Abstract
Background:
Mutated circulating cell-free DNA (cfDNA) has been suggested as a surrogate marker of tumour burden and aggressiveness of disease. We examined the association between the level of plasma mutant cfDNA and metabolic tumour burden (MTB) measured by 18F-fluoro-D-glucose positron emission tomography/computed tomography (18F-FDG PET/CT). Furthermore, the presence of mutant cfDNA was correlated with patient survival.
Methods:
Forty-six advanced non-small cell lung cancer (NSCLC) patients were included. At the time of inclusion, blood sampling and a PET/CT scan were performed. cfDNA was isolated and next-generation sequencing (NGS) was performed (Ion AmpliSeq Colon and Lung Cancer panel v2). MTB was defined by a volumetric PET parameter.
Results:
NGS succeeded in 41 patients. Mutations were detected in the blood of 24 patients. A significant correlation between the allele frequency of the most frequent mutation and MTB was found (P=0.001). Patients with detectable mutated cfDNA had a significantly shorter median overall survival compared with patients without (3.7 versus 10.6 months, P=0.019). This impact on survival was independent of the MTB.
Conclusions:
Level of mutated cfDNA tends to correlate with MTB in advanced-stage NSCLC patients. Patients with detectable mutant DNA in plasma had an inferior survival, indicating that this could be an important predictor of survival.
Keywords: circulating tumour DNA, circulating cell-free DNA, tumour volume, positron emission tomography, tumour lesion glycolysis, non-small cell lung cancer, next-generation sequencing, survival
Circulating cell-free DNA (cfDNA) is small DNA fragments circulating in the blood stream (Jahr et al, 2001). Elevated levels of cfDNA have been identified in cancer patients compared with healthy individuals (Leon et al, 1977). This elevation is believed to origin from both malignant tumour cells as well as from non-malignant cells constituting the tumour microenviroment (Mouliere and Thierry, 2012; Thierry et al, 2016). The precise mechanism for release of the DNA into the blood stream is not fully elucidated. It has been demonstrated that the amount of DNA in the circulation increases when cells are undergoing apoptosis or necrosis (Jahr et al, 2001). Other alternative mechanisms such as active secretion from tumour cells (Schwarzenbach et al, 2011) and release from circulating tumour cells (Bettegowda et al, 2014) have been suggested and may contribute to the total cfDNA level. In the recent years, it has been revealed that the contribution of cfDNA origin from tumour cells can be identified in the total cfDNA level by measuring cancer-specific mutations in the cfDNA also known to be present in the original tumour (Wan et al, 2017).
It has been proposed that the level of mutated cfDNA correlates with tumour burden. The level of tumour necrosis is thought to increase when tumour cells are rapidly proliferating as enlarging tumours are more likely to outgrow their blood supply (Leek et al, 1999). This causes tumour areas that are poorly vascularised and ischaemic, and if this ischaemia is sustained, ultimately necrosis will develop. As these necrotic areas are believed to release tumour DNA into the circulation, the amount of mutated cfDNA could be related to tumour burden (Diehl et al, 2005). Previous studies have supported this hypothesis by showing increased mutated cfDNA levels in advanced-stage patients compared with early-stage patients (Fleischhacker and Schmidt, 2007; Bettegowda et al, 2014; Newman et al, 2014). Moreover, correlations have been identified between changes in the amount of mutated cfDNA and changes in treatment response (Dawson et al, 2013; Newman et al, 2014; Sorensen et al, 2014) as well as relapse of disease (Tie et al, 2015). In addition to tumour volume, tumour aggressiveness may also be an important factor affecting the level of mutated cfDNA. An association between increasing necrosis grade and higher tumour grade has been identified, indicating that the amount of mutated cfDNA is associated with an aggressive tumour phenotype (Leek et al, 1999).
18F-fluoro-D-glucose (18F-FDG) positron emission tomography/computed tomography (PET/CT) is a well-established nuclear imaging modality for use in cancer patients. 18F-FDG is a radiolabelled glucose analogue accumulating in tissue with high glucose utilisation. FDG activity thereby reflects the metabolic activity and aggressiveness of a tumour. In addition, data on tumour volume can be assessed on an 18F-FDG-PET/CT scan. New volume-based PET parameters, as tumour lesion glycolysis (TLG), have been introduced capable of measuring the entire metabolic tumour burden (MTB) by incorporating both metabolic activity and volumetric data on all tumours in the patient (Larson et al, 1999). Thus TLG assessment on an 18F-FDG-PET/CT scan reflects both tumour burden and, to some extent, tumour aggressiveness of the disease and could be a more informative parameter for assessing the total active tumour burden in patients than tumour burden assessment on a CT scan.
Identification of oncogenic alterations in cfDNA is challenging due to the low concentration of target cfDNA in plasma. With the emergence of next-generation sequencing (NGS), it has become possible to assess a large amount of different mutations from a low input of DNA. NGS-based targeted panels directed against the most frequent alterations in cancer have been designed. In non-small cell lung cancer (NSCLC), a 22-gene NGS panel has been developed, which includes the most frequent mutations in NSCLC (Ion AmpliSeq Colon and Lung Cancer Panel v2). This panel has proven useful for mutation detection in plasma cfDNA (Frenel et al, 2015; Zonta et al, 2016).
In the present study, we evaluated the correlation between the amount of mutated cfDNA in plasma, and the MTB was measured by TLG in advanced-stage NSCLC patients. Moreover, we evaluated whether the presence of mutated cfDNA was correlated with patient survival.
Materials and methods
Patients
In this retrospective study, advanced-stage NSCLC patients prospectively enrolled in a single-centre study from April 2013 to August 2015 at the Department of Oncology, Aarhus University Hospital, Denmark were evaluated. Details on inclusion and exclusion criteria have been described previously (Winther-Larsen et al, 2016). At the time of inclusion, patients underwent an 18F-FDG-PET/CT scan and a blood sample was drawn. All patients were treated with erlotinib in a palliative setting. Testing for somatic epidermal growth factor receptor (EGFR) mutations had been performed on the diagnostic tumour biopsy in all adenocarcinoma patients as part of the routine diagnostic work-up (see Supplementary S1). For the purpose of this study, we included only patients from the total cohort who were EGFR mutation wild type (wt), had progression on first or second line of chemotherapy, and in whom both an 18F-FDG-PET/CT scan and a blood sample were available and evaluable. Selection of patients is illustrated in Supplementary S2. All patients gave informed written consent before inclusion and the study was approved by the Central Denmark Region Committees on Biomedical Research Ethics (no. 1-10-72-19-12). The study was reported to ClinicalTrial.gov (NCT02043002).
18F-fluoro-D-glucose positron emission tomography/computed tomography
All 18F-FDG-PET/CT scans were performed on a combined PET/CT scanner (Siemens Biograph TruePoint 40, Siemens Healthcare GMbH, Erlangen, Germany) at the Department of Nuclear Medicine and PET-Centre, Aarhus University Hospital, Denmark. The imaging protocol is described in Supplementary S1. Same scanner model, protocol for acquisition, and reconstruction software was used for all patients. An experienced nuclear medicine physician blinded to the patient outcome analysed all PET/CT scans using the Siemens Syngo.via software, Siemens Healthcare GMbH. In all scans, TLG was calculated for all evaluable lesions according to the Positron Emission Tomography Response Criteria in Solid Tumours 1.0 guideline (Wahl et al, 2009) (described in Supplementary S1 and illustrated in Supplementary S3). The sum of TLG for all evaluable lesions was defined as the MTB.
DNA extraction from blood
A peripheral blood sample of 10 ml was collected in an EDTA-containing tube for each patient. Within 2 h of withdrawal, samples were centrifuged (1400 g for 15 min) and plasma was isolated from the peripheral blood cells. Plasma was subsequently frozen at −80 °C until further analysis. cfDNA was purified from 2 ml of plasma by use of the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol and eluted in a volume of 100 μl TE buffer. The amount of cfDNA was quantified by measuring the beta-2-microglobulin gene by droplet digital PCR (Pallisgaard et al, 2015) as described in Supplementary S1.
Next-generation sequencing
NGS was carried out on all cfDNA samples. The Oncomine Solid Tumor DNA Kit was used to prepare libraries on 1.1–10 ng of cfDNA following the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). NGS was conducted using the Ion Chef Instrument and the Ion Personal Genome Machine (PGM) System (Thermo Fisher Scientific). Sequencing was performed using Ion 316 v2 BC chips containing eight cfDNA samples per chip. Primary data processing was carried out using the Torrent Suite Software (version 5.0.4) on a Torrent Server and the Ion AmpliSeq Colon and Lung Cancer panel v2 template (all Thermo Fisher Scientific). Variant calling was performed using the Ion Reporter Software (version 5.0) and the AmpliSeq CHPv2 peripheral/CTC/CF DNA single sample workflow (Thermo Fisher Scientific). Default settings were used except from the reference and hotspot BED files, which were supplied in the kit. Sequencing was considered successful if the mean sequencing depth was ⩾2000. If this criterion could not be met, the sample was disqualified. Called variants were only accepted if the allele frequency (AF) was ⩾1%. The Integrative Genomics Viewer (Broad Institute, Cambridge, MA, USA) was used for visualisation of variants (Thorvaldsdóttir et al, 2013).
Statistical analysis
Correlations between detection of mutated cfDNA and clinical characteristics were calculated using χ2 or Fisher’s exact tests, where appropriate. Kruskal–Wallis test was used to calculate the correlation between the presence of mutated cfDNA and the levels of TLG and total cfDNA. Correlation between TLG and concentration of mutated cfDNA was evaluated using the Pearson’s correlation coefficient after log-transformation of both variables. Follow-up time was calculated using the reverse Kaplan–Meier method. Overall survival (OS) was measured from the start of enrolment until death of any cause or last follow-up date (19 April 2016). If patients were still alive on the last follow-up date, they were censored at that day. Estimates of median OS were calculated by Kaplan–Meier method and compared by the log-rank test. The Cox proportional hazards model was used to calculate crude and adjusted hazard ratios (HRs). Clinical variables were dichotomised except for age (continuous). All tests were two-sided, and P-values <0.05 were considered to be statistically significant. Statistical analyses were performed using SPSS statistics version 20.0 for windows (IBM SPSS Statistics, Chicago, IL, USA). STATA version 13 (Stata Corporation, College Station, TX, USA) was used for preparation of Kaplan–Meier curve and GraphPad Prism 7.0 (GraphPad Software Inc., San Diego, CA, USA) for preparation of scatter plot.
Results
Patients
A total of 46 patients were analysed. Patient characteristics are shown in Table 1. All patients had stage IV disease except for three patients with stage III disease. The majority of patients had progression on first-line chemotherapy. No patients were lost to follow-up. The median time interval between blood sampling and PET/CT scanning was 2 days (range: 0–9 days). After a median follow-up time of 20.9 months (95% CI: 11.2–30.6 month), five patients were still alive. The median OS in all patients was 5.9 months (95% CI: 2.9–8.9 months).
Table 1. Patient and tumour characteristics (N=46).
| Characteristics | N (%) |
|---|---|
|
Age | |
| Median years (range) | 67 (48–81) |
|
Gender | |
| Female | 20 (43) |
| Male | 26 (57) |
|
Performance status, ECOG | |
| 0–1 | 38 (83) |
| 2 | 8 (17) |
|
Smoking status | |
| Never or formera | 33 (72) |
| Current | 12 (26) |
| Unknown | 1 (2) |
|
Stage | |
| III | 3 (7) |
| IV | 43 (93) |
|
Histology | |
| Adenocarcinoma | 38 (83) |
| Squamous cell carcinoma | 8 (17) |
|
Progression on | |
| First-line chemotherapy | 37 (80) |
| Second-line chemotherapy | 9 (20) |
|
Prior treatment | |
| First line | |
| Carboplatin/vinorelbineb | 26 (56) |
| Carboplatin/vinorelbine/bevacizumabc | 20 (44) |
| Second line | |
| Pemetrexed | 5 (56) |
| Docetaxel | 4 (44) |
Abbreviation: ECOG=Eastern Cooperative Oncology Group.
Former smoker was defined as having stopped smoking at the time of diagnosis.
Carboplatin day 1 (AUC 5) and vinorelbine day 1 and day 8 (60–80 mg m−2 (p.o.)) every 3 weeks for a maximum of four cycles.
Bevacizumab (7.5 mg m−2 i.v. day 1) was given in combination with chemotherapy. Patients with disease control received subsequent maintenance therapy every 3 weeks until progression or toxicity.
Correlation between the level of mutated cfDNA and MTB
cfDNA isolation was successful in all patients and a median value of 4587 copies ml−1 (range: 297–170 720 copies ml−1) was measured. Sequencing of cfDNA succeeded in 41 out of 46 patients (89%). A least one mutation was detected in 24 patients (59%). The number of identified mutations in each patient ranged from 1 to 4 with a median of 1 mutation. In total, 43 mutations were identified. The most frequently mutated genes were: TP53 (18 patients), KRAS (12 patients), and EGFR (2 patients; Table 2). The median AF of the most frequent mutation in each patient was 2.7% (range: 1.1–62.5%).
Table 2. Mutations identified in circulating cell-free DNA by use of next-generation sequencing (N=24).
| ID | Gene | Coding | Protein | Coverage | Mutation allele coverage | Allele frequency (%) | COSMIC |
|---|---|---|---|---|---|---|---|
| 4 | TP53 | c.833C>G | p.Pro278Arg | 1536 | 27 | 1.76 | COSM10887 |
| 6 | KRAS | c.34G>A | p.Gly12Ser | 3201 | 40 | 1.25 | COSM517 |
| 9 | TP53 | c.714_715insT | p.Asn239Ter | 5734 | 339 | 5.91 | COSM45870 |
| KRAS | c.34G>A | p.Gly12Ser | 3387 | 86 | 2.54 | COSM517 | |
| 10 | TP53 | c.313G>T | p.Gly105Cys | 2800 | 32 | 1.14 | COSM44481 |
| 12 | KRAS | c.34G>T | p.Gly12Cys | 3084 | 66 | 2.14 | COSM516 |
| 14 | SMAD4 | c.1051G>A | p.Asp351Asn | 4554 | 612 | 13.44 | COSM32910 |
| TP53 | c.313G>T | p.Gly105Cys | 2750 | 343 | 12.47 | COSM44481 | |
| KRAS | c.34G>A | p.Gly12Ser | 3003 | 153 | 5.09 | COSM517 | |
| 15 | STK11 | c.766G>T | p.Glu256Ter | 6164 | 168 | 2.73 | COSM5731897 |
| TP53 | c.799C>T | p.Arg267Trp | 3961 | 82 | 2.07 | COSM11183 | |
| 21 | KRAS | c.34G>A | p.Gly12Ser | 8580 | 1913 | 22.30 | COSM517 |
| KRAS | c.35G>A | p.Gly12Asp | 6666 | 426 | 6.39 | COSM521 | |
| TP53 | c.478A>G | p.Met160Val | 5363 | 105 | 1.96 | COSM44328 | |
| TP53 | c.491A>C | p.Lys164Thr | 5420 | 95 | 1.75 | COSM44387 | |
| 23 | KRAS | c.34_35delGGinsTT | p.Gly12Phe | 3432 | 45 | 1.31 | COSM512 |
| 26 | MET | c.3029C>T | p.Thr1010Ile | 5643 | 1994 | 35.34 | COSM707 |
| TP53 | c.578A>G | p.His193Arg | 2674 | 662 | 24.76 | COSM10742 | |
| 29 | TP53 | c.641A>G | p.His214Arg | 4574 | 1214 | 26.54 | COSM43687 |
| EGFR | c.2235_2249delGGAATTAAGAGAAGC | p.Glu746_Ala750del | 4149 | 140 | 3.37 | COSM6223 | |
| EGFR | c.2240T>C | p.Leu747Ser | 4178 | 60 | 1.44 | COSM26704 | |
| 30 | TP53 | c.730G>T | p.Gly244Cys | 5693 | 393 | 6.90 | COSM11524 |
| 34 | KRAS | c.34G>T | p.Gly12Cys | 1517 | 51 | 3.36 | COSM516 |
| TP53 | c.716A>G | p.Asn239Ser | 4539 | 112 | 2.47 | COSM44094 | |
| ERBB4 | c.512G>T | p.Trp171Leu | 2605 | 61 | 2.34 | COSM5369807 | |
| 35 | TP53 | c.711G>A | p.Met237Ile | 4353 | 60 | 1.38 | COSM10834 |
| 36 | TP53 | c.404G>A | p.Cys135Tyr | 2745 | 1716 | 62.51 | COSM10801 |
| PIK3CA | c.1624G>C | p.Glu542Gln | 9973 | 2250 | 22.56 | COSM17442 | |
| 37 | TP53 | c.743G>A | p.Arg248Gln | 8221 | 129 | 1.57 | COSM10662 |
| 38 | KRAS | c.34G>T | p.Gly12Cys | 3081 | 40 | 1.30 | COSM516 |
| 40 | TP53 | c.742C>T | p.Arg248Trp | 7006 | 256 | 3.65 | COSM10656 |
| KRAS | c.34G>A | p.Gly12Ser | 3743 | 71 | 1.90 | COSM517 | |
| TP53 | c.578A>G | p.His193Arg | 4570 | 60 | 1.31 | COSM10742 | |
| TP53 | c.830G>T | p.Cys277Phe | 3210 | 37 | 1.15 | COSM10749 | |
| 44 | TP53 | c.469G>T | p.Val157Phe | 3714 | 48 | 1.29 | COSM10670 |
| 45 | KRAS | c.34G>T | p.Gly12Cys | 2020 | 100 | 4.95 | COSM516 |
| TP53 | c.747G>T | p.Arg249Ser | 6103 | 121 | 1.98 | COSM10817 | |
| 49 | EGFR | c.2234_2245delAGGAATTAAGAG | p.Glu746_Ala750del | 6360 | 91 | 1.43 | COSM6223 |
| 51 | KRAS | c.34G>T | p.Gly12Cys | 4237 | 46 | 1.09 | COSM516 |
| 54 | KRAS | c.34G>A | p.Gly12Ser | 3214 | 86 | 2.68 | COSM517 |
| TP53 | c.488A>G | p.Tyr163Cys | 2946 | 50 | 1.70 | COSM10808 | |
| KRAS | c.47A>G | p.Lys16Arg | 3208 | 47 | 1.47 | COSM1163765 | |
| 64 | TP53 | c.844C>G | p.Arg282Gly | 1814 | 53 | 2.92 | COSM10992 |
Abbreviation: COSMIC=Catalogue of Somatic Mutations in Cancer.
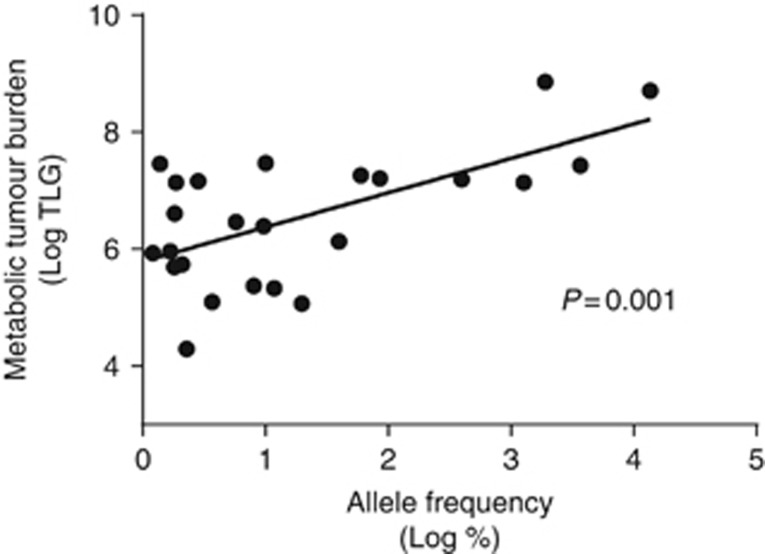
TLG was assessable in all FDG-PET/CT. A significant correlation between the AF of the most frequent mutation (%) and the level of TLG was found as shown in Figure 1 (r=0.628, P=0.001). By contrast, no significant correlation could be identified between MTB and the number of mutations identified in each patient (data not shown).
Figure 1.
Scatter plot showing the correlation between the allele frequency of the most frequent mutation and the level of TLG in patients with circulating mutated cell-free DNA present in plasma (N=24). The P-value was calculated using the Pearson’s correlation coefficient.
Evaluation of patients with and without detectable mutated cfDNA
No mutations could be identified in the cfDNA of 17 patients. A comparison of clinical and tumour characteristics between the mutated cfDNA-positive and the mutated cfDNA-negative group was performed, and no significant differences were identified (Supplementary S4). In addition, no significant difference in the level of total cfDNA was found between the mutated cfDNA-positive (5170 copies ml−1, range 1518–170 720) and the mutated cfDNA-negative (4510 copies ml−1, range 1232–39 160) patients (χ2 (2)=0.018, P=0.895). However, a significant higher median MTB was found in the mutated cfDNA-positive patients (689 (range 73–7034)) compared with the mutated cfDNA-negative patients (269 (range 67–3958)) (P=0.047).
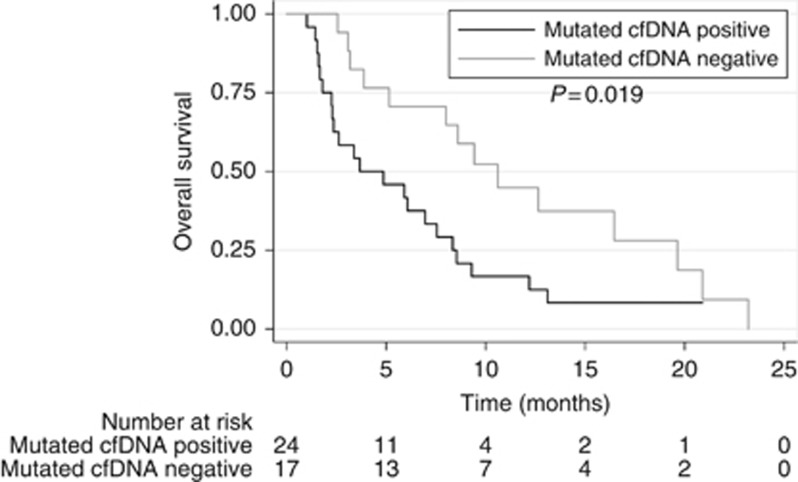
When evaluating OS, patients with mutated cfDNA presence in the blood had a significant shorter median OS (3.7 months (95% CI: 0.0–7.6)) compared with mutated cfDNA-negative patients (10.6 months (95% CI: 7.1–14.1), P=0.019; Figure 2). In a univariate Cox regression analysis, the presence of mutated cfDNA together with high level of MTB was found to be associated with decreased OS (Table 3). To evaluate the independent impact of the presence of mutated cfDNA on survival, a multivariate Cox regression analysis was performed. Being mutated cfDNA positive remained an independent predictor of shorter OS with an adjusted HR of 2.11 (95% CI: 1.02–4.38) (Table 3).
Figure 2.
Kaplan–Meier curve for overall survival (OS) according to the presence or absence of circulating mutated cell-free DNA (cfDNA) in plasma. A median OS of 3.7 months (95% confidence interval (CI): 0–7.6 months) was found in patients with detectable mutated cfDNA, while a median OS of 12.6 months (95% CI: 8.4–16.9 months) was found in patients without detectable mutated cfDNA. The difference between the groups was calculated using the log-rank test.
Table 3. Univariate and multivariate Cox regression analyses of overall survival (N=46).
| Variables | HR (95% CI) | P-value | Adjusted HR (95% CI) | P-value |
|---|---|---|---|---|
|
Age | ||||
| Continuous | 0.99 (0.95–1.04) | 0.690 | ||
|
Gender | ||||
| Female | 0.84 (0.41–1.74) | 0.842 | ||
| Male | 1.00 | |||
|
Histology | ||||
| Adenocarcinoma | 0.36 (0.12–1.10) | 0.073 | ||
| Squamous cell | 1.00 | |||
|
Smoking | ||||
| Never or former | 1.37 (0.68–3.01) | 0.400 | ||
| Current | 1.00 | |||
|
Performance status, ECOG | ||||
| 0–1 | 0.60 (0.20–1.74) | 0.344 | ||
| 2 | 1.00 | |||
|
Stage | ||||
| IV | 0.51 (0.15–1.76) | 0.287 | ||
| III | 1.00 | |||
|
Erlotinib treatment | ||||
| Second line | 0.69 (0.27–1.75) | 0.432 | ||
| Third line | 1.00 | |||
|
Presence of mutated cfDNA | ||||
| Mutated cfDNA positive | 2.26 (1.13–4.55) | 0.022 | 2.11 (1.02–4.38) | 0.044 |
| Mutated cfDNA negative | 1.00 | 1.00 | ||
|
TLGa | ||||
| >420 | 2.80 (1.41–5.55) | 0.003 | 3.08 (1.46–6.47) | 0.003 |
| ⩽420 | 1.00 | 1.00 | ||
Abbreviations: cfDNA=cell-free DNA; CI=confidence interval; ECOG=Eastern Cooperative Oncology Group; HR=hazard ratio; TLG=tumour lesion glycolysis.
Divided by the median value.
No difference in OS was found between mutated cfDNA-positive patients, with only one mutation identified compared with a higher number of mutations (P=0.355). Neither was any difference in survival found between mutated cfDNA-positive patients with an AF above or below the median 2.7% (P=0.949).
Discussion
Our study shows a significant correlation between the frequency of mutated cfDNA and the MTB measured by PET/CT in advanced-stage NSCLC patients. The study thereby supports the hypothesis of mutated cfDNA as a surrogate marker of tumour burden and tumour aggressiveness. In addition, we observed that mutated cfDNA-positive patients had a higher median MTB than patients without mutated cfDNA present in plasma and could confirm from previous reports showing that mutated cfDNA positivity is correlated to an inferior survival.
To our knowledge, only one previous study has, similar to ours, compared the exact level of mutated cfDNA with radiological tumour burden in NSCLC patients. Newman et al (2014) evaluated the correlation between amount of mutated cfDNA and tumour volume, defined as visible tumour on a CT scan, in nine newly diagnosed or recurrent NSCLC patients with different stages (I–IV). In line with ours, they found a significant correlation. In a study by Pécuchet et al (2016), the concentration of mutated cfDNA found in 75 advanced-stage NSCLC patients was divided in tertiles and correlated to the tumour burden defined as the sum of the RECIST target lesions. A significant higher median tumour burden was found in the groups with high and intermediate concentration of mutated cfDNA compared with the low group. In addition, there was also a significantly higher median tumour burden when comparing the group with high concentration of mutated cfDNA to the group with intermediate concentration. Both of these studies used a CT-defined tumour volume and no measurement of tumour aggressiveness was therefore incorporated. In contrast, we measured the MTB by use of the PET parameter TLG. This parameter not only measures the entire tumour burden by incorporating all measurable tumours in the patient (and not only RECIST target lesions) but also integrates the metabolic activity of each tumour. Thereby, the parameter becomes a measurement of both tumour burden and aggressiveness of the tumour. MTB has previously been used for assessment of tumour burden in a study evaluating the correlation between tumour burden and total amount of total cfDNA in 53 advanced NSCLC patients (Nygaard et al, 2014). No correlation could be found in this study indicating that total cfDNA is a more unspecific marker of tumour burden than mutated cfDNA. The majority of cfDNA originates from non-malignant tissue. Hence, the total amount of cfDNA not simply reflects tumour burden and tumour aggressiveness but rather reflect much more complicated biological mechanisms yet not fully understood.
Additionally, we found that patients with detectable mutated cfDNA in plasma had an inferior median survival compared with patients without mutated cfDNA present in plasma. A prognostic value of mutated cfDNA has previously been evaluated in several types of cancers, such as colorectal (Lecomte et al, 2002), breast (Dawson et al, 2013; Bettegowda et al, 2014), pancreatic (Hadano et al, 2016), and gastric (Gao et al, 2016). In NSCLC patients, a study of EGFR mutation-positive patients treated in the EURTAC trail found that the subgroup of L858R-positive patients with detectable mutations in plasma had a significant shorter OS as compared with L858R-positive patients with no plasma mutations, also supporting our finding (Karachaliou et al, 2015). However, the importance of mutated cfDNA positivity in EGFR mutation wt patients is less investigated. A recent study performed NGS of cfDNA and tumour samples in 105 unselected advanced NSCLC patients using the same NGS panel as used in the present study (Pécuchet et al, 2016). Mutations in cfDNA were identified in 71% of patients and a significantly reduced OS was seen in the mutated cfDNA-positive patients. Though, 43% of the patients harboured an EGFR mutation either in the tumour or in the cfDNA making the patient cohort heterogeneous. In our cohort, all patients had tumours that were EGFR wt and only 2 patients (2%) showed EGFR mutations in plasma. Owing to the homogenous nature of our cohort, we could show that mutated cfDNA positivity is a valuable tool for predicting survival in advanced EGFR mutation wt patients. The prognostic value of mutated cfDNA positivity could in theory be explained by the close correlation to tumour burden. However, we found that the impact of mutated cfDNA on survival was independent of the MTB in the multivariate analysis, indicating that mutated cfDNA positivity is an independent marker of survival.
Our study is the first of its kind and strengthened by the prospective nature of the original study allowing the PET scanning and the blood sampling to be performed in very close proximity to each other. The same PET scanner model, protocol for acquisition, and reconstruction software was used for all patients, thereby reducing the possible interindividual variability of the scans. Additionally, we had a homogeneous patient cohort receiving the same treatment and with complete clinical data in all patients. On the other hand, our study had some limitations to consider. Even though our study is the largest in the field, the number of patients is limited and our results are primarily hypothesis generating. Furthermore, we performed a targeted sequencing using a 22-gene panel with the risk that rare lung cancer mutations would not be detected. As we used the AF for quantification of mutated DNA, a possibility exist that a mutation could have been rejected (if the AF<1%) owing to a high amount of non-mutated cfDNA in the sample. Finally, we chose to define the most frequently mutation as the mutation defining the level of mutated cfDNA. This may underestimate the true level, but it would be more problematic to combine the frequency of different mutation types as it is not possible to determine whether different mutations exist in the same clone or arise from several different clones contributing their individual mutations.
In conclusion, we showed for the first time that a correlation exists between the level of mutated cfDNA and MTB in EGFR mutation wt NSCLC patients with advanced-stage disease. This indicates that mutated plasma cfDNA can be used as an indirect measure of tumour biology. Furthermore, we observed an inferior survival in patients with mutated cfDNA present in plasma compared with patients without. This correlation was independent of tumour burden and, if further validated, mutated cfDNA positivity could be a valuable tool for prediction of survival in patients with advanced NSCLC in future.
Acknowledgments
We are thankful to Birgit Westh Mortensen for laboratory help.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong S-M, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SKN, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih L-M, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang T-L, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6: 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B (2005) Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci 102: 16368–16373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker M, Schmidt B (2007) Circulating nucleic acids (CNAs) and cancer—a survey. Biochim Biophys Acta 1775: 181–232. [DOI] [PubMed] [Google Scholar]
- Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, Nava-Rodrigues D, Smith A, Leux C, Garcia-Murillas I, Ferraldeschi R, Lorente D, Mateo J, Ong M, Yap TA, Banerji U, Gasi Tandefelt D, Turner N, Attard G, de Bono JS (2015) Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 21: 4586–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang K, Xi H, Cai A, Wu X, Cui J, Li J, Qiao Z, Wei B, Chen L (2016) Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget 8: 6330–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadano N, Murakami Y, Uemura K, Hashimoto Y, Kondo N, Nakagawa N, Sueda T, Hiyama E (2016) Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer 115: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R (2001) DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 61: 1659–1665. [PubMed] [Google Scholar]
- Karachaliou N, Mayo-de las Casas C, Queralt C, de Aguirre I, Melloni B, Cardenal F, Garcia-Gomez R, Massuti B, Sánchez JM, Porta R, Ponce-Aix S, Moran T, Carcereny E, Felip E, Bover I, Insa A, Reguart N, Isla D, Vergnenegre A, de Marinis F, Gervais R, Corre R, Paz-Ares L, Morales-Espinosa D, Viteri S, Drozdowskyj A, Jordana-Ariza N, Ramirez-Serrano JL, Molina-Vila MA, Rosell R Spanish Lung Cancer Group (2015) Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol 1: 149–157. [DOI] [PubMed] [Google Scholar]
- Larson SM, Erdi Y, Akhurst T, Mazumdar M, Macapinlac HA, Finn RD, Casilla C, Fazzari M, Srivastava N, Yeung HWD, Humm JL, Guillem J, Downey R, Karpeh M, Cohen AE, Ginsberg R (1999) Tumor treatment response based on visual and quantitative changes in global tumor glycolysis using PET-FDG imaging. The visual response score and the change in total lesion glycolysis. Clin Positron Imaging 2: 159–171. [DOI] [PubMed] [Google Scholar]
- Lecomte T, Berger A, Zinzindohoué F, Micard S, Landi B, Blons H, Beaune P, Cugnenc P-H, Laurent-Puig P (2002) Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J cancer 100: 542–548. [DOI] [PubMed] [Google Scholar]
- Leek RD, Landers RJ, Harris AL, Lewis CE (1999) Necrosis correlates with high vascular density and focal macrophage infiltration in invasive carcinoma of the breast. Br J Cancer 79: 991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon SA, Shapiro B, Sklaroff DM, Yaros MJ (1977) Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 37: 646–650. [PubMed] [Google Scholar]
- Mouliere F, Thierry AR (2012) The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin Biol Ther 12(12 Suppl 1): S209–S215. [DOI] [PubMed] [Google Scholar]
- Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW, Alizadeh AA, Diehn M (2014) An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 20: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard AD, Holdgaard PC, Spindler K-LG, Pallisgaard N, Jakobsen A (2014) The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer 110: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallisgaard N, Spindler K-LG, Andersen RF, Brandslund I, Jakobsen A (2015) Controls to validate plasma samples for cell free DNA quantification. Clin Chim Acta 446: 141–146. [DOI] [PubMed] [Google Scholar]
- Pécuchet N, Zonta E, Didelot A, Combe P, Thibault C, Gibault L, Lours C, Rozenholc Y, Taly V, Laurent-Puig P, Blons H, Fabre E (2016) Base-Position error rate analysis of next-generation sequencing applied to circulating tumor DNA in non-small cell lung cancer: a prospective study. PLOS Med 13: e1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DSB, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11: 426–437. [DOI] [PubMed] [Google Scholar]
- Sorensen BS, Wu L, Wei W, Tsai J, Weber B, Nexo E, Meldgaard P (2014) Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 120: 3896–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M (2016) Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 35: 347–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, Tacey M, Wong R, Singh M, Karapetis CS, Desai J, Tran B, Strausberg RL, Diaz LA, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P (2015) Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 26: 1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl RL, Jacene H, Kasamon Y, Lodge MA (2009) From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J Nucl Med 50(Suppl 1): 122S–150S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N (2017) Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17: 223–238. [DOI] [PubMed] [Google Scholar]
- Winther-Larsen A, Fledelius J, Sorensen BS, Meldgaard P (2016) Metabolic tumor burden as marker of outcome in advanced EGFR wild-type NSCLC patients treated with erlotinib. Lung Cancer 94: 81–87. [DOI] [PubMed] [Google Scholar]
- Zonta E, Garlan F, Pécuchet N, Perez-Toralla K, Caen O, Milbury C, Didelot A, Fabre E, Blons H, Laurent-Puig P, Taly V (2016) Multiplex detection of rare mutations by picoliter droplet based digital PCR: sensitivity and specificity considerations. PLoS One 11: e0159094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.