Supplemental Digital Content is available in the text.
Keywords: brain injuries, cerebrospinal fluid, microRNAs, real-time polymerase chain reaction, stroke, subarachnoid hemorrhage
Abstract
Background and Purpose—
Delayed cerebral ischemia (DCI) accounts for a major part of the morbidity and mortality after aneurysmal subarachnoid hemorrhage (SAH). MicroRNAs (miRNAs) are pathophysiologically involved in acute cerebral ischemia. This study compared miRNA profiles in cerebrospinal fluid from neurologically healthy patients, as well as SAH patients with and without subsequent development of DCI.
Methods—
In a prospective case–control study of SAH patients treated with external ventricular drainage and neurologically healthy patients, miRNA profiles in cerebrospinal fluid were screened and validated using 2 different high-throughput real-time quantification polymerase chain reaction techniques. The occurrence of DCI was documented in patient charts and subsequently reviewed independently by 2 physicians.
Results—
MiRNA profiles from 27 SAH patients and 10 neurologically healthy patients passed quality control. In the validation, 66 miRNAs showed a relative increase in cerebrospinal fluid from SAH patients compared with neurologically healthy patients (P<0.001); 2 (miR-21 and miR-221) showed a relative increase in SAH patients with DCI compared with those without (P<0.05) in both the screening and validation.
Conclusions—
SAH is associated with marked changes in the cerebrospinal fluid miRNA profile. These changes could be associated to the development of DCI.
Clinical Trial Registration—
URL: http://www.clinicaltrials.gov. Unique identifier: NCT01791257.
Among patients who survive to hospital admission after aneurysmal subarachnoid hemorrhage (SAH), clinical deterioration caused by delayed cerebral ischemia (DCI) accounts for a major part of the morbidity and mortality.1,2 Current prevention and therapy of DCI are primarily based on a hypothesis of large-vessel vasospasm. Despite a marked reduction in the prevalence of vasospasm, intravenous clazosentan neither reduced the incidence of DCI nor improved outcome in multicenter randomized controlled trials.3 Furthermore, nimodipine, the only agent with a documented effect on the risk of DCI,4 does not reduce cerebral vasospasm and, thus, seems to exert its effect through different mechanisms.5 Consequently, vasospasm should no longer be viewed as the single important mediator of DCI. Instead, alternative mechanisms leading to DCI should be explored.
MicroRNAs (miRNAs) are 22 nucleotides long, noncoding RNA molecules involved in post-transcriptional regulation of complementary mRNA targets.6 MiRNAs are highly tissue-specific. They act intracellularly but are transported outside the cells in exosomes and exist in stable forms in body fluids.7 Specific miRNAs are involved in the development of neuronal apoptosis after experimental acute cerebral ischemia6,8,9 and are clinically associated with the severity of traumatic brain injury.10 In the present explorative study, we compared miRNA profiles in cerebrospinal fluid (CSF) between neurologically healthy patients and SAH patients with and without subsequent development of DCI. Moreover, we compared profiles obtained with 2 different commonly used commercial high-throughput real-time quantification polymerase chain reaction (RT-qPCR) platforms. We hypothesized that the relative concentration of specific miRNAs would be altered between SAH and neurological healthy patients, as well as between SAH patients with and without DCI.
Materials and Methods
This was a prospective case–control study designed primarily to study miRNA in SAH and neurologically healthy patients. The protocol was approved by the Danish Scientific Ethics Committee of the Capital Region (file No. H-3-2013-009), as well as the Danish Data Protection Agency (file No. RH-2013-30-0935), and registered on clinicaltrials.gov (file No. NCT01791257). According to Danish law, informed consent was obtained from either the patient or from their next of kin and general practitioner.
Enrollment
SAH Patients
From February 2013 to January 2014, patients admitted to the neurointensive care unit at Rigshospitalet, Copenhagen, with spontaneous aneurysmal SAH were screened for inclusion. Eligible participants were patients >18 years of age who had an external ventricular drain placed within 5 days after ictus. Exclusion criteria were the following: (1) inability of patient or next of kin to speak and understand Danish; (2) >24 hours’ uncertainty of the time of ictus; and (3) inability to diagnose DCI because of a persistent Glasgow Coma Score <7 as a result of initial brain injury, a known complication (eg, rebleeding, surgical/endovascular trauma), or deep sedation.
Neurologically Healthy Control Patients
From September to October 2013, all patients admitted to the orthopedic department at Frederiksberg Hospital, Copenhagen, for elective lower limb surgery and planned for spinal anesthesia were screened for inclusion. Eligible participants had no clinically overt neurological disease or diagnosis, were >18 years of age, and had an American Society of Anesthesiologists physical status classification ≤2.
Sampling of CSF
Using a strict protocol, including sterile precautions, CSF was sampled from the external ventricular drain on day 5 after ictus in SAH patients and prior to administration of spinal anesthesia for orthopedic surgery in neurologically healthy patients. All CSF samples were put on ice immediately and spun within 15 minutes at 500g for 10 minutes; the supernatant was stored at −80°C until use.
Primary Outcome Measures
All cases of SAH were diagnosed by computed tomography scanning and computed tomographic angiography. Patients were continually evaluated for signs of DCI as described by Vergouwen et al.2 Because DCI is a clinical diagnosis based on the exclusion of other causes (eg, rebleeding, surgical complications, hydrocephalus, and infection), confirmation or exclusion will be subjective in some cases.2 To minimize error, 2 physicians (S. Bache and R. Rasmussen) reviewed all clinical, biochemical, and radiological data, obtained 3 weeks from ictus, and independently assessed if cases of clinical deterioration fulfilled the criteria of DCI. Patients were categorized as definite DCI, probable DCI, possible DCI, probably not DCI, or definitely not DCI. Disagreements were discussed, and all decisions were made in consensus. Only patients with a priori agreement between the 2 physicians could be categorized as definitely DCI or definitely not DCI. According to the protocol, inclusion was stopped when at least 12 patients had been categorized as either definitely DCI or probably DCI and at least 12 patients were categorized as either probably not DCI or definitely not DCI. Only patients categorized as definite DCI and definitely not DCI were compared in the final analysis.
Secondary Outcome Measures
To relate the CSF miRNA profiles to initial and secondary injury, as well as outcome, SAH patients were dichotomized according to the following:
Clinical status on admission based on World Federation of Neurosurgical Societies grading scale (WFNS) with good grade (WFNS score 1–3) versus poor grade (WFNS score 4–5)
Early brain injury (EBI) defined by our protocol according to the Glasgow Come Score at 24 hours after aneurysm closure as no or minor EBI (Glasgow Coma Score 13–15 and no major focal deficit) versus major EBI (Glasgow Coma Score 3–12 and major focal deficit)
Presence or absence of angiographic vasospasm diagnosed by computed tomography or magnetic resonance angiographic sequences between 3 and 14 days from ictus
Delayed cerebral infarction as defined by Vergouwen et al2
Outcome at 3 months according to the modified Rankin Scale and classified as good (modified Rankin Scale score 0–2) versus poor (modified Rankin Scale score 3–6).
Spectrophotometric Measures of Hemolysis
Spectrophotometry was performed on each CSF sample; the net absorbance was calculated for hemoglobin and oxyhemoglobin using a standardized method11 and correlated against the non-normalized average cycle quantification (Cq) value of all detected miRNAs.
MiRNA Profiling
CSF miRNA profiles were screened in individual patients using 1 high-throughput RT-qPCR platform, which is based on the stem-loop primer technique (TaqMan, Thermo Fisher, Cambridge, MA). Subsequently, the miRNAs that were detected in all SAH patients were validated using another high-throughput RT-qPCR platform, which is based on the locked nuclear acid technique (LNA, Exiqon, Vedbaek, Denmark).
Screening: RNA Isolation
Total RNA was isolated from 200 μL of each sample of CSF according to the manufacturer’s protocol (Total RNA isolation kit, Appendix B; Norgen Biotek, Thorold, Canada). As proposed by the manufacturer, β-mercaptoethanol was added to the lysis solution.
Screening: Real-Time Quantification
We screened for a total of 754 miRNAs. Reverse transcription and RT-qPCR was performed as previously described.12 Briefly, for each sample, a fixed volume of 6 μL eluted RNA sample was mixed on ice with 9 μL of a reverse transcription reaction containing 1.6 μL of 10× RT buffer, 1.8 μL of MgCl2, 0.2 μL of RNase-inhibitor (20 U/μL), 0.4 μL of dNTPs with dTTP (100 mmol/L), 3.0 μL of Multiscribe Reverse Transcriptase (50 U/μL), 1.6 μL of Megaplex RT primers (×10), and 0.4 μL of nuclease free water. Reverse transcription was performed using 40 cycles of 16°C, 2 minutes; 42°C, 1 minute; and 50°C, 1 s followed by 85°C for 5 minutes and hold at 4°C. For each sample, a preamplification reaction comprising 62.5 μL of TaqMan PreAmp Master Mix (2×), 12.5 μL of Megaplex PreAmp Primers (10×), 37.5 μL of nuclease-free water, and 12.5 μL of RT product were mixed on ice. After 10 minutes at 95°C, 2 minutes at 55°C, and 2 minutes at 72°C, the cDNA was subjected to 12 cycles of 15 s at 95°C and 4 minutes at 60°C followed by 10 minutes at 99.9°C and hold at 4°C. The preamplified product was diluted with 375 μL of 0.1 TE buffer. Subsequently, 450 μL of diluted preamplified product was mixed on ice with 450 μL of TaqMan Fast Advanced Master Mix and 800 μL loaded on a TaqMan Human MicroRNA Array Card. RT-qPCR was performed using a Viia 7 real-time PCR system (Applied Biosystems, Foster City, CA). The above steps were performed using Human Pools A v.2.1 and repeated using Human Pools B v.3.0 for a total screen of 754 specific miRNAs. Reagents and array cards were all purchased from Invitrogen, Carlsbad, CA.
Validation: RNA Isolation
Total RNA was isolated from 200 μL of each sample of CSF using miRCURY RNA Isolation Kit Biofluids. This included the addition of MS2 carrier RNA (Roche, Switzerland) and spike-ins, UniSp2, UniSp4, and UniSp5, according to the manufacturer’s protocol (Exiqon, Vedbæk, Denmark).
Validation: Real-Time Quantification
The validation was tested for the presence of 151 miRNAs of the 155 miRNAs originally detected in all SAH patients during screening analysis because 4 of the original targets were excluded either because they represent mRNA or tRNA fragments rather than miRNA or because they were unavailable from the manufacturer. Reverse transcription was performed using the Universal cDNA synthesis kit II and included the addition of spike-ins, UniSp6, and cel-miR-39, according to the manufacturer’s protocol (Exiqon, Vedbæk, Denmark). Based on our own optimization studies, we used 9.6 μL of template RNA for a total volume of 24 μL. After incubation, cDNA was diluted 100× in nuclease-free water and ExiLENT SYBR Green master mix according to the manufacturers protocol (Exiqon, Vedbæk, Denmark) with addition of a ROX reference dye 50× (Invitrogen, Carlsbad, CA). Pick & Mix microRNA PCR panels (Exiqon, Vedbæk, Denmark) were loaded with 10 μL in each of 384 wells, and RT-qPCR was performed using a Viia 7 real-time PCR system (Applied Biosystems, Foster City, CA), with settings suggested by the manufacturer including a melting curve analysis (Exiqon).
Sample Size Calculations
Using a web-based sample size calculator for microarray studies13 and an SD of 0.5 Cq values based on in-house RT-qPCR measurements of biological replicates of CSF, we estimated that 8 patients in each group would be necessary to detect a 2-fold change. To allow for dropouts, exclusion of patients difficult to categorize as DCI, and missing data in RT-qPCR profiling, we planned to include 12 patients in each of the 3 groups (ie, SAH with and without DCI, as well as neurologically healthy patients).
Preprocessing and Quality Control
For all targets, the RT-qPCR data were preprocessed by setting cycle threshold to 0.1 for screening and 0.01 for validation; the baseline was set from 3 to 15 for screening and calculated automatically by the instrument software for validation. Screened miRNA profiles with U6snRNA replicate SDs >0.5 Cq values were excluded from further analysis. Validated miRNA profiles were subjected to interplate calibration using the synthetic DNA spike-in, UniSp3, and internal amplification control using the synthetic RNA spike-ins UniSp2, -4, and -6 and cel-miR-39-3p in GenEx v.6.0.1 (Multid, Göteborg, Sweden). MiRNA targets with Cq values >32 in the screening study and 36 in the validation study were discarded.
Normalization and Statistical Analysis
The average Cq value of all detected miRNAs in each sample differed between SAH and neurologically healthy patients. Thus, non-normalized data were used to compare these groups. Otherwise, Cq values were normalized to the sample mean.14 GenEx v.6.0.1 (Multid, Göteborg, Sweden) was used for preprocessing, and fold changes were calculated using the formula 2(−ΔΔCq). Statistical analyses were performed using Qlucore Omics Explorer 3.2. Continuous variables were tested for normality, and groups were compared using Student’s t test. One-way hierarchically clustered heat maps were generated using Euclidean distance and average linkage. Significance criteria were defined as P<0.001 for non-normalized data and P<0.05 for normalized data. As an estimate of the false discovery rate, corresponding q values were calculated using the Benjamini & Hochberg procedure where applicable.15 In this context, a q value of 0.05 equals one false-positive discovery in 20 significant discoveries, and lower q values equal lower risk of a false-positive discovery.
Results
Patients
During 11 months, 54 adult SAH patients treated with external ventricular drainage were screened for participation in the study. Thirty-six patients were included, of whom 7 were later excluded. The remaining 29 patients were grouped as follows: definitely DCI (N=8), probably DCI (N=5), possible DCI (N=3), probably not DCI (N=4), and definitely not DCI (N=9). The miRNA profiles for 2 patients failed quality control after RT-qPCR, leaving miRNA profiles from a total of 27 SAH patients for final analyses.
During October and November 2013, 14 neurologically healthy control patients were included; 2 were subsequently excluded as CSF could not be sampled. Two miRNA profiles failed quality control, leaving 10 miRNA profiles for final analyses.
Please see Figure 1 and Table 1 for details on patient recruitment and clinical characteristics.
Figure 1.
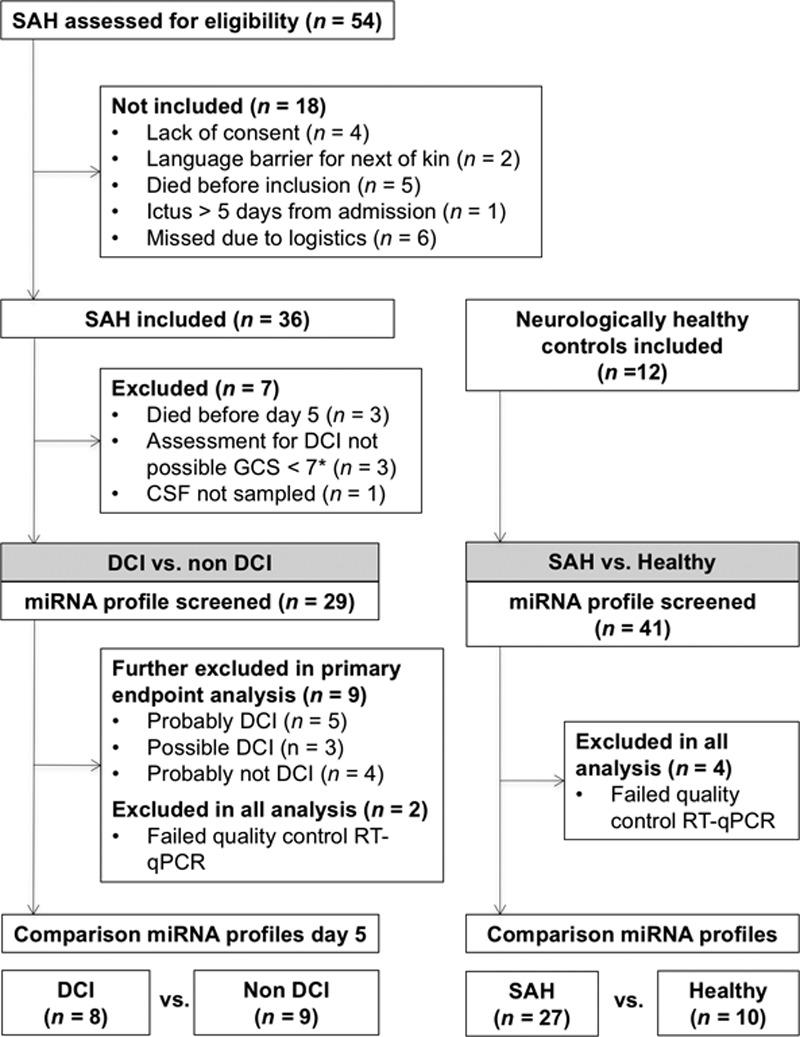
Flow diagram of patient inclusion. CSF indicates cerebrospinal fluid; DCI, delayed cerebral ischemia; GCS, Glasgow Coma Scale; RT-qPCR, real-time quantification polymerase chain reaction; and SAH, subarachnoid hemorrhage. *GCS <7 (2 because of poor neurological performance, 1 needed heavy sedation for cardiopulmonary complication, and 1 had a thrombus of a posterior inferior cerebellar artery at day 3).
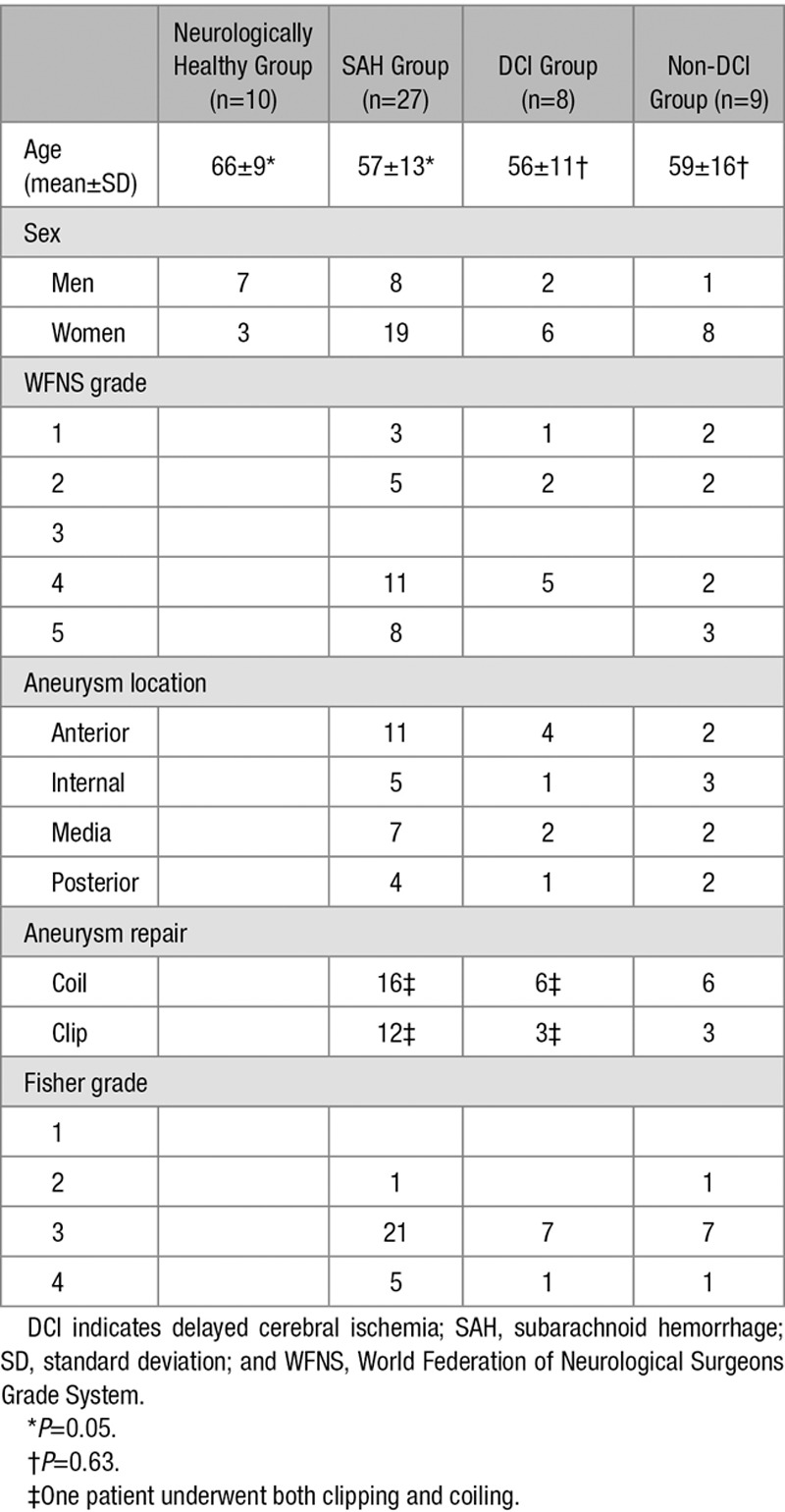
Table 1.
Clinical Characteristics of Included Patients
Screening and Validation miRNA Analyses
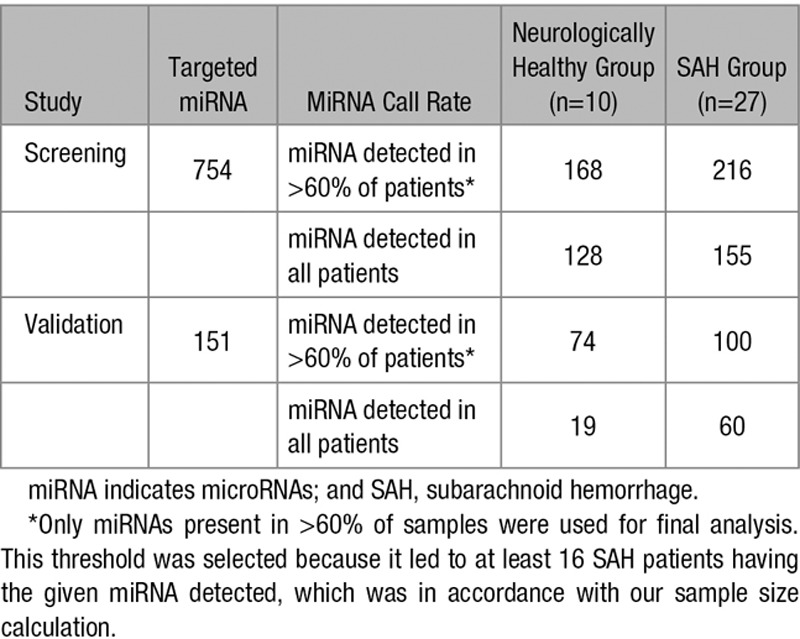
Detection rates of miRNAs for each samples are presented in Table 2. Both the screening and validation study detected an increased number of miRNAs in CSF from SAH patients compared with neurologically healthy patients. Also the average Cq value for all miRNAs in each sample was higher in SAH patients (data not shown). Standard deviations of the Cq values for each miRNA within samples were similar between the 2 techniques and increased as expected when Cq values increased consistent with lower copy numbers. Conversely, raw and normalized Cq values of the 66 miRNAs detected with both techniques in all SAH patients correlated well between screening and validation. For raw and normalized Cq values for all miRNAs in all samples for both studies, see related data in the online-only Data Supplement:.
Table 2.
Overall Detected miRNAs in the Screening and the Validation Studies
Correlation of miRNA to Hemolysis
In profiling studies of circulating miRNA, quantification of miR-16-5p and -451a have been recommended to evaluate serum and plasma samples for hemolysis.16 In agreement with this, we found that Cq values of miR-16-5p and -451a, as well as the average Cq value for all miRNAs in each sample, correlated well to the net absorbance of oxyhemoglobin and hemoglobin in the CSF of SAH patients (Figure I and Table I in the online-only Data Supplement).
SAH Versus Healthy
Of the 151 miRNAs that were detected in the screening study and targeted during validation, 66 exhibited an increase in SAH compared with neurologically healthy patients, both in the screening (P<0.001 and q<0.002) and validation study (P<0.001 and q<0.0009); and none exhibited a decrease. Figure 2 shows the distribution of the 66 increased miRNAs for each patient in each study. Fold change, as well as P and q values are presented for each of these miRNAs in Table II in the online-only Data Supplement.
Figure 2.
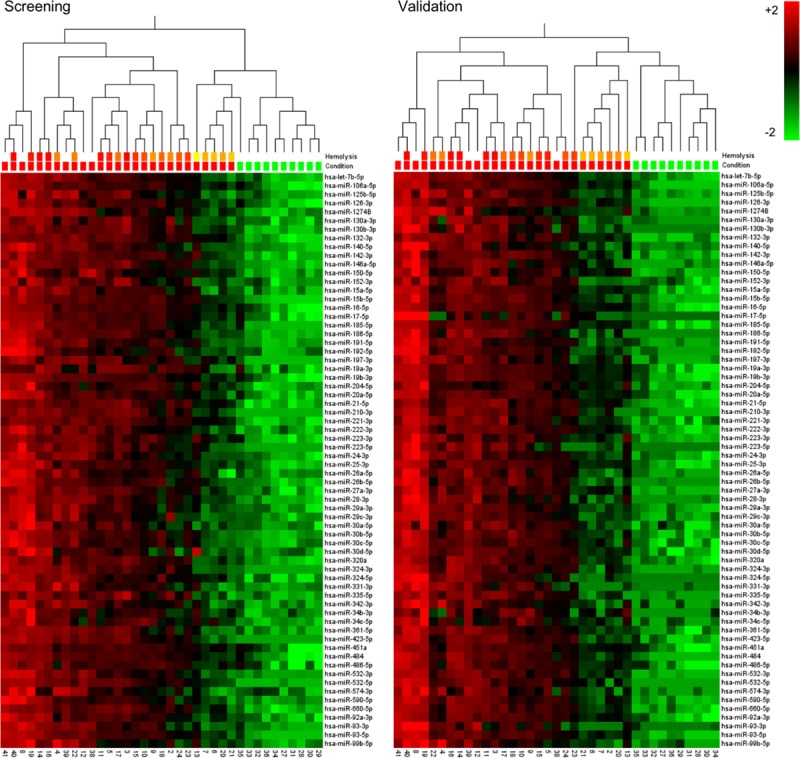
One-way hierachically clustered relative differences of non-normalized Cq values of microRNAs, which differ in cerebrospinal fluid between subarachnoid hemorrhage (SAH; red) and neurologically healthy patients (green) in both the screening (P<0.001, q<0.002) and the validation study (P<0.001, q<0.0009). Downregulation shown in green and upregulation in red. The line “Hemolysis” indicates the degree of hemolysis as measured by the net absorbance of oxyhemoglobin and colored from red (high) to yellow (low); the white color represents a value that is either out of range on the spectrophotometer (SAH) or not available (Healthy).
Occurrence of DCI
In the screening, a relative increase in miR-10b-3p, 21-5p, -132-3p, -146a-5p, 193a-5p, and -221-3p and a relative decrease in miR-208a-3p, -490-3p, -520h and -553 and -643 was found in CSF from SAH patients with DCI, compared with those without DCI (P<0.05 and q=0.74). Validation confirmed a relative increase in 2 of these, namely, miR-21-5p and -221-3p in patients with DCI compared with SAH patients without DCI (P<0.05 and q=0.38). Still, as can be deducted from the q values, none of these remained significant after correction for multiple testing. For the remaining miRNAs, none showed significant change in both the screening and the validation study. Figure 3 shows fold change and sample by sample correlations between screening and validation analyses for miR-21-5p and -221-3p.
Figure 3.
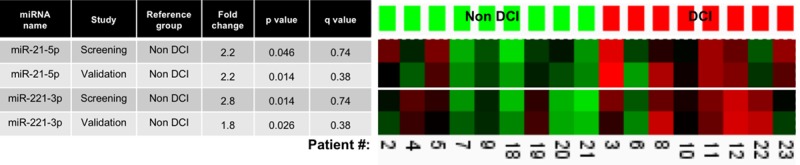
MicroRNAs (miRNAs) associated with delayed cerebral ischemia (DCI) in both screening and validation studies. Table (left) shows fold changes, significance levels (P value), and estimated false discovery rates (q value) for each miRNA in each study. Heat maps (right) depict sample by sample correlation between screening and validation of relative increases (red) or decreases (green) in each specific miRNA.
Secondary End Points
For the secondary end points, only results for miRNAs that differed in both screening and validation are given. We found a relative increase in miR-486-5p in patients with a poor neurological admission status (WFNS score 4–5) compared with those with a good status (WFNS score 1–3). Angiographic vasospasm was associated with a relative increase in miR-132-3p, -19b-3p, -210-3p, -221-3p, and -484. MiR-221-3p, which was also associated with DCI, showed a 3.3-fold increase in the screening study and a 2.6-fold increase in the validation study in SAH patients with angiographic vasospasm compared with patients without. These associations were strengthened by the fact that miR-221-3p was even lower in the neurologically healthy patients. No miRNAs differed between SAH patients with and without delayed cerebral infarction in both the screening and the validation. Finally, we observed a relative increase of miR-9-3p both in patients with major signs of EBI after aneurysm repair and in patients with a poor outcome 3 months after SAH, compared with patients without these injuries. Figure 4 shows fold change and sample by sample correlations between screening and validation analyses for secondary end points.
Figure 4.
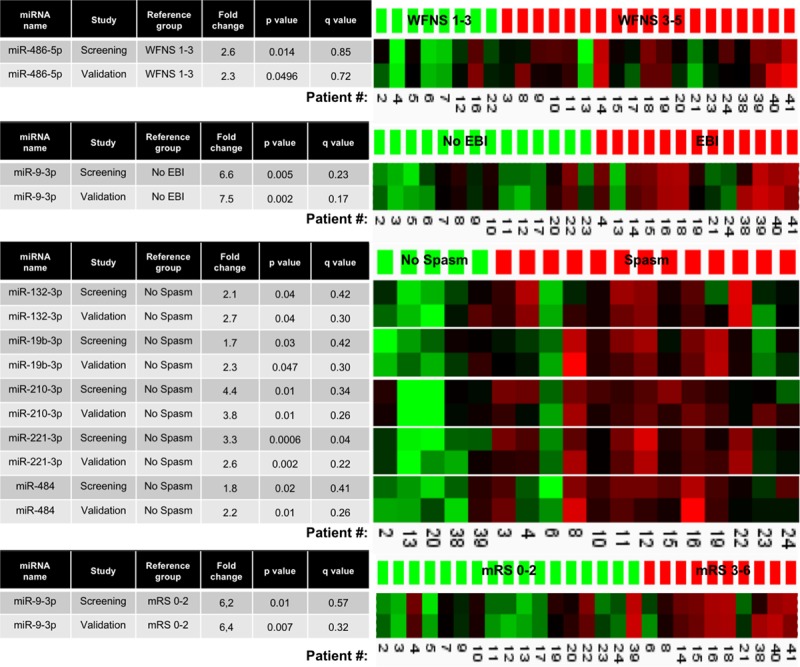
MicroRNAs (miRNAs) associated with neurological condition on admission (WFNS), early brain injury (EBI), angiographic vasospasm (Spasm), and outcome after 3 months (mRS) in both screening and validation studies. Tables (left) show fold changes, significance levels (P value), and estimated false discovery rates (q value) for each miRNA in each study. Heat maps (right) depict sample by sample correlation between screening and validation of relative increases (red) or decreases (green) in each specific miRNA. mRS indicates modified Rankin Scale; and WFNS, World Federation of Neurosurgical Societies SAH grading scale.
Discussion
According to our results, considerable changes occur in the miRNA composition of CSF after SAH, indicating a highly altered intrathecal miRNA environment at this time. On average, 48 additional miRNAs were present in CSF from SAH patients, and the 66 most differentially expressed miRNAs changed by a median of 34-fold (median P=1×10–8).
Experimental manipulation of levels of intrathecal miRNA with mimics or inhibitors has shown regulation of central pathways of apoptosis and neuronal survival after acute cerebral ischemia.17–19 Although inhibitory in nature, both up- and downregulated miRNAs could be of relevance depending on whether they target mRNA related to proapoptotic8 or antiapoptotic pathways.9 Extensive research into the origin of angiographic vasospasm and DCI has proposed that activation of various pathways are essential for their development.5,20 Our study is the first that aims to associate cerebral changes in miRNA with DCI, as well as with outcome after SAH.
We found a relative increase of miR-21-5p and miR-221-3p 5 days after ictus in CSF from patients who developed DCI and of miR-221-3p in those with angiographic vasospasm. Experimentally, miR-21 has been studied extensively in many organs.6,21 In the context of the present study, miR-21 was increased after neointimal lesion of the carotid artery in rats. In the same study, inhibition of miR-21 reduced neointima formation, decreased proliferation, and increased apoptosis of cells related to the injured vessel walls,22 indicating that miR-21 promoted reparatory mechanisms in this model. In contrast, another experimental study after middle cerebral artery occlusion showed reduced infarction after intrathecal inhibition of miR-24 but not miR-21.17 Recently, Stylli et al23 studied miRNA profiles in CSF from 19 SAH patients and 4 healthy patients. As in the present study, the most pronounced miRNA differences were found between SAH and control patients, but neither miR-21-5p nor miR-221-3p differed significantly between the reported groups.23 However, methodological differences, such as a variation of time from ictus to CSF sampling, the choice of instrument for miRNA detection, and a primary focus on angiographic vasospasm, make direct comparisons difficult between the study by Stylli et al23 and our findings. Recently, 2 other groups have studied circulating miRNAs after SAH. One study reported that a combination of plasma levels of miR-1290, -4463, -4532, and -4793 on day 7 could differentiate between SAH patients with and without delayed cerebral infarction.24 The other study found the serum levels of miR-502-5p and -1297 to be upregulated 72 hours after SAH in patients with a poor compared with a good outcome, as well as in SAH patients compared with healthy volunteers.25 However, none of the miRNAs reported in these studies differed significantly between groups in the present study. To our knowledge, hitherto no study has compared intrathecal and circulating miRNA profiles in SAH, but it would be interesting to see if an increase in circulating brain-specific miRNA could be associated with brain injury, as previously proposed for S100B and neuron specific enolase.26,27 In another clinical study of acute ischemic stroke, increased miR-221 was also reported in CSF28; the same 2 miRNAs, miR-21-5p and miR-221-3p, have been proposed as circulating biomarkers in ischemic stroke patients.29
We also found a >6-fold relative increase of miR-9-3p in patients with EBI or a poor outcome after 3 months, or both. Although the significance of this miRNA in the pathophysiology of SAH needs to be further established, the finding does suggest that future dynamic studies of CSF miRNAs in the early phase may help to reveal the extent of brain injury and, thereby, aid prognostication.
A strength of this study is the use of 2 different miRNA profiling platforms. Despite rapid development of new miRNA detection systems, RT-qPCR remains the gold standard for reliable detection of extracellular miRNA. As considerable analytic variability has been demonstrated between RT-qPCR platforms,30 we elected to conduct the screening part with the widely used and probably most sensitive30 stem-loop primer technique. This was followed up by validation using the more specific30 locked nuclear acid technique. The validation method includes melting curve analyses and extensive use of spike-ins integrating quality control after RNA extraction, reverse transcription, and RT-qPCR. The results indicate that the previously reported differences between RT-qPCR techniques30 also apply to studies of CSF and stresses the need for caution when interpreting results from either technique in isolation. Even so, the fact that we are able to measure this large number of miRNAs reliably in CSF with a low analytic variation may hold some promise to explore miRNAs in the pathophysiology of other types of acute brain injury.
For each CSF sample, the net absorbance of oxyhemoglobin correlated well to the mean Cq values for each sample. Therefore, comparisons between subgroups of SAH patients, where the mean normalization approach was used, provide Cq values relative to the mean Cq value for each miRNA in each sample. This strategy partly compensates for the contamination of CSF with blood. Consequently, a miRNA that is found to be relatively downregulated between groups may well exhibit an unchanged or increased absolute concentration.
The present findings apply only to SAH patients requiring external ventricular drainage. However, one could speculate that SAH patients without this need are more likely to have a miRNA profile resembling those SAH patients in our study with less severe blood contamination and hemolysis in the CSF. Furthermore, the association of mir-21-5p and -221-3p to DCI was not significant when corrected for multiple comparisons. Nevertheless, in our opinion, the reproducibility when using another RT-qPCR technique justifies reporting these associations, although their use should be restricted to hypothesis generation. Also, the strict criteria for DCI, with the exclusion of any patient who was difficult to categorize, increases the likelihood that a true association reflect processes related to DCI and not for instance poor neurological status on admission or EBI. Further, miRNA profiles in CSF were screened at a fixed time, 5 days after ictus. In contrast, DCI occurred variably during Day 4 to 14, and it could be argued that any changes in miRNA associated with DCI are more likely to occur at a specific point up until or after the occurrence of DCI and not in relation to ictus. Additionally, although we excluded patients in whom time of ictus could not be estimated with an error margin of <24 hours, we cannot exclude that a preceding minor hemorrhage or errors at estimating the time of ictus might have skewed the average miRNA profile because CSF was not truly sampled at 5 days after ictus. Finally, the neurologically healthy patients differed slightly from SAH patients regarding age and sex. Nonetheless, age and sex did not drive any of the reported large differences between SAH and neurologically healthy patients (data not shown). Also, we cannot exclude that the neurologically healthy patients had a nondiagnosed neurological disease. To interpret these results, one also needs to consider that the diagnosis of DCI is to a large extent based on clinical signs and exclusion of other diagnoses. Therefore, DCI may well constitute the final common pathway of many activated or deactivated physiological or pathophysiological pathways. Moreover, the present study is one of association only and does not imply causality.
We have previously reported that certain miRNAs may be detected in cerebral microdialysate,12 although quantification was performed solely with the highly sensitive protocol reserved for screening in the present study. Regarding our present finding of marked changes in the intrathecal miRNA profile after SAH, its pathophysiological implications are to date unknown. With our current understanding of the role of miRNAs, extracellular miRNA levels are important only inasmuch as they reflect the intracellular levels. In vitro studies indicate that an energy-dependent equilibrium exists between intracellular and extracellular miRNA concentrations,31 that is, the extracellular levels may reflect intracellular levels. However, potential additional mechanisms, such as cell lysis, and the kinetics of these mechanisms await further studies.
Summary
In patients with SAH, the miRNA profile in CSF differs markedly from that in normal CSF. Intrathecal changes in miRNA could be associated to the development of DCI.
Sources of Funding
Dr Bache was salaried by a grants from the Research Board at Copenhagen University Hospital, Rigshospitalet, and The Lundbeck Foundation. Reagents for miRNA profiling were funded from Grosserer Jakob Ehrenreich & Hustru Grete Ehrenreichs Fond, Brødrene Hartmanns Fond, Torben & Alice Frimodts Fond, Grosserer L. F. Foghts Fond and Aase & Ejnar Danielsens Fond.
Disclosures
None.
Supplementary Material
Footnotes
The online-only Data Supplement is available with this article at http://stroke.ahajournals.org/lookup/suppl/doi:10.1161/STROKEAHA.117.017804/-/DC1.
References
- 1.van Gijn J, Kerr RS, Rinkel GJ. Subarachnoid haemorrhage. Lancet. 2007;369:306–318. doi: 10.1016/S0140-6736(07)60153-6. doi: 10.1016/S0140-6736(07)60153-6. [DOI] [PubMed] [Google Scholar]
- 2.Vergouwen MD, Vermeulen M, van Gijn J, Rinkel GJ, Wijdicks EF, Muizelaar JP, et al. Definition of delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage as an outcome event in clinical trials and observational studies: proposal of a multidisciplinary research group. Stroke. 2010;41:2391–2395. doi: 10.1161/STROKEAHA.110.589275. doi: 10.1161/STROKEAHA.110.589275. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Li YM, Li WQ, Huang CG, Lu YC, Hou LJ. Effect of clazosentan in patients with aneurysmal subarachnoid hemorrhage: a meta-analysis of randomized controlled trials. PLoS One. 2012;7:e47778. doi: 10.1371/journal.pone.0047778. doi: 10.1371/journal.pone.0047778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorhout Mees SM, Rinkel GJ, Feigin VL, Algra A, van den Bergh WM, Vermeulen M, et al. Calcium antagonists for aneurysmal subarachnoid haemorrhage. Cochrane Database Syst Rev. 2007:CD000277. doi: 10.1002/14651858.CD000277.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowland MJ, Hadjipavlou G, Kelly M, Westbrook J, Pattinson KT. Delayed cerebral ischaemia after subarachnoid haemorrhage: looking beyond vasospasm. Br J Anaesth. 2012;109:315–329. doi: 10.1093/bja/aes264. doi: 10.1093/bja/aes264. [DOI] [PubMed] [Google Scholar]
- 6.Saugstad JA. MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J Cereb Blood Flow Metab. 2010;30:1564–1576. doi: 10.1038/jcbfm.2010.101. doi: 10.1038/jcbfm.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Z, Zhang C, Wu R, Hu W. Tumor suppressor p53 meets microRNAs. J Mol Cell Biol. 2011;3:44–50. doi: 10.1093/jmcb/mjq040. doi: 10.1093/jmcb/mjq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi G, Liu Y, Liu T, Yan W, Liu X, Wang Y, et al. Upregulated miR-29b promotes neuronal cell death by inhibiting Bcl2L2 after ischemic brain injury. Exp Brain Res. 2012;216:225–230. doi: 10.1007/s00221-011-2925-3. doi: 10.1007/s00221-011-2925-3. [DOI] [PubMed] [Google Scholar]
- 10.Redell JB, Moore AN, Ward NH, 3rd, Hergenroeder GW, Dash PK. Human traumatic brain injury alters plasma microRNA levels. J Neurotrauma. 2010;27:2147–2156. doi: 10.1089/neu.2010.1481. doi: 10.1089/neu.2010.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruickshank A, Auld P, Beetham R, Burrows G, Egner W, Holbrook I, et al. UK NEQAS Specialist Advisory Group for External Quality Assurance of CSF Proteins and Biochemistry. Revised national guidelines for analysis of cerebrospinal fluid for bilirubin in suspected subarachnoid haemorrhage. Ann Clin Biochem. 2008;45(pt 3):238–244. doi: 10.1258/acb.2008.007257. doi: 10.1258/acb.2008.007257. [DOI] [PubMed] [Google Scholar]
- 12.Bache S, Rasmussen R, Rossing M, Hammer NR, Juhler M, Friis-Hansen L, et al. Detection and quantification of microRNA in cerebral microdialysate. J Transl Med. 2015;13:149. doi: 10.1186/s12967-015-0505-1. doi: 10.1186/s12967-015-0505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu W, Lee ML. SPCalc: A web-based calculator for sample size and power calculations in micro-array studies. Bioinformation. 2006;1:251–252. doi: 10.6026/97320630001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 16.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59:S1–S6. doi: 10.1016/j.ymeth.2012.09.015. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Chen X, Zhang Y. Effects of microRNA-21 and microRNA-24 inhibitors on neuronal apoptosis in ischemic stroke. Am J Transl Res. 2016;8:3179–3187. [PMC free article] [PubMed] [Google Scholar]
- 18.Tao Z, Zhao H, Wang R, Liu P, Yan F, Zhang C, et al. Neuroprotective effect of microRNA-99a against focal cerebral ischemia-reperfusion injury in mice. J Neurol Sci. 2015;355:113–119. doi: 10.1016/j.jns.2015.05.036. doi: 10.1016/j.jns.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 19.Ni J, Wang X, Chen S, Liu H, Wang Y, Xu X, et al. MicroRNA let-7c-5p protects against cerebral ischemia injury via mechanisms involving the inhibition of microglia activation. Brain Behav Immun. 2015;49:75–85. doi: 10.1016/j.bbi.2015.04.014. doi: 10.1016/j.bbi.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Vergouwen MD, Vermeulen M, Coert BA, Stroes ES, Roos YB. Microthrombosis after aneurysmal subarachnoid hemorrhage: an additional explanation for delayed cerebral ischemia. J Cereb Blood Flow Metab. 2008;28:1761–1770. doi: 10.1038/jcbfm.2008.74. doi: 10.1038/jcbfm.2008.74. [DOI] [PubMed] [Google Scholar]
- 21.Xu X, Kriegel AJ, Jiao X, Liu H, Bai X, Olson J, et al. miR-21 in ischemia/reperfusion injury: a double-edged sword? Physiol Genomics. 2014;46:789–797. doi: 10.1152/physiolgenomics.00020.2014. doi: 10.1152/physiolgenomics.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 23.Stylli SS, Adamides AA, Koldej RM, Luwor RB, Ritchie DS, Ziogas J, et al. miRNA expression profiling of cerebrospinal fluid in patients with aneurysmal subarachnoid hemorrhage. J Neurosurg. 2016;126:1131–1139. doi: 10.3171/2016.1.JNS151454. doi: 10.3171/2016.1.JNS151454. [DOI] [PubMed] [Google Scholar]
- 24.Lu G, Wong MS, Xiong MZQ, Leung CK, Su XW, Zhou JY, et al. Circulating microRNAs in delayed cerebral infarction after aneurysmal subarachnoid hemorrhage. J Am Heart Assoc. 2017;6:e005363. doi: 10.1161/JAHA.116.005363. doi: 10.1161/JAHA.116.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai NS, Zhang JQ, Qin FY, Sheng B, Fang XG, Li ZB. Serum microRNAs are non-invasive biomarkers for the presence and progression of subarachnoid haemorrhage. Biosci Rep. 2017;37:BSR20160480. doi: 10.1042/BSR20160480. doi: 10.1042/BSR20160480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kacira T, Kemerdere R, Atukeren P, Hanimoglu H, Sanus GZ, Kucur M, et al. Detection of caspase-3, neuron specific enolase, and high-sensitivity C-reactive protein levels in both cerebrospinal fluid and serum of patients after aneurysmal subarachnoid hemorrhage. Neurosurgery. 2007;60:674–679; discussion 679. doi: 10.1227/01.NEU.0000255394.77538.BB. doi: 10.1227/01.NEU.0000255394.77538.BB. [DOI] [PubMed] [Google Scholar]
- 27.Hajduková L, Sobek O, Prchalová D, Bílková Z, Koudelková M, Lukášková J, et al. Biomarkers of brain damage: S100B and NSE concentrations in cerebrospinal fluid–a normative study. Biomed Res Int. 2015;2015:379071. doi: 10.1155/2015/379071. doi: 10.1155/2015/379071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sørensen SS, Nygaard AB, Nielsen MY, Jensen K, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Transl Stroke Res. 2014;5:711–718. doi: 10.1007/s12975-014-0364-8. doi: 10.1007/s12975-014-0364-8. [DOI] [PubMed] [Google Scholar]
- 29.Tsai PC, Liao YC, Wang YS, Lin HF, Lin RT, Juo SH. Serum microRNA-21 and microRNA-221 as potential biomarkers for cerebrovascular disease. J Vasc Res. 2013;50:346–354. doi: 10.1159/000351767. doi: 10.1159/000351767. [DOI] [PubMed] [Google Scholar]
- 30.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 31.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]


