Abstract
This study was performed to investigate the concordance in terms of molecular characteristics and antimicrobial susceptibility between colonizing and clinical Staphylococcus aureus isolates obtained from children in Korea, where ST72 is the major genotype.
This was a prospective observational descriptive study of culture-confirmed S aureus infections obtained from children ≤18 years old admitted to Asan Medical Center Children's Hospital in Seoul, Korea, from March 2014 to April 2015. Molecular studies including multilocus sequence typing (MLST), SCCmec typing, polymerase chain reaction amplification of the Panton-Valentine leukocidin (PVL) genes, and antibiotic susceptibility tests were performed on S aureus isolates obtained from nares and clinical specimens.
During the study period, 126 clinically significant S aureus infections were identified. Nasal swab cultures were made from 113 of the 126 children, and 46.0% (52/113) showed S aureus colonization. The overall concordance between colonizing and clinical isolates by methicillin susceptibility was 94.2% (49/52); all 3 discordant cases were HA-MSSA cases with nasal MRSA. Among the 37 pairs of colonizing and clinical S aureus isolates included in the genotyping analysis, ST72-SCCmec type IV was the most prevalent clone and the PVL genes were positive in 2 patients. Among the 31 pairs of healthcare-associated cases, concordance rates by methicillin susceptibility and sequence type (ST) were 90.3% (28/31) and 84% (26/31), respectively. For the 6 pairs of community-associated (CA) S aureus including 3 CA-MRSA cases, 100% concordance was observed by methicillin susceptibility and ST.
The concordance between isolates obtained from children who required medical services was relatively high in Korean children where ST72-SCCmec type IV is the predominant clone as the colonizer and the pathogen. It is suggested that decolonization and continuous care to prevent transmission could be effective in managing and preventing both HA- and CA-SA infections in our setting.
Keywords: children, colonization, concordance, Korea, MRSA, S aureus, ST72
1. Introduction
Staphylococcus aureus is a leading cause of skin and soft tissue infections (SSTIs) throughout the world and can lead to serious invasive infections, such as bone and joint infection, pneumonia, bacteremia, and infective endocarditis, which are associated with significant morbidity and mortality.[1,2]S aureus normally colonizes the anterior nares[3]; 38.1% of healthy Korean preschool children attending a day care center were found to be colonized by S aureus strains, a quarter of which were methicillin-resistant (MRSA).[4] Recently, an increasing rate of MRSA colonization has been noted throughout the world, including the United States.[5]
It is well known that endogenous S. aureus colonization is closely related to nosocomial infection[6] and is associated with healthcare-associated bacteremia, increased risk of wound infection after surgery, and mortality among long-term healthcare facility patients.[6,7] However, it is relatively unclear whether any community-associated S aureus infections are of endogenous origin. Only 59% of S aureus clinical and colonizing isolates obtained from children were identical according to methicillin susceptibility tests and pulsed field gel electrophoresis (PFGE).[1] Even in colonized individuals, more than one-third of community-associated S aureus (CA-SA) infections were caused by strains different from the colonizing form.[8] These findings suggest that CA-SA infections in children are not always of endogenous origin. However, most previous studies measuring the concordance between colonizing and clinical isolates were based on SSTIs caused by the USA300 (sequence type [ST]8) strain, which is the major genotype in community settings in the United States,[1,9] and there have been few studies of isolates from other geographic regions such as Asia, where the prevalent S aureus genotype is different—for examples, ST59 in China and Taiwan; ST8 and 30 in Japan[10]; and ST72 in Korea.[10–12] The relationship between colonizing and clinical isolates obtained from children in regions where ST72 is the major genotype has not been much studied. In addition, few studies have focused on not only SSTIs but also diverse clinical entities such as bacteremia and pneumonia in children.
The purpose of this study was to examine the concordance between the genotypes of colonizing and clinical S aureus isolates in children in Korea, where ST72 is the predominant clone in healthcare as well as community settings. In addition, we aimed to evaluate the nasal colonization rate in clinically significant S aureus infections in children.
2. Methods
2.1. Study populations
This was a prospective observational descriptive study. We prospectively enrolled children ≤18 years of age with culture-confirmed S aureus infections who were hospitalized or visited the pediatric emergency department or outpatient clinic at Asan Medical Center Children's Hospital, Seoul, Korea, over 13 consecutive months from March 2014 to April 2015. We used the electronic databases of the bacterial surveillance system at our institute to collect S aureus isolates obtained from children with clinical infections and exclude those obtained for surveillance from specimens such as urine, eye discharges, and skin swabs, which were considered to be without clinical significance. The isolates with clinical significance were kept at −70°C.
Nasal swab cultures for screening both methicillin-susceptible (MS) and MR S aureus colonization were performed as soon as possible during each clinical episode in cases of CA-SA infections including SSTIs, suppurative lymphadenitis, abscesses, and bone and joint infections that were mainly caused by S aureus. Surveillance cultures of nasal swabs for detecting MRSA carriage were made at the outset of periods of stay in pediatric units or neonatal intensive care units (ICU), and contact precautions were applied to the patients colonized by MRSA as a standard of care in our institute; in cases of healthcare-associated S aureus infections such as ventilator-associated pneumonia, hospital-acquired pneumonia, and catheter-related infections, nasal swab cultures were made to detect only MRSA colonizers.
The colonizing S aureus isolates were also kept at −70°C. Only the first S aureus isolates detected during a single clinical episode were included in the analysis, and duplicates from the same patient were excluded. Demographic data and clinical information including primary site of infection, hospitalization, and underlying medical condition were abstracted from the electronic medical records. This study was approved by the Institutional Review Board of Asan Medical Center (IRB No. 2014-0175).
2.2. Definitions
Healthcare-associated S aureus (HA-SA) infection was defined as an S aureus infection with risk factors for healthcare-associated infection (e.g., hospitalized within the previous year, recent surgery, residence in a long-term care facility, dialysis patient, indwelling catheter at time of admission).[13,14] A CA-SA infection was defined as a case in which the isolate was obtained from a patient with no risk factors for healthcare-associated infections including prior hospitalization in the 12 months preceding culture.
2.3. Strain identification and antimicrobial susceptibility testing
S aureus isolates were identified and their antimicrobial susceptibilities established using a MicroScan WalkAway96 with a MicroScan–Pos BP Combo Panel Type 28 (Siemens, West Sacramento, CA). Eight antibiotics were tested: gentamycin, ciprofloxacin, trimethoprim/sulfamethoxazole, rifampin, tetracyclin, clindamycin, linezolid, and vancomycin. Intermediate resistance and full resistance were both considered resistance. The in vitro macrolide–lincosamide–streptogramin B (MLSB)-inducible phenotype was detected by the D-zone test (double-disk diffusion test).
Nasal swab specimens were obtained from both anterior nares with Amies agar gel double plastic swabs (Copan Diagnostics, Breschia, Italy) after soaking the rayon bulbs of the swabs in sterile normal saline, and inoculated onto MRSASelect (BioRad, Redmond, WA) agar the same day. After incubation at 35°C for 18–24 hours, small pink colonies were selected for gram staining. For screening MSSA colonization, clustered gram-positive cocci were tested with a MicroScan PBC 28 for identification and susceptibility testing.
2.4. Molecular analysis of colonizing and clinical isolates
For the cases with concomitant clinical and colonizing S aureus isolates obtained from March to December 2014, molecular analysis including multilocus sequence typing (MLST), SCCmec typing, and identification of the Panton–Valentine leukocidin (PVL) genes by PCR was conducted as described by Lee et al.[12]Staphylococcus aureus genotypes were designated by their ST and their SCCmec type, as proposed by Enright et al.[15]
3. Results
3.1. Patient characteristics
During the study period, a total of 548 cases of S aureus infection were reported in children ≤18 years old (Fig. 1). After excluding isolates obtained from eye discharge, urine, and simple surveillance cultures, 126 patients with clinically significant S aureus infections were enrolled. Nasal swab culture was performed on 113 of the 126 enrolled children, and 46.0% (52/113) were confirmed as having S aureus colonization of the nares (MRSA colonization, n = 46; and MSSA colonizaion, n = 6). Of 113 S aureus infections, 96 (85.0%) and 17 (15.0%) were HA-SA and CA-SA infections, of which 45.8% (44/96) and 47.1% (8/17) cases occurred in conjunction with S aureus colonization, respectively.
Figure 1.
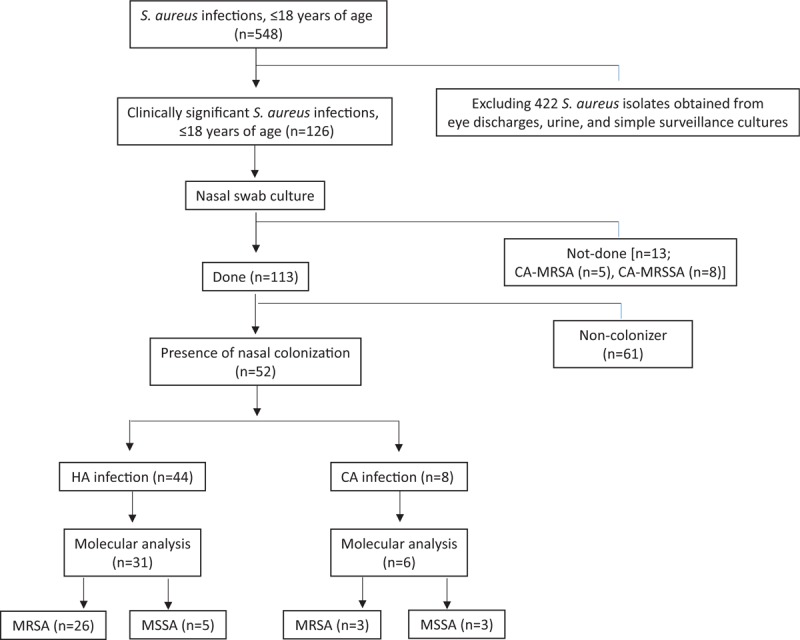
Flow-chart of the study. CA = community-associated, HA = healthcare-associated, MR = methicillin-resistant, MS = methicillin-susceptible, SA = S aureus.
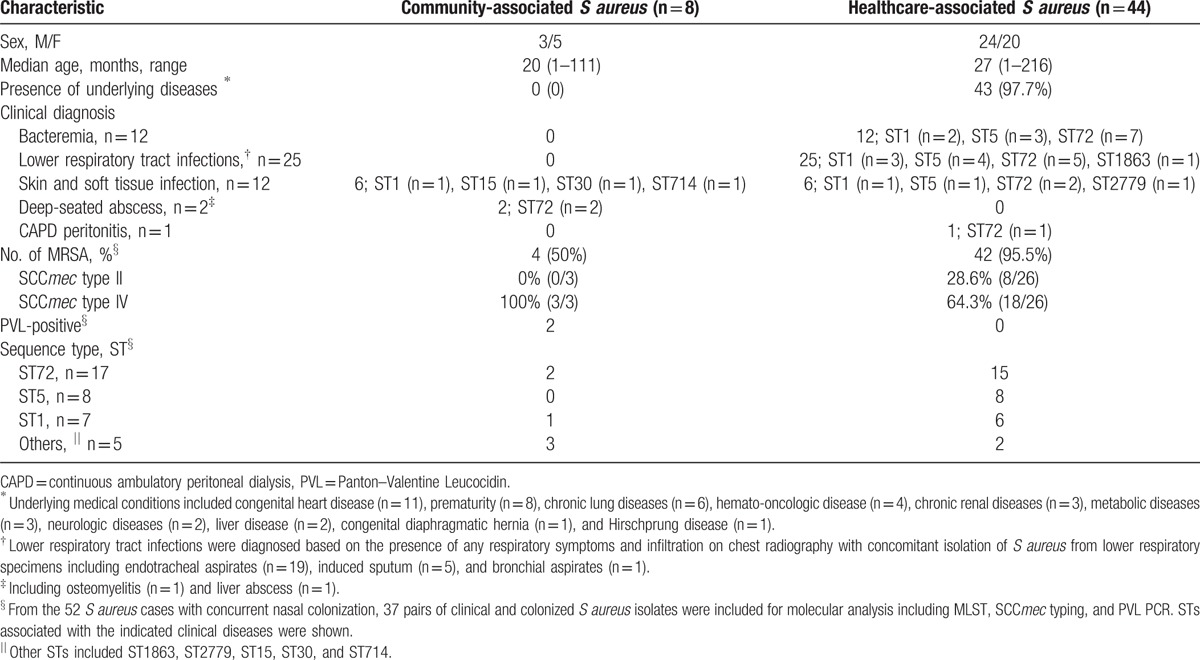
The demographic characteristics of the total of 52 S aureus cases with concurrent nasal colonization are shown in Table 1. Median age was 26 months (range, 1–216 months), and 44 cases were HA-SA infections. The most common underlying medical condition of the patients with HA-SA infections was congenital heart disease (11/44, 25%), followed by prematurity (7/44, 15.9%), chronic lung diseases (6/44, 13.6%), and hemato-oncologic diseases (4/44, 9.1%). The most common source of the HA-SA isolates was respiratory specimens (25/44, 56.8%) including endotracheal sputum and bronchial aspirates obtained from children with pneumonia, followed by blood (12/44, 27.3%). The 8 CA-SA cases presented with diagnoses of SSTI (n = 6) and deep-seated abscess (n = 2). There were no fatal cases related to S aureus infection during the study period.
Table 1.
Demographic and clinical characteristics of 52 S aureus infections with concurrent nasal colonization.
3.2. Molecular characterization of the S aureus isolates
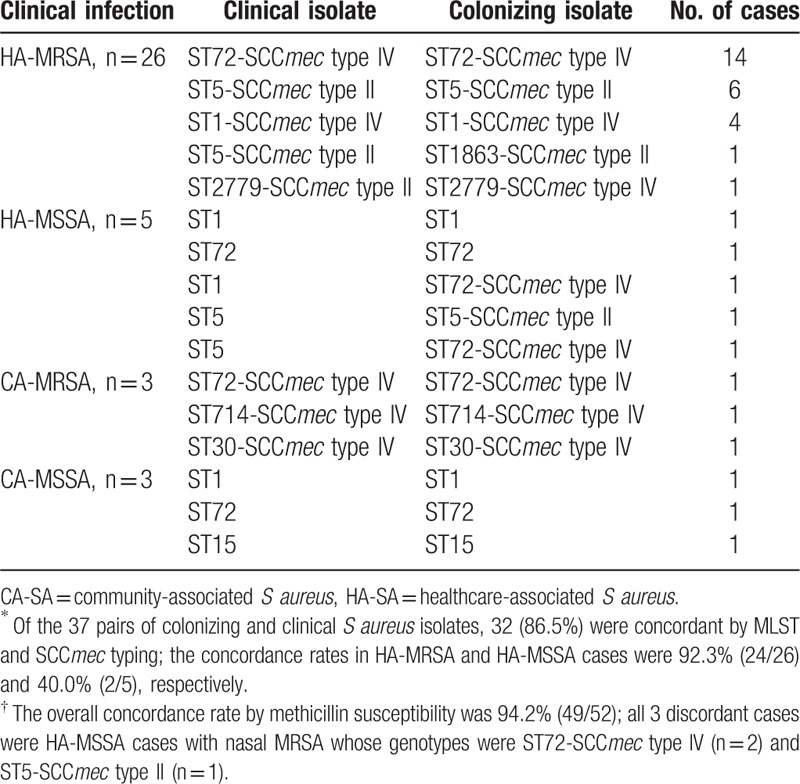
From among the 52 S aureus cases with concurrent nasal colonization, 37 pairs of clinical and colonizing S aureus isolates could be submitted to molecular analysis of MLST, and SCCmec typing (Fig. 1). STs associated with the indicated clinical diseases were shown in Table 1.
Among the total of 74 isolates, the most common ST was ST72 (n = 36), followed by ST5 (n = 16) and ST1 (n = 13) (Table 2). Among the total of 58 clinical and colonizing MRSA isolates obtained from the 3 cases of CA-MRSA and the 26 of HA-MRSA, the most common genotype was ST72-SCCmec type IV (n = 30), followed by ST5-SCCmec type II (n = 13), and ST1-SCCmec type IV (n = 8). The STs of the 8 clinical and 5 colonizing MSSA isolates were: ST1 (n = 5), ST72 (n = 4), ST5 (n = 2), and ST15 (n = 2). The 3 discordant pairs of HA-MSSA infections with nasal MRSA were as follows: 1 case of ST1-MSSA bacteremia with nasal ST72-SCCmec type IV-MRSA, and 2 cases of respiratory infections caused by either ST5-MSSA with nasal ST5-SCCmec type II-MRSA or ST5-MSSA with nasal ST72-SCCmec type IV, respectively.
Table 2.
Concordance between colonizing and clinical isolates by genotype∗ and methicillin-susceptibility†.
The PVL genes were detected in 2 CA-MRSA infections involving an ST714-SCCmec type IV and an ST30-SCCmec type IV strain, respectively; these isolates were both obtained from patients with SSTIs.
3.3. Antibiotic susceptibility profiles
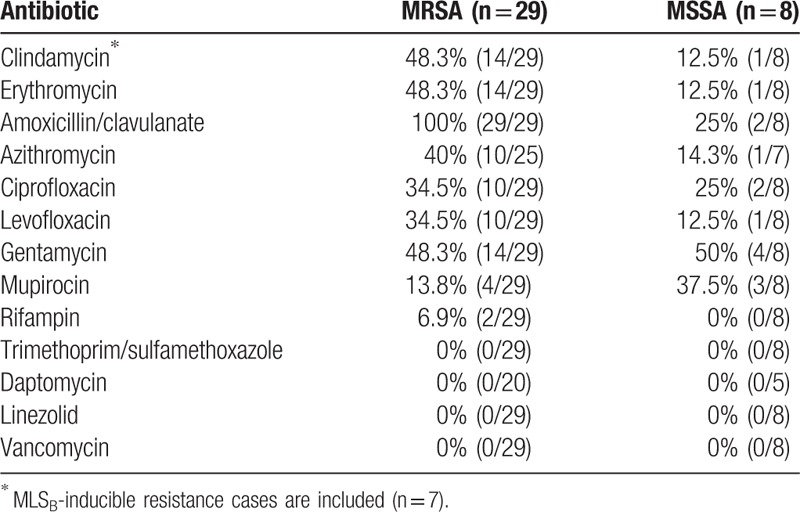
The antimicrobial susceptibility patterns of the 37 clinical isolates included in the molecular analysis are presented in Table 3. All the 29 MRSA isolates were fully susceptible to vancomycin and trimethoprim/sulfamethoxazole. The double-disk diffusion test was positive in 43.8% (7/16) of the erythromycin-resistant and clindamycin-susceptible MRSA isolates; all 7 of these isolates had MLSB-inducible phenotypes. Considering both inducible and constitutive phenotypes, 48.3% of the MRSA isolates were clindamycin-resistant.
Table 3.
Antibiotic resistance of clinical S aureus isolates.
3.4. Concordance between colonizing and clinical isolates by genotype and methicillin susceptibility
The overall concordance rate by methicillin susceptibility was 94.2% (49/52); all 3 discordant cases were HA-MSSA cases with nasal MRSA (Table 2). Of the 31 pairs of HA-SA cases and 6 pairs of CA-SA infections, the concordance rates by methicillin susceptibility were 90.3% (28/31) and 100% (6/6), respectively.
Of the 29 pairs of colonizing and clinical MRSA isolates, 27 (93.1%) were concordant by both ST and SCCmec type; the concordance rates in HA-MRSA and CA-MRSA cases were 92.3% (24/26) and 100% (3/3), respectively. Of the 8 MSSA cases, the concordance rates by ST in HA-MSSA and CA-MSSA cases were 60% (3/5) and 100% (3/3), respectively.
4. Discussion
In this prospective study, we found a high concordance rate between colonizing and clinical S aureus isolates from both healthcare and community settings in Korea where ST72 is the predominant clone: the infecting strain in 94.2% and 86.5% of the patients was concordant with the colonizing strain by methicillin susceptiblity and genotype, respectively. Active surveillance showed that at least 46.5% of the pediatric patients with clinically significant S aureus infections had nasal S aureus colonization.
Colonization pressure is a predictive factor for nosocominal transmission of MRSA,[16,17] and MRSA colonization is a risk factor for subsequent MRSA infection in children admitted to a pediatric intensive care unit.[18] In a neonatal care unit where MRSA infection was endemic, colonized infants had a significantly higher rate of MRSA infection than those without colonization, and the genotypes of the clinical and colonizing isolates were indistinguishable in >90% of episodes.[19] However, among children in a community setting, there was no relationship between baseline S aureus household colonization pressure and subsequent SSTI.[20] The striking difference in the concordance rates of HA and CA cases might be explained by differences in their transmission dynamics. In a hospital setting, most individual contacts are limited to healthcare staff, whereas in the community, routes of transmission are more diverse because the colonizing strain can come not only from within the household but also from external sources.[20,21]
Compared to the reports from the United States, where the USA 300 strain is the major genotype in SSTI outbreaks, we found relatively diverse genotypes, including ST72, among our CA-SA isolates, but no ST8 strains. Because routine screening is not performed in cases of CA-SA infection in our institute, all the cases that failed to undergo nasal screening were CA infections, irrespective of methicillin susceptibility. Even though only a small number of CA-SA cases could be included in our molecular analysis, the infecting and colonizing isolates in all 6 cases were indistinguishable by ST, SCCmec type, and methicillin susceptibility. Among the 26 HA-MRSA cases, more than 80% were caused by the 2 most common strains, ST72 (54%) and ST5 (27%); colonizing and infecting strains were indistinguishable in 24 of these cases. We suggest that decolonization and taking continuous precautions might be effective in managing and preventing both HA- and CA-SA infections in our setting.
Clinical outcomes and bacterial virulence factors may differ depending on genotype; rapid spread of the USA 300 strain has been reported, which is highly contagious and is now the most common cause of global epidemics.[22,23] Freitas et al[23] reported that individuals colonized with USA 300 were more likely to have SSTIs than those colonized with non-USA 300 strains (P = .02); however, no significant difference between USA 300 and non-USA 300 strains was observed in other types of clinical infection. The ST72 strain was considered as a community-associated strain, found frequently in Korea; however, it is now the most common genotype in HA-SA infections,[10,25] replacing ST239-SCCmec type III and ST5-SCCmec type II.[26,27] Although Uhlemann et al report that the ST72 strain is the most prevalent in the Dominican Republic, its spa type seems to be different from the predominant Korean spa type.[28] ST72-SCCmec type IV is associated with lower mortality than ST5-SCCmec type II in adult patients with MRSA bacteremia.[29] Since the majority of previous studies were conducted on patients with SSTIs in a region where the USA 300 clone was the major genotype,[21,24] it would be inappropriate to extrapolate the relationship between colonizing and clinical isolates observed in those studies to other clinical manifestations and bacterial genotypes.
PVL is associated with CA infections causing necrotic lesions involving the skin or mucosa, such as pyogenic skin lesions or necrotizing pneumonia.[30,31] Recently, 10% of pediatric patients with CA-MRSA infections were found to be positive for the PVL gene in Korea.[11] We found 2 examples of CA infections due to PVL gene positive strains; 1 strain was ST714-SCCmec type IV, a genotype not previously reported in Korea.[32]
There are several limitations to our study. First, it was based on S aureus infections observed in a single tertiary university hospital in Seoul, Korea, and the results cannot be extrapolated to the general pediatric population. Because some patients were already on antibiotic treatment when nasal swabs were obtained, the colonization rate might be underestimated. In addition, significant conclusions regarding the concordance rate among pediatric CA-SA infections and control measures cannot be drawn owing to the paucity of CA-SA cases included in the molecular analysis. Further studies focusing on CA-SA infections would be helpful for understanding the relationship between colonization and clinical infection in non-outbreak settings. Nevertheless, this study could suggest the clinical significance of colonizing S aureus isolates in the circumstances where most S aureus strains were ST72 clones without PVL genes.
In conclusion, most colonizing and clinical S aureus isolates in pediatric S aureus infections in Korea where ST72 is the major genotype as an endemic strain in both healthcare and community settings were indistinguishable from each other. Precautions to minimize the transmission, and decolonization, might be useful to reduce S aureus infections in Korean children, and their effects should be studied.
Acknowledgments
The authors are very grateful to Jung-Hwa Kim for assistance with an experiment during the course of this research.
Footnotes
Abbreviations: CA = community-associated, HA = healthcare-associated, MLST = multilocus sequence typing, MR = methicillin-resistant, MS = methicillin-susceptible, PVL = Panton-Valentine leukocidin, SA = S aureus, SCCmec = staphylococcal cassette chromosome mec, SSTIs = skin and soft tissue infections, ST = sequence type.
Funding: This work was supported by a grant from Asan Institute for Life Sciences (grant No. 2014-0288). There is no conflict of interest to declare.
The authors have no conflicts of interest to disclose.
References
- [1].Chen AE, Cantey JB, Carroll KC, et al. Discordance between Staphylococcus aureus nasal colonization and skin infections in children. Pediatr Infect Dis J 2009;28:244–6. [DOI] [PubMed] [Google Scholar]
- [2].Daum RS. Skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus. N Engl J Med 2007;357:380–90. [DOI] [PubMed] [Google Scholar]
- [3].Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 1997;10:505–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kim YM, Oh CE, Kim SH, et al. Nasal carriage of Staphylococcus aureus from healthy children attending day care center. Korean J Pediatr Infect Dis 2010;17:9–15. [Google Scholar]
- [5].Gorwitz RJ, Moran DK, McAllister SK, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis 2008;197:1226–34. [DOI] [PubMed] [Google Scholar]
- [6].von Eiff C, Becker K, Machka K, et al. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 2001;344:11–6. [DOI] [PubMed] [Google Scholar]
- [7].Vendrell E, Capdevila JA, Barrufet P, et al. Mortality among methicillin-resistant Staphylococcus aureus carriers in long-term care facilities. Rev Esp Quimioter 2015;28:92–7. [PubMed] [Google Scholar]
- [8].Fritz SA, Hogan PG, Hayek G, et al. Household versus individual approaches to eradication of community-associated Staphylococcus aureus in children: a randomized trial. Clin Infect Dis 2012;54:743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gonzalez BE, Martinez-Aguilar G, Hulten KG, et al. Severe Staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics 2005;115:642–8. [DOI] [PubMed] [Google Scholar]
- [10].Chuang Y-Y, Huang Y-C. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis 2013;13:698–708. [DOI] [PubMed] [Google Scholar]
- [11].Sung JY, Lee J, Choi EH, et al. Changes in molecular epidemiology of community-associated and health care-associated methicillin-resistant Staphylococcus aureus in Korean children. Diagn Microbiol Infect Dis 2012;74:28–33. [DOI] [PubMed] [Google Scholar]
- [12].Lee J, Sung JY, Kim YM, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus obtained from the anterior nares of healthy Korean children attending daycare centers. Int J Infect Dis 2011;15:1–6. [DOI] [PubMed] [Google Scholar]
- [13].David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 2010;23:616–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fridkin SK, Hageman JC, Morrison M, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med 2005;352:1436–44. [DOI] [PubMed] [Google Scholar]
- [15].Enright MC, Robinson DA, Randle G, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci USA 2002;99:7687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Williams VR, Callery S, Vearncombe M, et al. The role of colonization pressure in nosocomial transmission of methicillin-resistant Staphylococcus aureus. Am J Infect Control 2009;37:106–10. [DOI] [PubMed] [Google Scholar]
- [17].Bloemendaal AL, Fluit AC, Jansen WM, et al. Acquisition and cross-transmission of Staphylococcus aureus in European intensive care units. Infect Control Hosp Epidemiol 2009;30:117–24. [DOI] [PubMed] [Google Scholar]
- [18].Milstone AM, Goldner BW, Ross T, et al. Methicillin-resistant Staphylococcus aureus colonization and risk of subsequent infection in critically Ill children: importance of preventing nosocomial methicillin-resistant Staphylococcus aureus transmission. Clin Infect Dis 2011;53:853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang YC, Chou YH, Su LH, et al. Methicillin-resistant Staphylococcus aureus colonization and its association with infection among infants hospitalized in neonatal intensive care units. Pediatrics 2006;118:469–74. [DOI] [PubMed] [Google Scholar]
- [20].Rodriguez M, Hogan PG, Krauss M, et al. Measurement and impact of Staphylococcus aureus colonization oressure in households. J Pediatr Infect Dis Soc 2013;2:147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rodriguez EA, Correa MM, Ospina S, et al. Differences in epidemiological and molecular characteristics of nasal colonization with Staphylococcus aureus (MSSA-MRSA) in children from a university hospital and day care centers. PLoS One 2014;9:e101417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].DeLeo FR, Otto M, Kreiswirth BN, et al. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 2010;375:1557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Freitas EA, Harris RM, Blake RK, et al. Prevalence of USA300 strain type of methicillin-resistant Staphylococcus aureus among patients with nasal colonization identified with active surveillance. Infect Control Hosp Epidemiol 2010;31:469–75. [DOI] [PubMed] [Google Scholar]
- [24].Eko KE, Forshey BM, Carrel M, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization and infection isolates in a Veterans Affairs hospital. Antimicrob Resist Infect Control 2015;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Song J-H, Hsueh P-R, Chung DR, et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother 2011;66:1061–9. [DOI] [PubMed] [Google Scholar]
- [26].Soo Ko K, KIM YS, Song J-H, et al. Genotypic diversity of methicillin-resistant Staphylococcus aureus isolates in Korean hospitals. Antimicrob Agents Chemother 2005;49:3583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ko KS, Lee J-Y, Suh JY, et al. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J Clin Microbiol 2005;43:421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Uhlemann A-C, Dumortier C, Hafer C, et al. Molecular characterization of Staphylococcus aureus from outpatients in the Caribbean reveals the presence of pandemic clones. Eur J Clin Microbiol Infect Dis 2011;31:505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Park KH, Chong YP, Kim SH, et al. Community-associated MRSA strain ST72-SCCmecIV causing bloodstream infections: clinical outcomes and bacterial virulence factors. J Antimicrob Chemother 2014;70:1–8. [DOI] [PubMed] [Google Scholar]
- [30].Boan P, Tan H-L, Pearson J, et al. Epidemiological, clinical, outcome and antibiotic susceptibility differences between PVL positive and PVL negative Staphylococcus aureus infections in Western Australia: a case control study. BMC Infect Dis 2015;15:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lina G, Piémont Y, Godail- Gamot F, et al. Involvement of Panton–Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 1999;29:1128–32. [DOI] [PubMed] [Google Scholar]
- [32].Reenar Yoo, Seohee Kim, Lee J. ST714-SCCmec type IV CA-MRSA isolated from a child with recurrent skin and soft tissue infections in South Korea: a case report. Pediatr infect Vaccine 2016;23:62–6. [Google Scholar]


