Abstract
Background
An electronic medical record (EMR) database of a large unselected population who received screening colonoscopies may minimize sampling error and represent real-world estimates of risk for screening target lesions of advanced colorectal neoplasia (CRN). Our aim was to develop and validate a prediction model for assessing the probability of advanced CRN using a clinical data warehouse.
Methods
A total of 49,450 screenees underwent their first colonoscopy as part of a health check-up from 2002 to 2012 at Samsung Medical Center, and the dataset was constructed by means of natural language processing from the computerized EMR system. The screenees were randomized into training and validation sets. The prediction model was developed using logistic regression. The model performance was validated and compared with existing models using area under receiver operating curve (AUC) analysis.
Results
In the training set, age, gender, smoking duration, drinking frequency, and aspirin use were identified as independent predictors for advanced CRN (adjusted P < .01). The developed model had good discrimination (AUC = 0.726) and was internally validated (AUC = 0.713). The high-risk group had a 3.7-fold increased risk of advanced CRN compared to the low-risk group (1.1% vs. 4.0%, P < .001). The discrimination performance of the present model for high-risk patients with advanced CRN was better than that of the Asia-Pacific Colorectal Screening score (AUC = 0.678, P < .001) and Schroy’s CAN index (AUC = 0.672, P < .001).
Conclusion
The present 5-item risk model can be calculated readily using a simple questionnaire and can identify the low- and high-risk groups of advanced CRN at the first screening colonoscopy. This model may increase colorectal cancer risk awareness and assist healthcare providers in encouraging the high-risk group to undergo a colonoscopy.
Introduction
Colorectal cancer (CRC) is the third most common cancer in the world [1]. A colonoscopy is considered the preferred CRC screening modality [2]; however, adherence is generally not sufficient [3]. One of the barriers to CRC screening is a lack of perceived risk among the patients and primary care providers [4]. Risk stratification provides a rational strategy for facilitating appropriate CRC screening and can improve the distribution of resources. A prerequisite for this risk stratification approach is the accessibility of a precise risk assessment tool.
Although several risk prediction models for screening target lesions, CRC, or advanced colorectal neoplasia (CRN) have been developed [4–14], previous models have some limitations, such as the lack of inclusion of possible risk factors [4, 5, 7, 9–11, 13]. In the current healthcare system, electronic medical records (EMRs) encompass a plethora of data related with patients, such as demographics, vital signs, medical history, medication, laboratory test results, results from laboratory and imaging studies. The use of medical data mining and correlational studies using EMRs could serve as a valuable resource to aid the determination of unrevealed risk factors under deductive assumptions to establish a real-world prediction model for advanced CRN.
However, EMR data contains unstructured data, such as endoscopic and pathology reports, which requires laborious efforts for transforming text to numerical data [15]. Recent advanced natural language processing algorithms, such as the Concept Extraction-based Text Analysis System (CETAS), are able to transform information from endoscopic and pathology reports to a numerical dataset. In this study, we constructed a database using the EMR data from 49,450 patients who underwent their first screening colonoscopy as part of routine health check-up examinations by means of the CETAS and developed a risk prediction model to identify individuals at high risk of advanced CRN.
Methods
Study population
The Center for Health Promotion of Samsung Medical Center, Seoul, Republic of Korea, provides regular health screening examinations that include a colonoscopy [16]. We included consecutive subjects who underwent a screening colonoscopy during health screening examinations at the Center for Health Promotion between January 2003 and December 2012. Regular routine health screening is very common in Korea due to the Industrial Safety and Health Law [16, 17]. The health screening examinations were performed as described previously [16]. All participants completed a questionnaire and received a detailed physical examination as part of the screening program. Self-administered questionnaire data were used to identify current smoking status, alcohol drinking frequency, physical activity, family history of colon cancer, history of colorectal polyps/cancer, comorbidities, and regular use of aspirin. Participants were asked to fast for at least 12 hours and to avoid smoking on the morning of the examination. Blood samples were collected on the day of the colonoscopy. Serum biochemical tests were carried out using an automatic analyzer at the Department of Laboratory Medicine at Samsung Medical Center.
Screening colonoscopies
All colonoscopies were performed by board-certified endoscopists. During colonoscopy, the location, size, number, and appearance of CRN were recorded. The location was assessed by the endoscopists, and the size was estimated using open biopsy forceps. The gross appearance of each lesion was classified using Paris endoscopic classification [18]. All of the colorectal lesions were histologically evaluated and classified according to the World Health Organization classification [19]. However, because the colonoscopy and pathology reports were described by the performing endoscopists and pathologists, the natural language for describing the lesions was different in each report despite using standardized terms. For example, even though the information was the same, the endoscopists used different units, such as cm or mm, and various modifiers, such as elevated, raised, upraised, protruded, and bulged. Therefore, the reports were considered to contain unstructured data, and it was difficult to extract unified forms of variables in real practice.
Data collection
This study used only de-identified medical records that were collected for administrative or clinical purposes as part of routine health screening examinations in the Center for Health Promotion of Samsung Medical Center. The Center for Health Promotion provides researchers de-identified information for biomedical research, which was approved by the Institutional Review Board of Samsung Medical Center for studies that investigate decision-making and the relationships and potential patterns between disease progression and management. The EMRs included both structured and unstructured data. Structured data refer to information that was organized in a row-column database including demographics, physical measurement, smoking, alcohol drinking, physical activity, co-morbidities, aspirin use, and laboratory biochemical measurements. Unstructured data refers to information that does not reside in a traditional row-column database including the free text from colonoscopy and pathology reports. This study was approved by the Institutional Review Board of Samsung Medical Center, which waived the requirement for informed consent because the researchers only obtained de-identified routinely collected data from the institution's clinical data warehouse.
Unstructured text data analysis: Concept Extraction-based Text Analysis System (CETAS)
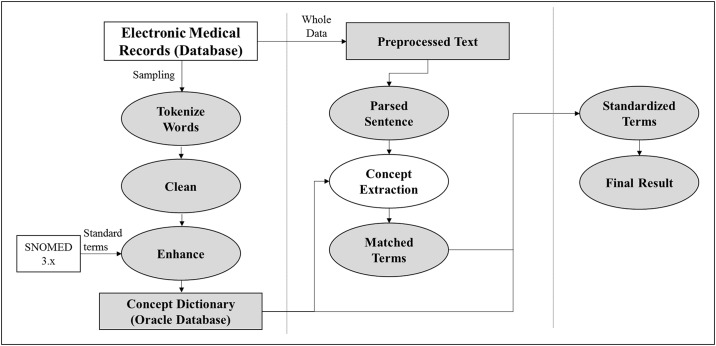
Among the data collected in this study, we obtained the data about the number and size related to CRN from the free text of the colonoscopy reports, and the histology and dysplasia grade related to CRN from the free text of the pathology reports. Unstructured data were transformed from text to numerical data by the CETAS. The CETAS is based on SAS Enterprise Contents Categorization 12.2 (SAS Institution; Cary, NC, USA), and it does not have add-on modules, such as text mining. SAS ECC 12.2 is an NLP solution that is separate from SAS Base; since it has a built-in LITI (Language Interface Taxonomy Interface) for performing Concept Extraction in a simple and effective manner in the text, it offers a solution that enables rule-based construction of the matching of terms and extraction (Fig 1) [15, 20].
Fig 1. Process diagram of a Concept Extraction-based Text Analysis System.
1. Concept dictionary
In order to create a Concept Dictionary that underlies the configuration of the Concept Extraction Rule, by extracting about 500 colonoscopy tests and pathologic result reports in a random sampling manner, the terms that represent the information for the colorectal polyps which is the number, location, size, histology, dysplasia grade through a natural language processing methodology are organized and cleansed. In this process, in order to determine, the standard terminology of non-standard terms included in the EMR database, a Concept Dictionary was configured by referencing the SNOMED 3.x.
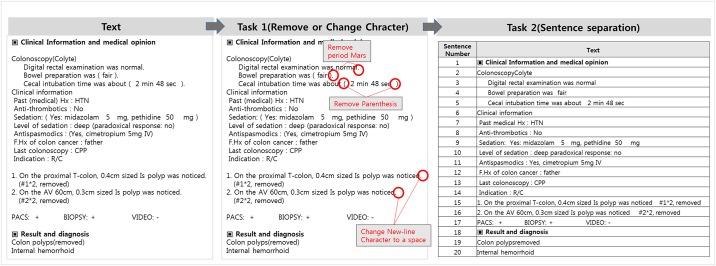
2. Preprocess
A pretreatment process for changing the colonoscopy reports created in different sentence structures by each endoscopists into coherent sentence structures is constructed (Fig 2). The Preprocessing is comprised of two operations. Task1 is composed of functions that delete or change the special symbols that became non-standardized special symbols, such as Bullet Mark, comma, line breaks, spacing, etc.; through which, a base that can perform Concept Extraction based on the special symbols in the natural language processing steps was created. Task2 performs the task of dividing the text into sentences or syntactical units in accordance with the period, line breaks, conjunctions, prepositions, etc. With this, in the case where the text about organs and lesions is expressed through a number of sentences and paragraphs, the error of generating results in conjunction with the information of sentence 1 and sentence 2 that are not related in terms of processing the natural language can be reduced.
Fig 2. Text pre-processing.
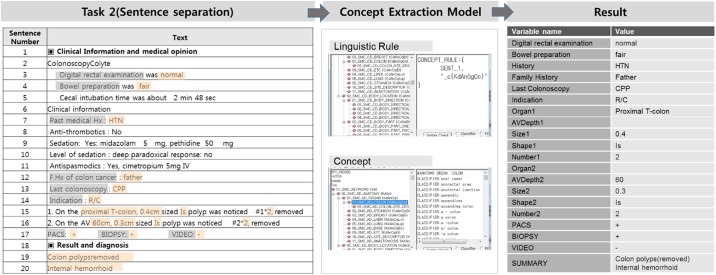
3. Concept extraction
As a natural language processing step, categories are configured in accordance with the hierarchical structure of the colorectal structures and lesions, and the Concept Extraction Rule was developed for each category. The effectiveness for Rule-based natural language processing has already been frequently proven in the previous studies.[21, 22] Concept Extraction is performed through the Rule that extracts the terms stored in the Concept Dictionary that is within the particular words expressed in sentences or within the Keyword Count specified by special symbols. As such, the researchers did not have to change the Rule when adding/removing terms; they simply had to change the Concept Dictionary and that automatically changed the results of the Concept Extraction (Fig 3).
Fig 3. Concept extraction process.
4. Validation
Through a random sampling of 500 colonoscopy and pathology result reports from the clinical data warehouse of Samsung Medical Center, the first ever Concept Extraction Rule was created, and by expanding the sample size to 2,000, the precision of the Rule was increased. After the Concept Extraction Rule was created, the accuracy was verified by comparing the Concept Extraction results and the results manually generated. The accuracy can be verified by the following two indicators.
Precision means the ability to find the correct data when it is in the colonoscopy and pathology result reports, and Recall means the ability to find only the correct data when it is in the colonoscopy and pathology result reports to the meaning. In order to verify the accuracy of the Concept Extraction Rule built in the CETAS, finally, Precision and Recall were calculated through a random sampling of 50 specimens from the 3 months, and the Precision and Recall were each 99.27% and 99.83%, respectively (Table 1). After validation, unstructured data from remaining 48950 colonoscopy and pathology was transformed text to numerical data.
Table 1. Comparison of concept extraction results and manual data extraction.
| Category | Precision (%) | Recall (%) | Category | Precision (%) | Recall (%) |
|---|---|---|---|---|---|
| PAST MEDICAL HISTORY | 100.00 | 100.00 | LESION | 100.00 | 100.00 |
| ANTITHROMBOTICS | 100.00 | 100.00 | ABNORMALITY | 100.00 | 100.00 |
| FAMILY HISTORY OF CANCER | 100.00 | 100.00 | HISTOLOGICAL CLASSIFICATIONADJ | 100.00 | 100.00 |
| LAST COLONOSCOPY | 100.00 | 100.00 | HISTOLOGIC TYPE | 100.00 | 100.00 |
| INDICATION | 100.00 | 97.75 | TUMOR GRADING | 100.00 | 100.00 |
| SEDATION | 100.00 | 100.00 | SIZE | 92.05 | 98.78 |
| MIDAZOLAM | 100.00 | 100.00 | NUMBER | 100.00 | 100.00 |
| PETHIDINE | 100.00 | 100.00 | SHAPE | 100.00 | 100.00 |
| LEVEL OF SEDATION | 100.00 | 98.88 | COLOR | 100.00 | 100.00 |
| PARADOXICAL RESPONSE | 100.00 | 100.00 | VIDEO | 95.51 | 100.00 |
| ANTISPASMODICS | 100.00 | 100.00 | SLIDE | 95.51 | 100.00 |
| CIMETROPIUM | 100.00 | 100.00 | ORGAN | 100.00 | 100.00 |
| DIGITAL RECTAL EXAMINATION | 96.63 | 100.00 | BIOPSY | 100.00 | 100.00 |
| BOWEL PREPARATION | 100.00 | 100.00 | BIOPSY STATUS | 98.88 | 100.00 |
| CECAL INBUTIONTIME | 100.00 | 97.75 | BIOPSY METHOD | 93.26 | 100.00 |
| WITHDRAWAL TIME | 100.00 | 97.75 | SUBMUCOSAL INJECTION | 100.00 | 100.00 |
| INSERTED UPTO | 100.00 | 98.88 | HEMOSTASIS | 100.00 | 100.00 |
| ORGAN | 100.00 | 100.00 | DIAGNOSIS | 100.00 | 100.00 |
| SITE | 98.88 | 100.00 | IMPRESSION | 100.00 | 100.00 |
| Total | 99.27 | 99.83 |
Study design
We performed a cross-sectional analysis of patients ≥ 20 years of age who underwent their first screening colonoscopy. The exclusion criteria were as follows: 1) incomplete colonoscopy, 2) poor (semisolid stool that could not be suctioned or washed away and less than 90% of surface seen) and inadequate (repeat preparation and colonoscopy needed) bowel preparation, 3) incomplete colonoscopy report about the number and size related to CRN, 4) incomplete pathology report about the histology and dysplasia grade related to CRN, 5) history of previous colonoscopy, 6) history of colorectal polyps, cancer, or surgery, and 7) inflammatory bowel disease.
Definition of outcome measurement
An advanced CRN was defined as a cancer or adenoma that was at least 10 mm in diameter and had high-grade dysplasia, villous or tubulovillous histological characteristics, or any combination thereof [23]. For patients with multiple neoplasms, the size and appearance of the neoplasms with advanced pathology or of the largest polyp were reported. The main outcome measurement in this study is an advanced CRN detected by means of a colonoscopy and evaluated pathologically.
Prediction model
Structured data and unstructured data transformed from text to numerical data using the CETAS were used as the input variables of the prediction model. The enrolled subjects were randomly partitioned into a training set and a validation set using a 50–50 allocation. Candidate predictors with P < .10 in univariate analyses were included in the multivariable logistic regression. Backward selection was used to remove variables with not significant (P < .05) contributions to the multivariable model fit. Two prediction models were fitted. The first one used both inquiry and lab variables, and the second only used inquiry variables.
Model performance and calibration
A two-sided alpha of 5% was used as insertion and deletion criteria of the two-stage variable selection in fitting a prediction model (i.e., training). The prediction score from the fitted prediction model was applied to the validation set, and the performance of the prediction model was evaluated using area under receiver operating curve (AUC) analysis. Models with a AUC near 1 suggest excellent predictive ability, and an AUC near 0.5 indicates hardly any predictive ability. The calibration is a measure of how accurately the predicted probabilities of advanced CRN inferred from the training set match the subsequently observed event rate in the validation set. The negative predictive value (NPV) is the probability that a patient who is termed “no disease” by the risk score really has no disease. We want this probability to be very high (at least 99%) so as not to miss any significant disease. A cutoff value for the trained risk score was identified and shown to have over 99% negative predictive value when applied to the test set and the combined data set.
Results
Study population
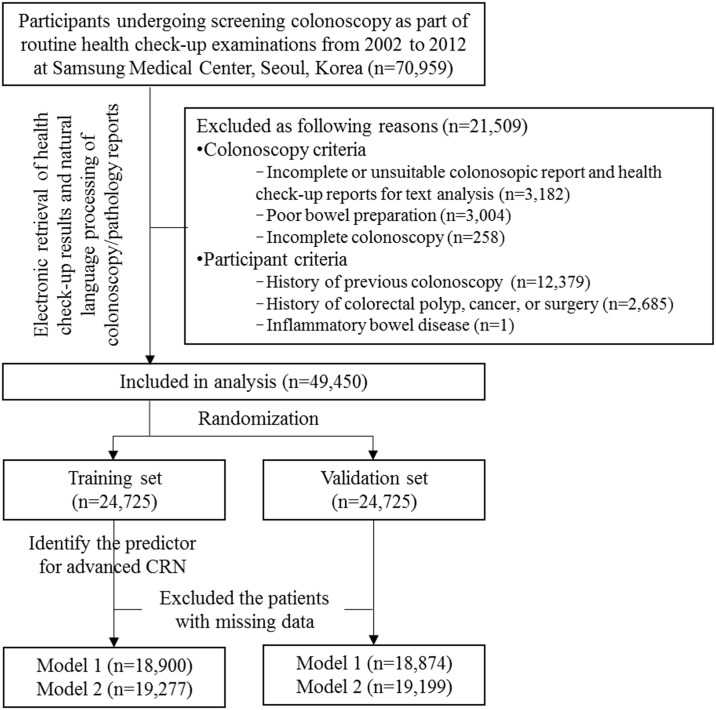
A total of 70,959 consecutive subjects underwent screening colonoscopy during health screening examinations at the Center for Health Promotion. We excluded 21,509 subjects who had incomplete or unsuitable reports for text analysis; poor bowel preparation; incomplete colonoscopy; or history of previous colonoscopy, colorectal polyps, cancer, or surgery, or inflammatory bowel disease. For subjects who underwent multiple colonoscopies, we selected the first colonoscopy for the present analysis. Finally, this study used only de-identified data from 49,450 participants who underwent their first screening colonoscopy and a health check-up. A flow diagram of the study population is shown in Fig 4. Of the eligible 49,450 patients who underwent their first screening colonoscopy, 27,688 were male (55.99%) and 21,762 were female (44.01%), all were Korean, and the mean age was 49.86 ± 9.33 years. One or more colorectal adenomas were found in 14,716 (29.8%) patients, 1,025 (2.1%) of whom had advanced adenoma, and 92 of whom had invasive cancer (0.2%). The overall prevalence of advanced CRN was 2.3%. The clinical characteristics of the enrolled participants are listed in Table 2. Enrolled participants were randomly divided into training and validation sets using a 50–50 allocation.
Fig 4. Flow diagram of the study population.
Table 2. Clinical characteristics of enrolled subjects.
| Variable | Total | Training set | Validation set | p | |||
|---|---|---|---|---|---|---|---|
| N | Value | N | Value | N | Value | ||
| Demographics | |||||||
| Age (years), mean ± SD | 49,450 | 49.9 ± 9.3 | 24,726 | 49.9 ± 9.4 | 24,724 | 49.8 ± 9.3 | 0.160 |
| Sex | 49,450 | 24,726 | 24,724 | 0.008 | |||
| Female, n (%) | 21,762 (44.0) | 10,735 (43.4) | 11,027 (44.6) | ||||
| Male, n (%) | 27,688 (56.0) | 13,991 (56.6) | 13,697 (55.4) | ||||
| Family history of colorectal cancer | 45,583 | 22,759 | 21,706 | 0.694 | |||
| Yes, n (%) | 2,251 (4.9) | 1,133 (5.0) | 1,118 (4.9) | ||||
| No, n (%) | 43,332(95.1) | 21,626 (95.0) | 21,706 (95.1) | ||||
| Physical measurement | |||||||
| Body mass index, mean ± SD | 44,581 | 23.7 ± 3.1 | 22,275 | 23.7 ± 3.1 | 22,306 | 23.6 ± 3.1 | 0.410 |
| Waist circumference (cm), mean ± SD | 44,145 | 83.4 ± 44.3 | 22,057 | 83.7 ± 62.0 | 22,088 | 83.2 ± 9.1 | 0.229 |
| Body fat percentage* (%), mean ± SD | 49,058 | 25.5 ± 6.6 | 24,517 | 25.4 ± 6.5 | 24,541 | 25.5 ± 6.6 | 0.813 |
| Cigarette smoking | |||||||
| Smoking status | 42,579 | 21,271 | 21,308 | 0.319 | |||
| Non-smoker, n (%) | 23,841 (56.0) | 11,838 (55.7) | 12,003 (56.4) | ||||
| Ex-smoker, n (%) | 5,260 (12.3) | 2,631 (12.3) | 2,629 (12.3) | ||||
| Current smoker, n (%) | 13,478 (31.7) | 6,802 (32.0) | 6,676 (31.3) | ||||
| Smoking duration (year), mean ± SD | 43,108 | 9.9 ± 12.8 | 21,526 | 10.0 ± 12.9 | 21,582 | 9.8 ± 12.7 | 0.143 |
| Smoking amount (pack/day), mean ± SD | 43,107 | 0.9 ± 1.1 | 21,534 | 0.9 ± 1.1 | 21,573 | 0.9 ± 1.1 | 0.238 |
| Alcohol drinking | |||||||
| Regular alcohol drinking | 43,777 | 21,868 | 21,909 | 0.719 | |||
| Yes, n (%) | 19,814 (45.3) | 9,879 (45.2) | 9,935 (45.4) | ||||
| No, n (%) | 23,963 (54.7) | 1,1989 (54.8) | 11,974 (54.6) | ||||
| Drinking duration (year), mean ± SD | 26,395 | 23.8 ± 10.1 | 13,250 | 23.8 ± 10.2 | 13,145 | 23.8 ± 10.0 | 0.551 |
| Drinking frequency (/week), mean ± SD | 40,171 | 20,096 | 20,075 | ||||
| No drinking, n (%) | 19,814 (49.3) | 9,879 (49.2) | 9,935 (49.5) | 0.843 | |||
| Once a week, n (%) | 3,268 (8.1) | 1,671 (8.3) | 1,597 (8.0) | ||||
| 2–3 times per month, n (%) | 5,792 (14.4) | 2,897 (14.4) | 2,895 (14.4) | ||||
| 1–2 times per week, n (%) | 6,730 (16.8) | 3,344 (16.6) | 3,386 (16.9) | ||||
| 3–4 times per week, n (%) | 3,436 (8.6) | 1,737 (8.6) | 1,699 (8.5) | ||||
| 5–6 times per week, n (%) | 779 (1.9) | 411 (2.0) | 368 (1.8) | ||||
| Everyday, n (%) | 352 (0.9) | 157 (0.8) | 195 (1.0) | ||||
| Drinking amount at one (bottle), mean ± SD | 40,027 | 1.2 ± 1.4 | 20,028 | 1.2 ± 1.4 | 19,999 | 1.2 ± 1.4 | 0.782 |
| Physical activity | |||||||
| Type of physical activities† | 29,150 | 14,558 | 14,592 | 0.132 | |||
| Strenuous activities, n (%) | 2,148 (7.4) | 1,097 (7.5) | 1,051 (7.2) | ||||
| Moderate activities, n (%) | 6,719 (23.1) | 3,299 (22.7) | 3,420 (23.4) | ||||
| Mild activities, n (%) | 17,895 (61.4) | 8,930 (61.3) | 8,965 (61.4) | ||||
| None, n (%) | 2,388 (8.2) | 1,232 (8.5) | 1,156 (7.9) | ||||
| Physical activity frequency (/week), mean ± SD | 28,510 | 2.8 ± 0.9 | 14,267 | 2.79 ± 0.88 | 14,243 | 2.80 ± 0.87 | 0.121 |
| Physical activity duration (minutes), mean ± SD | 28,663 | 36.8 ± 11.6 | 14,337 | 36.74 ± 11.62 | 14,326 | 36.82 ± 11.49 | 0.562 |
| Co-morbidity | |||||||
| Hypertension, n (%) | 6,545 (13.2) | 3,325 (13.5) | 3,220 (13.0) | 0.166 | |||
| Diabetes, n (%) | 1,917 (3.9) | 932 (3.8) | 985 (5.0) | 0.216 | |||
| Hyperlipidemia, n (%) | 1,941 (3.9) | 932 (3.8) | 1,009 (4.1) | 0.355 | |||
| Aspirin use | |||||||
| Regular use, n (%) | 2,612 (5.3) | 1,336 (5.4) | 1,276 (5.2) | 0.229 | |||
| No use, n (%) | 46,838 (94.7) | 23,390 (94.6) | 23,448 (94.8) | ||||
| Laboratory measurement | |||||||
| Hemoglobin, mean ± SD | 49,136 | 14.3 ± 31.5 | 24,556 | 14.3 ± 1.6 | 24,580 | 14.3 ± 1.5 | 0.601 |
| Hematocrit, mean ± SD | 49,136 | 42.4 ± 34.2 | 24,556 | 42.4 ± 4.2 | 24,580 | 42.4 ± 4.2 | 0.463 |
| Platelet, mean ± SD | 49,136 | 234.9 ± 52.3 | 24,556 | 234.7 ± 51.9 | 24,580 | 235.1 ± 52.8 | 0.421 |
| Prothrombine time (INR) | 46,820 | 1.0 ± 0.1 | 23,404 | 1.0 ± 0.1 | 23,416 | 1.0 ± 0.1 | 0.537 |
| Total_protein | 49,137 | 7.1 ± 0.4 | 24,559 | 7.1 ± 0.4 | 24,578 | 7.1 ± 0.4 | 0.051 |
| Albumin | 49,137 | 4.5 ± 0.3 | 24,559 | 4.5 ± 0.3 | 24,578 | 4.5 ± 0.3 | 0.419 |
| Total bilirubin, mean ± SD | 49,137 | 0.9 ± 0.4 | 24,559 | 0.9 ± 0.4 | 24,578 | 0.9 ± 0.4 | 0.507 |
| Aspartate transaminase | 49,142 | 26.1 ± 16.1 | 24,561 | 26.2 ± 16.6 | 24,581 | 26.0 ± 15.6 | 0.233 |
| Alanine transaminase | 49,142 | 26.4 ± 24.6 | 24,561 | 26.5 ± 24.6 | 24,581 | 26.3 ± 24.5 | 0.292 |
| Alkaline phosphatase | 49,137 | 63.3 ± 18.3 | 24,558 | 63.6 ± 18.7 | 24,579 | 62.9 ± 17.9 | 0.001 |
| γ-glutamyltransferase, mean ± SD | 48,603 | 33.9 ± 44.8 | 24,295 | 34.1 ± 47.0 | 24,308 | 33.7 ± 42.6 | 0.268 |
| Uric acid, mean ± SD | 49,129 | 5.2 ± 1.4 | 24,554 | 5.2 ± 1.4 | 24,575 | 5.2 ± 1.4 | 0.001 |
| Blood urea nitrogen | 49,135 | 13.3 ± 3.4 | 24,559 | 13.3 ± 3.4 | 24,576 | 13.3 ± 3.4 | 0.499 |
| Creatinine | 49,138 | 0.9 ± 0.2 | 24,560 | 0.9 ± 0.2 | 24,578 | 0.9 ± 0.2 | 0.182 |
| Fasting glucose, mean ± SD | 49,146 | 93.7 ± 18.0 | 24,563 | 93.7 ± 17.7 | 24,583 | 93.8 ± 18.3 | 0.625 |
| Hemoglobin a1c, mean ± SD | 47,575 | 5.6 ± 0.7 | 23,790 | 5.6 ± 0.7 | 23,785 | 5.6 ± 0.7 | 0.916 |
| Insulin | 34,601 | 7.4 ± 4.4 | 17,337 | 7.4 ± 4.5 | 17,264 | 7.3 ± 4.3 | 0.206 |
| C-peptide | 34,602 | 1.7 ± 0.8 | 17,337 | 1.7 ± 0.8 | 17,265 | 1.7 ± 0.8 | 0.174 |
| Total cholesterol, mean ± SD | 49,153 | 196.5 ± 34.7 | 24,569 | 196.7 ± 34.8 | 24,584 | 196.3 ± 34.5 | 0.194 |
| Triglyceride, mean ± SD | 48,757 | 119.0 ± 76.0 | 24,377 | 119.2 ± 75.6 | 24,380 | 118.7 ± 76.5 | 0.501 |
| HDL-cholesterol, mean ± SD | 48,755 | 55.9 ± 14.6 | 24,376 | 55.4 ± 14.7 | 24,379 | 55.6 ± 14.6 | 0.120 |
| LDL- cholesterol, mean ± SD | 48,759 | 123.9 ± 31.1 | 24,378 | 124.2 ± 31.2 | 24,381 | 123.7 ± 31.0 | 0.117 |
| C-reactive protein, mean ± SD | 43,613 | 0.1 ± 0.3 | 21,800 | 0.1 ± 0.3 | 21,813 | 0.1 ± 0.3 | 0.073 |
| Calcium, mean ± SD | 49,128 | 9.2 ± 0.4 | 24,554 | 9.2 ± 0.4 | 24,574 | 9.2 ± 0.4 | 0.755 |
| Ferritin, mean ± SD | 40,343 | 121.1 ± 120.2 | 20,211 | 122.4 ± 126.5 | 20,132 | 119.7 ± 113.6 | 0.089 |
| Colonoscopic and pathologic finding of enrolled patients | |||||||
| No adenoma | 34,734 | 70.2% | 17,377 | 70.3% | 17,357 | 70.2% | 0.855 |
| Serrated polyp | 5,868 | 11.9% | 2,916 | 11.8% | 2,952 | 11.9% | 0.614 |
| Any adenoma | 14,716 | 29.8% | 7,349 | 29.7% | 7,367 | 29.8% | 0.855 |
| Number of adenomas | |||||||
| 1 or 2 | 12,251 | 24.8% | 6,084 | 24.6% | 6,167 | 24.9% | 0.319 |
| ≥3 | 2,465 | 5.0% | 1,265 | 5.1% | 1,200 | 4.9% | |
| Size of the largest adenoma | |||||||
| ≤10 mm | 14,002 | 28.3% | 6,994 | 28.3% | 7,008 | 28.3% | 0.976 |
| >10 mm | 714 | 1.4% | 355 | 1.4% | 359 | 1.5% | |
| Histology of adenoma | |||||||
| Tubular adenoma | 14,586 | 29.5% | 7,279 | 29.4% | 7,307 | 29.6% | 0.659 |
| Tubulovillous or villous adenoma | 130 | 0.3% | 70 | 0.3% | 60 | 0.2% | |
| Dysplasia grade | |||||||
| Low-grade dysplasia | 14,511 | 29.3% | 7,251 | 29.3% | 7,260 | 29.4% | 0.928 |
| High-grade dysplasia | 125 | 0.3% | 59 | 0.2% | 66 | 0.1% | |
| Non-advanced adenoma | 13,691 | 27.7% | 6,836 | 27.7% | 6,855 | 27.7% | 0.981 |
| Advanced adenoma§ - | 989 | 2.0% | 495 | 2.0% | 494 | 2.0% | 0.975 |
| Invasive cancer | 92 | 0.2% | 45 | 0.2% | 47 | 0.2% | 0.834 |
| Advanced neoplasia¶ | 1,025 | 2.1% | 513 | 2.1% | 512 | 2.1% | 0.976 |
*Measured by bioelectrical impedance device
†Type of physical activities have done for the last 7 days including recreation, exercise, sports activities, activities at the work
- strenuous activities—ex) labor, aerobics, fast running bicycle, jogging, soccer- moderate activities—ex) a quick step, swimming, mountain climbing, four-up tennis- mild activities—ex) walking, golf, household-chores- none—I do not even walk for 10 m
§Advanced adenoma was defined as adenoma with villous histology, high-grade dysplasia, or size >10 mm
¶Advanced neoplasia was referred to advanced adenoma and invasive cancer
Identifying risk predictors and developing a candidate risk prediction model
To identify the patients with advanced CRN among the individuals who underwent their first colonoscopy, a stepwise logistic regression using all available variables listed in Table 1 was conducted for the imputed training set. We identified age, gender, diabetes, aspirin use, smoking duration, alcohol drinking frequency, drinking duration, uric acid, and γ-glutamyltransferase as the potential predictors (Table 3). Predictors for advanced CRN were refined using the complete data from the training set and excluded drinking duration and uric acid due to P-values > 0.3. Finally, age, gender, smoking duration, alcohol drinking frequency, aspirin use, and γ-glutamyltransferase were included in the prediction model (model 1). The prediction score from the refined prediction model 1 was determined by the following equation:
Table 3. Stepwise logistic regression for predicting patients with advanced colorectal neoplasia among individuals who underwent their first colonoscopy.
| 1. Predictor selection for advanced neoplasia using the imputed training set | |||
| Parameter | Estimate | Standard Error | P |
| Intercept | -8.282 | 0.394 | < .001 |
| Uric acid | 0.062 | 0.039 | .110 |
| γ-Glutamyltransferase | 0.001 | 0.001 | .035 |
| Smoking duration | 0.015 | 0.004 | < .001 |
| Drinking duration | 0.010 | 0.007 | .131 |
| Drinking frequency | 0.082 | 0.030 | .007 |
| Aspirin use | -0.299 | 0.096 | .002 |
| Diabetes | 0.225 | 0.110 | .041 |
| Gender | 0.122 | 0.069 | .075 |
| Age | 0.065 | 0.006 | < .001 |
| 2. Predictor refining for advanced neoplasia using the complete training set: variables with a P-value > 0.3 were excluded | |||
| Parameter | Estimate | Standard Error | P |
| Intercept | -8.720 | 0.515 | < .001 |
| Uric acid | 0.073 | 0.049 | .140 |
| γ-Glutamyltransferase | 0.001 | 0.001 | .026 |
| Smoking duration | 0.015 | 0.005 | .002 |
| Drinking duration | 0.005 | 0.009 | .538 |
| Drinking frequency | 0.089 | 0.035 | .011 |
| Aspirin use | -0.192 | 0.111 | .082 |
| Gender | 0.138 | 0.123 | .261 |
| Age | 0.071 | 0.009 | < .001 |
| Parameter | Estimate | Standard Error | P |
| Intercept | -8.710 | 0.432 | <001 |
| Uric acid | 0.050 | 0.044 | .253 |
| γ-Glutamyltransferase | 0.001 | 0.001 | .024 |
| Smoking duration | 0.015 | 0.004 | .001 |
| Drinking frequency | 0.095 | 0.031 | .002 |
| Aspirin use | -0.288 | 0.104 | .006 |
| Gender | 0.153 | 0.083 | .064 |
| Age | 0.074 | 0.006 | < .001 |
| 3. Prediction model for advanced neoplasia (Model 1) | |||
| Parameter | Estimate | Standard Error | P |
| Intercept | -8.428 | 0.354 | < .001 |
| γ-Glutamyltransferase | 0.002 | 0.001 | .016 |
| Smoking duration | 0.015 | 0.004 | .001 |
| Drinking frequency | 0.094 | 0.031 | .002 |
| Aspirin use | -0.286 | 0.104 | .006 |
| Gender | 0.190 | 0.076 | .012 |
| Age | 0.074 | 0.006 | < .001 |
| Parameter | Estimate | Standard Error | P |
| Intercept | -8.390 | 0.350 | <. 001 |
| Smoking duration | 0.015 | 0.004 | < .001 |
| Drinking frequency | 0.100 | 0.031 | .001 |
| Aspirin use | -0.289 | 0.104 | .006 |
| Gender | 0.205 | 0.075 | .007 |
| Age | 0.074 | 0.006 | < .001 |
Among the identified predictors, γ-glutamyltransferase was the only laboratory parameter that requires blood sampling and laboratory costs. When γ-glutamyltransferase was removed from the prediction model, all predictors could be obtained from a simple questionnaire, and a simple 5-item risk index could be readily determined from the questionnaire clinical data. The final prediction model was constructed with age, gender, smoking duration, alcohol drinking frequency, and aspirin use (model 2). The prediction score from the refined prediction models 1 and 2 was determined by the following equation:
Evaluating the performance of the prediction model
Discrimination refers to the ability to separate the variables with events from those without events. Using the prediction models 1 and 2, AUC values were calculated and used to evaluate the discrimination power of the prediction models. The AUC for prediction model 1 was 0.716 for the training set and 0.701 for the validation set (Fig 5A), whereas the AUC for prediction model 2 was 0.726 for the training set and 0.713 for the validation set (Fig 5B). Model 2 showed slightly higher discriminatory ability than model 1, although the risk factors were eliminated. The reason why model 2 was superior to model 1 was that the number of participants included in the calculation was larger in model 2 (training set: n = 18,874, validation set: n = 19,199) than model 1 (training set: n = 18,900, validation set: n = 19,277).
Fig 5. Model performance.
Area under the receiver operating curve (AUC) was calculated to evaluate the discrimination power between the training set (line) and validation set (dot) in prediction model 1 (A) and model 2 (B).
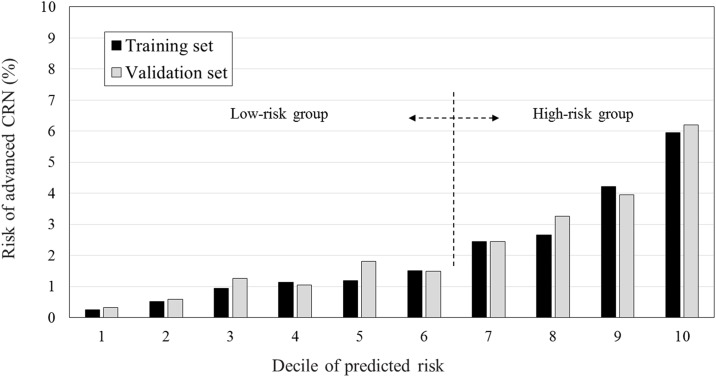
Prediction model 2 was selected as the final prediction model for advanced CRN. The calibration is a measure of how accurately the predicted probabilities of advanced CRN inferred from the training set match the subsequently observed event rate in the validation set. The individuals included in the training set were divided into deciles according to predicted risk for advanced CRN. Then, the predicted rate of the training set and observed rates of the validation set in each category were compared (Fig 6), indicating good calibration performance. To improve clinical utilization, cut-off values were set at the point of discrimination between the high- and low-risk group for advanced CRN in simulated calibration charts. Between the sixth and seventh deciles, the risk of advanced CRN increased from 1.51% to 2.45% in the training set and 1.50% to 2.45% in the validation set. The cut-off value of -4.195 was set at this point between the sixth and seventh deciles (Table 4).
Fig 6. Model calibration.
Cut-off values to discriminate between the high- and low-risk groups for advanced colorectal neoplasia were set at the point between the sixth and seventh deciles based on the risk of advanced colorectal neoplasia.
Table 4. Model calibration and estimation of cut-off value for discrimination between high- and low-risk for advanced colorectal neoplasia (CRN).
| Decile of predicted risk | Training set | Validation set | Risk group | |||
|---|---|---|---|---|---|---|
| N | Prevalence of advanced CRN (%) | N | Prevalence of advanced CRN (%) | |||
| 1 | 1922 | 0.260 | 1901 | 0.316 | 1.067 | Low-risk group |
| 2 | 1922 | 0.520 | 2064 | 0.581 | ||
| 3 | 1922 | 0.937 | 1983 | 1.261 | ||
| 4 | 1922 | 1.145 | 2079 | 1.058 | ||
| 5 | 1922 | 1.197 | 1715 | 1.808 | ||
| 6 | 1922 | 1.509 | 1871 | 1.497 | ||
| 7 | 1922 | 2.445 | 1960 | 2.449 | 3.955 | High-risk group |
| 8 | 1922 | 2.653 | 1867 | 3.267 | ||
| 9 | 1922 | 4.214 | 1873 | 3.951 | ||
| 10 | 1929 | 5.962 | 1885 | 6.207 | ||
Discrimination of the low-risk group from the high-risk group for advanced CRN
Based on the cut-off value, a simplified prediction model for discrimination of the low-risk group from the high-risk group for advanced CRN was constructed (Table 5). The high-risk group had a 3.7-fold increased risk of advanced CRN compared to the low-risk group (1.1% vs. 4.0%, P < .001). In the training set, the sensitivity, specificity, accuracy, PPV, and negative predictive value (NPV) of the simplified prediction model were 73.3%, 61.0%, 61.3%, 3.9%, and 99.1%, respectively. In the validation set, the sensitivity, specificity, accuracy, PPV, and NPV were 70.8%, 61.2%, 61.4%, 4.0%, and 98.9%, respectively.
Table 5. Discrimination ability of the low-risk group from the high-risk group for advanced colorectal neoplasia.
| Advanced CRN (-), n | Advanced CRN (+), n | p | Sensitivity, % (95% CI) |
Specificity, % (95% CI) |
Accuracy, % (95% CI) |
PPV, % (95% CI) |
NPV, % (95% CI) |
|
|---|---|---|---|---|---|---|---|---|
| Training set | ||||||||
| Low-risk group, n | 11491 | 107 | <.001 | 73.3 (69.0–77.6) |
61.0 (60.3–61.7) |
61.3 (60.6–62.0) |
3.9 (3.4–4.3) |
99.1 (98.9–99.3) |
| High-risk group, n | 7335 | 294 | ||||||
| Validation set | ||||||||
| Low-risk group, n | 11487 | 124 | <.001 | 70.8 (66.4–75.1) |
61.2 (60.5–61.9) |
61.4 (60.7–62.1) |
4.0 (3.5–4.4) |
98.9 (98.7–99.1) |
| High-risk group, n | 7288 | 300 | ||||||
| Total dataset | ||||||||
| Low-risk group, n | 22978 | 231 | <.001 | 72.0 (68.9–75.1) |
61.1 (60.6–61.6) |
61.3 (60.9–61.8) |
3.9 (3.6–4.2) |
99.0 (98.9–99.1) |
| High-risk group, n | 14623 | 594 | ||||||
CRN, colorectal neoplasia; PPV, positive predictive value; NPV, negative predictive value.
Comparison of the discrimination performance of the final model with previous published prediction models for advanced CRN
In the validation set, the discrimination performance of the final model was compared with that of the advanced CRN (ACN) index [14] and Asia-Pacific Colorectal Screening score (APCS) [12] using the AUC (Fig 7). The AUC of the final model was 0.716 (95% CI, 0.691–0.741), whereas that of the ACN index was 0.672 (95% CI, 0.645–0.699), and that of the APCS was 0.678 (95% CI, 0.651–0.705). The discrimination performance of the developed model for high-risk patients with advanced CRN was better than that of the ACN index (P < .001) or APCS (P < .001).
Fig 7. Comparison of the discrimination performance of the final model with previous published prediction models for advanced colorectal neoplasia.
Discussion
Big data can improve health by providing insights into public health, such as enhanced disease prediction and prevention. Using a big data analytics algorithm, we explored a large health screening examination database. The refined database with structured and unstructured data contained first screening colonoscopy and comprehensive health examination data from 49,450 patients. Big data can not only be applied for verifying alleged associations, but can also be used as a hypothesis-generating machine [24]. In this study, we generated a prediction model for advanced CRN, which might be the first trial for utilization of big data analytics in the field of gastroenterology. The final simplified prediction model was shown to have acceptable discriminative power for patients with advanced CRN. Our simple risk score using easily available information from the patient's clinical questionnaire stratified asymptomatic patients into low- and high-risk groups for advanced CRN before a screening colonoscopy was performed. The discrimination performance of the developed model for high-risk patients with advanced CRN was better than that of existing models.
Based on our results, it is deemed to be inefficient to undergo colonoscopy screening for patients in the low-risk group due to the low probability of advanced CRN as well as the cost and risk associated with colonoscopy. The specificity of our prediction model was not sufficiently high, but the NPVs in this prediction model were as high as 99%. Since this study was populated by asymptomatic individuals who underwent health check-ups, and not symptomatic patients, our objective was to develop and validate a prediction model for estimating the probability of having advanced CRN. We hope to apply this proposed prediction model for the purpose of identifying patients who may not need to undergo a colonoscopy.
There were many studies reporting different risk scoring system for CRC; however, almost none of them can be translated into clinical practice. It is possible because the fecal occult blood test is in fact very convenient, the result is straightforward, and the cost is low. In Korea, a national CRC screening has been in place using fecal immunochemical testing (FIT). The limitation of a stool-based test such as FIT is that it is a diagnostic tool only for the early detection of CRC. Recent guideline grouped the CRC screening tests into cancer prevention and cancer detection tests [25]. The benefits of cancer prevention test can eliminate advanced CRN and prevent CRC. Cancer prevention tests are preferred over detection tests. The goal of CRC screening shifted from “screening detection to prevention by polypectomy [26].” As such, the present study aimed to develop and validate a prediction model for estimating the probability of having advanced CRN and not CRC. Therefore, we think it is difficult to directly compare the predictive model based on FIT with a colonoscopy.
The issue of developing a prediction model for advanced CRN is not novel and several other models already exist. Our study has implemented a predictive model using varied clinical variables acquired in real-world clinical practice. Our prediction model showed more effective prediction for advanced CRN than previous proposed advanced CRN prediction models. We chose to compare our prediction model to studies by Schroy et al [14]. and Yeoh et al [12]. The reason we chose the studies by Schroy et al [14]. and Yeoh et al [12]. is because both studies evaluated advanced CRN predictability and were well designed. The study by Imperidale et al. was also a well-designed study [10], but the outcome measurement was advanced proximal advanced CRN. Therefore, we thought Imperidale’s study was inappropriate for comparison with our model.
Our study was performed with a large population who underwent their first colonoscopy and a comprehensive health screening examination, which may minimize sampling error and represent real-world practice and enhances its usefulness in facilitating shared decision-making for individuals who need CRC screening. The use of EMR systems among healthcare providers has spread widely over the past decade [15]. Using text from EMR system, we applied NLP and the CETAS method to demonstrate the replicability of manual chart review. Previous studies have revealed the utility of NLP in extracting information from clinical text [20–22]. In addition, our risk prediction models use extensive independent variables to estimate the probability of having or developing advanced CRN. Therefore, the discrimination performance of our model for high-risk patients with advanced CRN was better than that of existing models.
Our study had some limitations. External validation could not be performed, so there are concerns about overfitting and generalizability. In addition, the model was developed using a database of patients willing to undergo screening colonoscopy (It is a selected population of 70,959 subjects who underwent colonoscopy screening. It is furthermore selected once more because 21,509 subjects are excluded from the analysis); on that account, it is unclear whether our model can apply to the patients unable or unwilling to undergo colonoscopy. Our study population was quite young for routine screening colonoscopies. The mean age of study population was 50 years old and this may explain why the overall rate of advanced CRN of 2.3% in this study. However, all included subjects underwent colonoscopy as a part of their health check-up. So, even though the patients were young, they did not have symptoms or a family history of CRC. Furthermore, given the long time needed for an adenoma to progress to a carcinoma, the increased number of cases of CRC diagnosed in this age group may originate from adenomas present in individuals in their 40s or earlier [17]. These cancers may be prevented by colonoscopy with polypectomy of premalignant lesions in the preceding decade. Despite this theoretical argument for screening individuals in their 40s or earlier, we included patients who underwent colonoscopies at any age and analyzed the age as continuous variables to develop a prediction model. In addition, we used the mean substitution technique as imputation to deal with missing predictor values in training set. Mean substitution has the benefit of not changing the sample mean for that variable, however mean imputation attenuates any correlations involving the variables that are imputed. The mean imputation has some attractive properties for univariate analysis but becomes problematic for multivariate analysis. Although we used the dataset applied mean substitution technique during univariate logistic regression for the identification of predictors and not used during multivariate logistic regression, the uncertainty in the imputation can lead to overly precise results and errors in our prediction model [27, 28].
Despite these weak points, our model can serve as a clinically useful tool for facilitating shared decision-making related to select the screening modalities for early detection and prevention of CRC, especially when the provider and patient preferences differ. If physicians could predict which patients are at increased risk before colonoscopy, it is possible that they might make better decisions about screening. We developed a simple risk scoring model easily available by questionnaire and precisely identified low- and high-risk groups for advanced CRN at the first screening colonoscopy. This model may increase CRC risk awareness and help healthcare providers encourage the high-risk group to undergo colonoscopy. Furthermore, by identifying the patients with a high risk of advanced CRN, the present model may help to target primary prevention interventions. Once it has been externally validated, the model will be useful to facilitate more effective shared decision-making for CRC screening.
Data Availability
Due to ethical restrictions related to participant confidentiality imposed by the Samsung Medical Center Institutional Data Access / Ethics Committee, the data underlying this study are available upon request. Interested researchers may submit queries related to data access to the Samsung Medical Center Institutional Data Access / Ethics Committee by contacting Dr. Seil Jang (+82-2-3410-2972; sei.jang@samsung.com).
Funding Statement
The authors received no specific funding for this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer Journal international du cancer. 2015. March 1;136(5):E359–86. doi: 10.1002/ijc.29210 . [DOI] [PubMed] [Google Scholar]
- 2.Lee BI, Hong SP, Kim SE, Kim SH, Kim HS, Hong SN, et al. Korean guidelines for colorectal cancer screening and polyp detection. The Korean journal of gastroenterology. 2012. February;59(2):65–84. . [DOI] [PubMed] [Google Scholar]
- 3.Khalid-de Bakker C, Jonkers D, Smits K, Mesters I, Masclee A, Stockbrugger R. Participation in colorectal cancer screening trials after first-time invitation: a systematic review. Endoscopy. 2011. December;43(12):1059–86. doi: 10.1055/s-0031-1291430 . [DOI] [PubMed] [Google Scholar]
- 4.Kaminski MF, Polkowski M, Kraszewska E, Rupinski M, Butruk E, Regula J. A score to estimate the likelihood of detecting advanced colorectal neoplasia at colonoscopy. Gut. 2014. July;63(7):1112–9. doi: 10.1136/gutjnl-2013-304965 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin A, Joo J, Yang HR, Bak J, Park Y, Kim J, et al. Risk prediction model for colorectal cancer: National Health Insurance Corporation study, Korea. PloS one. 2014;9(2):e88079 doi: 10.1371/journal.pone.0088079 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park Y, Freedman AN, Gail MH, Pee D, Hollenbeck A, Schatzkin A, et al. Validation of a colorectal cancer risk prediction model among white patients age 50 years and older. Journal of clinical oncology. 2009. February 10;27(5):694–8. doi: 10.1200/JCO.2008.17.4813 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colditz GA, Atwood KA, Emmons K, Monson RR, Willett WC, Trichopoulos D, et al. Harvard report on cancer prevention volume 4: Harvard Cancer Risk Index. Risk Index Working Group, Harvard Center for Cancer Prevention. Cancer causes & control. 2000. July;11(6):477–88. . [DOI] [PubMed] [Google Scholar]
- 8.Freedman AN, Slattery ML, Ballard-Barbash R, Willis G, Cann BJ, Pee D, et al. Colorectal cancer risk prediction tool for white men and women without known susceptibility. Journal of clinical oncology. 2009. February 10;27(5):686–93. doi: 10.1200/JCO.2008.17.4797 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei EK, Colditz GA, Giovannucci EL, Fuchs CS, Rosner BA. Cumulative risk of colon cancer up to age 70 years by risk factor status using data from the Nurses' Health Study. American journal of epidemiology. 2009. October 1;170(7):863–72. doi: 10.1093/aje/kwp210 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imperiale TF, Wagner DR, Lin CY, Larkin GN, Rogge JD, Ransohoff DF. Using risk for advanced proximal colonic neoplasia to tailor endoscopic screening for colorectal cancer. Annals of internal medicine. 2003. December 16;139(12):959–65. . [DOI] [PubMed] [Google Scholar]
- 11.Driver JA, Gaziano JM, Gelber RP, Lee IM, Buring JE, Kurth T. Development of a risk score for colorectal cancer in men. The American journal of medicine. 2007. March;120(3):257–63. . [DOI] [PubMed] [Google Scholar]
- 12.Yeoh KG, Ho KY, Chiu HM, Zhu F, Ching JY, Wu DC, et al. The Asia-Pacific Colorectal Screening score: a validated tool that stratifies risk for colorectal advanced neoplasia in asymptomatic Asian subjects. Gut. 2011. September;60(9):1236–41. doi: 10.1136/gut.2010.221168 . [DOI] [PubMed] [Google Scholar]
- 13.Cai QC, Yu ED, Xiao Y, Bai WY, Chen X, He LP, et al. Derivation and validation of a prediction rule for estimating advanced colorectal neoplasm risk in average-risk Chinese. American journal of epidemiology. 2012. March 15;175(6):584–93. doi: 10.1093/aje/kwr337 . [DOI] [PubMed] [Google Scholar]
- 14.Schroy PC, Wong JB, O'Brien MJ, Chen CA, Griffith JL. A Risk Prediction Index for Advanced Colorectal Neoplasia at Screening Colonoscopy. The American journal of gastroenterology. 2015. July;110(7):1062–71. doi: 10.1038/ajg.2015.146 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strauss JA, Chao CR, Kwan ML, Ahmed SA, Schottinger JE, Quinn VP. Identifying primary and recurrent cancers using a SAS-based natural language processing algorithm. Journal of the American Medical Informatics Association. 2013. Mar-Apr;20(2):349–55. doi: 10.1136/amiajnl-2012-000928 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rampal S, Yang MH, Sung J, Son HJ, Choi YH, Lee JH, et al. Association between markers of glucose metabolism and risk of colorectal adenoma. Gastroenterology. 2014. July;147(1):78–87.e3. doi: 10.1053/j.gastro.2014.03.006 . [DOI] [PubMed] [Google Scholar]
- 17.Hong SN, Kim JH, Choe WH, Han HS, Sung IK, Park HS, et al. Prevalence and risk of colorectal neoplasms in asymptomatic, average-risk screenees 40 to 49 years of age. Gastrointestinal endoscopy. 2010. September;72(3):480–9. doi: 10.1016/j.gie.2010.06.022 . [DOI] [PubMed] [Google Scholar]
- 18.The Paris endoscopic classification of superficial neoplastic lesions: esophagus, stomach, and colon: November 30 to December 1, 2002. Gastrointestinal endoscopy. 2003. December;58(6 Suppl):S3–43. . [DOI] [PubMed] [Google Scholar]
- 19.Han DS, Sohn JH, Byeon JS, Choi H, Kim JM. Diagnostic Coding for Intramucosal Carcinoma and Neuroendocrine Tumor in the Colorectum: Proposal for Avoiding Confusing Coding in Korea. Clin Endosc. 2015. May;48(3):216–20. doi: 10.5946/ce.2015.48.3.216 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazlehurst B, Sittig DF, Stevens VJ, Smith KS, Hollis JF, Vogt TM, et al. Natural language processing in the electronic medical record: assessing clinician adherence to tobacco treatment guidelines. American Journal of Preventive Medicine. 2005. December;29(5):434–9. doi: 10.1016/j.amepre.2005.08.007 . [DOI] [PubMed] [Google Scholar]
- 21.Al-Haddad MA, Friedlin J, Kesterson J, Waters JA, Aguilar-Saavedra JR, Schmidt CM. Natural language processing for the development of a clinical registry: a validation study in intraductal papillary mucinous neoplasms. HPB (Oxford). 2010. December;12(10):688–95. doi: 10.1111/j.1477-2574.2010.00235.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedlin J, Overhage M, Al-Haddad MA, Waters JA, Aguilar-Saavedra JJ, Kesterson J, et al. Comparing methods for identifying pancreatic cancer patients using electronic data sources. Annual Symposium proceedings / AMIA Symposium. 2010. November 13;2010:237–41. . [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2012. September;143(3):844–57. doi: 10.1053/j.gastro.2012.06.001 . [DOI] [PubMed] [Google Scholar]
- 24.Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo clinic NLP system for patient smoking status identification. Journal of the American Medical Informatics Association. 2008. Jan-Feb;15(1):25–8. doi: 10.1197/jamia.M2437 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM. American College of Gastroenterology guidelines for colorectal cancer screening 2009. The American journal of gastroenterology. 2009. March;104(3):739–50. doi: 10.1038/ajg.2009.104 . [DOI] [PubMed] [Google Scholar]
- 26.Moiel D, Thompson J. Early detection of colon cancer-the kaiser permanente northwest 30-year history: how do we measure success? Is it the test, the number of tests, the stage, or the percentage of screen-detected patients? The Permanente journal. 2011. Fall;15(4):30–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham JW. Missing data analysis: making it work in the real world. Annual review of psychology. 2009;60:549–76. doi: 10.1146/annurev.psych.58.110405.085530 . [DOI] [PubMed] [Google Scholar]
- 28.Fox-Wasylyshyn SM, El-Masri MM. Handling missing data in self-report measures. Research in nursing & health. 2005. December;28(6):488–95. doi: 10.1002/nur.20100 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Due to ethical restrictions related to participant confidentiality imposed by the Samsung Medical Center Institutional Data Access / Ethics Committee, the data underlying this study are available upon request. Interested researchers may submit queries related to data access to the Samsung Medical Center Institutional Data Access / Ethics Committee by contacting Dr. Seil Jang (+82-2-3410-2972; sei.jang@samsung.com).