Melanocytic tumors arising in the central nervous system are also known as primary leptomeningeal melanocytic tumors (PLMTs). They frequently show a benign behavior and are designated as ‘melanocytomas’ in the World Health Organization classification of central nervous system tumors (Brat et al., 2016). Distinction of PLMTs from melanomas metastatic to the central nervous system can be challenging.
Recent studies have demonstrated that PLMTs have a very characteristic genetic profile, frequently harboring GNAQ and GNA11 mutations (Gessi et al., 2014, Koelsche et al., 2015, Kusters-Vandevelde et al., 2010a, Kusters-Vandevelde et al., 2015, Kusters-Vandevelde et al., 2010b, Murali et al., 2012). These mutations are very rare in cutaneous melanoma (Cancer Genome Atlas, 2015), but frequent in uveal melanoma (Van Raamsdonk et al., 2009, Van Raamsdonk et al., 2010). The close molecular relationship between PMLTs and uveal melanomas was further demonstrated by the finding that EIF1AX, SF3B1 and BAP1 mutations, previously identified in uveal melanomas, can also occur in PMLT (Kusters-Vandevelde et al., 2016, van de Nes et al., 2016a, van de Nes et al., 2016b). Similar to uveal melanomas, a proportion of PLMTs are not found to harbor activating mutations in GNAQ or GNA11.
Recent studies have identified activating CYSLTR2 and PLCB4 mutations in uveal melanomas, occurring at the L129 and D630 hotspots, respectively (Johansson et al., 2016, Moore et al., 2016). CYSLTR2 mutations have also been detected in blue nevi (Moller et al., 2016). These mutations always occurred in tumors lacking GNAQ and GNA11 mutations, and have not yet been reported in cutaneous melanomas (Cancer Genome Atlas, 2015). To determine whether CYSLTR2 and PLCB4 mutations also occur in PLMTs, we analyzed our previously published cohort of tumors (van de Nes et al., 2016a, van de Nes et al., 2016b), using a next-generation sequencing gene panel covering the mutational hot-spots in CYSLTR2 and PLCB4 as well as other gene mutations reported in uveal melanoma (described in Supplemental Material and previously reported (Moller et al., 2016)).
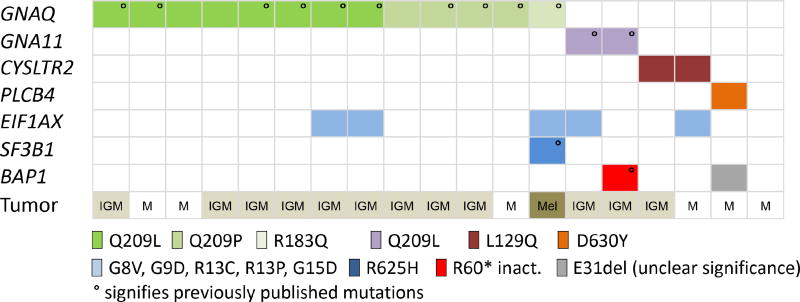
In 19 PLMTs, we found two tumors with activating L129Q (c.386T>A) mutations in CYSLTR2 and one tumor harboring an activating D630Y (c.1888G>T) mutation in PLCB4 (Figure 1, Table 1). These mutations were identified at the same hotspots previously described in uveal melanoma and predicted to be activating (Johansson et al., 2016, Moore et al., 2016). In our PLMT cohort, CYSLTR2 and PLCB4 mutations were found to be mutually exclusive of GNAQ and GNA11 mutations. Together, mutations in GNAQ, GNA11, CYSLTR2 or PLCB4 were identified in 18/19 (94.7%) PLMTs.
Figure 1. Distribution of mutations in primary leptomeningeal melanocytic tumors.
Distribution of mutations identified in primary leptomeningeal melanocytic tumors (PLMT). The histological diagnosis is demonstrated in the lowest row (M = melanocytoma, IGM = intermediate grade melanocytoma, Mel = primary leptomeningeal melanoma). ° signifies previously pu blished mutations (van de Nes et al., 2016a, van de Nes et al., 2016b)
Table 1.
Clinicopathological and mutation information of primary leptomeningeal melanocytic tumors
| Nr. | Age | Sex | Location | Diag. | Sample | GNAQ | GNA11 | PLCB4 | CYSLTR2 | EIF1AX | SF3B1 | BAP1 | Status | FU |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 46 | f | cavernous sinus | M | P | - | - | - | - | - | - | - | NK | NK |
| 2 | 48 | f | spinal | M | P | Q209L° | - | - | - | - | - | - | NK | NK |
| 3 | 41 | f | frontal | M | P | Q209P° | - | - | - | - | - | - | NK | NK |
| 4 | 73 | m | spinal C2-3 | M | P | Q209L | - | - | - | - | - | - | NK | NK |
| 5 | 73 | f | temporal right | M | P | - | - | D630Y | - | - | - | E31del | NK | NK |
| 6 | 77 | f | spinal T7-8 | M | P | - | - | - | L129Q | R13C | - | - | NK | NK |
| 7 | 34 | m | spinal T10-12 | IGM | P | Q209P | - | - | - | - | - | - | A | 7y |
| 8 | 44 | m | spinal C1 | IGM | P | Q209L° | - | - | - | - | - | - | A | 5y |
| 9 | 79 | m | spinal T4-5 | IGM | R | Q209L° | - | - | - | - | - | - | NK | NK |
| 10 | 35 | f | spinal C5-6 | IGM | P | Q209P° | - | - | - | - | - | - | A | 8y |
| 11 | 65 | m | spinal C3 | IGM | P | Q209L° | - | - | - | G8V | - | - | A | 3y |
| 12 | 69 | f | spinal T8 | IGM | P | Q209L° | - | - | - | G9D | - | - | NK | NK |
| 13 | 64 | m | spinal C5 | IGM | P | Q209L° | - | - | - | - | - | - | NK | NK |
| 14 | NK | NK | NK | IGM | P | Q209L | - | - | - | - | - | - | NK | NK |
| 15 | 20 | m | frontal | IGM | P | Q209P° | - | - | - | - | - | - | A, R# | 5y |
| 16 | 67 | m | spinal T9-10 | IGM | P | - | - | - | L129Q | - | - | - | NK | NK |
| 17 | 48 | f | spinal T8-9 | IGM | R | - | Q209L° | - | - | R13P | - | - | A | 6y |
| 18 | 78 | f | tentorium | IGM | P | - | Q209L° | - | - | - | - | R60*° | R§ | 1y |
| 19 | 66 | m | brain | Mel | P | R183Q° | - | - | - | G15D | R625H° | - | D | 1y |
green = activating mutations; blue = mutations expected to alter protein function; red = loss of function mutation; grey = mutation of unclear functional significances; Nr. = number; Diag. = diagnosis; m = male; f = female; NK = not known; P = primary; R = recurrence; M = melanocytoma; IGM = intermediate grade melanocytoma; Mel = melanoma; del. = deletion; FU = follow-up period; NK = not known; A = alive; R# = recurred after 3 years (mutation profile was identical to the primary); R§ = recurred, then lost to follow-up; D = dead (disease related)
signifies mutations previously published (van de Nes et al., 2016a, van de Nes et al., 2016b)
The hotspot mutations in GNAQ and GNA11 (R183 and Q209), PLCB4 (D630) and CYSLTR2 (L129) result in activation of the corresponding proteins and increased stimulation of a common downstream signaling pathway (Johansson et al., 2016, Moore et al., 2016, Van Raamsdonk et al., 2009, Van Raamsdonk et al., 2010). CYSLTR2 codes for the CYSLTR2 receptor, a seven transmembrane receptor which signals through the products of the GNAQ and GNA11 genes, the highly homologous heterotrimeric G proteins Gαq and Gα11. Gαq and Gα11 directly activate the PLCB4 gene product, phospholipase-C β4, which hydrolyses PIP2 (phosphatidylinositol 4,5-bisphosphate) releasing DAG (diacylglyerol) and IP3 (inositol-1,4,5-trisphosphate), subsequently releasing Ca+ (Suh et al., 2008, Waldo et al., 2010). The similar functional consequences of these gene mutations is demonstrated by the occurrence of these mutations in a mutually exclusive fashion.
The increased coverage and sensitivity of our sequencing assay identified multiple additional mutations in other genes, including EIF1AX (Figure 1), as was recently reported (Kusters-Vandevelde et al., 2016). One EIF1AX mutation (R13C, c.37C>T) was identified in a tumor that also harbored an activating CYSLTR2 L129Q mutation (Supplemental Figure 1). In addition to a known SF3B1 R625H (c.1874G>A) and inactivating BAP1 R60* (c.178C>T) mutation (van de Nes et al., 2016a, van de Nes et al., 2016b), our targeted next-generation sequencing approach detected a BAP1 E31del (c.91_93delGAG) mutation in the PLMT sample also harboring a PLCB4 D630Y mutation (Supplemental Figure 1). The functional relevance of this BAP1 in-frame one amino acid deletion is unclear. BAP1 immunohistochemistry showed retained nuclear protein expression (Supplemental Figure 2). However, copy number analysis (Supplemental Figure 3) demonstrated loss of chromosome 3 including the other wild-type BAP1 allele which means the mutation would be highly relevant if it resulted in loss or impairment of protein function. The relevance of EIF1AX, SF3B1 and BAP1 mutations in terms of clinical behavior and prognosis needs to be fully elucidated in future larger studies.
In summary, our study demonstrates the occurrence of activating CYSLTR2 or PLCB4 mutations in PLMTs lacking GNAQ and GNA11 mutations. Activating mutations in these genes are exceedingly rare in other melanomas, with the exception of uveal melanomas. The diagnosis of a PLMT requires exclusion of a more frequently occurring CNS metastasis of a non-CNS melanoma (most commonly of cutaneous origin). Presence of a PLCB4, CYSLTR2, GNAQ or GNA11 mutation is strong evidence in favor of a PLMT, but a rare CNS uveal melanoma metastasis should also be considered.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brat D, Perry A, Wesseling P, Bastian B. Melanocytic tumours. In: Louis D, Ohgaki H, Wiestler O, Cavanee W, editors. WHO Classification of Tumours of the Central Nervous System. Lyon: IARC Press; 2016. pp. 266–70. [Google Scholar]
- Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessi M, van de Nes J, Griewank K, Barresi V, Buckland ME, Kirfel J, et al. Absence of TERT promoter mutations in primary melanocytic tumours of the central nervous system. Neuropathology and applied neurobiology. 2014;40(6):794–7. doi: 10.1111/nan.12138. [DOI] [PubMed] [Google Scholar]
- Johansson P, Aoude LG, Wadt K, Glasson WJ, Warrier SK, Hewitt AW, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7(4):4624–31. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelsche C, Hovestadt V, Jones DT, Capper D, Sturm D, Sahm F, et al. Melanotic Tumors of the Nervous System are Characterized by Distinct Mutational, Chromosomal and Epigenomic Profiles. Brain pathology. 2015;25(2):202–8. doi: 10.1111/bpa.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters-Vandevelde HV, Creytens D, van Engen-van Grunsven AC, Jeunink M, Winnepenninckx V, Groenen PJ, et al. SF3B1 and EIF1AX mutations occur in primary leptomeningeal melanocytic neoplasms yet another similarity to uveal melanomas. Acta neuropathologica communications. 2016;4:5. doi: 10.1186/s40478-016-0272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters-Vandevelde HV, Klaasen A, Kusters B, Groenen PJ, van Engen-van Grunsven IA, van Dijk MR, et al. Activating mutations of the GNAQ gene: a frequent event in primary melanocytic neoplasms of the central nervous system. Acta neuropathologica. 2010a;119(3):317–23. doi: 10.1007/s00401-009-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters-Vandevelde HV, van Engen-van Grunsven IA, Coupland SE, Lake SL, Rijntjes J, Pfundt R, et al. Mutations in g protein encoding genes and chromosomal alterations in primary leptomeningeal melanocytic neoplasms. Pathology oncology research : POR. 2015;21(2):439–47. doi: 10.1007/s12253-014-9841-3. [DOI] [PubMed] [Google Scholar]
- Kusters-Vandevelde HV, van Engen-van Grunsven IA, Kusters B, van Dijk MR, Groenen PJ, Wesseling P, et al. Improved discrimination of melanotic schwannoma from melanocytic lesions by combined morphological and GNAQ mutational analysis. Acta neuropathologica. 2010b;120(6):755–64. doi: 10.1007/s00401-010-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller I, Murali R, Muller H, Wiesner T, Jackett LA, Scholz SL, et al. Activating cysteinyl leukotriene receptor 2 (CYSLTR2) mutations in blue nevi. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2016 doi: 10.1038/modpathol.2016.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AR, Ceraudo E, Sher JJ, Guan Y, Shoushtari AN, Chang MT, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nature genetics. 2016;48(6):675–80. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali R, Wiesner T, Rosenblum MK, Bastian BC. GNAQ and GNA11 mutations in melanocytomas of the central nervous system. Acta neuropathologica. 2012;123(3):457–9. doi: 10.1007/s00401-012-0948-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh PG, Park JI, Manzoli L, Cocco L, Peak JC, Katan M, et al. Multiple roles of phosphoinositide- specific phospholipase C isozymes. BMB reports. 2008;41(6):415–34. doi: 10.5483/bmbrep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- van de Nes J, Gessi M, Sucker A, Moller I, Stiller M, Horn S, et al. Targeted next generation sequencing reveals unique mutation profile of primary melanocytic tumors of the central nervous system. Journal of neuro-oncology. 2016a;127(3):435–44. doi: 10.1007/s11060-015-2052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Nes J, Wrede K, Ringelstein A, Stiller M, Horn S, Sucker A, et al. Diagnosing a Primary Leptomeningeal Melanoma by Gene Mutation Signature. The Journal of investigative dermatology. 2016b;136(7):1526–8. doi: 10.1016/j.jid.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457(7229):599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. The New England journal of medicine. 2010;363(23):2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo GL, Ricks TK, Hicks SN, Cheever ML, Kawano T, Tsuboi K, et al. Kinetic scaffolding mediated by a phospholipase C-beta and Gq signaling complex. Science. 2010;330(6006):974–80. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.