (0.44 pmol) d7-N-acetylserotonin [α, α, β, β, acetyl-D7] and 100 pg (0.49 pmol) of d3-N-acetyltryptamine [acetyl-d3] was added. Two volumes of acetonitrile were then added, the tubes were vortexed and placed on ice for 10 min, and centrifuged (10,000 × g, 15 min). The supernatant was collected, dried by vacuum centrifugation, and brought up in 0.5 mL of 2% ammonium hydroxide in water. The resulting sample was applied (30 mg/1 mL) to a Strata-X 33µ polymeric solid phase extraction (SPE) column (Phenomenex) that had been pre-equilibrated (0.5 mL of methanol followed by 2 × 0.5 mL 2% ammonium hydroxide in water). The column was washed (2 × 0.5 mL 2% ammonium hydroxide water:methanol (95:5)); bound compounds were eluted (0.5 mL of 2% acetic acid in methanol), dried by vacuum centrifugation, and dissolved in 50 µL 0.1% formic acid in water:methanol (1:1).
Chromatography
A 10 µL volume of the sample eluted from the SPE column was injected onto an Acquity UPLC BEH C18 (1.7 µm) column (2.1 × 150 mm) (Waters Corporation, Milford, MA) equilibrated with Solvent A (deionized water/0.1% formic acid) at 45 °C. Compounds were eluted over 15 min at a flow rate of 0.3 mL/min, using a gradient from 0 to 5% Solvent B (acetonitrile/0.1% formic acid) from 1 to 2 min, 5 to 30% B from 2 to 12 min. Solvent B was held at 30% from 12 to 13 min and returned to 0% B in a linear gradient from 13 to 15 min and then re-equilibrated for 2 min.
MS/MS detection
Compounds were ionized by electrospray ionization in positive ion mode using an Agilent Jet Stream source (350 °C) and detected using an Agilent 6460 QQQ mass spectrometer (Agilent Technologies, Santa Clara, CA). Data was collected by MRM scans, using the following transitions, fragmented at a constant collision energy of 20 V: N-acetyltryptamine (203.2/144), and d3-N-acetyltryptamine (206.2/144), N-acetylserotonin (219.2/160), d7-N-acetylserotonin (226.2/164), melatonin (233.2/174), d4-melatonin (237.2/178). Under these conditions, N-acetyltryptamine eluted at 12.68 min, N-acetylserotonin at 7.68 and melatonin at 12.1 min. The area under the curve for each transition was calculated, and the ratio of the peak areas of N-acetyltryptamine and melatonin to their respective internal standards determined. The concentration was calculated from the measured peak ratios, using a standard curve generated from known amounts authentic standard added to a fixed amount of deuterated standard. All values presented represent triplicate analytical determinations, 97% of which were within 10% of the mean.
Assays of plasma tryptophan, kynurenine, cortisol, dehydroepiandrosterone sulfate (DHEAS) and testosterone
Plasma tryptophan and kynurenine were measured using LC/MS-MS (Drug Study Unit, Department of Bioengineering and Therapeutics Sciences, University of California, San Francisco, CA 94143 (Huang, Louie et al. 2013); tryptophan was also measured by Mayo Medical Laboratories, Mayo Clinic, Rochester, MN). Plasma cortisol, dehydroepiandrosterone sulfate (DHEAS), and testosterone were assayed using a chemiluminescence-based automatic clinical platform (Roche Cobas e411, Roche Diagnostics, Indianapolis, IN), as previously described (Urbanski, Sorwell et al. 2014); these assays show intra-assay coefficients of variation of < 6% for nonhuman primates (Lemos, Downs et al. 2009).
Determination of acetylation of arylamines and arylalkylamines by NAT1 and NAT2
Nomenclature according to (Vatsis, Weber et al. 1995)
Enzyme preparation
Human NAT1 and NAT2 were recombinantly expressed in yeast cells. Briefly, the coding regions of the NAT1*4 (high NAT1 activity), NAT1*14B (low NAT1 activity), NAT2*4 (rapid NAT2 acetylator), and NAT2*5B (slow NAT2 acetylator) were amplified by polymerase chain reaction (PCR) using previously constructed plasmids as previously described (Leff, Fretland et al. 1999; Fretland, Doll et al. 2001; Fretland, Leff et al. 2001). The yeast vector pESP-3 and PCR products (Stratagene, La Jolla, CA) was digested with NdeI and AscI at 37° C overnight. Purified PCR products and 80 ng of digested plasmid were ligated overnig 16 ° C with T4 DNA ligase (New England Biolabs, Inc, Beverly, MA). Ligated plasmids were transformed into XL-10 Gold Ultracompetent Escherichia coli (Stratagene). Plasmids were isolated from cultures grown from selected colonies using the Qiagen Plasmid Midi kit (Qiagen, Valencia, CA) and sequenced. Constructs were then transformed into competent Schizosaccaromyces pombe and expressed following the manufacturer’s instructions (Stratagene). Mock transformed yeast used pESP-3 vector with no NAT insert. Total cell lysates were prepared by vigorous agitation of yeast in 20 mM sodium phosphate, 1 mM EDTA and 0.2% triton-X-100, pH 7.4 containing acid-washed glass beads (Stratagene) for ten min at 4° C. Liquid fractions were collected from the lysed cells and centrifuged at 13,000 × g for twenty min. Supernatants were collected, aliquoted, and stored at −80° C until utilized for enzymatic assays.
Catalytic Assays
Yeast lysates containing recombinant human NAT1*4, NAT1*14B, NAT2*4 or NAT2*5B were added to reactions containing p-aminobenzoic acid (reference substrate with selectivity for human NAT1), sulfamethazine (reference substrate with selectivity for human NAT2) serotonin or tryptamine (300 µM) in combination with acetyl coenzyme A (1 mM). Total protein in cell lysate was measured by the Bradford assay using the Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA). All N-acetyltransferase assays were conducted at 37° C at protein concentrations and over time periods in which the reactions were linear. Negative controls substituted water for acetyl coenzyme A and substituted buffer for enzyme. N-Acetyltransferase activities were measured using high performance liquid chromatography (HPLC) to separate N-acetyltryptamine product from tryptamine substrate. HPLC assays for the N-acetylation of p-aminobenzoic acid and sulfamethazine were performed as described previously. The HPLC method was modified for the separation of tryptamine and N-acetyl-tryptamine using a linear gradient of 98:02 sodium perchlorate pH 2.5: acetonitrile to 75:25 sodium perchlorate pH 2.5: acetonitrile over 5 min. The retention times of tryptamine and N-acetyltryptamine were 6.2 and 9.3 min respectively. HPLC separation of serotonin and N-acetylserotonin was achieved using a linear gradient of 98:02 sodium perchlorate pH 2.5: acetonitrile to 50:50 sodium perchlorate pH 2.5: acetonitrile over 5 min. The retention times of serotonin and N-acetylserotonin were 8.8 and 9.8 min, respectively.
Genotyping of human NAT2
Genomic DNA was isolated from frozen whole blood samples using the QIAamp DNA Mini Kit (Qiagen, Valencia CA) according to manufacturer’s instructions. NAT2 haplotypes, genotypes, and deduced phenotypes were determined as previously described (Doll and Hein 2001). Briefly, single nucleotide polymorphism (SNP)-specific polymerase chain reaction (PCR) primers and fluorogenic probes were designed using Primer Express (Applied Biosystems, Foster City, CA). The fluorogenic probes are labeled with a reporter dye (either FAM or VIC) and are specific for one of the two possible bases identified at seven SNPs in the NAT2 coding region. Controls (no DNA template) were run to ensure that there was no amplification of contaminating DNA. Subjects were classified as rapid, intermediate, and slow acetylator phenotypes. Individuals possessing two of the NAT2 alleles associated with rapid acetylation activity (NAT2*4, NAT2*12, and NAT2*13) were classified as rapid acetylators; individuals possessing one of these alleles and one allele associated with slow acetylation (NAT2*5, NAT2*6, NAT2*7, and NAT2*14) were classified as intermediate acetylators, and those individuals that possessed two slow acetylation alleles were classified as slow acetylators.
RNA preparation, sequencing and data analysis
RNA Preparation
Total RNA was extracted from pineal or retinal homogenates, individual pineal glands or pieces of other tissues (2 to 4 mm3 pieces) with TRIzol reagent (Invitrogen, Carlsbad, CA), followed by clean-up using an RNeasy Micro Kit with on-column DNase treatment as per the manufacture’s protocol (Qiagen, Valencia, CA). The amount and quality of RNA were determined using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RIN values were > 9 for all tissues. Two mixed tissue RNA samples were prepared (Day, male; Night, female) by adding equal amounts of RNA extracted from sixteen tissues, not including the pineal gland or retina.
Library construction
RNA-Seq libraries were constructed from 0.7 to 1 µg total RNA using the TruSeq Stranded Total RNA Sample Prep Kits (Illumina cat. no. RS-122-2301) following the manufacturer’s instructions. Insert sizes were approximately 175bp. Unique barcode adapters were applied to each library. Equal volumes of individual libraries were pooled and run on a MiSeq instrument (Illumina, San Diego, CA). The libraries were then repooled based on the MiSeq demultiplexing results. The pooled libraries were sequenced on a HiSeq2000 (Illumina, San Diego, CA) using version 3 chemistry.
RNA-Seq Alignment
Reads were aligned with the RNA-STAR aligner (v. 2.3.0e), using a rhesus rheMac3 genome build. Quality control metrics were calculated and visualized using FastQC and in-house written scripts; GC artifacts and k-mer abnormalities were not found. The mean alignment rate was 89.19 % for retina with 85.78 % and for the pineal gland. Unique aligned reads were kept for further analysis, and reads mapped to more than one locus were discarded. The average mean GC content was 43% (SD = 10). Of the aligned read-pairs, 33.1% mapped at least partially to the exon of a known gene, 11% were mapped to 3’UTRs, and 4% to 5’ UTRs. Of the remaining aligned read-pairs: 44% mapped to intronic regions of known genes, while 8 % mapped to intergenic regions. All processed reads were mapped to the correct strand by use of strand-specific alignment and read assignment. The large proportion of reads mapping to intronic regions is typical of sequencing results derived from libraries that were ribodepleted, which captures mature and immature transcripts (Li, Tighe et al. 2014). The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo (GEO series accession number GSE78165) and are accessible at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=epkvmssmvhmrlmf&acc=GSE78165.
Differential expression analysis
Gene read counts were provided by R subreads package, (featureCounts function) with parameters specifying: paired-end reads, and reverse-complement for the second strand. Annotation for assigning the reads was derived from Gencode v.19 human annotation. Gene coordinates were lifted over using RheMac3/hg19 liftover chains, and only sequences that shared no less than 95% homology were kept, resulting in a total of 50,308 genes. Differential expression analysis was performed using DESeq2. Fold-change estimates were calculated via DESeq2 using the parameter estimates from the model fit. All p-values were adjusted for multiple testing using the FDR method of Benjamini and coworkers (Benjamini and Hochberg 1995; Benjamini and Yekutieli 2001). Outlier detection was performed using Cook’s distance, a measure of the difference between fitted coefficients with and without each individual sample (Cook 1977). The pineal- and the retina- vs-mixed-tissue analyses followed a slightly different methodology. Two separate comparisons were run: all 12 pineal replicates vs mixed-tissue (2 replicates), as well as comparison of 12 retina samples replicates vs 2 mixed-tissue replicates. The raw data and differential expression analysis results are available in the Supporting Information file 2 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=epkvmssmvhmrlmf&acc=GSE78165).
RESULTS
LC-MS/MS detection of N-acetyltryptamine, N-acetylserotonin and melatonin
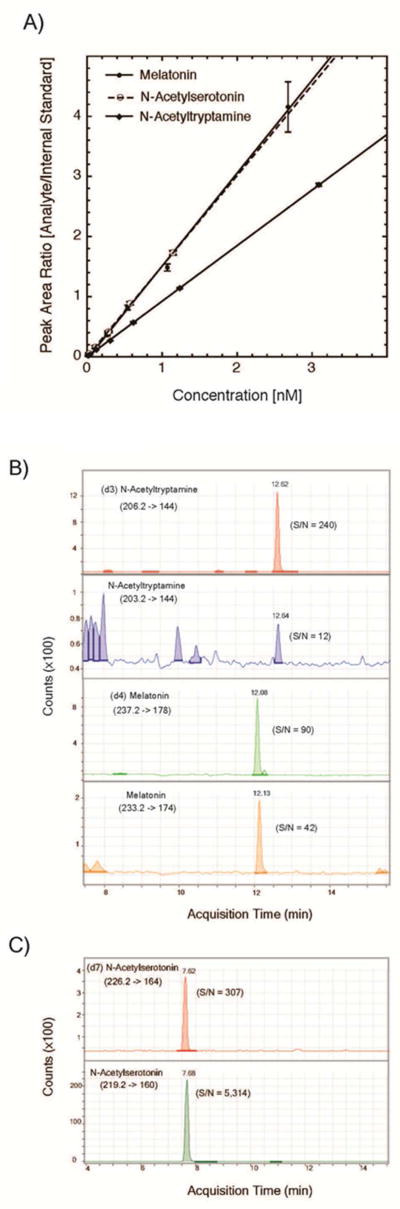
Calibration curves were generated adding known amounts of each compound to a fixed amount of labeled internal standard (Figure 2A). A linear response was obtained for all compounds with lower limit of quantitation values for amounts injected on the column between 1 fmol and 0.8 fmol for these compounds. The responses were linear from 0.02 to 10 nM (Coefficient of determination = 0.9996, Sum of squares of residuals = 0.0505 for N-acetyltryptamine, 0.9992 and 0.0172 for N-acetylserotonin and 0.9971 and 1.89 for melatonin).
Figure 2. Detection of N-acetyltryptamine, N-acetylserotonin and melatonin by liquid chromatography/tandem mass spectroscopy.
The SPE extract (10 µL of 50 µL total) from 1 mL of human plasma was injected onto the UPLC RP column and eluted as described. N-Acetyltryptamine, N-acetylserotonin, melatonin, and the internal deuterated standards were analyzed by MRM mass spectrometry, monitoring the indicated transitions. (A.) Calibration curves for N-acetyltryptamine, N-acetylserotonin and melatonin. The indicated concentrations of N-acetyltryptamine, N-acetylsertonin and melatonin (from 0.01 to 6 nM) were added to a constant concentration of deuterated internal standards (d3-N-acetyltryptamine and, d7-N-acetylsertonin and d4-melatonin). The integrated peak of MRM signal intensity was quantified and the ratio of analyte to internal standard was calculated. Each point indicates the mean ± SD (n=3), and a linear fit to the data points was calculated for each compound. (B.) N-Acetyltryptamine and melatonin in human plasma: Top: MRM signal for internal standard (98 fmol) d3-N-acetyltryptamine. Second from top: MRM signal for plasma N-acetyltryptamine (arrow, 12.64 min). Second from bottom: MRM signal for the internal standard (85 fmol) d4-melatonin. Bottom; MRM signal for plasma melatonin (peak at 12.13 min). (C.) N-Acetylserotonin in rhesus pineal tissue: Top: MRM signal for internal standard (198 fmol) d7-N-acetylserotonin. Bottom; MRM signal for N-acetylserotonin (peak at 7.68 min) in extracts from rhesus pineal gland (1.5 mg tissue) collected at dawn. Further details appear in the Materials and Methods section.
Recovery of the labeled standards ranged from 80 to 90%. When human plasma extracts were analyzed, peaks were observed eluting from the reversed-phase LC column with the same retention time and specific MRM transitions as authentic N-acetyltryptamine and melatonin (Figure 2B). Analysis of pineal extracts revealed a peak eluting with the same retention time and specific MRM transitions as authenic N-acetylseronin (Figure 2C). Within-day variance of triplicate determinations for N-acetyltryptamine and melatonin in a plasma pool were −5 to 12% and −13 to 16%, respectively; and were 4 to 8 % for N-acetylserotonin in a pineal gland extract.
Daytime plasma N-acetyltryptamine and melatonin
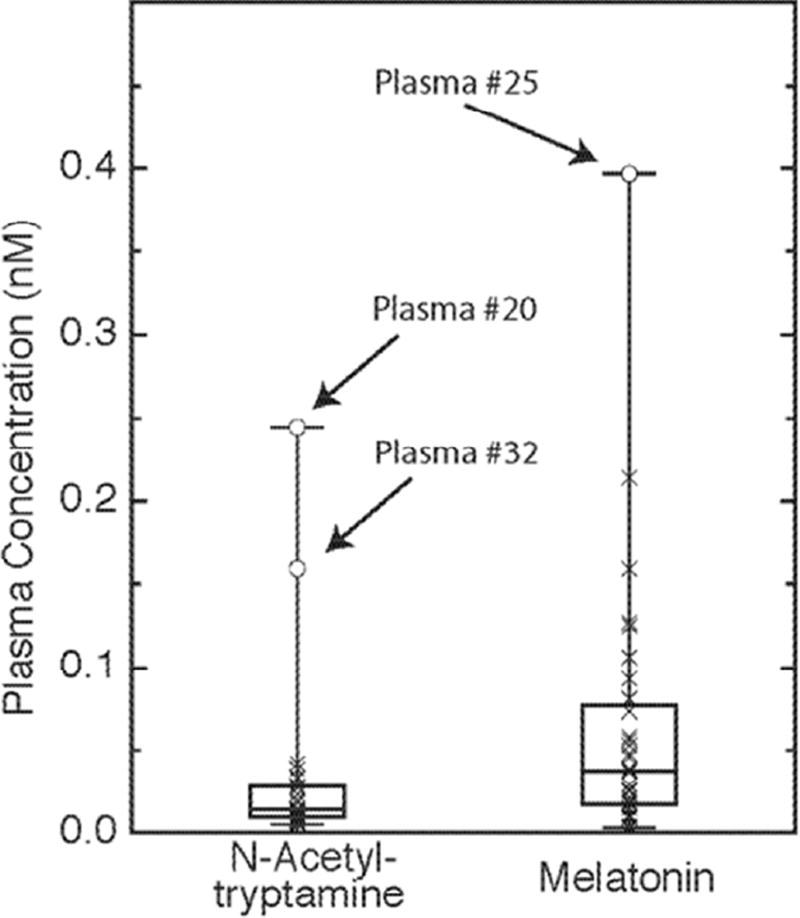
N-Acetyltryptamine and melatonin were detected in daytime rat, rhesus macaque and human plasma; N-acetylserotonin was not (Table 1). The amounts of N-acetyltryptamine and melatonin in daytime rat and rhesus plasma ranged from 0.05 to 0.5 nM. The average human plasma N-acetyltryptamine and melatonin values obtained from 32 healthy donors (22 to 69 years of age; 24 males and 8 females; admitted between 07:30 h to 14:00 h) were 0.03 and 0.06 nM, respectively (Figure 3, Table S1). Human daytime plasma N-acetyltryptamine values ranged from a minimum of 0.005 nM to a maximum of 0.25 nM (median = 0.014 nM), 1st quartile = 0.01 nM and 3rd quartile = 0.028 nM. Two of the donors were identified as outliers for plasma N-acetyltryptamine, with values 5- and 8-fold higher than the average. Plasma melatonin levels had a minimum concentration of 0.003 nM and a maximum of 0.4 nM (median = 0.037 nM, 1st quartile = 0.017 nM and 3rd quartile = 0.079 nM. No correlation was observed between N-acetyltryptamine and melatonin values. Tryptophan and kynurenine levels in the outlier’s plasma were within the range of the population studied; an association of plasma N-acetyltryptamine levels with NAT2 phenotype was not apparent (Table S1).
Table 1. Daytime levels of N-acetyltryptamine and melatonin in rat, human and rhesus macaque plasma.
Data represent the mean ± SEM for plasma N-acetyltryptamine and melatonin from the indicated number of individuals; individual values were calculated from the mean of three or more determinations of the sample. Rat plasma samples were drawn between 13:00 and 14:00 h; human plasma samples between 07:00 h and 14:00 h and the rhesus samples between 09:00 and 15:00 h. Additional experimental details appear in the Materials and Methods section.
| Plasma Concentration | |||
|---|---|---|---|
| Source | N-Acetyltryptamine [nM] | Melatonin [nM] | N-Acetyltryptamine/ Melatonin |
| Rat (N=5) | 0.29 ± 0.05 | 0.17 ± 0.01 | 1.7 |
| Human (N=32) | 0.03 ± 0.01 | 0.06 ± 0.01 | 0.5 |
| Rhesus (N=4) | 0.5 ± 0.2 | 0.17 ± 0.06 | 2.9 |
Figure 3. Human plasma N-acetyltryptamine and melatonin.
Box plot of daytime plasma N-acetyltryptamine and melatonin values in 32 healthy donors (22 to 69 years of age; 24 males and 8 females). Samples were collected between 07:30 and 14:00 h. Outliers are indicated by subject identifying numbers. Table S1 provides subject details including age, sex, weight and genotype and plasma values for N-acetyltryptamine, melatonin, tryptophan and kynurenine. Further details appear in the Materials and Methods section.
Plasma N-acetyltryptamine exhibits a 24-hour rhythm in the rhesus macaque
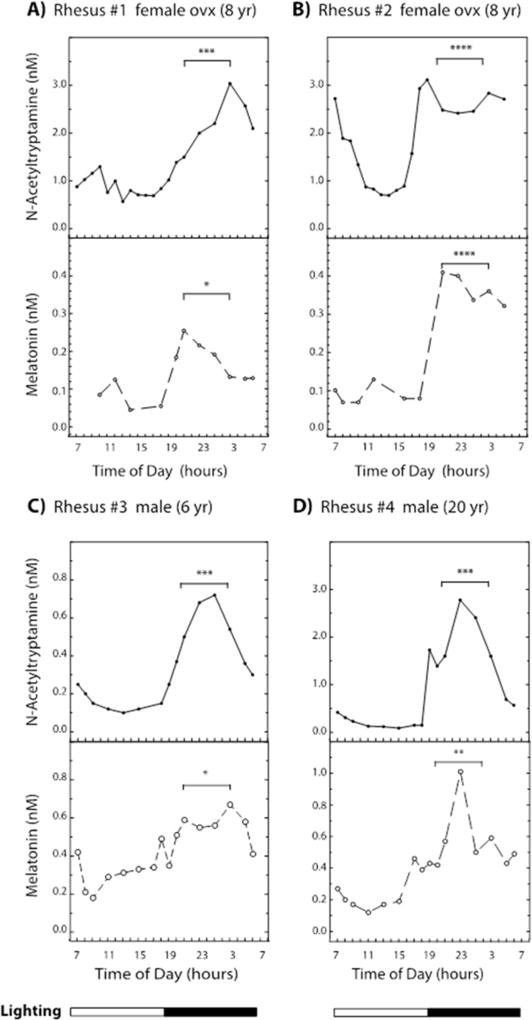
Plasma was prepared from blood obtained over a 24-hour period from four unrestrained rhesus macaques (Urbanski 2011) (Figure 4). The absolute concentrations of N-acetyltryptamine and melatonin at similar times of the day varied between individual animals. Specifically, the levels of both N-acetyltryptamine and melatonin increased in the dark, with significant differences observed between six-hour “day” (09:00 to 15:00 h) and “night” periods (21:00 to 03:00 h). N-Acetyltryptamine and melatonin levels were 2- to 15- fold and 2- to 4- fold higher at night, respectively (P>0.001). The individual-to-individual variation and nocturnal increase in melatonin are consistent with published reports on melatonin in the cerebral spinal fluid of restrained rhesus macaques (Reppert, Perlow et al. 1979; Reppert, Perlow et al. 1979; Perlow, Reppert et al. 1981).
Figure 4. Daily changes in plasma N-acetyltryptamine and melatonin in the rhesus macaque.
Blood was obtained continuously in ten to thirty minute fractions over a 25 hour period remotely from animals that were undisturbed, conscious and unrestrained (Urbanski 2011). Animals were exposed to 12 h of light followed by 12 h of dark over the 24 hour period, with lighting on from 07:00 h to 19:00 h. N-Acetyltryptamine and melatonin levels were determined for two ovariectomized (OVX) females (A & B) and two males (C & D); sequential day samples were pooled to increase the melatonin signal strength to detectable levels. Mean plasma values during 6-hour periods of the night (21:00 h to 03:00 h) and day (09:00 h to 15:00 h) were significantly different (P values: * = 0.01 to 0.05, ** = 0.001 to 0.01, *** = 0.0001 to 0.001, **** < 0.0001). Additional experimental details appear in the Materials and Methods section.
Plasma cortisol, DHEAS, and testosterone levels during a 24-hour period
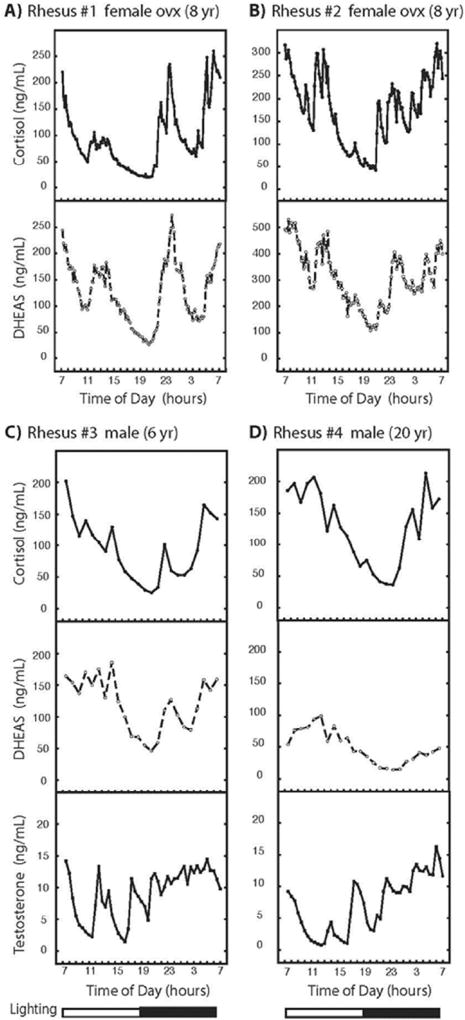
Cortisol, DHEAS, and testosterone in the plasma samples used in Figure 4 were determined (Figure 5) and found to exhibit 24-hour patterns that were generally similar to those previously reported for unrestrained rhesus macaques (Jacoby, Sassin et al. 1974; Perlow, Reppert et al. 1981; Lemos, Downs et al. 2009; Sitzmann, Brown et al. 2014; Sorwell, Kohama et al. 2014; Urbanski, Sorwell et al. 2014). This provides evidence of the physiological normalcy of the circadian system of the animals used in the present study.
Figure 5. Plasma cortisol, DHEAS and testosterone over 24 h in rhesus macaques.
Aliquots of the plasma samples used in Figure 4 for N-acetyltryptamine and melatonin determinations were also analyzed for levels of cortisol, DHEAS (A–D), and testosterone (in males, C & D). Steroid levels were measured in samples obtained at 10- or 30-minute intervals. Additional experimental details appear in the Materials and Methods section.
Expression of melatonin synthesis-related genes in the rhesus macaque pineal gland and retina
Limited data is available on the 24-hour pattern of expression of genes in the melatonin/N-acetyltryptamine synthesis pathway in the rhesus pineal gland and retina (Coon, Del Olmo et al. 2002). This was expanded here using RNA sequencing data obtained at four times of the day from the pineal gland, retina and a mixture of tissues. As expected, TPH1, DDC, AANAT and ASMT (Figure 1) transcripts were detectable at all four times in the pineal gland and retina (Table 2). AANAT transcripts were similarly abundant in both tissues. However, there were marked differences in the relative expression of some transcripts between the two tissues (Table 2). Specifically, the transcripts required to encode enzymes involved in O-methylation of tryptophan, TPH1 and ASMT, were ~200- and ~30-fold less abundant, respectively, in retina compared to the pineal gland. It was also observed that TPH2 transcript levels were more than 200-fold lower than those of TPH1 in the pineal gland and retina, indicating that TPH2 is unlikely to contribute significantly to tryptophan hydroxylation in these tissues. NAT1 and NAT2 transcript abundances in both tissues were insignificant relative to those of AANAT and to NAT1 and NAT2 in a mixture of tissues.
Table 2. Expression of selected genes in the rhesus macaque pineal gland and retina.
Each pineal and retina values are the mean ± S.E.M. (n = 3) in tissues removed at the indicated times of day. The expression in single samples of a mixture of RNA (Mixed tissue) from 12 tissues removed during the day or night are given for comparison. Differential analysis was performed using DESeq2 software package (Love, Huber et al. 2014), and P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg (Benjamini and Hochberg 1995).
| Abundance (FPKM) | |||||
|---|---|---|---|---|---|
| Gene | Tissue | Dawn | Midday | Dusk | Midnight |
| TPH1 | Pineal gland | 513 ± 62 | 612 ± 103 | 611± 39 | 601± 73 |
| Retina | 11 ± 3 | 15 ± 3 | 11 ± 1 | 15 ± 5 | |
| Mixed tissue | 1 ± 0.3 | 2 ± 0.3 | |||
| TPH2 | Pineal gland | 2.3 ± 1.3 | 0.53 ± 0.18 | 0.21± 0.04 | 1.49± 0.17 |
| Retina | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.02 | 0.01 ± 0.01 | |
| Mixed tissue | 0.16 ± 0.03 | 0.22 ± 0.03 | |||
| DDC | Pineal gland | 141 ± 20 | 79 ± 16 | 47± 6** | 107± 9** |
| Retina | 0.3 ± 0.07 | 0.2 ± 0.1 | 0.4 ± 0.1 | 0.2 ± 0.09 | |
| Mixed tissue | 39 ± 3 | 34 ± 3 | |||
| AANAT | Pineal gland | 54 ± 4 | 60 ± 1 | 47± 4 | 48 ± 3 |
| Retina | 71 ± 9 | 60 ± 5 | 49 ± 4 | 54 ± 2 | |
| Mixed tissue | 0.2 ± 0.06 | 0.1 ± 0.06 | |||
| ASMT | Pineal gland | 140 ± 6 | 120 ± 23 | 167 ± 14 | 168 ± 15 |
| Retina | 5 ± 2 | 2 ± 1 | 1 ± 1 | 6 ± 1* | |
| Mixed tissue | 0.1 ± 0.03 | 0.2 ± 0.03 | |||
| NAT1 | Pineal gland | 0.3 ± 0.1 | 0.37 ± 0.08 | 0.46 ± 0.01 | 0.34 ± 0.05 |
| Retina | 0.2 ± 0.04 | 0.17 ± 0.01 | 0.23 ± 0.05 | 0.2 ± 0.03 | |
| Mixed tissue | 1.1 ± 0.15 | 1.4 ± 0.15 | |||
| NAT2 | Pineal gland | 0.18 ± 0.06 | 0.06 ± 0.03 | 0.34 ± 0.11 | 0.31 ± 0.05 |
| Retina | 0.14 ± 0.07 | 0.12 ± 0.03 | 0.1 ± 0.02 | 0.26 ± 0.03 | |
| Mixed tissue | 2.04 ± 0.13 | 1.78 ± 0.13 | |||
Differentially expressed relative to midday value, adjusted-P <0.002*;
differentially expressed relative to midday and dawn values, adjusted-P <0.001. Additional experimental details appear in the Materials and Methods section.
Moreover, time-of-day changes in AANAT mRNA were not observed, confirming previous findings (Coon, Del Olmo et al. 2002), consistent with the view that the 24-hour rhythm in AANAT activity in the rhesus macaque does not involve transcriptional regulation but rather is dependent on posttranslational modifications. The time-of-day dependent changes in the abundance of DDC transcripts in the pineal gland and ASMT transcripts in the retina were observed, which argues for a role of these enzymes in controlling daily changes in the melatonin/N-acetyltryptamine pathways.
Indoleamines are poor substrates of NAT1 and NAT2
The broad distribution of NAT1 and NAT2 in the body and the extant evidence that arylamine N-acetyltransferase acetylates the arylalkylamine serotonin (Steinberg, Cohen et al. 1969) pointed to the possibility that these enzyme might contribute to the circulating N-acetyltryptamine. This led us to study indoleamine acetylation using recombinant human NAT1, NAT2 and variants of these enzymes expressed in yeast (Table 3). Both enzymes were found to acetylate tryptamine and serotonin. Genetic polymorphisms were exhibited towards the N-acetylation of tryptamine and serotonin by recombinant human NAT2, as indicated by the observation that N-acetylation rates were significantly (p<0.005) higher when catalyzed by the reference NAT2*4 than those associated with slow acetylator phenotype, i.e., NAT4*5B. Furthermore, rates of tryptamine N-acetylation catalyzed by NAT1 4 were significantly (p<0.005) higher than catalyzed by NAT1*14B. Although the rates of N-acetylation by recombinant human NAT1 and NAT2 are not relevant to absolute rates of acetylation in human tissues, it is noteworthy that the rates of N-acetylation of tryptamine and serotonin by recombinanant human NAT1*4 were 28,000-fold and 100,000-fold lower, respectively, than that for p-aminobenzoic acid. Similarly, the rates of tryptamine and serotonin N-acetylation catalyzed by recombinant human NAT2*4 were 275- and 1000-fold lower, respectively, than that for sulfamethazine. Thus, it is clear that whereas arylamines are preferred substrates of NAT1 and NAT2, these enzymes can also acetylate arylalkylamines, albeit at levels that are orders of magnitude lower.
Table 3. N-Acetylation of arylamines and arylalklylamines by NAT1 and NAT2.
Table values represent the mean ± S.E.M. (n = 3). Controls included no enzyme, no substrate and no acetyl coenzyme A each of which yielded no production of product. All assays were run at 300 µM substrate and 1 mM acetyl coenzyme A at conditions linear with respect to time and enzyme concentration. Differences were tested for significance by Student t test (unpaired). Additional experimental details appear in the Materials and Methods section.
| Substrate | |||||
|---|---|---|---|---|---|
| N-Acetylated product formation (nmol/min/mg protein) | |||||
|
Recombinant human gene |
Acetylator phenotype |
Sulfamethazine | p-Amino-benzoic acid | Tryptamine | Serotonin |
| NAT1*4 | High | NDc | 11,500 ± 1400 | 0.412 ± 0.038 | 0.115 ± 0.064 |
| NAT1*14Ba | Low | ND | 2,170 ± 159 | 0.0753±0.0341 | 0.0361 ± 0.018 |
| NAT2*4 | Rapid | 545 ± 74 | ND | 1.98 ± 0.25 | 0.484 ± 0.045 |
| NAT2*5Bb | Slow | 84.5 ± 15.2 | ND | 0.412±0.079 | 0.0741± 0.0492 |
Rates catalyzed by NAT1*14B were significantly (p<0.005) lower than catalyzed by NAT1*4 towards the N-acetylation of p-aminobenzoic acid and tryptamine.
Rates catalyzed by NAT2*5B were significantly (p<0.005) lower than catalyzed by NAT2*4 towards the N-acetylation of sulfamethazine, tryptamine, and serotonin.
ND; Not determined
N-Acetyltryptamine, N-acetylserotonin and melatonin in pineal and retinal extracts
Analysis of extracts of the pineal gland and retina revealed that melatonin and N-acetylserotonin were >1000 fold higher in pineal extracts than in retinal extracts (Table 4); for presentation purposes, units of pmol/ µg RNA are used for the pineal gland and fmol/ µg RNA for the retina. N-Acetyltryptamine was detected in both tissues, with pineal levels (mean ~ 10 fmol/ µg RNA) over 10-fold higher than those in the retina (mean ~ .6 fmol/µg RNA); significant time-of-day differences were not detected. The levels of N-acetyltryptamine in the pineal gland (mean ~ 0.01 pmol/ µg RNA) were approximately 1000-fold lower than those of melatonin (mean ~11 pmol/ µg), whereas in the retina they were relatively similar for N-acetyltryptamine (mean ~ 0.6 fmol/µg) and melatonin (mean ~ 0.9 fmol/µg RNA). N-Acetylserotonin was detected in the pineal gland but not retinal extracts (Table 4). Nighttime values were statistically greater than midday values in the pineal gland as previously reported for the rat pineal gland (Mefford, Chang et al. 1983).
Table 4. Amounts of N-acetyltryptamine, melatonin, and N-acetylserotonin in rhesus macaque pineal and retinal extracts.
Metabolite values are the mean ± S.E.M. for extracts from three tissues removed at the indicated times of day; metabolite values were normalized to the µg of total RNA in each sample.
| A) Pineal gland | |||||
|---|---|---|---|---|---|
| Piineal Concentration (pmol/µg RNA)a | |||||
| Compound | Dawn | Midday | Dusk | Midnight | Mean† |
| N-Acetyltryptamine | 0.010 ± 0.004 | 0.012 ± 0.001 | 0.010 ± 0.003 | 0.008 ± 0.001 | 0.010 ± 0.001 |
| N-Acetylserotonin | 5.60 ± 4.1 | N. D. | 3.4 ± 0.8 | 16.0 ± 13 | 4.6 ± 2.5 |
| Melatonin | 11.9 ± 7.6 | 4.4 ± 1.7 | 11.7 ± 5.0 | 17.1 ± 5.1 | 11.2 ±2.6 |
| B) Retina | |||||
|---|---|---|---|---|---|
| Retinal Concentration (fmol/ µg RNA)a | |||||
| Compound | Dawn | Midday | Dusk | Midnight | Meanc |
| N-Acetyltryptamine | 1.4 ± 0.5 | 0.35 ± 0.1 | 0.30 ± 0.02 | 0.29b | 0.51 ± 0.20 |
| N-Acetylserotonin | N. D. | N. D. | N. D. | N. D. | N.D. |
| Melatonin | 0.42 + 0.09 | 2.6b | N. D. | 0.64 + 0.13 | 0.48 ± 0.21 |
Pineal and retinal values are presented using different units.
N.D. Compound not detected in assay.
Compound was detected in one of three tissue extracts.
Calculation of mean value is based on 12 determinations; zero was assigned if the compound was not detected.
DISCUSSION
This report provides a new perspective on N-acetyltryptamine by establishing that it is a physiological constituent of blood in mammals including man, and that it exhibits a daily rhythm based on studies in the rhesus macaque. The method presented here can be readily established in facilities where the required equipment is available. The use of internal deuterated standards enhances the reliability and accuracy of determinations. LC-MS/MS methodology essentially eliminates interference, which is a common concern with assays based on immunodetection.
It is reasonable to suspect that the rhythm in N-acetyltryptamine is a common feature of vertebrate biology, a possibility that deserves further study to determine whether it, like melatonin and cortisol, might be a useful circadian marker. The rhythm generally follows the outlines of the melatonin rhythm, characterized by a nocturnal increase, pointing to the possibility that it contributes to circadian signalling. As discussed below, this advance raises a number of issues, including the source(s) of circulating N-acetyltryptamine, the mechanisms controlling its biosynthesis and the possible physiological roles it might play. In addition, the discovery of N-acetyltryptamine outliers in humans and the question of whether N-acetyltryptamine interferes with radioimmunoassay detection of melatonin are also addressed.
Source of N-acetyltryptamine
The observations that N-acetyltryptamine can be synthesized from tryptamine catalyzed by AANAT, NAT1 and NAT2 raises the possibility that there are multiple potential sources. However, there are several reasons to ignore NAT1 and NAT2 in this regard. First, the affinity of these enzymes for tryptamine is significantly lower than that for arylamines, their preferred substrates. Second, the finding that there was no association between plasma N-acetyltryptamine levels and NAT@ acetylator genotype in humans argues against a significant contribution by human NAT2. Rather, the rhythmic nature of plasma levels provides reason to suspect that it might be produced in either or both of the two tissues known to have daily rhythms in AANAT activity, the pineal gland (Klein and Weller 1970; Coon, Roseboom et al. 1995; Klein, Coon et al. 1997; Klein 2007) and retina (Bernard, Iuvone et al. 1997; Iuvone, Tosini et al. 2005; Pozdeyev, Taylor et al. 2006; Tosini, Chaurasia et al. 2006; Haque, Chong et al. 2011).
The view that the retina might contribute to circulating N-acetyltryptamine in the rhesus macaque is supported by the findings reported here and elsewhere that the AANAT transcript is abundant in the retina and that there is a daily rhythm in AANAT activity in the retina (Coon, Del Olmo et al. 2002); in addition retinal and pineal levels of AANAT mRNA and activity are similar. The very low levels of transcripts encoding enzymes required for melatonin synthesis, TPH1 and ASMT, and the low levels of melatonin in the retina argue that AANAT is devoted to N-acetyltryptamine synthesis in the retina, whereas the reverse is true in the pineal gland.
Other sites of AANAT expression should not be ignored, however, as potential contributors to circulating N-acetyltryptamine. These include the pituitary gland and brain (Gaudet, Palkovits et al. 1991; Gaudet and Namboodiri 1993; Coon, Roseboom et al. 1995; Coon, Mazuruk et al. 1996; Fleming, Barrett et al. 1999; Paulin, Cazamea-Catalan et al. 2015). Further investigation is required to answer the question of whether activity increases at night in these tissues.
Additional support for the view that the retina is a source of N-acetyltryptamine comes from the finding that it was detected in retinal tissue obtained at four times of the day in that tissue. However, the average value for midnight pineal N-acetyltryptamine was not statistically higher than other averages. This might reflect individual-to-individual variation in N-acetyltryptamine producton, as seen with plasma values, where daily rhythms in indivudals were obvious, although night values varied considerably between individuals. Another possible contributing factor is that retinal N-acetyltryptamine may have been extracted during the 0.5 to 1 minute wash that preceded freezing.
Physiological role of N-acetyltryptamine
N-Acetyltryptamine is a candidate for a role as a chronobiological signal based on the finding that there is a marked rhythm in the circulation and that it is a melatonin receptor mixed agonist/antagonist (Dubocovich 1983; Dubocovich 1984; Dubocovich, Delagrange et al. 2010). However, the low relative affinity of N-acetyltryptamine for the melatonin receptor (Dubocovich, Delagrange et al. 2010) and plasma levels that are similar to those of melatonin make it seem unlikely that it acts as a second melatonergic hormone. Rather, it seems more likely that it could act as a paracrine or autocrine signal at sites of synthesis where it could be released at sufficiently high levels so as to interact locally with melatonin receptors. In this light, circulating N-acetyltryptamine would be an indication of overflow from sites where it acts as a local regulator. Such a local role of N-acetyltryptamine is consistent with the colocalization of melatonin receptors and AANAT in the retina and pituitary gland (Vanecek 1988; Vanecek 1998; Fleming, Barrett et al. 1999; Klein 2007; Dubocovich, Delagrange et al. 2010), where they might mediate N-acetyltryptamine-based cell: cell signaling.
Similarly, it is possible that N-acetyltryptamine interactions with melatonin receptors in the vertebrate brain might also be involved in local signaling (Dubocovich 1991; Brooks and Cassone 1992; Davies, Hannah et al. 1994; Dubocovich, Delagrange et al. 2010; Paulin, Cazamea-Catalan et al. 2015). One could envision such N-acetyltryptamine-mediated communication within discrete brain regions where abundance would be regulated through circadian clock driven cyclic AMP-dependent mechanisms similar to those which operate in the pineal gland and retina to control AANAT activity (Bernard, Klein et al. 1997; Bolliet, Begay et al. 1997; Iuvone, Tosini et al. 2005). This leads to the possibility that the temporal pattern of N-acetyltryptamine production in discrete brain regions is controlled by peripheral clocks or by other signaling pathways that control cyclic AMP, or by a combination of the two.
A non-melatoninergic mode of action of N-acetyltryptamine might also exist, as suggested by recent pharmacological studies by Iuvone and coworkers who reported that N-acetylserotonin (Figure 1) influences neuroprotection in the retina through a direct interaction with BDNF receptors (Iuvone, Boatright et al. 2014). Based on structural similarity, it is reasonable to suspect that N-acetyltryptamine might play this role alone or in combination with N-acetylserotonin. The former is supported by the finding that retinal extracts contain N-acetyltryptamine but not detectable levels of N-acetylserotonin.
Human N-acetyltryptamine outliers
Our analysis of human daytime plasma revealed the existence of two outliers among the 32 healthy individuals studied (Figure 3, Table S1). The reason for this was not revealed in our studies, which failed to find a strong association of high N-acetyltryptamine with high levels of plasma tryptophan or low kynurenine levels. This points to mechanisms other than those involving tryptophan availability and tryptophan conversion to kynurenine. Similarly, high levels of N-acetyltryptamine were not found in individuals with a rapid acetylator phenotype, which point away from human NAT2 as an explanation. Moreover, we did not find that high levels of N-acetyltryptamine were associated with high levels of melatonin, suggesting that a mechanism or a combination of mechanisms specific for production or degradation of N-acetyltryptamine, but not impacting melatonin, is involved. The basis of the outlier observation remains a mystery; perhaps the solution lies in the levels of expression of AANAT in the retina or other tissue; or a shift in the timing of the rhythmic 24-hour pattern of AANAT activity in the source of N-acetyltryptamine relative to that in the pineal gland. Of practical importance is whether outlier values of N-acetyltryptamine are useful biomarkers, perhaps for diseases impacting retinal biology.
Interference by N-acetyltryptamine with immunoassay of melatonin
The question of whether N-acetyltryptamine interferes with detection of melatonin in the circulation is important because of the wide use of melatonin assays based on anti-melatonin antisera. Whereas interference has been reported (Kennaway, Frith et al. 1977), there is no evidence that the antisera used for detection of melatonin in commercial kits are more than 1/100 as sensitive towards N-acetyltryptamine than melatonin, based on product literature, communications with sources and publications (Kennaway, Gilmore et al. 1982). Accordingly, the potential for interference seems unlikely. However, the possibility that this might occur when plasma melatonin is low and N-acetyltryptamine levels are exceedingly high cannot be ignored, especially in view of our finding of the existence of outliers with high values of N-acetyltryptamine (Figure 3, Table S1). This issue is of special relevance in studies where accurate determinations of low levels of melatonin play an important role, i.e., the diagnostic use of melatonin to describe dynamic changes in the central circadian clock and chronotype (Lewy and Sack 1989; Lewy 1999; Lack, Bailey et al. 2009).
Final Comment
The evidence that N-acetyltryptamine is present in the circulation and undergoes a 24-hour rhythm in abundance leads to the important question of the role it plays in physiology independent of melatonin. This encourages reexamination of melatonergic signaling including special consideration of the proposal that N-acetyltryptamine has a paracrine or autocrine role.
Supplementary Material
Supplemental Information File S1. Expression analysis results are available on the following link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=epkvmssmvhmrlmf&acc=GSE78165. Full tables with expression values (in FPKM) for mixed tissue, retina and pineal gland are provided. Additionally, differential expression tables for both retina and pineal gland are available at the same GEO link.
Table S1. Individual human plasma N-acetyltryptamine and melatonin values.
Acknowledgments
This work was supported by funds from the Intramural Research Program of Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (P.B., S.L.C, D.C.K); by NIH grants AG-036670, AG-029612 and OD-011092 (H.F.U); and by the Bert L and N Kuggie Vallee Foundation, the WorldQuant Foundation, NASA (NNX14AH50G, 15-15Omni2-0063), and the Bill and Melinda Gates Foundation (OPP1151054) (C.E.M. and M.B.). The expert animal management of Daniel Abebe (NICHD) is greatly appreciated
References
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57(1):289–300. [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29(4):1165–1188. [Google Scholar]
- Bernard M, Iuvone PM, et al. Avian melatonin synthesis: photic and circadian regulation of serotonin N-acetyltransferase mRNA in the chicken pineal gland and retina. J Neurochem. 1997;68(1):213–224. doi: 10.1046/j.1471-4159.1997.68010213.x. [DOI] [PubMed] [Google Scholar]
- Bernard M, Klein DC, et al. Chick pineal clock regulates serotonin N-acetyltransferase mRNA rhythm in culture. Proc Natl Acad Sci U S A. 1997;94(1):304–309. doi: 10.1073/pnas.94.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolliet V, Begay V, et al. Photoreceptor cells of the pike pineal organ as cellular circadian oscillators. Eur J Neurosci. 1997;9(4):643–653. doi: 10.1111/j.1460-9568.1997.tb01413.x. [DOI] [PubMed] [Google Scholar]
- Brooks DS, Cassone VM. Daily and circadian regulation of 2-[125I]iodomelatonin binding in the chick brain. Endocrinology. 1992;131(3):1297–1304. doi: 10.1210/endo.131.3.1324157. [DOI] [PubMed] [Google Scholar]
- Carter MD, Calcutt MW, et al. Quantitation of melatonin and n-acetylserotonin in human plasma by nanoflow LC-MS/MS and electrospray LC-MS/MS. J Mass Spectrom. 2012;47(3):277–285. doi: 10.1002/jms.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong NW, Sugden D. Thermodynamic analysis of agonist and antagonist binding to the chicken brain melatonin receptor. Br J Pharmacol. 1994;111(1):295–301. doi: 10.1111/j.1476-5381.1994.tb14059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RD. Detection of Influential Observation in Linear-Regression. Technometrics. 1977;19(1):15–18. [Google Scholar]
- Coon SL, Del Olmo E, et al. Melatonin synthesis enzymes in Macaca mulatta: focus on arylalkylamine N-acetyltransferase (EC 2.3.1.87) The Journal of clinical endocrinology and metabolism. 2002;87(10):4699–4706. doi: 10.1210/jc.2002-020683. [DOI] [PubMed] [Google Scholar]
- Coon SL, Del Olmo E, et al. Melatonin synthesis enzymes in Macaca mulatta: focus on arylalkylamine N-acetyltransferase (EC 2.3.1.87) J Clin Endocrinol Metab. 2002;87(10):4699–4706. doi: 10.1210/jc.2002-020683. [DOI] [PubMed] [Google Scholar]
- Coon SL, Mazuruk K, et al. The human serotonin N-acetyltransferase (EC 2.3.1.87) gene (AANAT): structure, chromosomal localization, and tissue expression. Genomics. 1996;34(1):76–84. doi: 10.1006/geno.1996.0243. [DOI] [PubMed] [Google Scholar]
- Coon SL, Roseboom PH, et al. Pineal serotonin N-acetyltransferase: expression cloning and molecular analysis. Science. 1995;270(5242):1681–1683. doi: 10.1126/science.270.5242.1681. [DOI] [PubMed] [Google Scholar]
- Davies B, Hannah LT, et al. Central melatonin binding sites in rainbow trout (Onchorhynchus mykiss) Gen Comp Endocrinol. 1994;96(1):19–26. doi: 10.1006/gcen.1994.1155. [DOI] [PubMed] [Google Scholar]
- Desilva SO, Snieckus V. Indole N-Alkylation of Tryptamines Via Dianion and Phthalimido Intermediates - New Potential Indolealkylamine Haptens. Canadian Journal of Chemistry-Revue Canadienne De Chimie. 1978;56(12):1621–1627. [Google Scholar]
- Doll MA, Hein DW. Comprehensive human NAT2 genotype method using single nucleotide polymorphism-specific polymerase chain reaction primers and fluorogenic probes. Anal Biochem. 2001;288(1):106–108. doi: 10.1006/abio.2000.4892. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML. Melatonin is a potent modulator of dopamine release in the retina. Nature. 1983;306(5945):782–784. doi: 10.1038/306782a0. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML. N-Acetyltryptamine antagonizes the melatonin-induced inhibition of [3H]dopamine release from retina. Eur J Pharmacol. 1984;105(1–2):193–194. doi: 10.1016/0014-2999(84)90668-x. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML. Characterization of a retinal melatonin receptor. J Pharmacol Exp Ther. 1985;234(2):395–401. [PubMed] [Google Scholar]
- Dubocovich ML. Melatonin receptors in the central nervous system. Adv Exp Med Biol. 1991;294:255–265. doi: 10.1007/978-1-4684-5952-4_23. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML, Delagrange P, et al. International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharmacol Rev. 2010;62(3):343–380. doi: 10.1124/pr.110.002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon J, Coon SL, et al. Drastic neofunctionalization associated with evolution of the timezyme AANAT 500 Mya. Proc Natl Acad Sci U S A. 2014;111(1):314–319. doi: 10.1073/pnas.1312634110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JV, Barrett P, et al. Ovine arylalkylamine N-acetyltransferase in the pineal and pituitary glands: differences in function and regulation. Endocrinology. 1999;140(2):972–978. doi: 10.1210/endo.140.2.6496. [DOI] [PubMed] [Google Scholar]
- Fretland AJ, Doll MA, et al. Functional characterization of nucleotide polymorphisms in the coding region of N-acetyltransferase 1. Pharmacogenetics. 2001;11(6):511–520. doi: 10.1097/00008571-200108000-00006. [DOI] [PubMed] [Google Scholar]
- Fretland AJ, Leff MA, et al. Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Pharmacogenetics. 2001;11(3):207–215. doi: 10.1097/00008571-200104000-00004. [DOI] [PubMed] [Google Scholar]
- Gaudet S, Palkovits M, et al. Regional distribution of arylamine and arylalkylamine N-acetyltransferase activities in the rat brain. Brain Res. 1991;539(2):355–357. doi: 10.1016/0006-8993(91)91645-h. [DOI] [PubMed] [Google Scholar]
- Gaudet SJ, Namboodiri MA. Characterization of the brain family of aromatic amine N-acetyltransferases. Mol Cell Neurosci. 1993;4(4):310–318. doi: 10.1006/mcne.1993.1041. [DOI] [PubMed] [Google Scholar]
- Haque R, Chong NW, et al. Melatonin synthesis in retina: cAMP-dependent transcriptional regulation of chicken arylalkylamine N-acetyltransferase by a CRE-like sequence and a TTATT repeat motif in the proximal promoter. J Neurochem. 2011;119(1):6–17. doi: 10.1111/j.1471-4159.2011.07397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein DW. N-acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opin Drug Metab Toxicol. 2009;5(4):353–366. doi: 10.1517/17425250902877698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heward CB, Hadley ME. Structure-activity relationships of melatonin and related indoleamines. Life Sci. 1975;17(7):1167–1177. doi: 10.1016/0024-3205(75)90340-9. [DOI] [PubMed] [Google Scholar]
- Huang Y, Louie A, et al. A simple LC-MS/MS method for determination of kynurenine and tryptophan concentrations in human plasma from HIV-infected patients. Bioanalysis. 2013;5(11):1397–1407. doi: 10.4155/bio.13.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuvone PM, Boatright JH, et al. N-acetylserotonin: circadian activation of the BDNF receptor and neuroprotection in the retina and brain. Adv Exp Med Biol. 2014;801:765–771. doi: 10.1007/978-1-4614-3209-8_96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuvone PM, Tosini G, et al. Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog Retin Eye Res. 2005;24(4):433–456. doi: 10.1016/j.preteyeres.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Jacoby JH, Sassin JF, et al. Patterns of spontaneous cortisol and growth hormone secretion in rhesus monkeys during the sleep-waking cycle. Neuroendocrinology. 1974;14(3):165–173. doi: 10.1159/000122256. [DOI] [PubMed] [Google Scholar]
- Kennaway DJ, Frith RG, et al. A specific radioimmunoassay for melatonin in biological tissue and fluids and its validation by gas chromatography-mass spectrometry. Endocrinology. 1977;101(1):119–127. doi: 10.1210/endo-101-1-119. [DOI] [PubMed] [Google Scholar]
- Kennaway DJ, Gilmore TA, et al. Effect of melatonin feeding on serum prolactin and gonadotropin levels and the onset of seasonal estrous cyclicity in sheep. Endocrinology. 1982;110(5):1766–1772. doi: 10.1210/endo-110-5-1766. [DOI] [PubMed] [Google Scholar]
- Klein DC. The 2004 Aschoff/Pittendrigh lecture: Theory of the origin of the pineal gland--a tale of conflict and resolution. J Biol Rhythms. 2004;19(4):264–279. doi: 10.1177/0748730404267340. [DOI] [PubMed] [Google Scholar]
- Klein DC. Evolution of the vertebrate pineal gland: the AANAT hypothesis. Chronobiol Int. 2006;23(1–2):5–20. doi: 10.1080/07420520500545839. [DOI] [PubMed] [Google Scholar]
- Klein DC. Arylalkylamine N-acetyltransferase: "the Timezyme". J Biol Chem. 2007;282(7):4233–4237. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- Klein DC, Coon SL, et al. The melatonin rhythm-generating enzyme: molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res. 1997;52:307–357. discussion 357-308. [PubMed] [Google Scholar]
- Klein DC, Weller JL. Indole metabolism in the pineal gland: a circadian rhythm in N-acetyltransferase. Science. 1970;169(3950):1093–1095. doi: 10.1126/science.169.3950.1093. [DOI] [PubMed] [Google Scholar]
- Lack L, Bailey M, et al. Chronotype differences in circadian rhythms of temperature, melatonin, and sleepiness as measured in a modified constant routine protocol. Nat Sci Sleep. 2009;1:1–8. doi: 10.2147/nss.s6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff MA, Fretland AJ, et al. Novel human N-acetyltransferase 2 alleles that differ in mechanism for slow acetylator phenotype. Journal of Biological Chemistry. 1999;274(49):34519–34522. doi: 10.1074/jbc.274.49.34519. [DOI] [PubMed] [Google Scholar]
- Lemos DR, Downs JL, et al. Photoperiodic modulation of adrenal gland function in the rhesus macaque: effect on 24-h plasma cortisol and dehydroepiandrosterone sulfate rhythms and adrenal gland gene expression. J Endocrinol. 2009;201(2):275–285. doi: 10.1677/JOE-08-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewy AJ. The dim light melatonin onset, melatonin assays and biological rhythm research in humans. Biol Signals Recept. 1999;8(1–2):79–83. doi: 10.1159/000014573. [DOI] [PubMed] [Google Scholar]
- Lewy AJ, Markey SP. Analysis of melatonin in human plasma by gas chromatography negative chemical ionization mass spectrometry. Science. 1978;201(4357):741–743. doi: 10.1126/science.675255. [DOI] [PubMed] [Google Scholar]
- Lewy AJ, Sack RL. The dim light melatonin onset as a marker for circadian phase position. Chronobiol Int. 1989;6(1):93–102. doi: 10.3109/07420528909059144. [DOI] [PubMed] [Google Scholar]
- Li S, Tighe SW, et al. Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study (vol 32, pg 915, 2014) Nature Biotechnology. 2014;32(11):1166–1166. doi: 10.1038/nbt.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R, Brooks A. The electrophysiological effects of melatonin and a putative melatonin antagonist (N-acetyltryptamine) on rat suprachiasmatic neurones in vitro. Neurosci Lett. 1988;95(1–3):296–301. doi: 10.1016/0304-3940(88)90674-x. [DOI] [PubMed] [Google Scholar]
- Mefford IN, Chang P, et al. Reciprocal day/night relationship between serotonin oxidation and N-acetylation products in the rat pineal gland. Endocrinology. 1983;113(5):1582–1586. doi: 10.1210/endo-113-5-1582. [DOI] [PubMed] [Google Scholar]
- Paulin CH, Cazamea-Catalan D, et al. Subfunctionalization of arylalkylamine N-acetyltransferases in the sea bass Dicentrarchus labrax: two-ones for one two. J Pineal Res. 2015;59(3):354–364. doi: 10.1111/jpi.12266. [DOI] [PubMed] [Google Scholar]
- Pavlicek J, Sauzet S, et al. Evolution of AANAT: expansion of the gene family in the cephalochordate amphioxus. BMC Evol Biol. 2010;10:154. doi: 10.1186/1471-2148-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlow MJ, Reppert SM, et al. Daily rhythms in cortisol and melatonin in primate cerebrospinal fluid. Effects of constant light and dark. Neuroendocrinology. 1981;32(4):193–196. doi: 10.1159/000123157. [DOI] [PubMed] [Google Scholar]
- Pozdeyev N, Taylor C, et al. Photic regulation of arylalkylamine N-acetyltransferase binding to 14-3-3 proteins in retinal photoreceptor cells. J Neurosci. 2006;26(36):9153–9161. doi: 10.1523/JNEUROSCI.1384-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Perlow MJ, et al. Diurnal Melatonin Rhythm in Primate Cerebrospinal-Fluid. Endocrinology. 1979;104(2):295–301. doi: 10.1210/endo-104-2-295. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Perlow MJ, et al. Photic Regulation of the Melatonin Rhythm - Distinct Difference between Man and Monkey. Pediatr Res. 1979;13(4):362–362. [Google Scholar]
- Sitzmann BD, Brown DI, et al. Impact of moderate calorie restriction on testicular morphology and endocrine function in adult rhesus macaques (Macaca mulatta) Age (Dordr) 2014;36(1):183–197. doi: 10.1007/s11357-013-9563-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, March J. March's advanced organic chemistry : reactions, mechanisms, and structure. Hoboken, N.J.: Wiley-Interscience; 2007. [Google Scholar]
- Sorwell KG, Kohama SG, et al. Testosterone increases circulating dehydroepiandrosterone sulfate levels in the male rhesus macaque. Front Endocrinol (Lausanne) 2014;5:101. doi: 10.3389/fendo.2014.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MS, Cohen SN, et al. Acetylation of 5-hydroxytryptamine by isoniazid N-acetyltransferase. Biochim Biophys Acta. 1969;184(1):210–212. doi: 10.1016/0304-4165(69)90119-6. [DOI] [PubMed] [Google Scholar]
- Sugden D, Chong NW, et al. Structural requirements at the melatonin receptor. Br J Pharmacol. 1995;114(3):618–623. doi: 10.1111/j.1476-5381.1995.tb17184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosini G, Chaurasia SS, et al. Regulation of arylalkylamine N-acetyltransferase (AANAT) in the retina. Chronobiol Int. 2006;23(1–2):381–391. doi: 10.1080/07420520500482066. [DOI] [PubMed] [Google Scholar]
- Tosini G, Pozdeyev N, et al. The circadian clock system in the mammalian retina. Bioessays. 2008;30(7):624–633. doi: 10.1002/bies.20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyce GM, Flock EV, et al. Uptake and Metabolism of 5-Hydroxytryptamine by Isolated Perfused Rat Liver. American Journal of Physiology. 1968;215(3):611-&. doi: 10.1152/ajplegacy.1968.215.3.611. [DOI] [PubMed] [Google Scholar]
- Tyce GM, Flock EV, et al. Metabolism of serotonin by the isolated perfused rat liver--effect of glucuronyl transferase deficiency or monoamine oxidase inhibition. Biochem Pharmacol. 1968;17(8):1543–1552. doi: 10.1016/0006-2952(68)90213-x. [DOI] [PubMed] [Google Scholar]
- Urbanski HF. Circadian Variation in the Physiology and Behavior of Humans and Nonhuman Primates. Animal Models of Behavioral Analysis. 2011;50:217–235. [Google Scholar]
- Urbanski HF. Role of circadian neuroendocrine rhythms in the control of behavior and physiology. Neuroendocrinology. 2011;93(4):211–222. doi: 10.1159/000327399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanski HF, Sorwell KG, et al. Androgen supplementation during aging: development of a physiologically appropriate protocol. Rejuvenation Res. 2014;17(2):150–153. doi: 10.1089/rej.2013.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanecek J. Melatonin binding sites. J Neurochem. 1988;51(5):1436–1440. doi: 10.1111/j.1471-4159.1988.tb01108.x. [DOI] [PubMed] [Google Scholar]
- Vanecek J. Cellular mechanisms of melatonin action. Physiol Rev. 1998;78(3):687–721. doi: 10.1152/physrev.1998.78.3.687. [DOI] [PubMed] [Google Scholar]
- Vatsis KP, Weber WW, et al. Nomenclature for N-Acetyltransferases. Pharmacogenetics. 1995;5(1):1–17. doi: 10.1097/00008571-199502000-00001. [DOI] [PubMed] [Google Scholar]
- Wood S, Loudon A. Clocks for all seasons: unwinding the roles and mechanisms of circadian and interval timers in the hypothalamus and pituitary. J Endocrinol. 2014;222(2):R39–59. doi: 10.1530/JOE-14-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information File S1. Expression analysis results are available on the following link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=epkvmssmvhmrlmf&acc=GSE78165. Full tables with expression values (in FPKM) for mixed tissue, retina and pineal gland are provided. Additionally, differential expression tables for both retina and pineal gland are available at the same GEO link.
Table S1. Individual human plasma N-acetyltryptamine and melatonin values.