Abstract
Background:
Evidence on height and prostate cancer risk is mixed, however, recent studies with large data sets support a possible role for its association with the risk of aggressive prostate cancer.
Methods:
We analysed data from the PRACTICAL consortium consisting of 6207 prostate cancer cases and 6016 controls and a subset of high grade cases (2480 cases). We explored height, polymorphisms in genes related to growth processes as main effects and their possible interactions.
Results:
The results suggest that height is associated with high-grade prostate cancer risk. Men with height >180 cm are at a 22% increased risk as compared to men with height <173 cm (OR 1.22, 95% CI 1.01–1.48). Genetic variants in the growth pathway gene showed an association with prostate cancer risk. The aggregate scores of the selected variants identified a significantly increased risk of overall prostate cancer and high-grade prostate cancer by 13% and 15%, respectively, in the highest score group as compared to lowest score group.
Conclusions:
There was no evidence of gene-environment interaction between height and the selected candidate SNPs.
Our findings suggest a role of height in high-grade prostate cancer. The effect of genetic variants in the genes related to growth is seen in all cases and high-grade prostate cancer. There is no interaction between these two exposures.
Keywords: height, SNPs, gene and environment interaction, prostate cancer
Prostate cancer is the second most common cancer in men worldwide. Approximately 1.1 million men were diagnosed with prostate cancer in 2012 and almost 70% of the cases occur in more developed regions (IARC, 2014). The established risk factors include are age, ethnicity, family history, and over 100 common genetic variants. There are however other risk factors with less conclusive evidence including height (Key et al, 1997; Hayes et al, 1999; Villeneuve et al, 1999; Hsing et al, 2000; Norrish et al, 2000; Stattin et al, 2000). Height is a phenotypic trait determined by a combination of genetics and environmental factors. The relationship between height and prostate cancer risk has been proposed to act through possible factors including pre-adult nutritional status, androgen and insulin-like growth factor-I (IGF-I; Giovannucci et al, 1997; Calle, 2000; Willett, 2000; Freeman et al, 2001; Emerging Risk Factors Collaboration, 2012; Travis et al, 2016).
Height per se is not a cause of cancer but it is a marker for other exposures. It has also been suggested that taller stature may indicate increased risk of a number of cancers. The most consistent evidence has been found in relation to breast cancer (Willett, 2000; Gunnell et al, 2001).
In 2008, findings from a large nested case–control study (ProtecT) and meta-analysis (58 studies) suggested a positive association of height with high-grade prostate cancer (OR: 1.23; 95% CI: 1.06–1.43; Zuccolo et al, 2008). In this article, we present results from the international collaboration, the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome consortium (PRACTCAL; http://practical.ccge.medschl.cam.ac.uk/). The aim was to explore the effects of height on prostate cancer risk. We were also interested to see if selected candidate SNPs related to height were associated with prostate cancer risk. Finally, we explored possible interactions between the selected SNPs and height.
Materials and methods
PRACTICAL consortium
The PRACTICAL consortium consists of 78 study groups around the world. The consortium was established in September 2008. The co-ordination of PRACTICAL is funded by Cancer Research UK and data have been contributed to the Collaborative Oncology Gene-environment Study (COGS), a project funded by the European Commission and 7th Framework Programme and the NIH grant. Each study with relevant data contributed an epidemiological data set and blood samples. Data on epidemiological factors for each study were provided in accordance with an assembled data dictionary. We performed quality control checks for each study before merging the data into one combined database. The majority of the samples are of European ancestry (95%). Since we investigated height as our main exposure we only analysed studies that contained subjects with European ancestry in order to minimise variation of height potentially influenced by different ethnic groups.
Blood derived DNA samples were genotyped for 211,155 SNPs on a custom Illumina array (iCOGS) in 25 074 prostate cancer cases and 24 272 controls. Details of genotyping and quality control analysis can be found in previous publication (Eeles et al, 2013).
Analysis of height exposure
During the QC process, any subjects with outlier values were checked directly with the individual study group and subsequently either corrected or excluded. Height data were available in 10 out of 15 studies that submitted data on epidemiological factors. The inclusion criterion for this particular analysis is subjects with European ancestry. The total number of prostate cancer cases and controls were 6207 cases and 6016 controls. The list of studies included in the height exposure analyses are listed in Supplementary Table 1. Meta-analysis was performed using Meta-Analyst software (Wallace et al, 2009). We performed analysis in all PCA cases and high grade cases as compared to controls. The latter is defined by Gleason grade ⩾7. Out of 6207 cases, 2480 cases are high grade cases. Meta-analysis was carried out in 9 studies as one of the studies had no controls. Height was fitted as a continuous variable and study heterogeneity was explored. We also performed analysis whereby height was categorised into quartiles using control height values to determine the ranges. Results suggest study homogeneity hence results from a fixed effect model are reported. Pooled analysis was also performed. Tests for trend were carried out to assess possible dose-response relationships. Analyses were performed using IBM SPSS Statistics version 20.0. All analyses were adjusted for age, family history of prostate cancer, and study sites. As the data were derived from various studies with differing sample sizes, the analyses were therefore adjusted for study site to avoid possible confounding effects.
SNPs analyses
We explored the effects of candidate SNPs related to growth factors on prostate cancer risk. We identified 168 candidate SNPs in IGF-I, GH-1, SHOX, FMR1, GHITM, and GHRHR genes related to human growth based on evidence from the literature and these SNPs were genotyped within a custom Illumina array (iCOGS). The full list of 168 candidate SNPs and associated relative risk estimates are shown in Supplementary Table 2. To evaluate effect sizes of these SNPs, we created a data set consisting of individual subjects whose IDs appeared in both the genotype and epidemiological data sets by matching the IDs between the two sets. We included only Caucasian subjects. This resulted in 13 123 controls and 9424 cases. PLINK software was used to explore minor allele frequency (MAF) and Hardy-Weinberg equilibrium (HWE; Purcell et al, 2007). MAF ranges were from 0.017 to 0.496. Out of 155 SNPs, 168 SNPs met HWE (P>0.05). STATA (version14) was used to obtain risk estimates and R-square (LDscore; Cheng et al, 2006). To quantify risk, the log-additive model was used by including a single variable coded as 0, 1, or 2 based additively on the number of minor alleles. Multiple logistic regression analyses were carried out to obtain the odd ratios of all 168 SNPs. Variables included in the model were age, family history of prostate cancer, study sites, principal components for European ancestry, and SNPs. Twelve SNPs showed significant associations (P-value <0.05). We then computed the R-square value for these 12 SNPs (Table 1). The results showed that these SNPs fell into 4 regions. SNPs were excluded if r2 value was >0.8 among them and we kept the most informative SNP based on association and P-value in each region. R-squared values for these 8 SNPs were less than 0.26. After this process, eight SNPs were selected for further analysis. Among these significant SNPs, only two yielded odds ratios (ORs) above 1.15.
Table 1. R-square for 8 SNPs.
| SNPs | rs11630647 | rs11831436 | rs13317803 | rs2229765 | rs2871864 | rs35767 | rs5742612 | rs6503691 |
|---|---|---|---|---|---|---|---|---|
| rs11630647 | 1 | |||||||
| rs11831436 | 0.0000 | 1 | ||||||
| rs13317803 | 0.0001 | 0.0000 | 1 | |||||
| rs2229765 | 0.0000 | 0.0001 | 0.0000 | 1 | ||||
| rs2871864 | 0.2013 | 0.0001 | 0.0000 | 0.0030 | 1 | |||
| rs35767 | 0.0000 | 0.0484 | 0.0000 | 0.0000 | 0.0001 | 1 | ||
| rs5742612 | 0.0000 | 0.2629 | 0.0000 | 0.0000 | 0.0001 | 0.2080 | 1 | |
| rs6503691 | 0.0002 | 0.0001 | 0.0001 | 0.0001 | 0.0000 | 0.0000 | 0.0001 | 1 |
Abbreviation: SNP=single-nucleotide polymorphism.
Gene and environment interaction analyses
We carried out gene and environment (GE) analyses in 6207 cases and 6016 controls. These are subjects with data on genotype and height. We applied two type of analyses based on the effect sizes of the SNP analyses.
For the 8 SNPs that were significantly associated with prostate cancer risk, individual standardised genetic score was computed. First, we multiplied coefficient for each SNP derived from multiple logistic regression (as explained above) with individual risk allele of that particular SNPs. To obtain total genetic risk score, we summed results from each SNP. To compute standardised score, the total score was divided with s.d. value from control group. First, genetic risk scores were analysed as for main effect by comparing subjects in the second and third tertile to the referent category. For GE analysis, both height and genetic risk score were then compared as binary variables. We classified both variables into tertiles with lowest tertile as reference group and highest tertiles as exposed group. We applied empirical-Bayes (EB) method proposed by Mukherjee et al (Mukherjee et al, 2008). Results for all PCA and high grade cases are presented.
We also employed the general multifactor dimensionality reduction (GMDR) method (Chen et al, 2011). For this we included the top 2 SNPs with effect sizes >1.15 and fitted these into the model at the same time. This procedure is not possible in the conventional GE methods. Height was fitted as a binary variable. We included subjects with height in the reference (lowest tertile) and top third tertile. Analyses were carried out for all PCA and high grade cases. Age and family history of PCA were fitted as covariates.
Results
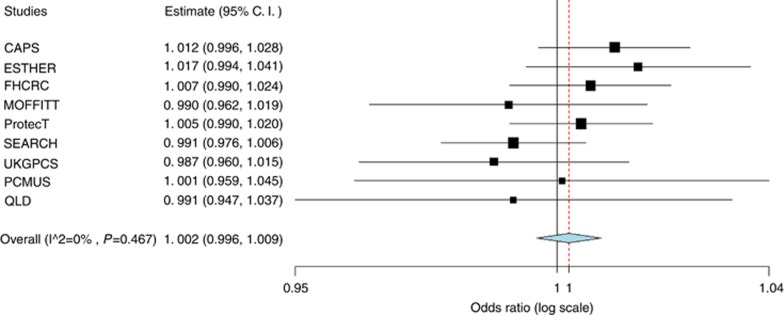
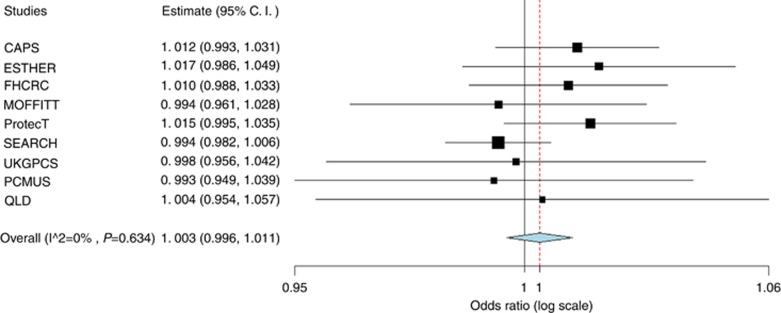
Subject characteristics are displayed in Table 2. Family history of prostate cancer is associated with prostate cancer risk. Subjects with a positive family history of prostate cancer had a 12% increase in prostate cancer risk. Mean height for cases and controls was 176.3 and 176.8 cms, respectively. The Student’s t-test suggests a significant difference in the means between the two groups (P-value <0.05). Results from a meta-analysis of height are presented in Figures 1 and 2. ORs were adjusted for age, family history of prostate cancer, and study site. In all cases and high grade cases, point risk estimates of each study are very similar and are close to 1. None of the estimated relative risks is statistically significant. The heterogeneity P-value of 0.467 in all cases and 0.634 in high grade cases suggests that studies are homogenous. ORs of fixed effect model in all cases and high grade cases are 1.002 (95% CI 0.996–1.009) and 1.003 (95% CI 0.996–1.011) respectively.
Table 2. Demographic data.
|
95% CI |
||||||
|---|---|---|---|---|---|---|
| Variables | Case | Control | OR | Lower | Upper | P-value |
|
Age (years) | ||||||
| Number | 6207 | 6016 | ||||
| Mean±s.d. | 63±7 | 60±7 | <0.001a | |||
|
Family history of PCAb | ||||||
| No | 4051 | 3594 | 1.00 | |||
| Yes | 904 | 831 | 1.12 | 1.00 | 1.24 | <0.05 |
|
Height (cm)- all cases | ||||||
| Mean±s.d. | 176.3±7.0 | 176.8±7.1 | <0.001a | |||
| Number | 2480 | 6016 | ||||
|
Height (cm)- aggressive cases | ||||||
| Mean±s.d. | 176.3±7.0 | 176.8±7.1 | <0.05a | |||
Abbreviations: CI=confidence interval; OR=odds ratio; PCA=prostate cancer.
P-value of Student t-test.
Adjusted for age.
Figure 1.
Forest plot (all prostate cancer cases).
Figure 2.
Forest plot (high grade cases).
Results from pooled analysis yielded similar risk estimates with OR 1.004, 95% CI 0.996–1.012 in all cases and OR 1.007, 95% CI 0.999–1.015 in high grade cases. We also analysed height as a categorical variable. Results are presented in Table 3. Results also suggest no overall association between height and prostate cancer risk comparing all cases with controls. In the high-grade case group, however, significant results were observed in the fourth quartile as compared to the first quartile (OR 1.22, 95% CI 1.014–1.477).
Table 3. Height as quartiles and prostate cancer risk.
|
All cases |
High grade cases |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
95% CI |
95% CI |
|||||||||
| Height (cm) | Number of subjects (all cases+controls) | ORa | Lower | Upper | P-value | Number of subjects (High grade cases+controls) | ORa | Lower | Upper | P-value |
| Q1 (⩽173.0) | 2949 | 1.00 | 2000 | 1.00 | ||||||
| Q2 (173.1–177.9) | 3210 | 1.16 | 0.99 | 1.35 | 0.064 | 2196 | 1.20 | 0.99 | 1.46 | 0.069 |
| Q3 (178.0–180.0) | 1865 | 1.07 | 0.89 | 1.28 | 0.467 | 1331 | 1.19 | 0.94 | 1.49 | 0.150 |
| Q4 (>180.0) | 4199 | 1.11 | 0.96 | 1.28 | 0.173 | 2969 | 1.22 | 1.01 | 1.48 | 0.035 |
Abbreviations: CI=confidence interval; OR=odds ratio.
All cases P for trend 0.407.
High-grade cases P for trend 0.075.
Adjusted for age, family history and study sites.
Table 4 shows the ORs of candidate SNPs with statistically significant results. ORs range from 0.90 to 1.32 with P-value from 10−2 to 10−3. One SNPs in the IGF-I gene had the highest ORs (1.32).
Table 4. Candidate SNPs with significant associations.
|
95% CI |
||||||
|---|---|---|---|---|---|---|
| SNP | Minor allele | Genes | Odds ratiosa | Lower | Upper | P-value |
| rs6503691 | A | GHDC:STAT5B:STAT5A | 0.90 | 0.82 | 0.99 | 0.036 |
| rs13317803 | G | GHSR:TNFSF10 | 1.08 | 1.01 | 1.14 | 0.016 |
| rs11831436 | A | IGF1 | 1.19 | 1.01 | 1.41 | 0.040 |
| rs35767 | A | IGF1 | 1.12 | 1.03 | 1.22 | 0.006 |
| rs5742612 | G | IGF1 | 1.32 | 1.13 | 1.55 | 0.001 |
| rs11630647 | A | IGF1R | 1.08 | 1.01 | 1.15 | 0.035 |
| rs2871864 | C | IGF1R | 1.11 | 1.01 | 1.22 | 0.031 |
| rs2229765 | A | IGF1R:PGPEP1L | 1.08 | 1.02 | 1.15 | 0.013 |
Abbreviations: CI=confidence interval; SNP=single-nucleotide polymorphism.
Multiple logistic regression adjusted for age, family history of prostate cancer, study site and Principal Components of EU ancestry.
Table 5 shows the ORs of genetic risk scores and prostate cancer risk. A significant result was observed in the third tertile as compared to reference tertile (OR 1.13 with 95% CI 1.03–1.23) when all prostate cancer cases were included. The P-value for trend is also statistically significant. In the high grade cases, similar results were observed. There is also a trend of increasing risk with increasing genetic risk scores in all prostate cancer cases and in high grade cases.
Table 5. Estimated risk of genetic risk scores (standardised score) and prostate cancer risk.
|
All PCA |
High grade PCA |
|||||||
|---|---|---|---|---|---|---|---|---|
|
95% CI |
95% CI |
|||||||
| Genetic risk score | Odds ratioa | Lower | Upper | P-value | Odds ratioa | Lower | Upper | P-value |
| Reference | 1.00 | 1.00 | ||||||
| 2nd tertile | 1.06 | 0.97 | 1.16 | 0.186 | 1.03 | 0.92 | 1.16 | >0.05 |
| 3rd tertile | 1.13 | 1.04 | 1.23 | 0.006 | 1.55 | 1.03 | 1.29 | <0.05 |
Abbreviations: CI=confidence interval; PCA=prostate cancer. All cases P for trend 0.006, high-grade cases P for trend 0.014.
Adjusted for height.
The interaction results between height and genetic risk scores suggest that there is no GE interaction between height and genetic risk score (Table 6) regardless of type of cases.
Table 6. GE interaction result (Bayesian method)–interaction between Height and genetic risk scores.
|
G=0 |
G=1 |
95% CI |
||||||
|---|---|---|---|---|---|---|---|---|
| Group | E=0 | E=1 | E=0 | E=1 | Total | Estimated interaction OR | Lower | Upper |
| Control | 666 | 743 | 670 | 725 | 2804 | |||
| All PCA case | 738 | 629 | 766 | 743 | 2876 | 1.14 | 0.98 | 1.33 |
| High-grade PCA case | 293 | 248 | 305 | 305 | 1151 | 1.18 | 0.94 | 1.49 |
Abbreviations: CI=confidence interval; PCA=prostate cancer.
G=0-subjects with genetic risk score in the first tertile, G=1-subjects with genetic risk score in the third tertile.
E=0-subjects with height in the first tertile, E=1-subjects with height in the third tertile.
Results of the GE analyses by GMDR method are depicted in Table 7. We fitted 2 SNPs with effect sizes >1.15 into the model and adjusted for covariates (age and family history of PCA). None of the models yield significant ORs regardless of case type. This is confirmed by cross-validation consistency. Both all and high grade cases, the extended models show consistency across testing sets.
Table 7. GE with 2 IGF-I pathway SNPs by GMDR method.
| Group | Best model | Testing accuracy | Testing sensitivity | Testing odds ratioa | Testing χ2 | Cross-validation consistency |
|---|---|---|---|---|---|---|
| All PCA cases | Height | 0.51 | 0.51 | 1.07 (95% CI 0.72–1.59) | 0.69 (P=0.408) | 10/10 |
| Height, rs5742612 | 0.51 | 0.51 | 1.12 (95% CI 0.75–1.66) | 0.84 (P=0.358) | 10/10 | |
| Height, rs5742612, rs11831436 | 0.51 | 0.51 | 1.09 (95% CI 0.73–1.61) | 0.75 (P=0.387) | 10/10 | |
| High grade cases | Height | 0.51 | 0.54 | 1.16 (95% CI 0.63–2.11) | 0.63 (P=0.429) | 10/10 |
| Height, rs11831436 | 0.51 | 0.52 | 1.06 (95% CI 0.58–1.94) | 0.22 (P=0.636) | 10/10 | |
| Height, rs5742612, rs11831436 | 0.51 | 0.53 | 1.12 (95% CI 0.62–2.05) | 0.44 (P=0.507) | 10/10 |
Abbreviations: GE=gene and environment; GMDR=general multifactor dimensionality reduction; IGF=insulin-like growth factor; PCA=prostate cancer; SNP=single-nucleotide polymorphism.
Testing odds ratios adjusted for age and family history of prostate cancer.
Discussion
This study investigated the effect of height and its possible interaction with selected SNPs from the PRACTICAL consortium in 6207 cases and 6016 controls. The consortium is an international collaboration on PCA and it has had notable successes for example in identifying 100 new genetic loci (Eeles et al, 2008, 2009, 2013; Al Olama et al, 2009, 2012, 2014). These loci confer small to medium risks with highly significant P-values of ⩽10−7 (GWAS significance).
There are, however, many polymorphisms with estimated risks less statistically significant which could still play an important role, particularly in the presence of environmental exposure. We therefore created a data set (subjects with epidemiological data and genotype data) which allowed us to investigate such a hypothesis.
Out of the 6207 cases, 2480 cases (40%) are high grade cases defined by Gleason grade ⩾7. One of the limitations of defining high grade cases is that we did not have data on Gleason grade breakdown hence we have to use combined score data of 7 rather than (4+3 or 3+4). Age and family history of PCA are confirmed risk factors in our study (Table 2). We investigated height in 3 ways. First, we explored height phenotype as a main exposure. Second, we investigated genetic profile (candidate SNPs) related to height, and third, we determined if there are any potential interactions between the selected SNPs and height. SNPs were deemed ‘related to height’ because they are found in candidate genes for height but they have not necessarily been identified in GWAS as underlying the variability of the height phenotype. We present results for all PCA cases and high grade cases as compared to controls. Although mean height values were very similar between cases and controls the mean difference was statistically significant and is in the opposite direction to that expected. In a multivariate analysis adjusted for age, family history of PCA and study sites, height as a continuous variable did not show associations with PCA risk in either all PCA cases or high grade PCA cases. However, height categorised in quartiles did show significantly increased risk in high grade cases. Subjects with a height >180 cm are at 22% increased risk compared with subjects with height <173 cm. We did not observe any association between height and low grade cases ((Gleason grade <7) results are not presented in the paper). Our findings suggest taller subjects are at increased risk of high grade PCA risk. A previous report from a large nested case–control study (ProtecT) reported the OR of prostate-specific antigen–detected high-grade PCA per 10 cm increase in height was 1.23; 95% CI: 1.06–1.43. In a meta-analysis of 58 studies, a smaller effect was reported (random-effects OR: 1.12; 95% CI: 1.05–1.19) (Zuccolo et al, 2008). Findings from The Early Stage Prostate Cancer Cohort Study which looked at the relationship between height and prostate cancer grade in various subpopulations of men with potentially different risk of high-grade PCA also suggested that participants in the highest quartile of height were more than twice as likely to have a Gleason score ⩾7 (4+3) at biopsy than participants in the lowest quartile of height (OR 2.14 (95% CI 1.11, 4.14); Farwell et al, 2011). Two other studies presented results exclusively on cases with advanced stage PCA and both supported a positive association between height and PCA risk (Hayes et al, 1999; Norrish et al, 2000). Hayes and colleagues observed a two-fold increased risk in white men with height>1.75 metres compared to height<1.67 metres. The association was absent among black men (Hayes et al, 1999). Norrish and colleagues investigated the role of height and PCA risk in both sporadic cancer cases and familial cancer cases. The study used the Gleason grading score to characterise the cases. Advanced PCA cases were defined by combined Gleason score ⩾7 and localised PCA cases by combined Gleason score ⩽6. Results on sporadic advanced cancer showed an indication of risk increasing across the quintiles (p for trend=0.07) which is similar to our high grade cases. Moreover the risk was greater among those with a positive family history of PCA (OR for height >179 cm compared to <170 cm=7.41, 95% CI 1.68–32.67, p for trend=0.02). A null association was reported in localised cases. Not only is height potentially associated with PCA risk but it also shows association with PCA mortality. A recent publication including more than 1 million subjects investigated adult height and the risk of cause-specific death and vascular morbidity suggested that hazard ratios per 6.5 cm greater height were 1.04 (1.03–1.06) for death from cancers and 1.07 (1.02–1.11) for death from PCA (Emerging Risk Factors Collaboration, 2012). In contrast, the results form a large cohort of 10 501 PCA cases and 10 831 controls within the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) suggested that height was not associated with PCA risk both as a continuous variable (OR: 1.001, 95% CI: 1.000–1.002 per cm increase, P=0.12) or as in tertiles (OR: 1.02, 95% CI: 0.99–1.06, P=0.24) (Lindstrom et al, 2011). A null association was reported in the study also using PRACTICAL genotype data set and investigated the effect of height and prostate cancer incidence and mortality using Mendelian randomisation approach (Davies et al, 2015). The authors analysed genetic variants associated with height from published genome-wide association studies and reported that these genetic variants are strong instrument for the variable. There are some limitations in that GWA studies will not explain a majority of the estimated 80% contribution of genetic factors to variation in height (Lango Allen et al, 2010).
Human height is well known as a polygenic trait with a number of genes that contribute to height (Chial, 2008). Recent GWAS studies have identified strong and moderate effects of genes related to human height (Weedon and Frayling, 2008; McEvoy and Visscher, 2009). Single SNPs with small effects in aggregate form can be applied to assign individuals to their height distribution (Lettre, 2009). We applied a candidate SNPs approach and identified SNPs in genes that had been genotyped in our consortium that were related to growth processes. These SNPs were in the genes IGF-I, GH-1, SHOX, FMR1, GHITM, and GHRHR (Gunnell, 2000; Ellis et al, 2001; Gunnell et al, 2001). Twelve SNPs in these genes show significant associations. We computed r2 and kept the 8 SNP based on association and P-value in each region. Only one SNP (rs6503691) showed a small protective effect. This SNP is reported to associate with significantly decreased risk of breast cancer (Johansson et al, 2007; Zhao et al, 2015). Polymorphisms in the IGF signalling pathway have been shown to associate with PCA mortality (Cao et al, 2014). Other studies reported null associations (Gu et al, 2010; Tsilidis et al, 2013). We also explored association between aggregated SNPs score as main effect; results support that individuals with genetic risk scores in the third tertiles are at increased risk of high grade PCA at 15% and of all PCA cases at 13% as compared with the lowest tertile. A test for trend also supports a dose-response relationship (P-value <0.05 in both case groups). These findings support that a genetic risk score in the growth pathway are associated with high grade PCA. IGF genes have been previously linked with PCA (Cheng et al, 2006; Johansson et al, 2007; Cao et al, 2014; Gan et al, 2014; Qian et al, 2014; Takeuchi et al, 2014; Travis et al, 2016). GHSR genes are also previously reported to associate with prostate cancer risk (Dressen, 2007). We also investigated possible gene-environment interactions using two approaches. The first approach uses combined genetic risk scores and a binary variable of height with the first tertile as the reference group and the third tertile as the ‘exposed’ group. Analyses were done in both PCA and high grade case group using the Bayesian method proposed by (Mukherjee et al, 2008). Results of the GE analyses however suggested no interaction between genetic risk scores and height. In the second approach, we selected the top 2 SNPs with the strongest effect sizes and fitted a model using the GMDR method (Chen et al, 2011). The GMDR method allows adjustment for discrete and quantitative covariates and is applicable to both dichotomous and continuous phenotypes. The GMDR with covariate adjustment had a power of >80% in a case–control design with a sample size of ⩾2000. We applied the GMDR method because it differs from the traditional GE method in that it allows more than 1 SNP in the model (traditional method-based on the concept of single-factor–based approaches; Lou et al, 2007). The results also showed no interactions. None of the main effect (height) and extended models showed any significant results.
In summary, our findings suggest that height and genetic variants related to the human growth pathway are associated with high grade PCA risk. Taller men of >1.80 m are at increased risk of high grade PCA. Genetic variants in genes that relate to growth pathways are associated with prostate cancer risk. The estimated risk is evident amongst subjects in the highest score group when combined genetic risk scores were used. There is, however, no GE interaction between selected genetic variants and height.
Acknowledgments
This study would not have been possible without the contributions of the following: Per Hall (COGS); Douglas F Easton, Paul Pharoah, Kyriaki Michailidou, Manjeet K Bolla, Qin Wang (BCAC), Andrew Berchuck (OCAC), Rosalind A Eeles, Douglas F Easton, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Georgia Chenevix-Trench, Antonis Antoniou, Lesley McGuffog, Fergus Couch and Ken Offit (CIMBA), Joe Dennis, Alison M Dunning, Andrew Lee, and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Javier Benitez, Anna Gonzalez-Neira, and the staff of the CNIO genotyping unit, Jacques Simard and Daniel C Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, Stig E Bojesen, Sune F Nielsen, Borge G Nordestgaard, and the staff of the Copenhagen DNA laboratory, and Julie M Cunningham, Sharon A Windebank, Christopher A Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility. Funding for the iCOGS infrastructure came from: the European Community’s Seventh Framework Programme under grant agreement no 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, and C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112–the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. Funding for ICEP (‘This work was also supported by CRUK (grant number C18281/A19169)’). Funding for the CRUK study and PRACTICAL consortium: this work was supported by the Canadian Institutes of Health Research, European Commission’s Seventh Framework Programme grant agreement no 223175 (HEALTH-F2-2009-223175), Cancer Research UK Grants C5047/A7357, C1287/A10118, C5047/A3354, C5047/A10692, C16913/A6135, and The National Institute of Health (NIH) Cancer Post-Cancer GWAS initiative grant: No. 1 U19 CA 148537-01 (the GAME-ON initiative). We acknowledge support from the NIHR to the Biomedical Research Centre at The Institute of Cancer Research and Royal Marsden NHS Foundation Trust.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Contributor Information
The PRACTICAL consortium:
Johanna Schleutker, BØrge G Nordestgaard, Fredrik Wiklund, Ruth C Travis, Christopher A Haiman, Stephen N Thibodeau, Christiane Maier, Vogel Walther, William J Blot, Adam S Kibel, Cezary Cybulski, Lisa Cannon-Albright, Hardev Pandha, Manuel R Teixeira, Margaret Cook, Koveela Govindasami, Michelle Guy, Daniel Leongamornlert, Emma J Sawyer, Rosemary Wilkinson, Angela Morgan, Cyril Fisher, Edward J Saunders, Malgorzata Tymrakiewicz, Naomi Livni, Steve Hazel, Tokhir Dadaev, Angela Cox, Anne George, Athene Lane, Gemma Marsden, Michael Davis, Paul Brown, John Pedersen, John L Hopper, Ami Karlsson, Carin Cavalli-Bjoerkman, Jan Adolfson, Jan-Erik Johansson, Michael Broms, Paer Stattin, Suzanne Kolb, Christa Stegmaier, Babu Zachariah, Hyun Park, James Haley, Julio Pow-Sang, Maria Rincon, Selina Radlein, Aleksandrina Vlahova, Atanaska Mitkova, Darina Kachakova, Elenko Popov, Svetlana Christova, Tihomir Dikov, Allison Eckert, Angus Collins, Glenn Wood, Greg Malone, Kimberly Alexander, Kris Kerr, Mary-Anne Kedda, Megan Turner, Pamela Saunders, Peter Heathcote, Srilakshmi Srinivasan, Tracy Omara, Trina Yeadon, and Felicity Lose
Supplementary Material
References
- Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, Benlloch S, Hazelett DJ, Wang Z, Saunders E, Leongamornlert D, Lindstrom S, Jugurnauth-Little S, Dadaev T, Tymrakiewicz M, Stram DO, Rand K, Wan P, Stram A, Sheng X, Pooler LC, Park K, Xia L, Tyrer J, Kolonel LN, Le Marchand L, Hoover RN, Machiela MJ, Yeager M, Burdette L, Chung CC, Hutchinson A, Yu K, Goh C, Ahmed M, Govindasami K, Guy M, Tammela TL, Auvinen A, Wahlfors T, Schleutker J, Visakorpi T, Leinonen KA, Xu J, Aly M, Donovan J, Travis RC, Key TJ, Siddiq A, Canzian F, Khaw KT, Takahashi A, Kubo M, Pharoah P, Pashayan N, Weischer M, Nordestgaard BG, Nielsen SF, Klarskov P, Roder MA, Iversen P, Thibodeau SN, McDonnell SK, Schaid DJ, Stanford JL, Kolb S, Holt S, Knudsen B, Coll AH, Gapstur SM, Diver WR, Stevens VL, Maier C, Luedeke M, Herkommer K, Rinckleb AE, Strom SS, Pettaway C, Yeboah ED, Tettey Y, Biritwum RB, Adjei AA, Tay E, Truelove A, Niwa S, Chokkalingam AP, Cannon-Albright L, Cybulski C, Wokolorczyk D, Kluzniak W, Park J, Sellers T, Lin HY, Isaacs WB, Partin AW, Brenner H, Dieffenbach AK, Stegmaier C, Chen C, Giovannucci EL, Ma J, Stampfer M, Penney KL, Mucci L, John EM, Ingles SA, Kittles RA, Murphy AB, Pandha H, Michael A, Kierzek AM, Blot W, Signorello LB, Zheng W, Albanes D, Virtamo J, Weinstein S, Nemesure B, Carpten J, Leske C, Wu SY, Hennis A, Kibel AS, Rybicki BA, Neslund-Dudas C, Hsing AW, Chu L, Goodman PJ, Klein EA, Zheng SL, Batra J, Clements J, Spurdle A, Teixeira MR, Paulo P, Maia S, Slavov C, Kaneva R, Mitev V, Witte JS, Casey G, Gillanders EM, Seminara D, Riboli E, Hamdy FC, Coetzee GA, Li Q, Freedman ML, Hunter DJ, Muir K, Gronberg H, Neal DE, Southey M, Giles GG, Severi G, Cook MB, Nakagawa H, Wiklund F, Kraft P, Chanock SJ, Henderson BE, Easton DF, Eeles RA, Haiman CA (2014) A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet 46: 1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar S, Saunders E, Hopper JL, Southey MC, Muir KR, English DR, Dearnaley DP, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Sawyer E, Lophatananon A, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper C, Donovan JL, Hamdy FC, Neal DE, Eeles RA, Easton DF (2009) Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet 41: 1058–1060. [DOI] [PubMed] [Google Scholar]
- Al Olama AA, Kote-Jarai Z, Schumacher FR, Wiklund F, Berndt SI, Benlloch S, Giles GG, Severi G, Neal DE, Hamdy FC, Donovan JL, Hunter DJ, Henderson BE, Thun MJ, Gaziano M, Giovannucci EL, Siddiq A, Travis RC, Cox DG, Canzian F, Riboli E, Key TJ, Andriole G, Albanes D, Hayes RB, Schleutker J, Auvinen A, Tammela TL, Weischer M, Stanford JL, Ostrander EA, Cybulski C, Lubinski J, Thibodeau SN, Schaid DJ, Sorensen KD, Batra J, Clements JA, Chambers S, Aitken J, Gardiner RA, Maier C, Vogel W, Dork T, Brenner H, Habuchi T, Ingles S, John EM, Dickinson JL, Cannon-Albright L, Teixeira MR, Kaneva R, Zhang HW, Lu YJ, Park JY, Cooney KA, Muir KR, Leongamornlert DA, Saunders E, Tymrakiewicz M, Mahmud N, Guy M, Govindasami K, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English D, Virtamo J, Le Marchand L, Campa D, Kaaks R, Lindstrom S, Diver WR, Gapstur S, Yeager M, Cox A, Stern MC, Corral R, Aly M, Isaacs W, Adolfsson J, Xu J, Zheng SL, Wahlfors T, Taari K, Kujala P, Klarskov P, Nordestgaard BG, Roder MA, Frikke-Schmidt R, Bojesen SE, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Rinckleb A, Luedeke M, Herkommer K, Meyer A, Serth J, Marthick JR, Patterson B, Wokolorczyk D, Spurdle A, Lose F, McDonnell SK, Joshi AD, Shahabi A, Pinto P, Santos J, Ray A, Sellers TA, Lin HY, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V, Chanock S, Gronberg H, Haiman CA, Kraft P, Easton DF, Eeles RA (2012) A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet 22(2): 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EE (2000) Invited Commentary: Do Anthropometric Measures Predict risk of prostate cancer? Am J Epidemiol 151: 550–553. [DOI] [PubMed] [Google Scholar]
- Cao Y, Lindstrom S, Schumacher F, Stevens VL, Albanes D, Berndt S, Boeing H, Bueno-de-Mesquita HB, Canzian F, Chamosa S, Chanock SJ, Diver WR, Gapstur SM, Gaziano JM, Giovannucci EL, Haiman CA, Henderson B, Johansson M, Le Marchand L, Palli D, Rosner B, Siddiq A, Stampfer M, Stram DO, Tamimi R, Travis RC, Trichopoulos D, Willett WC, Yeager M, Kraft P, Hsing AW, Pollak M, Lin X, Ma J (2014) Insulin-like growth factor pathway genetic polymorphisms, circulating IGF1 and IGFBP3, and prostate cancer survival. J Natl Cancer Inst 106: dju085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GB, Xu Y, Xu HM, Li MD, Zhu J, Lou XY (2011) Practical and theoretical considerations in study design for detecting gene-gene interactions using MDR and GMDR approaches. PLoS One 6: e16981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng I, Stram DO, Penney KL, Pike M, Le Marchand L, Kolonel LN, Hirschhorn J, Altshuler D, Henderson BE, Freedman ML (2006) Common genetic variation in IGF1 and prostate cancer risk in the Multiethnic Cohort. J Natl Cancer Inst 98: 123–134. [DOI] [PubMed] [Google Scholar]
- Chial H (2008) Polygenic inheritance and gene mapping. Nat Educ 1: 17. [Google Scholar]
- Davies NM, Gaunt TR, Lewis SJ, Holly J, Donovan JL, Hamdy FC, Kemp JP, Eeles R, Easton D, Kote-Jarai Z, Al Olama AA, Benlloch S, Muir K, Giles GG, Wiklund F, Gronberg H, Haiman CA, Schleutker J, Nordestgaard BG, Travis RC, Neal D, Pashayan N, Khaw KT, Stanford JL, Blot WJ, Thibodeau S, Maier C, Kibel AS, Cybulski C, Cannon-Albright L, Brenner H, Park J, Kaneva R, Batra J, Teixeira MR, Pandha H, Lathrop M, Smith GD, Martin RM (2015) The effects of height and BMI on prostate cancer incidence and mortality: a Mendelian randomization study in 20 848 cases and 20 214 controls from the PRACTICAL consortium. Cancer Causes Control 26: 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressen AS (2007) Growth Hormone Genes And Prostate Cancer Risk. Master of Science, University of Pittsburgh: Pittsburgh. [Google Scholar]
- Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J, Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA, Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, Spurdle A, Clements J, Dickinson JL, Maier C, Vogel W, Dork T, Rebbeck TR, Cooney KA, Cannon-Albright L, Chappuis PO, Hutter P, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, English DR, Leongamornlert DA, Tymrakiewicz M, Morrison J, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Saunders EJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Southey MC, Lophatananon A, Liu JF, Kolonel LN, Le Marchand L, Wahlfors T, Tammela TL, Auvinen A, Lewis SJ, Cox A, FitzGerald LM, Koopmeiners JS, Karyadi DM, Kwon EM, Stern MC, Corral R, Joshi AD, Shahabi A, McDonnell SK, Sellers TA, Pow-Sang J, Chambers S, Aitken J, Gardiner RA, Batra J, Kedda MA, Lose F, Polanowski A, Patterson B, Serth J, Meyer A, Luedeke M, Stefflova K, Ray AM, Lange EM, Farnham J, Khan H, Slavov C, Mitkova A, Cao G, Easton DF (2009) Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet 41: 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF (2008) Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 40: 316–321. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock SJ, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah PD, Pashayan N, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle AB, Clements JA, Teixeira MR, Dicks E, Lee A, Dunning AM, Baynes C, Conroy D, Maranian MJ, Ahmed S, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As NJ, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatananon A, Cox A, Southey MC, Hopper JL, English DR, Aly M, Adolfsson J, Xu J, Zheng SL, Yeager M, Kaaks R, Diver WR, Gaudet MM, Stern MC, Corral R, Joshi AD, Shahabi A, Wahlfors T, Tammela TL, Auvinen A, Virtamo J, Klarskov P, Nordestgaard BG, Roder MA, Nielsen SF, Bojesen SE, Siddiq A, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Blot WJ, Zheng W, Cai Q, McDonnell SK, Rinckleb AE, Drake B, Colditz G, Wokolorczyk D, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Sellers TA, Lin HY, Slavov C, Mitev V, Lose F, Srinivasan S, Maia S, Paulo P, Lange E, Cooney KA, Antoniou AC, Vincent D, Bacot F, Tessier DC, Kote-Jarai Z, Easton DF (2013) Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 45: 385–391, e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JA, Stebbing M, Harrap SB (2001) Polymorphism of the androgen receptor gene is associated with male pattern baldness. J Invest Dermatol 116: 452–455. [DOI] [PubMed] [Google Scholar]
- Emerging Risk Factors Collaboration (2012) Adult height and the risk of cause-specific death and vascular morbidity in 1 million people: individual participant meta-analysis. Int J Epidemiol 41: 1419–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farwell WR, Lourenco C, Holmberg E, Hall RB, D’Avolio L, Lawler EV, Michael Gaziano J (2011) The association between height and prostate cancer grade in the Early Stage Prostate Cancer Cohort Study. Cancer Causes Control 22: 1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2013) GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer. Available from: http://globocan.iarc.fr (accessed on 12 June 2016).
- Freeman VL, Liao YL, Durazo-Arvizu R, Cooper RS (2001) Height and risk of fatal prostate cancer: findings from the National Health Interview Survey (1986 to 1994). Ann Epidemiol 11: 22–27. [DOI] [PubMed] [Google Scholar]
- Gan Y, Buckels A, Liu Y, Zhang Y, Paterson AJ, Jiang J, Zinn KR, Frank SJ (2014) Human GH receptor-IGF-1 receptor interaction: implications for GH signaling. Mol Endocrinol 28(11): 1841–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci E, Rimm EB, Stampfer MJ, Colditz GA, Willett WC (1997) Height, body weight, and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev 6: 557–563. [PubMed] [Google Scholar]
- Gu F, Schumacher FR, Canzian F, Allen NE, Albanes D, Berg CD, Berndt SI, Boeing H, Bueno-de-Mesquita HB, Buring JE, Chabbert-Buffet N, Chanock SJ, Clavel-Chapelon F, Dumeaux V, Gaziano JM, Giovannucci EL, Haiman CA, Hankinson SE, Hayes RB, Henderson BE, Hunter DJ, Hoover RN, Johansson M, Key TJ, Khaw KT, Kolonel LN, Lagiou P, Lee IM, LeMarchand L, Lund E, Ma J, Onland-Moret NC, Overvad K, Rodriguez L, Sacerdote C, Sanchez MJ, Stampfer MJ, Stattin P, Stram DO, Thomas G, Thun MJ, Tjonneland A, Trichopoulos D, Tumino R, Virtamo J, Weinstein SJ, Willett WC, Yeager M, Zhang SM, Kaaks R, Riboli E, Ziegler RG, Kraft P (2010) Eighteen insulin-like growth factor pathway genes, circulating levels of IGF-I and its binding protein, and risk of prostate and breast cancer. Cancer Epidemiol Biomarkers Prev 19: 2877–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnell D (2000) Height, insulin-like growth factors and cancer risk. Growth Horm IGF Res 10: S39–S40. [DOI] [PubMed] [Google Scholar]
- Gunnell D, Okasha M, Smith GD, Oliver SE, Sandhu J, Holly JMP (2001) Height, leg length, and cancer risk: a systematic review. Epidemiol Rev 23: 313–342. [DOI] [PubMed] [Google Scholar]
- Hayes RB, Ziegler RG, Gridley G, Swanson C, Greenberg RS, Swanson GM, Schoenberg JB, Silverman DT, Brown LM, Pottern LM, Liff J, Schwartz AG, Fraumeni JF Jr, Hoover RN (1999) Dietary factors and risks for prostate cancer among blacks and whites in the United States. Cancer Epidemiol Biomarkers Prev 8: 25–34. [PubMed] [Google Scholar]
- Hsing AW, Deng J, Sesterhenn IA, Mostofi FK, Stanczyk FZ, Benichou J, Xie T, Gao YT (2000) Body size and prostate cancer: a population-based case-control study in China. Cancer Epidemiol Biomarkers Prev 9: 1335–1341. [PubMed] [Google Scholar]
- Johansson M, McKay JD, Stattin P, Canzian F, Boillot C, Wiklund F, Adami HO, Balter K, Gronberg H, Kaaks R (2007) Comprehensive evaluation of genetic variation in the IGF1 gene and risk of prostate cancer. Int J Cancer 120: 539–542. [DOI] [PubMed] [Google Scholar]
- Key TJ, Silcocks PB, Davey GK, Appleby PN, Bishop DT (1997) A case-control study of diet and prostate cancer. Br J Cancer 76: 678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpelainen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Pare G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietilainen KH, Pouta A, Ridderstrale M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kahonen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimaki T, Melander O, Mosley TH Jr, Musk AW, Nieminen MS, O’Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tonjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Gronberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Volzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O’Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G (2009) Genetic regulation of adult stature. Curr Opin Pediatr 21: 515–522. [DOI] [PubMed] [Google Scholar]
- Lindstrom S, Schumacher F, Siddiq A, Travis RC, Campa D, Berndt SI, Diver WR, Severi G, Allen N, Andriole G, Bueno-de-Mesquita B, Chanock SJ, Crawford D, Gaziano JM, Giles GG, Giovannucci E, Guo C, Haiman CA, Hayes RB, Halkjaer J, Hunter DJ, Johansson M, Kaaks R, Kolonel LN, Navarro C, Riboli E, Sacerdote C, Stampfer M, Stram DO, Thun MJ, Trichopoulos D, Virtamo J, Weinstein SJ, Yeager M, Henderson B, Ma J, Le Marchand L, Albanes D, Kraft P (2011) Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers—results from BPC3. PLoS One 6: e17142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD (2007) A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet 80: 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy BP, Visscher PM (2009) Genetics of human height. Econ Hum Biol 7: 294–306. [DOI] [PubMed] [Google Scholar]
- Mukherjee B, Ahn J, Gruber SB, Rennert G, Moreno V, Chatterjee N (2008) Tests for gene-environment interaction from case-control data: a novel study of type I error, power and designs. Genet Epidemiol 32: 615–626. [DOI] [PubMed] [Google Scholar]
- Norrish AE, McRae CU, Holdaway IM, Jackson RT (2000) Height-related risk factors for prostate cancer. Br J Cancer 82: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Zhou H, Chen J, Ding Q, Cao Q, Qin C, Shao P, Li P, Cai H, Meng X, Ju X, Wang M, Zhang Z, Li J, Hua L, Yin C (2014) Genetic polymorphisms in IGF-I and IGFBP-3 are associated with prostate cancer in the Chinese population. PLoS One 9: e85609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stattin P, Bylund A, Rinaldi S, Biessy C, Dechaud H, Stenman UH, Egevad L, Riboli E, Hallmans G, Kaaks R (2000) Plasma insulin-like growth factor-I, insulin-like growth factor-binding proteins, and prostate cancer risk: a prospective study. J Natl Cancer Inst 92: 1910–1917. [DOI] [PubMed] [Google Scholar]
- Takeuchi A, Shiota M, Beraldi E, Thaper D, Takahara K, Ibuki N, Pollak M, Cox ME, Naito S, Gleave ME, Zoubeidi A (2014) Insulin-like growth factor-I induces CLU expression through Twist1 to promote prostate cancer growth. Mol Cell Endocrinol 384: 117–125. [DOI] [PubMed] [Google Scholar]
- Travis RC, Appleby PN, Martin RM, Holly JM, Albanes D, Black A, Bueno-de-Mesquita HB, Chan JM, Chen C, Chirlaque MD, Cook MB, Deschasaux M, Donovan JL, Ferrucci L, Galan P, Giles GG, Giovannucci EL, Gunter MJ, Habel LA, Hamdy FC, Helzlsouer KJ, Hercberg S, Hoover RN, Janssen JA, Kaaks R, Kubo T, Le Marchand L, Metter EJ, Mikami K, Morris JK, Neal DE, Neuhouser ML, Ozasa K, Palli D, Platz EA, Pollak MN, Price AJ, Roobol MJ, Schaefer C, Schenk JM, Severi G, Stampfer MJ, Stattin P, Tamakoshi A, Tangen CM, Touvier M, Wald NJ, Weiss NS, Ziegler RG, Key TJ, Allen NE (2016) A Meta-analysis of individual participant data reveals an association between circulating levels of IGF-I and prostate cancer risk. Cancer Res 76: 2288–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilidis KK, Travis RC, Appleby PN, Allen NE, Lindstrom S, Albanes D, Ziegler RG, McCullough ML, Siddiq A, Barricarte A, Berndt SI, Bueno-de-Mesquita HB, Chanock SJ, Crawford ED, Diver WR, Gapstur SM, Giovannucci E, Gu F, Haiman CA, Hayes RB, Hunter DJ, Johansson M, Kaaks R, Kolonel LN, Kraft P, Le Marchand L, Overvad K, Polidoro S, Riboli E, Schumacher FR, Stevens VL, Trichopoulos D, Virtamo J, Willett WC, Key TJ (2013) Insulin-like growth factor pathway genes and blood concentrations, dietary protein and risk of prostate cancer in the NCI Breast and Prostate Cancer Cohort Consortium (BPC3). Int J Cancer 133: 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve PJ, Johnson KC, Kreiger N, Mao Y (1999) Risk factors for prostate cancer: results from the Canadian National Enhanced Cancer Surveillance System. Cancer Causes Control 10: 355–367. [DOI] [PubMed] [Google Scholar]
- Wallace BC, Schmid CH, Lau J, Trikalinos TA (2009) Meta-Analyst: software for meta-analysis of binary, continuous and diagnostic data. BMC Med Res Methodol 9: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Frayling TM (2008) Reaching new heights: insights into the genetics of human stature. Trends Genet 24: 595–603. [DOI] [PubMed] [Google Scholar]
- Willett WC (2000) Diet and cancer. [Review] [152 refs]. Oncologist 5: 393–404. [DOI] [PubMed] [Google Scholar]
- Zhao H, Wang Z, Wu H, Xiao Q, Yao W, Wang E, Liu Y, Wei M (2015) STAT3 genetic variant, alone and in combination with STAT5b polymorphism, contributes to breast cancer risk and clinical outcomes. Med Oncol 32: 375. [DOI] [PubMed] [Google Scholar]
- Zuccolo L, Harris R, Gunnell D, Oliver S, Lane JA, Davis M, Donovan J, Neal D, Hamdy F, Beynon R, Savovic J, Martin RM (2008) Height and prostate cancer risk: a large nested case-control study (ProtecT) and meta-analysis. Cancer Epidemiol Biomarkers Prev 17: 2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.