Abstract
Background:
Chromosomal instability (CIN) has repeatedly been identified as a prognostic marker. Here we evaluated the percentage of aberrant genome per cell (PAG) as a measure of CIN in single disseminated tumour cells (DTC) isolated from patients with operable oesophageal adenocarcinoma (EAC), to assess the impact of CINhigh DTCs on prognosis.
Methods:
We isolated CK18positive DTCs from bone marrow (BM) or lymph node (LN) preparations of operable EAC patients. After whole-genome amplification, single DTCs were analysed for chromosomal gains and losses using metaphase-based comparative genomic hybridisation (mCGH). We calculated the PAG for each DTC and determined the critical threshold value that identifies high-risk patients by STEPP (Subpopulation Treatment Effect Pattern Plot) analysis in two independent EAC patient cohorts (cohort #1, n=44; cohort #2; n=29).
Results:
The most common chromosomal alterations observed among the DTCs were typical for EAC, but the DTCs showed a varying PAG between individual patients. Generally, LNDTCs displayed a significantly higher PAG than BMDTCs. STEPP analysis revealed an increasing PAG of DTCs to be correlated with an increased risk for short survival in two independent EAC cohorts as well as in the corresponding pooled analysis. In all three data sets (cohort #1, cohort #2 and pooled cohort), PAGhigh DTCs conferred an independent risk for a significantly decreased survival.
Conclusions:
The analysis of PAG/CIN in solitary marker-positive DTCs identifies operable EAC patients with poor prognosis, indicating a more aggressive minimal residual disease.
Keywords: disseminated tumour cells, single cell analysis, minimal residual disease, oesophageal cancer, chromosomal instability, heterogeneity, copy number alteration, prognosis
Oesophageal adenocarcinomas (EACs) represent 20–30% of all malignant oesophageal tumours, which are quite aggressive cancers with 5-year survival rates of <30% (Rustgi and El-Serag, 2014). Owing to frequent diagnosis at an advanced disease stage, only around one-third of the patients can be treated with curative intention (Shapiro et al, 2016). EACs tend to metastasise early and disseminate initially predominately into lymph nodes (LNs; Klein and Stoecklein, 2009; Dulak et al, 2012; Lee et al, 2013). As a result, 30–60% of patients with small primary lesions (pT1/2) already present with LN metastases (LNMET; Anderegg et al, 2016). However, the aggressive metastatic behaviour and the intrinsic therapeutic resistance of EAC currently restrains successful treatment of affected patients. In all, 48–80% of the selected patients in whom complete tumour resection can be achieved and who receive multimodal treatments develop lethal metastatic relapse within 5 years after surgery (Visser et al, 2016). The obvious starting point of those metastatic relapses are cancer cells that have disseminated to distant organs before the initial diagnosis and that were left behind after the surgical resection of the primary tumour (Polzer and Klein, 2013; Pantel and Alix-Panabieres, 2014). Current routine diagnostics fail to detect this early systemic spread. Among the accepted experimental approaches to trace such disseminated tumour cells (DTCs) are immuno-detection based methods targeting epithelial cells in the mesenchyme-derived indicator organs bone marrow (BM) and LNs. In operable oesophageal cancer, DTCs can be observed in 8–37% of BM samples (Thorban et al, 1996; Izbicki et al, 1997; Bonavina et al, 2001; Stoecklein et al, 2008) and in 10–71% of histopathologically tumour-free LN (Izbicki et al, 1997; Hosch et al, 2000; Mueller et al, 2000; Bandla et al, 2012). Although their prognostic value is not fully established in oesophageal cancer so far, DTCs are generally thought to comprise potential metastatic precursor cells (Klein, 2013). Therefore, their further molecular characterisation should provide a deeper insight into systemic disease progression and might represent or reveal new biomarkers eventually improving therapy.
Previous single-cell whole-genome analyses using metaphase-based comparative genomic hybridisation (mCGH) assessing copy number alterations (CNAs) of DTCs isolated from patients with different cancer types uncovered a highly varying extent of CNAs, reflecting different levels of chromosomal instability (CIN; Klein et al, 1999, 2002; Schmidt-Kittler et al, 2003; Stoecklein et al, 2008; Weckermann et al, 2009). Intriguingly, a common finding was that BMDTCs display significantly lower CIN levels than their matched primary tumours or LNDTCs (Schmidt-Kittler et al, 2003; Stoecklein et al, 2008; Weckermann et al, 2009). The presence of non-/low-aberrant DTCs indicated that cancer cells might disseminate into the BM very early during tumour development (Braun et al, 2000; Husemann et al, 2008). However, as only few cells of the DTC population eventually grow out into metastases, it is critical to understand the biology of the successful metastatic precursors. CIN is a known driver of tumour progression (Nowak et al, 2002; Bakhoum and Swanton, 2014; Tanaka and Hirota, 2016) and was frequently described as an early event in oesophageal cancer (Paulson et al, 2009; Nones et al, 2014). Therefore, we hypothesised that DTCs with an increased aberrant genome, reflecting elevated CIN levels, indicate more advanced systemic cancer and are more prone to metastasise, whereas DTCs with absent or few CNAs indicate retarded systemic progression or even a dead end and will never grow out during the lifespan of an EAC patient. To test our hypothesis, we isolated DTCs from BM and LN to cover both potential metastatic routes in operable EAC patients, assessed their CNAs by mCGH as a measure for CIN and correlated the extent of aberrant genome with patient outcome.
Materials and methods
Supplementary Figure S1 provides an overview of the complete experimental workflow.
Study patients and tissue samples
We processed preparations of BM aspirates and macroscopically tumour-free LN from 59 patients with EAC treated at the clinic for General, Visceral and Paediatric Surgery of the University Hospital Duesseldorf, Germany. From selected cases of this cohort, we also obtained formalin-fixed and paraffin-embedded (FFPE) tissue samples of primary tumours (OESPT) and LNMET. Written consent of the patients was obtained prior to the study and the study was approved by the ethics committee of the medical faculty of the Heinrich-Heine-University Duesseldorf (#2316 and #2655). The cohort in the present study is part of a larger collective described previously by Driemel et al (2013). The clinicopathological data of these patients (n=59) are presented in Table 1.
Table 1. Clinicopathological features from 59 EAC patients of cohort #1.
| Patients | n=59 | BMDTCpositive | BMDTCnegative | LNDTCpositive | LNDTCnegative |
|---|---|---|---|---|---|
|
Sex | |||||
| Male | 47 (ø=64 years) | 8 (19%) | 34 (81%) | 9 (43%) | 12 (57%) |
| Female | 12 (ø=65 years) | 5 (45%) | 6 (55%) | — | 1 (100%) |
|
Primary tumour | |||||
| pTx | 1 | — | 1 (100%) | — | — |
| pT1–2 | 26 | 3 (13%) | 20 (87%) | 5 (45%) | 6 (55%) |
| pT3–4 | 32 | 10 (34%) | 19 (66%) | 4 (36%) | 7 (64%) |
|
Lymph node statusa | |||||
| pN0 | 16 | — | 14 (100%) | 2 (40%) | 3 (60%) |
| pN1–3 | 43 | 13 (33%) | 26 (66%) | 7 (41%) | 10 (59%) |
|
Distant organ metastasis | |||||
| M0 | 48 | 12 (29%) | 30 (71%) | 8 (47%) | 9 (53%) |
| M1 | 11 | 1 (9%) | 10 (91%) | 1 (20%) | 4 (80%) |
|
Tumour grade | |||||
| Gx | 2 | — | 2 (100%) | — | — |
| G2 | 21 | 6 (32%) | 13 (68%) | 4 (57%) | 3 (43%) |
| G3 | 36 | 7 (22%) | 25 (78%) | 5 (33%) | 10 (67%) |
|
Resection margins | |||||
| R0 | 52 | 13 (28%) | 34 (72%) | 9 (43%) | 12 (57%) |
| R1 | 6 | — | 5 (100%) | — | 1 (100%) |
| R2 | 1 | — | 1 (100%) | — | — |
|
Neoadjuvant therapy | |||||
| 13 | 2 (15%) | 11 (85%) | 1 (25%) | 3 (75%) | |
Abbreviations: BMDTC=disseminated tumour cell from the bone marrow; EAC=oesophageal adenocarcinoma; LNDTC=disseminated tumour cell from the lymph node.
The presence of DTCs significantly correlated (P=0.03) to LN metastases.
Single-cell preparation
The preparation of single-cell suspensions from BM aspirates and LN samples was performed as previously described (Hosch et al, 2000; Klein et al, 2002). Approximately 20 ml BM from the upper iliac crest was aspirated into a heparinised syringe before the surgical procedure. Then the sample was mixed and washed with an equal volume Hank’s salt solution. Subsequently, the pellet was resuspended with 1 × PBS and peripheral blood mononuclear cells (PBMCs) were separated by Ficoll density-gradient centrifugation. In all, 2.5 × 105 cells in 1 ml 1 × PBS were seeded on each well of a two-well adhesion slide. After sedimentation for 45 min, the slides were dried overnight. Slides were stored at −20 °C until staining procedure. Macroscopically tumour-free LN samples were mechanically disaggregated with the MediMachine (Dako, Hamburg, Germany) to produce a single-cell solution. These samples were processed in the same manner as the PBMCs of the BM aspirates.
Double immunofluorescence staining
For the double immunofluorescence staining (IF) of 1 × 106 cells of the BM and LN samples, we applied the monoclonal mouse antibody BerEP4 against the epithelial cell adhesion molecule (EpCAM; Clone: M0804) together with a monoclonal rabbit antibody against Cytokeratin 18 (CK18; Clone: E431-1). The IF was performed as described by Driemel et al (2013). DTCs were defined as CK18positive non-granulated round cells located within the same focal plane of the adhesion slide as the peripheral blood lymphocytes. To isolate the positively stained DTCs from BM and LN samples with a micromanipulator from the adhesion slide, 300 μl of 0.5% Igepal solution in 1 × PBS was pipetted onto the slides. To exclude contaminations with unstained cells, the isolated single DTCs were transferred on a chamber slide from which the individual cell was isolated with a 1 μl pipette and finally transferred into a PCR reaction tube. Subsequently, the single cell was whole-genome amplified (WGA) by an adapter-linker/MseI-PCR, as described previously (Klein et al, 1999), which is now commercialised as Ampli1 WGA Kit (Silicon Biosystems, Bologna, Italy).
Laser capture microdissection (LCM)
For LCM, we cut serial slides with a microtome from FFPE tissues of OESPT and LNMET of the DTCpositive patients. On the first 5 μm section, we performed haematoxylin and eosin staining (HE) to evaluate the tumour areal for LCM and used the following 8 μm-thick sections adhered to Superfrost glass slides (Menzel, Braunschweig, Germany) for LCM. Afterwards, the slides were incubated for 20 min at 70 °C and overnight at 56 °C. The 8 μm FFPE sections were washed twice with xylene for 20 min and were subsequently rehydrated with a series of 100, 85 and 70% ethanol. Then the samples were stained with Gill’s haematoxylin and incubated for 5 min in xylene after a dehydrating ethanol series. Finally, the slides were dried overnight at room temperature under a fume hood. The tumour cells were isolated using the Arcturus PIXCELL II laser capture microscope (Applied Biosystems, Darmstadt, Germany; settings: Power 75 mW; Spotsize 15 μm; Duration 3.0 ms; Target 200 mV) and were then digested within 10 μl Proteinase K Digestion master mix overnight at 42 °C. Three microlitres of the digested sample were transferred into a 0.2 ml PCR reaction tube and Proteinase K inhibition was performed at 80 °C for 10 min. Afterwards the DNA was subjected to WGA by an adapter-linker/MseI-PCR as described previously (Klein et al, 1999), which is now commercialised as Ampli1 WGA Kit (Silicon Biosystems).
Metaphase-based comparative genomic hybridisation
mCGH of the WGA single-cell DNA and the LCM FFPE DNA was carried out as described previously (Stoecklein et al, 2002; Schmidt-Kittler et al, 2003). mCGH profiles were generated and evaluated with the Isis software (V 5.5.1; MetaSystems, Altlußheim, Germany) and mCGH karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN, Shaffer et al, 2009). For each sample, the number of alterations was counted and the mean number of alterations was calculated for each sample group. The percentage of aberrant genome per cell (PAG) was determined to obtain an integrated measure for CIN in each sample. For this analysis, we considered both alteration number and size by translating each alteration into megabase pairs (Mbp), calculated the total size of genomic alterations and divided them by the total genome size, 3.15 Gb, respectively. The number of alterations and PAG correlated well in our cohort (Supplementary Figure S2, linear regression, R2=0.90).
Computational and statistical analyses
A data matrix assigning distinct values (1=chromosomal gain, −1=chromosomal loss, 0=CNA neutral) to copy number changes was generated from the mCGH ISCN karyotypes (Baudis and Cleary, 2001; http://www.progenetix.net/), which was used for generating a penetrance plot to visualise overall CNAs for each sample group. For this analysis, mCGH karyotypes were joined in a composite karyotype for OESPT and LNMET, for which more than one sample was analysed, respectively.
Hierarchical cluster analysis was performed using the ‘rhclust’ function embedded in the Progenetix analysis software (Baudis and Cleary, 2001; http://www.progenetix.net/).
Mann–Whitney U-test was performed to calculate P-values for differences in CNA numbers or PAG between sample groups. Differences were considered as significant with a P-value of ⩽0.05.
For survival analysis, Log-rank statistics (Kaplan–Meier estimator) were applied to determine the prognostic impact of clinicopathological parameters, DTCs and PAG on patient survival during a follow-up period of 7.5 years (median=14 month, range=0–90 months). For multivariable analysis, Cox regression (Likelihood ratio test, backwards) analyses were performed to estimate the prognostic impact of the variables. Patients with an overall survival <3 months (n=6) as well as positive resection margins (⩾R1; n=7) were excluded from survival analysis. One patient was lost to follow-up and could therefore not be included in survival analysis. Four patients were DTCpositive, but no mCGH analysis could be performed, therefore these patients were as well excluded from survival analysis. In total, 44 patients were included in survival analyses.
For Subpopulation Treatment Effect Pattern Plot (STEPP) analysis, the R-package ‘STEPP’ (version 3.0–11; https://cran.r-project.org/web/packages/stepp/index.html; Bonetti and Gelber, 2004) was used together with R version 3.3.1 to calculate PAG-dependent hazard ratios (HRs) employing a tail-oriented subpopulation scheme. If a patient had more than one DTC, we selected for this patient the DTC with the highest PAG (PAGmax) for STEPP analysis, based on the hypothesis that the highest PAG is associated with the most deleterious effect. For STEPP analysis, we started from the lowest PAGmax value, limiting PAGmax values were successively increased and only patients with at least the specified value were included. This resulted in successively smaller first treatment groups. The second treatment groups were fixed (independent of PAG) and comprised all patients with no cells or only non-aberrant DTCs. From the ‘STEPP’ R-package, we selected the cumulative incidence method to calculate HRs. For obtaining 95% confidence intervals, pointwise critical normal quantiles were multiplied by the STEPP-derived s.e. estimates for the HR log ratios. Group-splitting PAG values for Kaplan–Meier survival analyses (Log-rank test) were determined according to the respective STEPP plot by choosing the PAG value that marks the onset of a HR plateau or continuous HR increase.
To validate our results in an independent cohort (cohort #2), we investigated 21 BMDTCs and 10 LNDTCs from 19 DTCpositive patients with operable EAC by mCGH and in total 29 patients could be included for survival analysis. This cohort is part of a larger collective described previously by Stoecklein et al (2008). The PAG was calculated and STEPP analysis and Log-rank statistics (Kaplan–Meier estimator) were performed to determine the prognostic impact of the PAG on patient survival also in this independent cohort.
Results
Detection of DTCs in EAC patients
To determine the specificity of our CK18-IF assay, we screened BM samples from 48 non-cancer patients with benign tumours or inflammatory diseases (see Supplementary Methods). We detected CK18positive cells in 6% (3 out of 48) of patients in this control cohort. In contrast, using the same CK18-IF assay on BM and LN preparations of 59 operable EAC patients (Table 1), we detected significantly more BMDTCs, that is, in 24% of samples (13 out of 53; P=0.01) as well as significantly more LNDTCs, that is, in 41% of investigated cases (9 out of 22; P<0.01) (Supplementary Figure S1).
mCGH analyses of single DTCs and corresponding OESPTs and LNMETs
Next, we used mCGH to analyse 43 BM/LNDTCs from 18 DTCpositive patients (31%) that were isolated and successfully subjected to WGA (Supplementary Figure S1; Supplementary Tables S1 and S2). In addition to the DTCs, we were able to obtain 67 corresponding whole-genome amplified OESPT/LNMET samples from 17 DTCpositive patients (Supplementary Figure S1; Supplementary Tables S1 and S2) to compare potential CNA differences between DTCs isolated from different organs and their matched primary tumours, as well as LNMET.
The common CNAs detected among all samples were typical for EAC (Baudis and Cleary, 2001; http://www.progenetix.net/), for example, gains of chromosomes 1q, 3q, 5p, 6p, 7p, 8q, 12, 17q, 19 and 20 and losses of chromosomes 4, 5q, 8p, 9p, 16q and 18q (Supplementary Figure S3; Supplementary Table S3). These typical EAC alterations were present in all OESPT and LNMET samples, in 12 out of 16 (75%) of LNDTCs and in 6 out of 16 (38%) of BMDTCs. This low percentage for BMDTCs did go along with a low number of altered chromosomes.
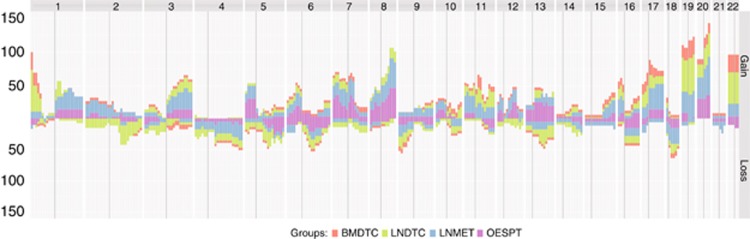
In the following step, we calculated the frequency of each alteration for the different sample types (BMDTCs, LNDTCs, OESPTs and LNMETs) and examined the data for shared alterations across the different sample sets. We observed several common alterations in LNDTCs and LNMETs, which could not be observed in the OESPTs (Figure 1; Supplementary Table S4). On the other hand, BMDTCs had similar alterations compared with the OESPTs (Figure 1; Supplementary Table S5).
Figure 1.
Penetrance plot from all mCGH samples of cohort #1. Horizontal view of chromosomes 1 to 22 on the x axis. The cumulative percentages are plotted on the y axis. Upward peaks denote chromosomal gains, whereas downward peaks represent chromosomal losses. OESPT=purple bars, LNMET=blue bars, LNDTC=green bars and BMDTC=red bars.
Unsupervised hierarchical cluster analysis revealed in the majority of patients (53%) with more than one aberrant DTC that CNAs were highly similar between the DTCs from the same patient (Supplementary Figure S4).
CIN in DTCs from EAC patients
Upon mCGH, we found 91% (21 out of 23) of the LNDTCs being aberrant, while only 50% of the BMDTCs (10 out of 20) displayed any CNA. We also noted a significant higher mean alteration number in the LNDTCs compared with the BMDTCs (8.78±6.52 vs 2.40±3.53; P<0.01; Figure 1; Table 2).
Table 2. Overview of results from the mCGH analyses of cohort #1.
| BMDTC | OESPT | LNDTC | LNMET | |
|---|---|---|---|---|
| Samples analysed (n=110) | 20 | 30 | 23 | 37 |
| Samples with chromosomal alterations | 10 | 29 | 21 | 34 |
| Mean number of chromosomal gains | 1.90 | 4.37 | 5.22 | 4.54 |
| Mean number of chromosomal losses | 0.50 | 1.70 | 3.57 | 2.32 |
| Mean number of alterations | 2.40 | 6.07 | 8.78 | 6.86 |
| Maximum percentage of aberrant genome | 25.39 | 30.57 | 42.83 | 62.22 |
| Mean percentage of aberrant genome | 3.89 | 10.04 | 16.26 | 12.49 |
| Median percentage of aberrant genome | 0.44 | 9.24 | 16.74 | 8.62 |
Abbreviations: BMDTC=disseminated tumour cell from the bone marrow; LNDTC=disseminated tumour cell from the lymph node; LNMET=lymph node metastasis; mCGH=metaphase-based comparative genomic hybridisation; OESPT=primary tumour.
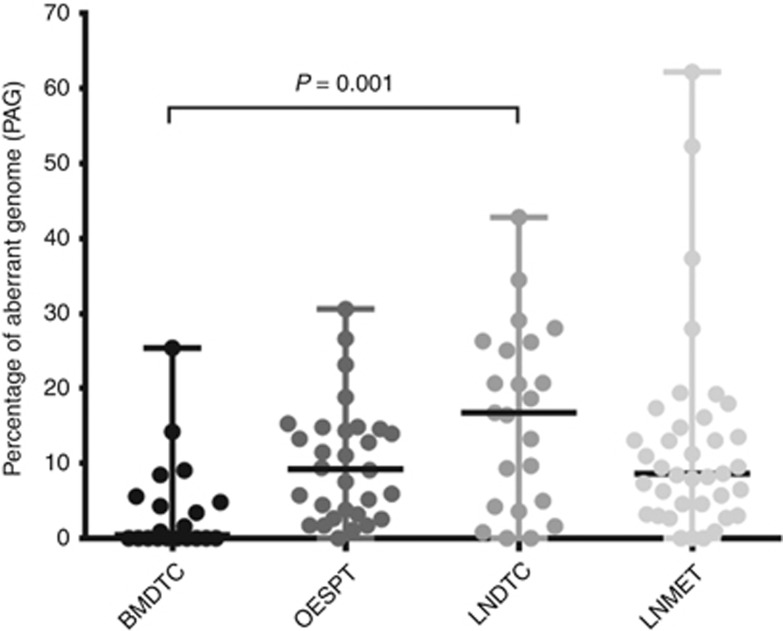
In a next step, we determined the extent of CIN in DTCs, corresponding OESPTs and LNMETs. For these analyses, we used the PAG (see Material and Methods section). Comparing the different sample groups, the BMDTCs displayed the lowest PAG (25.39) and LNMET the highest PAG (62.22) (Figure 2; Table 2). We then compared the PAG of the BMDTCs and LNDTCs to test whether DTCs from different organs show a different extent of CIN. Here we observed a significant difference in the median PAG of BMDTCs and LNDTCs (0.44 vs 16.74; P<0.01; Figure 2, Table 2), even if we excluded all non-aberrant cells from the analysis (5.20 vs 18.64; P=0.02).
Figure 2.
Overview of the PAG in cohort #1. Dot-plot diagram for samples analysed by mCGH. The black horizontal lines denote the median values. BMDTCs showed a significant lower PAG than LNDTCs (0.44, n=20 vs 17.74, n=23; P<0.01).
Prognostic value of DTC's PAG/CIN
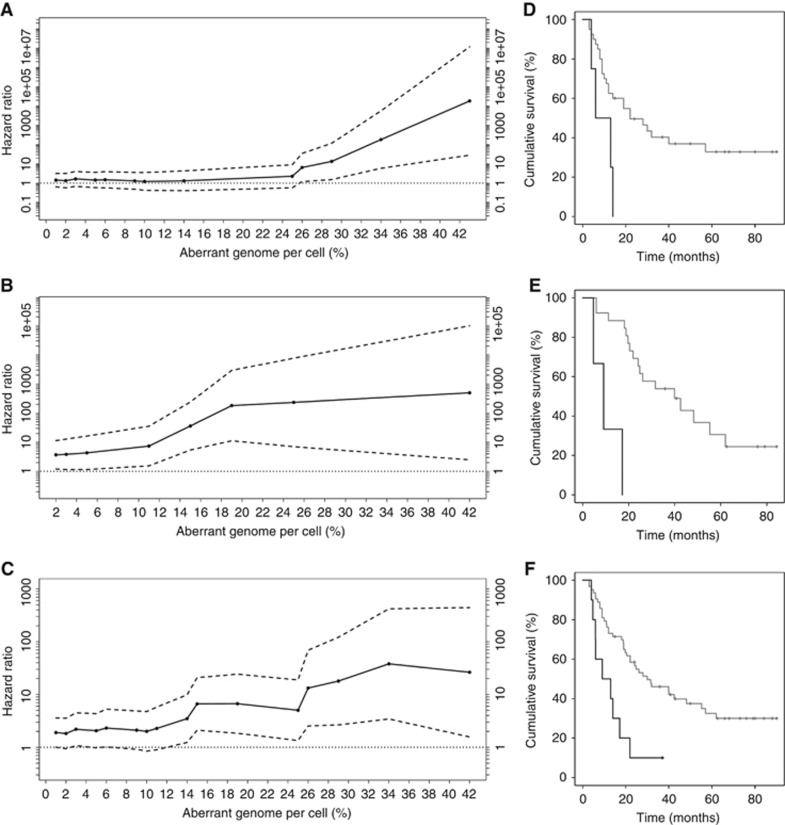
Having observed typical CNA patterns for EAC and highly variable PAG among individual DTCs, as reported in a previous study of an independent cohort (Stoecklein et al, 2008), we tested whether PAGhigh DTCs reflect a more aggressive (minimal residual) disease and correlated the PAG to clinical follow-up data. For this, we evaluated typical clinicopathological parameters in a first step and observed a significant prognostic impact of the presence of LNMET (pN0 vs pN1–3; P<0.01; Table 3). In a next step, we examined the prognostic value of the presence of DTCs in cohort #1 and noticed no influence on patient survival (P=0.38; Supplementary Figure S5A; Table 3). Subsequently, we used a STEPP analysis to assess the prognostic impact of the DTC's PAG in cohort #1. This revealed an increasing risk with advancing PAG (PAG⩾26; P=0.03; Figure 3A). Consistently, corresponding Kaplan–Meier analyses disclosed that DTCpositive patients with a PAG of ⩾26 (n=4) had a significantly reduced survival compared with patients with a PAG<26 or DTCnegative patients (P=0.02; Figure 3D; Table 3). Comparing only DTC-positive patients with high PAG to those with low PAG (CINhigh vs CINlow), we observed an analogous significant difference in survival when splitting the cohort at a PAG of 26 (P=0.04; Supplementary Figure S5B; Table 3). In both BMDTCs as well as LNDTCs, PAG values that were above the threshold contributed to shorter survival. In a multivariable analysis, the presence of LNMET (P=0.01) and a PAG of ⩾26 in DTCpositive patients (P=0.01) reached statistical significance (Table 3).
Table 3. Survival analysis of the patient cohorts included in DTC analyses.
| Univariate analysis |
Multivariable analysis |
||||
|---|---|---|---|---|---|
| Data set | Risk factor | Median survival (months) | P-value | Relative risk (95% confidence interval) | P-value |
| #1, n=44 | Age: ⩽65 (n=24) vs ⩾65 (n=20) | 31.0 vs 13.0 | 0.13 | — | NS |
| Sex: m (n=34) vs f (n=10) | 16.5 vs 31.0 | 0.62 | — | NS | |
| T-category: pT1–2 (n=22) vs pT3–4 (n=22) | 22.0 vs 13.0 | 0.27 | — | NS | |
| N-category: pN0 (n=14) vs pN1–3 (n=30) | NA vs 13.5 | <0.01 | 4.03 (1.40–11.63) | 0.01 | |
| M-category: M0 (n=38) vs M1 (n=6) | 22.0 vs 13.0 | 0.38 | — | NS | |
| Grading: G1–2 (n=18) vs G3–4 (n=26) | 30.0 vs 16.5 | 0.46 | — | NS | |
| DTCnegative (n=30) vs DTCpositive (n=14) | 22.0 vs 14.0 | 0.38 | — | NS | |
| DTCpositive PAG <26/DTCnegative (n=40) vs DTCpositive PAG⩾26 (n=4) | 22.0 vs 9.5 | 0.02 | — | NS | |
| DTCpositive PAG <26 (n=10) vs DTCpositive PAG⩾26 (n=4) | 26.0 vs 9.5 | 0.04 | 142.35 (4.05–5009.81) | 0.01 | |
| #2, n=29 | Age: ⩽62 (n=15) vs >62 (n=14) | 42.5 vs 20.1 | 0.21 | — | NS |
| Sex: m (n=27) vs f (n=2) | 26.1 vs 39.9 | 0.38 | — | NS | |
| T-category: pT1–2 (n=22) vs pT3–4 (n=7) | 42.5 vs 21.9 | 0.13 | 5.50 (1.55–19.55) | 0.01 | |
| N-category: pN0 (n=8) vs pN1–3 (n=21) | 62.0 vs 24.8 | 0.16 | — | NS | |
| M-category: M0 (n=25) vs M1 (n=4) | 39.9 vs 18.5 | 0.54 | — | NS | |
| Grading: G1–2 (n=12) vs G3–4 (n=17) | 39.9 vs 24.8 | 0.32 | — | NS | |
| DTCnegative (n=20) vs DTCpositive (n=9) | 42.5 vs 21.9 | 0.02 | 3.39 (1.03–11.15) | 0.04 | |
| DTCpositive PAG <19/DTCnegative (n=26) vs DTCpositive PAG⩾19 (n=3) | 39.9 vs 9.1 | <0.01 | 10.44 (1.60–68.07) | 0.01 | |
| DTCpositive PAG <19 (n=6) vs DTCpositive PAG⩾19 (n=3) | 25.1 vs 9.1 | 0.02 | — | NS | |
| #1 and #2, n=73 | Age: ⩽64 (n=38) vs >64 (n=35) | 39.9 vs 20.4 | 0.27 | — | NS |
| Sex: m (n=61) vs f (n=12) | 22.0 vs 36.0 | 0.44 | — | NS | |
| T-category: pT1–2 (n=44) vs pT3–4 (n=29) | 32.0 vs 19.7 | 0.10 | — | NS | |
| N-category: pN0 (n=22) vs pN1–3 (n=51) | NA vs 19.7 | <0.01 | 2.73 (1.26–5.94) | 0.01 | |
| M-category: M0 (n=63) vs M1 (n=10) | 28.0 vs 16.1 | 0.22 | — | NS | |
| Grading: G1–2 (n=30) vs G3–4 (n=43) | 31.6 vs 19.7 | 0.17 | — | NS | |
| DTCnegative (n=50) vs DTCpositive (n=23) | 32.0 vs 17.2 | 0.05 | — | NS | |
| DTCpositive PAG <15/DTCnegative (n=63) vs DTCpositive PAG⩾15 (n=10) | 30.0 vs 11.1 | <0.01 | 2.65 (1.24–5.66) | 0.01 | |
| DTCpositive PAG <15 (n=13) vs DTCpositive PAG⩾15 (n=10) | 26.1 vs 11.1 | 0.02 | 3.23 (1.15–9.10) | 0.03 | |
Abbreviations: DTC=disseminated tumour cell; f=female; m=male; M-category=distant metastases; NA=not available; N-category=lymph node metastases; NS=not significant in multivariable analysis; PAG=percentage of aberrant genome per cell; T-category=primary tumour size. P-values ⩽0.05 are presented in bold.
Figure 3.
Overview of results from STEPP and survival analyses for cohorts #1, #2 and the pooled data set (cohort #1 and cohort #2). (A–C) STEPP analyses of the effect of PAG on patient survival as measured by the HR. Solid black lines indicate HRs and dashed black lines indicate 95% upper and lower confidence intervals (CIs). CIs and P-values correspond to HR log ratios. A PAG⩾26 showed a significant effect in cohort #1 (n=44; HR=6.51; CI: 1.21–34.97; P=0.03; (A)). In cohort #2, PAG⩾19 demonstrated a significant effect (n=29; HR=180.77; CI: 11.20–2917.72; P<0.01; (B)). In the pooled data set (cohort #1 and cohort #2), a PAG⩾15 had a significant effect (n=73; HR=6.64; CI: 2.11–20.88; P<0.01; (C)). (D–F) Survival analyses by Kaplan–Meier estimator. Prognostic influence of PAG (⩾26) in DTCs from BM or LN in cohort #1 (Log-rank test; median survival 22.0 vs 9.5 months; P=0.02). Black line=PAG⩾26, grey line=PAG <26 (D). Prognostic influence of PAG (⩾19) in DTCs from BM or LN in cohort #2 (Log-rank test; median survival 39.9 vs 9.1 months; P<0.01). Black line=PAG⩾19, grey line=PAG<19 (E). Prognostic influence of PAG (⩾15) in DTCs from BM or LN in the pooled data set (Log-rank test; median survival 30.0 vs 11.1 months; P<0.01). Black line=PAG⩾15, grey line=PAG<15 (F).
Strikingly, we could validate this association when testing an independent, previously published mCGH data set consisting of informative 29 EAC patients (Stoecklein et al, 2008; cohort #2). The genomic data were overall comparable, also displaying a significantly lower median PAG in the BMDTCs compared with the LNDTCs (19.1 vs 20.04; P<0.01; Supplementary Figure S6A). Using the same clinical inclusion parameters, STEPP analysis disclosed an increasing risk with advancing PAG (PAG⩾19; P<0.01; Figure 3B) as in cohort #1, which as well was correlated to a shortened survival (P<0.01; Figure 3E; Table 3). Also in this data set, we found a significantly decreased survival in patients with CINhigh DTCs compared with patients with CINlow DTCs (P=0.02; Supplementary Figure S5C; Table 3). In a final analysis, we pooled the data from both independent cohorts (cohort #1 and cohort #2) to increase the number of patients (n=73). We noted a significantly lower PAG in the BMDTCs compared with the LNDTCs as already observed for the two separate cohorts (3.41 vs 18.64; P<0.01; Supplementary Figure S6B). Again, we could confirm an increasing risk with advancing PAG by STEPP analysis (PAG⩾15; P<0.01; Figure 3C) and a significantly shorter survival in patients with a higher PAG (P<0.01; Figure 3F; Table 3), even if we compared only CINhigh to CINlow DTCs (P=0.02; Supplementary Figure S5D; Table 3). As in the separate analysis of cohort #1, the prognostic impact of the presence of LNMET (P=0.01) and of an increased PAG (P=0.01) reached statistical significance in multivariable analysis (Table 3). Interestingly, we could also observe a trend for increasing risk with advancing PAG by STEPP analysis (PAG⩾15; P=0.07, Supplementary Figure S7A) when analysing BMDTCs only. This trend emerged also in survival analysis, showing a shorter survival in patients with BMDTCs with a higher PAG (P=0.06; Supplementary Figure S7B).
Discussion
Here we investigated whether increased CIN (using PAG as a measure) in solitary DTCs isolated from operable EAC patients is associated with poorer prognosis and therefore indicates a more aggressive minimal residual disease (MRD).
To obtain information on the genomic characteristics of the MRD cells in EAC patients, we used an anti-CK18/EpCAM IF to detect marker-positive DTCs. The methods for single-cell isolation, whole-genome amplification and mCGH were essentially performed as described in previous investigations (Klein et al, 2002; Schmidt-Kittler et al, 2003; Stoecklein et al, 2008; Weckermann et al, 2009). Using this workflow, we observed genomic alterations of the DTCs that were in line with CNA patterns typical for EAC (Goh et al, 2011; Dulak et al, 2012; Frankel et al, 2014; http://www.progenetix.net/). Despite this, CNA patterns frequently diverged between solitary DTCs and matched OESPT/LNMET, which can be expected from the known intratumoural heterogeneity of EAC (Maley et al, 2006; Stoecklein and Klein, 2010; Dulak et al, 2013; Cao et al, 2015; Hao et al, 2016) and other cancer types (e.g., Jamal-Hanjani et al, 2017). This heterogeneity is obviously more pronounced in our investigation, as single-cell DNA was compared with a DNA profile from thousands of laser microdissected cells. Nonetheless, LNDTCs shared alterations with LNMETs, which were not detected in OESPTs, while other alterations were exclusively shared by BMDTCs and OESPTs but not by established LNMETs, indicating site-specific CNAs in lymphatic metastases. In this context, it is interesting to highlight two observations confirming previous mCGH data of DTCs in oesophageal cancer (Stoecklein et al, 2008): (i) LNDTCs displayed a significantly higher PAG compared with BMDTCs; and (ii) If more than one aberrant LNDTC was present per patient, LNDTCs displayed highly similar CNAs, suggesting clonal expansion.
Careful interpretation is warranted concerning non-aberrant DTCs isolated from the BM. Our screening experiment of BM samples from a similarly large non-cancer patient cohort revealed a false-positive rate of 6%, indicating that some of the unaltered DTCs might be non-malignant cells. One possible explanation is unspecific staining (e.g., by antibody entrapment). However, it is notable that our control group did not consist of healthy controls but of patients with benign tumours and inflammatory diseases. For such a patient cohort, previous investigations on peripheral blood revealed higher detection rates of disseminated epithelial cells (7.5% in >200 individuals) when compared with healthy controls (Miller et al, 2010). Nevertheless, the significantly higher frequency of DTCs observed in EAC patients suggests that the majority of these cells was indeed of malignant origin, especially because we cannot exclude the existence of alterations <10 Mbp, which is the lower resolution level of our mCGH (Fuhrmann et al, 2008; Möhlendick et al, 2013). For example, Schardt et al (2005) could demonstrate cancer-relevant chromosomal alterations as well as subchromosomal DNA changes in breast cancer BMDTCs without mCGH alterations with higher-resolution methods (i.e., quantitative PCR for LOH).
Low-aberrant BMDTCs may represent early DTCs that homed to the BM niche (Klein, 2009). Once arrived, colonisation and metastatic outgrowth may have been impeded by the BM microenvironment, by inducing a dormant, non-proliferative state (Aguirre-Ghiso et al, 2013; Gužvić and Klein, 2013; Pantel and Alix-Panabieres, 2014; Sosa et al, 2014). Although our current study cannot prove or refute this hypothesis, we were intrigued by the varying levels of CIN, reflected by the different PAG observed in both DTC groups, the BMDTCs and the LNDTCs, respectively.
CIN is a hallmark of most human cancers and a prerequisite for genetic heterogeneity, tumour progression and metastasis (Nowak et al, 2002; Bakhoum and Swanton, 2014). It is generally accepted that higher numbers of alterations are directly linked to CIN (Thompson et al, 2010; Nicholson and Cimini, 2015), to disease progression (Chin et al, 2004; Li et al, 2014) and increased genomic heterogeneity (Jamal-Hanjani et al, 2017). In EAC development, the number of CNAs, and thus the extent of CIN, increases during progression through the different premalignant stages of Barrett’s oesophagus to invasive cancer and LN metastasis (Barrett et al, 1999; Maley et al, 2006; Pasello et al, 2009; Paulson et al, 2009; Merlo et al, 2010; Davison et al, 2014; Li et al, 2014). Recent data suggested that the risk of developing EAC is mediated by the acquisition of genetic instability early in Barrett lesion development (Martinez et al, 2016). For established oesophageal primary tumours, Pasello et al (2009) reported a significant correlation between CNA number and patient survival (12 aberrations; P=0.014). Accordingly, a study by Davison et al (2014) reported higher degrees of aneuploidy being directly linked to prognosis.
Motivated by published CNA data in EAC development, we hypothesised that non-/low-aberrant DTCs (CINlow) represent a dead end or at least retarded progression of MRD, whereas highly aberrant DTCs (CINhigh) indicate a more progressed and aggressive MRD. According to our hypothesis, patients with DTCs displaying high CIN levels should therefore exhibit shorter survival intervals than those with lower or absent alterations upon mCGH. In fact, our analyses revealed in two independent cohorts and in the pooled data set that patients with DTCs displaying higher aberrant genomes (PAG⩾26 cohort #1; PAG⩾19 cohort #2; PAG⩾15 pooled cohort) had a significantly worse prognosis compared with those patients with a lower PAG. It seems likely that only the high-aberrant LNDTCs contribute to the increased risk in our patient cohorts, but notably, DTCs from both organs contributed to the reduced survival of our patients. Additionally, we could observe a trend for an increased risk for shorter survival in the exclusive analysis of BMDTCs as well. Here a PAG of ⩾15 seems to reduce patient prognosis, reflecting the difference in CIN levels of BMDTCs and LNDTCs as well.
The clinical relevance of CIN and CNA-heterogeneity in early-stage cancers clearly emerged from recent large-scale next-generation sequencing studies in non-small cell lung cancer, showing that CIN is not only a significant driver of parallel evolution but also a predictor of poor outcome (Abbosh et al, 2017; Jamal-Hanjani et al, 2017). Importantly, primary tumour biopsies cannot provide all relevant genomic information of an individual disease, but there is hope that liquid biopsies provide better information. In this context, direct analysis of systemic cancer by DTC profiling therefore appears as a promising approach to identify high-risk patients to deliver the required intensified care and therapy, respectively. From a clinical point of view, it would be interesting to test whether circulating tumour cells (CTCs) could provide similar molecular information on the MRD state. As the CTC concentration is extremely low in the MRD situation, diagnostic leukapheresis might represent a feasible approach (Fischer et al, 2013; Stoecklein et al, 2016) to enable an informative liquid biopsy.
Acknowledgments
This work was supported by the German Research Foundation grants STO464/2-1 and STO464/2-2.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, Marafioti T, Kirkizlar E, Watkins TBK, McGranahan N, Ward S, Martinson L, Riley J, Fraioli F, Al Bakir M, Grönroos E, Zambrana F, Endozo R, Bi WL, Fennessy FM, Sponer N, Johnson D, Laycock J, Shafi S, Czyzewska-Khan J, Rowan A, Chambers T, Matthews N, Turajlic S, Hiley C, Lee SM, Forster MD, Ahmad T, Falzon M, Borg E, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Hafez D, Naik A, Ganguly A, Kareht S, Shah R, Joseph L, Marie Quinn A, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Oukrif D, Akarca AU, Hartley JA, Lowe HL, Lock S, Iles N, Bell H, Ngai Y, Elgar G, Szallasi Z, Schwarz RF, Herrero J, Stewart A, Quezada SA, Peggs KS, van Loo P, Dive C, Lin CJ, Rabinowitz M, Aerts HJWL, Hackshaw A, Shaw JA, Zimmermann BG, Swanton C (2017) Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545: 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Ghiso JA, Bragado P, Sosa MS (2013) Metastasis awakening: targeting dormant cancer. Nat Med 19: 276–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderegg MCJ, Lagarde SM, Jagadesham VP, Gisbertz SS, Immanuel A, Meijer SL, Hulshof MCCM, Bergman JJGHM, van Laarhoven HWM, Griffin SM, van Berge Henegouwen MI (2016) Prognostic significance of the location of lymph node metastases in patients with adenocarcinoma of the distal esophagus or gastroesophageal junction. Ann Surg 264: 847–853. [DOI] [PubMed] [Google Scholar]
- Bakhoum SF, Swanton C (2014) Chromosomal instability, aneuploidy, and cancer. Front Oncol 4: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandla S, Pennathur A, Luketich JD, Beer DG, Lin L, Bass AJ, Godfrey TE, Litle VR (2012) Comparative genomics of esophageal adenocarcinoma and squamous cell carcinoma. Ann Thorac Surg 93: 1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, Rabinovitch PS, Reid BJ (1999) Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet 22: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudis M, Cleary ML (2001) Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics 17: 1228–1229. [DOI] [PubMed] [Google Scholar]
- Bonavina L, Soligo D, Quirici N, Bossolasco P, Cesana B, Deliliers GL, Peracchia A (2001) Bone marrow—disseminated tumor cells in patients with carcinoma of the esophagus or cardia. Surgery 129: 15–22. [DOI] [PubMed] [Google Scholar]
- Bonetti M, Gelber RD (2004) Patterns of treatment effects in subsets of patients in clinical trials. Biostatistics 5: 465–481. [DOI] [PubMed] [Google Scholar]
- Braun S, Kentenich C, Janni W, Hepp F, de Waal J, Willgeroth F, Sommer H, Pantel K (2000) Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. J Clin Oncol 18: 80–86. [DOI] [PubMed] [Google Scholar]
- Cao W, Wu W, Yan M, Tian F, Ma C, Zhang Q, Li X, Han P, Liu Z, Gu J, Biddle FG (2015) Multiple region whole-exome sequencing reveals dramatically evolving intratumor genomic heterogeneity in esophageal squamous cell carcinoma. Oncogenesis 4: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo W-L, Ljung B-M, Chew K, Myambo K, Miranda M, Krig S, Garbe J, Stampfer M, Yaswen P, Gray JW, Lockett SJ (2004) In situ analyses of genome instability in breast cancer. Nat Genet 36: 984–988. [DOI] [PubMed] [Google Scholar]
- Davison JM, Yee M, Krill-Burger JM, Lyons-Weiler MA, Kelly LA, Sciulli CM, Nason KS, Luketich JD, Michalopoulos GK, LaFramboise WA (2014) The degree of segmental aneuploidy measured by total copy number abnormalities predicts survival and recurrence in superficial gastroesophageal adenocarcinoma. PLoS One 9: e79079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driemel C, Kremling H, Schumacher S, Will D, Wolters J, Lindenlauf N, Mack B, Baldus SA, Hoya V, Pietsch JM, Panagiotidou P, Raba K, Vay C, Vallböhmer D, Harréus U, Knoefel WT, Stoecklein NH, Gires O (2013) Context-dependent adaption of EpCAM expression in early systemic esophageal cancer. Oncogene 33: 4904–4915. [DOI] [PubMed] [Google Scholar]
- Dulak AM, Schumacher SE, van Lieshout J, Imamura Y, Fox C, Shim B, Ramos AH, Saksena G, Baca SC, Baselga J, Tabernero J, Barretina J, Enzinger PC, Corso G, Roviello F, Lin L, Bandla S, Luketich JD, Pennathur A, Meyerson M, Ogino S, Shivdasani RA, Beer DG, Godfrey TE, Beroukhim R, Bass AJ (2012) Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res 72: 4383–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, McKenna A, Carter SL, Cibulskis K, Sivachenko A, Saksena G, Voet D, Ramos AH, Auclair D, Thompson K, Sougnez C, Onofrio RC, Guiducci C, Beroukhim R, Zhou Z, Lin L, Lin J, Reddy R, Chang A, Landrenau R, Pennathur A, Ogino S, Luketich JD, Golub TR, Gabriel SB, Lander ES, Beer DG, Godfrey TE, Getz G, Bass AJ (2013) Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 45: 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JC, Niederacher D, Topp SA, Honisch E, Schumacher S, Schmitz N, Zacarias Fohrding L, Vay C, Hoffmann I, Kasprowicz NS, Hepp PG, Mohrmann S, Nitz U, Stresemann A, Krahn T, Henze T, Griebsch E, Raba K, Rox JM, Wenzel F, Sproll C, Janni W, Fehm T, Klein CA, Knoefel WT, Stoecklein NH (2013) Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proc Natl Acad Sci USA 110: 16580–16585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A, Armour N, Nancarrow D, Krause L, Hayward N, Lampe G, Smithers BM, Barbour A (2014) Genome-wide analysis of esophageal adenocarcinoma yields specific copy number aberrations that correlate with prognosis. Genes Chromosomes Cancer 53: 324–338. [DOI] [PubMed] [Google Scholar]
- Fuhrmann C, Schmidt-Kittler O, Stoecklein NH, Petat-Dutter K, Vay C, Bockler K, Reinhardt R, Ragg T, Klein CA (2008) High-resolution array comparative genomic hybridization of single micrometastatic tumor cells. Nucleic Acids Res 36: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh XY, Rees JRE, Paterson AL, Chin SF, Marioni JC, Save V, O'Donovan M, Eijk PP, Alderson D, Ylstra B, Caldas C, Fitzgerald RC (2011) Integrative analysis of array-comparative genomic hybridisation and matched gene expression profiling data reveals novel genes with prognostic significance in oesophageal adenocarcinoma. Gut 60: 1317–1326. [DOI] [PubMed] [Google Scholar]
- Gužvić M, Klein CA (2013) Cancer dormancy: time to explore its clinical relevance. Breast Cancer Res 15: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J-J, Lin D-C, Dinh HQ, Mayakonda A, Jiang Y-Y, Chang C, Jiang Y, Lu C-C, Shi Z-Z, Xu X, Zhang Y, Cai Y, Wang J-W, Zhan Q-M, Wei W-Q, Berman BP, Wang M-R, Koeffler HP (2016) Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet 48: 1500–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosch S, Kraus J, Scheunemann P, Izbicki JR, Schneider C, Schumacher U, Witter K, Speicher MR, Pantel K (2000) Malignant potential and cytogenetic characteristics of occult disseminated tumor cells in esophageal cancer. Cancer Res 60: 6836–6840. [PubMed] [Google Scholar]
- Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, Forni G, Eils R, Fehm T, Riethmuller G, Klein CA (2008) Systemic spread is an early step in breast cancer. Cancer Cell 13: 58–68. [DOI] [PubMed] [Google Scholar]
- Izbicki JR, Hosch SB, Pichlmeier U, Rehders A, Busch C, Niendorf A, Passlick B, Broelsch CE, Pantel K (1997) Prognostic value of immunohistochemically identifiable tumor cells in lymph nodes of patients with completely resected esophageal cancer. N Engl J Med 337: 1188–1194. [DOI] [PubMed] [Google Scholar]
- Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, Salm M, Horswell S, Escudero M, Matthews N, Rowan A, Chambers T, Moore DA, Turajlic S, Xu H, Lee S-M, Forster MD, Ahmad T, Hiley CT, Abbosh C, Falzon M, Borg E, Marafioti T, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Shah R, Joseph L, Quinn AM, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Dentro S, Taniere P, O'Sullivan B, Lowe HL, Hartley JA, Iles N, Bell H, Ngai Y, Shaw JA, Herrero J, Szallasi Z, Schwarz RF, Stewart A, Quezada SA, Le Quesne J, van Loo P, Dive C, Hackshaw A, Swanton C (2017) Tracking the evolution of non-small-cell lung cancer. N Engl J Med 376: 2109–2121. [DOI] [PubMed] [Google Scholar]
- Klein CA (2009) Parallel progression of primary tumours and metastases. Nat Rev Cancer 9: 302–312. [DOI] [PubMed] [Google Scholar]
- Klein CA (2013) Selection and adaptation during metastatic cancer progression. Nature 501: 365–372. [DOI] [PubMed] [Google Scholar]
- Klein CA, Blankenstein TJF, Schmidt-Kittler O, Petronio M, Polzer B, Stoecklein NH, Riethmüller G (2002) Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet 360: 683–689. [DOI] [PubMed] [Google Scholar]
- Klein CA, Schmidt-Kittler O, Schardt JA, Pantel K, Speicher MR, Riethmüller G (1999) Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci USA 96: 4494–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CA, Stoecklein NH (2009) Lessons from an aggressive cancer: evolutionary dynamics in esophageal carcinoma. Cancer Res 69: 5285–5288. [DOI] [PubMed] [Google Scholar]
- Lee L, Ronellenfitsch U, Hofstetter WL, Darling G, Gaiser T, Lippert C, Gilbert S, Seely AJ, Mulder DS, Ferri LE (2013) Predicting lymph node metastases in early esophageal adenocarcinoma using a simple scoring system. J Am Coll Surg 217: 191–199. [DOI] [PubMed] [Google Scholar]
- Li X, Galipeau PC, Paulson TG, Sanchez CA, Arnaudo J, Liu K, Sather CL, Kostadinov RL, Odze RD, Kuhner MK, Maley CC, Self SG, Vaughan TL, Blount PL, Reid BJ (2014) Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of Barrett's esophagus. Cancer Prev Res (Phila) 7: 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, Paulson TG, Blount PL, Risques R-A, Rabinovitch PS, Reid BJ (2006) Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 38: 468–473. [DOI] [PubMed] [Google Scholar]
- Martinez P, Timmer MR, Lau CT, Calpe S, Sancho-Serra MDC, Straub D, Baker A-M, Meijer SL, ten Kate FJW, Mallant-Hent RC, Naber AHJ, van Oijen AHAM, Baak LC, Scholten P, Bohmer CJM, Fockens P, Bergman JJGHM, Maley CC, Graham TA, Krishnadath KK (2016) Dynamic clonal equilibrium and predetermined cancer risk in Barrett's oesophagus. Nat Commun 7: 12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LMF, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, Maley CC (2010) A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 3: 1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Doyle GV, Terstappen LWMM (2010) Significance of circulating tumor cells detected by the cellsearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol 2010: 617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhlendick B, Bartenhagen C, Behrens B, Honisch E, Raba K, Knoefel WT, Stoecklein NH (2013) A robust method to analyze copy number alterations of less than 100 kb in single cells using oligonucleotide array CGH. PLoS One 8: e67031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JD, Stein HJ, Oyang T, Natsugoe S, Feith M, Werner M, Rudiger Siewert J (2000) Frequency and clinical impact of lymph node micrometastasis and tumor cell microinvolvement in patients with adenocarcinoma of the esophagogastric junction. Cancer 89: 1874–1882. [DOI] [PubMed] [Google Scholar]
- Nicholson JM, Cimini D (2015) Link between aneuploidy and chromosome instability. Int Rev Cell Mol Biol 315: 299–317. [DOI] [PubMed] [Google Scholar]
- Nones K, Waddell N, Wayte N, Patch A-M, Bailey P, Newell F, Holmes O, Fink JL, Quinn MCJ, Tang YH, Lampe G, Quek K, Loffler KA, Manning S, Idrisoglu S, Miller D, Xu Q, Waddell N, Wilson PJ, Bruxner TJC, Christ AN, Harliwong I, Nourse C, Nourbakhsh E, Anderson M, Kazakoff S, Leonard C, Wood S, Simpson PT, Reid LE, Krause L, Hussey DJ, Watson DI, Lord RV, Nancarrow D, Phillips WA, Gotley D, Smithers BM, Whiteman DC, Hayward NK, Campbell PJ, Pearson JV, Grimmond SM, Barbour AP (2014) Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun 5: 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Komarova NL, Sengupta A, Jallepalli PV, Shih I-M, Vogelstein B, Lengauer C (2002) The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci USA 99: 16226–16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K, Alix-Panabieres C (2014) Bone marrow as a reservoir for disseminated tumor cells: a special source for liquid biopsy in cancer patients. Bonekey Rep 3: 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasello G, Agata S, Bonaldi L, Corradin A, Montagna M, Zamarchi R, Parenti A, Cagol M, Zaninotto G, Ruol A, Ancona E, Amadori A, Saggioro D (2009) DNA copy number alterations correlate with survival of esophageal adenocarcinoma patients. Mod Pathol 22: 58–65. [DOI] [PubMed] [Google Scholar]
- Paulson TG, Maley CC, Li X, Li H, Sanchez CA, Chao DL, Odze RD, Vaughan TL, Blount PL, Reid BJ (2009) Chromosomal instability and copy number alterations in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res 15: 3305–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzer B, Klein CA (2013) Metastasis awakening: the challenges of targeting minimal residual cancer. Nat Med 19: 274–275. [DOI] [PubMed] [Google Scholar]
- Rustgi AK, El-Serag HB (2014) Esophageal carcinoma. N Engl J Med 371: 2499–2509. [DOI] [PubMed] [Google Scholar]
- Schardt JA, Meyer M, Hartmann CH, Schubert F, Schmidt-Kittler O, Fuhrmann C, Polzer B, Petronio M, Eils R, Klein CA (2005) Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell 8: 227–239. [DOI] [PubMed] [Google Scholar]
- Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein, Thomas J F, Kaufmann M, Diebold J, Arnholdt H, Muller P, Bischoff J, Harich D, Schlimok G, Riethmuller G, Eils R, Klein CA (2003) From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA 100: 7737–7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Slovak ML, Campbell LJ (eds). (2009) ISCN 2009: An International System for Human Cytogenetic Nomenclature (2009); Recommendations of the International Standing Committee on Human Cytogenetic Nomenclature. Karger: Basel, Switzerland. [Google Scholar]
- Shapiro J, van Klaveren D, Lagarde SM, Toxopeus ELA, van der Gaast A, Hulshof MCCM, Wijnhoven BPL, van Berge Henegouwen MI, Steyerberg EW, van Lanschot JJB (2016) Prediction of survival in patients with oesophageal or junctional cancer receiving neoadjuvant chemoradiotherapy and surgery. Br J Surg 103: 1039–1047. [DOI] [PubMed] [Google Scholar]
- Sosa MS, Bragado P, Aguirre-Ghiso JA (2014) Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer 14: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklein NH, Erbersdobler A, Schmidt-Kittler O, Diebold J, Schardt JA, Izbicki JR, Klein CA (2002) SCOMP is superior to degenerated oligonucleotide primed-polymerase chain reaction for global amplification of minute amounts of DNA from microdissected archival tissue samples. Am J Pathol 161: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklein NH, Fischer JC, Niederacher D, Terstappen LWMM (2016) Challenges for CTC-based liquid biopsies: low CTC frequency and diagnostic leukapheresis as a potential solution. Expert Rev Mol Diagn 16: 147–164. [DOI] [PubMed] [Google Scholar]
- Stoecklein NH, Hosch SB, Bezler M, Stern F, Hartmann CH, Vay C, Siegmund A, Scheunemann P, Schurr P, Knoefel WT, Verde PE, Reichelt U, Erbersdobler A, Grau R, Ullrich A, Izbicki JR, Klein CA (2008) Direct genetic analysis of single disseminated cancer cells for prediction of outcome and therapy selection in esophageal cancer. Cancer Cell 13: 441–453. [DOI] [PubMed] [Google Scholar]
- Stoecklein NH, Klein CA (2010) Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int J Cancer 126: 589–598. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hirota T (2016) Chromosomal instability: a common feature and a therapeutic target of cancer. Biochim Biophys Acta 1866: 64–75. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA (2010) Mechanisms of chromosomal instability. Curr Biol 20: R285–R295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorban S, Roder JD, Nekarda H, Funk A, Siewert JR, Pantel K (1996) Immunocytochemical detection of disseminated tumor cells in the bone marrow of patients with esophageal carcinoma. J Natl Cancer Inst 88: 1222–1227. [DOI] [PubMed] [Google Scholar]
- Visser E, Leeftink AG, van Rossum PSN, Siesling S, van Hillegersberg R, Ruurda JP (2016) Waiting time from diagnosis to treatment has no impact on survival in patients with esophageal cancer. Ann Surg Oncol 23: 2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckermann D, Polzer B, Ragg T, Blana A, Schlimok G, Arnholdt H, Bertz S, Harzmann R, Klein CA (2009) Perioperative activation of disseminated tumor cells in bone marrow of patients with prostate cancer. J Clin Oncol 27: 1549–1556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.