Abstract
Taxus spp. is a highly valuable medicinal plant with multiple pharmacological effects on various cancers. Cytochrome P450s (CYP450s) play important roles in the biosynthesis of active compounds in Taxus spp., such as the famous diterpenoid, Taxol. However, some specific CYP450 enzymes involved in the biosynthesis of Taxol remain unknown, and the systematic identification of CYP450s in Taxus has not been reported. In this study, 118 full-length and 175 partial CYP450 genes were identified in Taxus chinensis transcriptomes. The 118 full-length genes were divided into 8 clans and 29 families. The CYP71 clan included all A-type genes (52) belonging to 11 families. The other seven clans possessed 18 families containing 66 non-A-type genes. Two new gymnosperm-specific families were discovered, and were named CYP864 and CYP947 respectively. Protein sequence alignments revealed that all of the T. chinensis CYP450s hold distinct conserved domains. The expression patterns of all 118 CYP450 genes during the long-time subculture and MeJA elicitation were analyzed. Additionally, the expression levels of 15 novel CYP725 genes in different Taxus species were explored. Considering all the evidence, 6 CYP725s were identified to be candidates for Taxol biosynthesis. The cis-regulatory elements involved in the transcriptional regulation were also identified in the promoter regions of CYP725s. This study presents a comprehensive overview of the CYP450 gene family in T. chinensis and can provide important insights into the functional gene studies of Taxol biosynthesis.
Keywords: Taxus chinensis, cytochrome P450, transcriptome-wide identification, gene expression, Taxol biosynthesis
Introduction
Taxol (generic name paclitaxel), the main bioactive component of the Taxus species, is a highly effective anti-cancer agent widely used in the treatment of various sarcomas, melanomas, and carcinomas (Murphy et al., 1993). However, Taxol availability is restricted due to the insufficient natural supply; thus, Taxol supply and cost remain serious concerns because of its increasing requirements in chemotherapy (Cragg et al., 1993). In the near future, Taxol production and its potential precursors must rely on biological methods, either in Taxus tissues or in cell cultures (Ro et al., 2006; Ajikumar et al., 2010; Zhou et al., 2015). Therefore, understanding the biosynthetic pathway of Taxol and the enzymes that catalyze this series of reactions and their underlying molecular mechanism is essential.
The Taxol biosynthetic pathway starts with the cyclization of the universal diterpenoid precursor geranylgeranyl diphosphate to taxa-4(5),11(12)-diene. This taxane core is then decorated with a series of eight Cytochrome P450 (CYP450)-mediated oxidations, three CoA-dependent acylations, and several other transformations that lead to baccatin III, to which the C13-side chain is appended to afford Taxol (Jennewein et al., 2001; Croteau et al., 2006). Thus, CYP450s play a major role in Taxol biosynthesis (Rasool and Mohamed, 2016). Approximately half of the 20 enzymatic steps of the pathway are thought to be catalyzed by CYP450 oxygenases. The proposed order of the oxygenations on the taxane core begins with C5 and C10, then C13 and C9, and later C7 and C2. The final reactions include the epoxidation of the C4, C20-double bond (leading to the oxetane ring) and C1 oxygenation (Wheeler et al., 2001). In the past few years, studies have concentrated on the molecular biochemistry of Taxol biosynthesis (Chau et al., 2004; Wang et al., 2016). However, the CYP450 genes responsible for C1 hydroxylation, oxetane formation, C9 oxidation of the taxane core, and C2′-side-chain hydroxylation in Taxus remain unidentified.
CYP450s represent one of the largest gene families and play vital roles in many plant metabolic processes. They catalyze the oxidative modification of various substrates using oxygen and NAD(P)H (Chapple, 1998). Structurally, all plant CYP450s identified so far are membrane-bound enzymes. The vast majority of them are anchored on the endoplasmic reticulum by the hydrophobic signal sequence at the N-terminus (Williams et al., 2000). CYP450 protein sequences contain four unique structures, including the Heme-binding, PERF, and EXXR motifs, located in the K-helix and I-helix. The conserved cystein in the heme-binding motif (F–G-R-C-G) regulates the iron in their heme group. The R amino acid residue of the short string in the PERF motif, together with E and R amino acid residues in the K-helix, forms a salt bridge that is generally considered to be involved in locking the heme pocket into the corresponding position and ensuring the stability of the core structure (Hasemann et al., 1995). CYP450s for all organisms are named and classified by a P450 nomenclature committee (David Nelson: dnelson@uthsc.edu). To differentiate them from other species, plant CYP450s belong to families ranging from CYP71-CYP99, and then from CYP701-CYP999 (Durst and Nelson, 1995; Danielson, 2002). Moreover, CYP450s in plants are classified into two categories: A-type and non-A-type (Paquette et al., 2000). Based on the available sequences, plant CYP450s are further classified into 11 clans (Paquette et al., 2000). A-type CYP450s include only the CYP71 clan, whereas non-A-type CYP450s include 10 clans, namely, CYP51, CYP72, CYP74, CYP85, CYP86, CYP97, CYP710, CYP711, CYP727, and CYP746.
In recent years, the functional characterization of CYP450s involved in terpene biosynthesis has gathered attention. Many studies focusing on the transcriptome-wide identification of CYP450s for terpene biosynthesis have been done. An earlier research performed transcriptomic analyses based on 454 pyrosequencing data from Panax ginseng flowers, roots, stems, and leaves, thereby leading to the identitification of 326 potential CYP450s, including CYP716A47, which is related to the ginsenoside biosynthesis (Li C. et al., 2013). In addition, ~300 isotigs similar to CYP450s have been discovered from Salvia miltiorrhiza hairy roots using the RNA-Seq technology, of which six were further studied and the CYP76AH1 was functionally confirmed to catalyze the turnover of miltiradiene in tanshinones biosynthesis (Guo et al., 2013). Last year, 70 highly expressed CYP450s in the Xanthium strumarium trichomes were studied using an extensive analysis of transcriptomes. Among them, four CYP71 members (CYP71AV14, CYP71BL7, CYP71DD1, and CYP71AX30) were found to be the candidates involved in sesquiterpene lactone biosynthesis (Li et al., 2016). Transcriptome identification of genes for Taxol biosynthesis has been studied, however, no study has demonstrated interest in the CYP450 genes of Taxus (Wu et al., 2011; Sun et al., 2013). So, a systematic set of CYP450s and a standard nomenclature could be benefit to the functional identification of CYP450s, which might be involved in the biosynthesis of active ingredients in Taxus.
In this study, we established systematic information of CYP450s in Taxus chinensis by mining available transcriptome data. We first identified and classified putative full-length CYP450-encoding sequences. Phylogenetic analysis allowed us to identify groups of paralogs for further evaluation. Next, based on the KEGG database, we investigated the potential involvement in various biosynthesis pathways of these CYP450s, with emphasis on taxane biosynthesis. Moreover, the expression profiles in silico were characterized, some selected CYP450 genes were confirmed by qRT-PCR. And the expression levels of 15 novel CYP725 genes in different Taxus species were also explored. Lastly, the cis-regulatory elements involved in the transcriptional regulation were identified in the promoter regions of CYP725s. Our findings would contribute to advanced research and applications of CYP450 genes in Taxus.
Materials and methods
In silico mining of T. chinense CYP450s
All unigenes (containing contigs and singletons) of assembled T. chinense transcriptomes (Accession Numbers: SRR1343578, SRR1339474, and GSE28539) were obtained, then, they were clustered using CD-HIT software (Version 4.6), and the sequence identity >90% as the cutoff. The HMM model (PF00067) were retrieved from the Pfam database (http://pfam.sanger.ac.uk). After redundant sequences were removed, the HMMER program was used to identify the CYP450s, with an e-value cutoff of 1e-5. The nucleotide sequences of these selected unigenes were subjected to ORF Finder software (http://bioinf.ibun.unal.edu.co/servicios/sms/orf_find.html) for open reading frame (ORF) identification. The full-length CYP450 genes were identified as described by Chen et al. (2014). The search results were further consolidated with previously published genes in Genbank and other existing Taxus transcriptomes, including Taxus × media (SRR534003 and SRR534004), Taxus mairei (SRR350719), Taxus cuspidata (SRR032523).
Phylogenetic analysis
For comparison, a collection of CYP450s from Picea glauca and corresponding CYP names were obtained from Genbank. Multiple sequence alignment was performed with the ClustalW algorithm-based AlignX module in MEGA 7 software. The phylogenetic tree was constructed by Neighbour-Joining (NJ) method by p-distance in MEGA 7. The significance level for NJ analysis was examined by bootstrapping with 1,000 repeats.
Classification and characterization of full-length T. chinense CYP450s
All full-length CYP450 proteins were classified with reference sequences from an established P450 database (Nelson, 2009). Overall, 40, 55, and 97% sequence identities were used as cutoffs for family, subfamily, and allelic variants, respectively. The names of these CYP450 proteins were assigned by Prof. David Nelson. Theoretical iso-electric points (PI) and molecular weight (kDa) for each full-length CYP450 protein were predicted by ExPASy tool (http://www.expasy.org/tools/). Furthermore, CYP450 motifs were confirmed by Multiple Expectation Maximization for Motif Elicitation (MEME) program (http://alternate.meme-suite.org/). The subcellular locations were conducted using the TargetP1.1 server with specificity >0.95 (http://www.cbs.dtu.dk/services/TargetP/).
Functional annotation
Functional annotation was performed by searching T. chinensis CYP450s against the SWISS-PROT, NR, and NT databases, using BLAST with an E-value of 1e-5. The CYP450 genes were also mapped in Kyoto Encyclopedia of Genes and Genomes (KEGG) database to obtain reference metabolic pathways.
Expression pattern analysis
To analyze the expression levels of CYP450s in different Taxus cell lines, we used the reported Solexa sequencing libraries from cell lines CA (subcultured for 10 years) and NA (subcultured for 6 months), and Methyl Jasmonate (MeJA)-mediated Taxus cells harvested at 16 h after inoculation (Li et al., 2012; Zhang et al., 2015). Raw expression counts were calculated by FPKM method (reads of fragments per kilobase per million mapped). To identify the differentially expressed genes (DEGs), a greater than two-fold change and a false discovery rate (FDR) ≤ 0.001 were used to determine significant changes in expression. Heatmaps based on raw expression counts were generated with HemI 1.0 (Heatmap Illustrator software, version 1.0). The expression patterns of 17 randomly selected CYP450 genes in CA and NA were validated by qRT-PCR using the same RNA as the Solexa sequencing sample. The RNA were stored at −80°C.
The expression patterns of 15 novel CYP725 genes in different Taxus species, including T. chinensis, T. cuspidate, and T. media were detected using leaves from 5-year-old plants as sample.
Total RNAs was extracted using the RNAprep Pure Plant kit following manufacturer's protocal (TianGen, Beijing). RNA purity and concentration were detected using a NanoDrop_2000 UV-Vis spectrophotometer (Thermo, USA). Approximately 2 μg total RNA was reverse transcribed using the RevertAid™ First Strand cDNA Synthesis Kit (Thermo, USA). cDNA products were diluted 10-fold prior to use for real-time PCR reaction. Gene specific primers were designed by primer premier 5.0 software, and a housekeeping gene (actin) was chosen as the internal reference gene. All primer sequences were listed in Table S1. qRT-PCR reactions were performed in 10 μl volume containing 1 μl diluted cDNA, 0.3 μM forward primer, 0.3 μM reverse primer, and 5 μl 2 × SYBR Green PCR Master Mix (Applied Biosystems). The thermal conditions of the qRT-PCR reactions was 95°C for 5 min, and 40 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 15 s. Each experiment was performed with three biological and technical replicates. The relative expression levels were calculated using the 2−ΔΔCT method.
Cis-elements analysis of CYP725 genes
All promoter sequences (2 Kb upstream of initiation codon “ATG”) of CYP725 genes were extracted from the Taxus genome (Nystedt et al., 2013) by using GMAP (http://research-pub.gene.com/gmap/). Then, the on-line database PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) was used to identify the cis-acting elements of promoters for each gene (Yan et al., 2017).
Results
The construction of T. chinensis transcriptomes
To globally identify potential genes in T. chinensis transcriptomes were constructed from two Taxus cell lines CA and NA (Zhang et al., 2015), and the MeJA-treated Taxus cells for 16 h (Tm16) and those of mock-treated cells (Tm0; Li et al., 2012), resulting in 67,147 Unigenes with an average length of 910 bp and N50 of 1,552 bp (Table 1). The GC percentage of the unigenes was 41.37%.
Table 1.
Statistics of assembly of T. chinensis transcriptomes.
| Sample | Unigene | Total_length (bp) | GC (%) | N50 (bp) | Average (bp) |
|---|---|---|---|---|---|
| CA | 39,396 | 31,110,882 | 45.07 | 1428 | ND |
| NA | 43,009 | 31,758,961 | 45.63 | 1320 | ND |
| Tm0 | 39,176 | 29,459,951 | 45.69 | 812 | ND |
| Tm16 | 38,713 | 29,896,420 | 44.96 | 839 | ND |
| Total | 67,147 | 61,138,078 | 41.37 | 1552 | 910.51 |
N50, The largest length L such that 50% of all nucleotides are contained in unigenes of size at least L (de Jong et al., 2001). ND, no data.
Identification and classification of T. chinensis CYP450s
A total of 118 full-length and 175 partial CYP450 genes were identified in T. chinensis. The total number of T. chinensis CYP450 genes was 293, which is more than that of Arabidopsis thaliana (272). However, without the whole genome sequence, the strict criteria of all T. chinensis CYP450 genes were unmet. To further screen for full-length CYP450 genes, we performed BLAST search using standard CYP450 domains against other existing Taxus transcriptomes, such as T. media, T. mairei, and T. cuspidata. No new full-length CYP450 gene was discovered, because their sequencing size was highly limited. Then, the 175 partial CYP450s were assembled using previously known genes in Genbank, and no full-length CYP450s were obtained.
Classification of the 118 full-length CYP450 genes was executed by alignment with CYP450 database (Nelson, 2009) using standard sequence similarity cutoffs, specifically 97, 55, and 40% for allelic, subfamily, and family variants, respectively. Based on these cutoffs, the 118 full-length CYP450s were classified into 8 clans and 29 families and were divided into two categories: A-type (CYP71 clan) and non-A-type (all other clans; Table 2). Among them, only 6 CYP450s have been previously identified, and the other 112 CYP450s were obtained for the first time (Accession Numbers: MF448573-MF448684).
Table 2.
List of full-length CYP450s of T. chinensis identified in this study.
| Name | Type | Clan | Family | Subfamily | Length | Mol. wt (kDa) | Theoretical pI | TargetP |
|---|---|---|---|---|---|---|---|---|
| TcCYP51G1 | non-A | 51 | CYP51 | CYP51G | 497 | 56.56 | 8.81 | S |
| TcCYP73A170 | A | 71 | CYP73 | CYP73A | 501 | 57.50 | 8.55 | S |
| TcCYP73A171 | A | 71 | CYP73 | CYP73A | 515 | 59.07 | 8.68 | S |
| TcCYP74A73 | non-A | 74 | CYP74 | CYP74A | 525 | 58.84 | 9.1 | C |
| TcCYP74A74 | non-A | 74 | CYP74 | CYP74A | 474 | 53.37 | 6 | – |
| TcCYP74A75 | non-A | 74 | CYP74 | CYP74A | 475 | 53.58 | 5.95 | – |
| TcCYP75A77 | A | 71 | CYP75 | CYP75A | 504 | 56.27 | 9.33 | – |
| TcCYP75B115 | A | 71 | CYP75 | CYP75B | 516 | 57.26 | 6.78 | S |
| TcCYP76AA66 | A | 71 | CYP76 | CYP76AA | 512 | 58.05 | 8.57 | S |
| TcCYP76AA67 | A | 71 | CYP76 | CYP76AA | 506 | 57.43 | 6.58 | S |
| TcCYP76AA68 | A | 71 | CYP76 | CYP76AA | 505 | 57.57 | 9 | S |
| TcCYP76AA69 | A | 71 | CYP76 | CYP76AA | 507 | 57.39 | 8.71 | S |
| TcCYP76AA70 | A | 71 | CYP76 | CYP76AA | 513 | 58.57 | 8.77 | S |
| TcCYP76AA71 | A | 71 | CYP76 | CYP76AA | 523 | 59.13 | 7.58 | – |
| TcCYP76AA72 | A | 71 | CYP76 | CYP76AA | 505 | 57.68 | 8.73 | S |
| TcCYP76AA73 | A | 71 | CYP76 | CYP76AA | 523 | 59.71 | 9.03 | – |
| TcCYP76Z4 | A | 71 | CYP76 | CYP76Z | 511 | 57.63 | 9.08 | – |
| TcCYP77F31 | A | 71 | CYP77 | CYP77F | 508 | 57.18 | 9.03 | S |
| TcCYP77F32 | A | 71 | CYP77 | CYP77F | 503 | 57.05 | 8.54 | S |
| TcCYP77F33 | A | 71 | CYP77 | CYP77F | 525 | 59.16 | 9.06 | – |
| TcCYP77F34 | A | 71 | CYP77 | CYP77F | 461 | 51.55 | 8.62 | – |
| TcCYP77F35 | A | 71 | CYP77 | CYP77F | 503 | 56.85 | 8.77 | S |
| TcCYP78A233 | A | 71 | CYP78 | CYP78A | 554 | 61.89 | 8.66 | – |
| TcCYP78A234 | A | 71 | CYP78 | CYP78A | 548 | 61.33 | 8.93 | – |
| TcCYP86A149 | non-A | 86 | CYP86 | CYP86A | 523 | 59.27 | 8.93 | S |
| TcCYP86J5 | non-A | 86 | CYP86 | CYP86J | 540 | 61.85 | 8.58 | – |
| TcCYP86P14 | non-A | 86 | CYP86 | CYP86P | 491 | 55.59 | 9.32 | S |
| TcCYP90A54 | non-A | 85 | CYP90 | CYP90A | 487 | 55.26 | 8.64 | S |
| TcCYP94D79 | non-A | 86 | CYP94 | CYP94D | 519 | 59.13 | 8.87 | S |
| TcCYP94D80 | non-A | 86 | CYP94 | CYP94D | 522 | 59.50 | 8.85 | S |
| TcCYP94D81 | non-A | 86 | CYP94 | CYP94D | 517 | 59.23 | 7.59 | S |
| TcCYP94D82 | non-A | 86 | CYP94 | CYP94D | 514 | 58.80 | 8.43 | S |
| TcCYP94D83 | non-A | 86 | CYP94 | CYP94D | 515 | 58.21 | 8.59 | S |
| TcCYP94P4 | non-A | 86 | CYP94 | CYP94P | 501 | 57.22 | 7.1 | S |
| TcCYP94P5 | non-A | 86 | CYP94 | CYP94P | 510 | 57.80 | 9.31 | S |
| TcCYP94P6 | non-A | 86 | CYP94 | CYP94P | 530 | 60.00 | 6.63 | S |
| TcCYP94P7 | non-A | 86 | CYP94 | CYP94P | 518 | 58.50 | 9.11 | S |
| TcCYP97A57 | non-A | 97 | CYP97 | CYP97A | 673 | 75.86 | 8.96 | C |
| TcCYP97B52 | non-A | 97 | CYP97 | CYP97B | 584 | 65.81 | 8.26 | C |
| TcCYP701A59 | A | 71 | CYP701 | CYP701A | 507 | 58.32 | 6.97 | S |
| TcCYP703D4 | A | 71 | CYP703 | CYP703D | 524 | 59.78 | 9.23 | S |
| TcCYP704C10 | non-A | 86 | CYP704 | CYP704C | 510 | 59.12 | 7.28 | S |
| TcCYP710A78 | non-A | 710 | CYP710 | CYP710A | 499 | 57.33 | 8.05 | S |
| TcCYP715C54 | non-A | 72 | CYP715 | CYP715C | 532 | 60.09 | 9.01 | – |
| TcCYP715D11 | non-A | 72 | CYP715 | CYP715D | 525 | 59.67 | 8.65 | S |
| TcCYP716B29 | non-A | 85 | CYP716 | CYP716B | 484 | 54.52 | 8.46 | S |
| TcCYP716B30 | non-A | 85 | CYP716 | CYP716B | 475 | 53.70 | 8.54 | S |
| TcCYP716B31 | non-A | 85 | CYP716 | CYP716B | 475 | 53.56 | 8.64 | S |
| TcCYP718B1 | non-A | 85 | CYP718 | CYP718B | 482 | 55.20 | 8.61 | S |
| TcCYP720B23 | non-A | 85 | CYP720 | CYP720B | 485 | 56.18 | 8.48 | S |
| TcCYP720B24 | non-A | 85 | CYP720 | CYP720B | 485 | 55.99 | 8.83 | S |
| TcCYP720B25 | non-A | 85 | CYP720 | CYP720B | 486 | 56.12 | 8.67 | S |
| TcCYP720B26 | non-A | 85 | CYP720 | CYP720B | 486 | 55.87 | 8.64 | S |
| TcCYP725A1 | non-A | 85 | CYP725 | CYP725A | 497 | 56.55 | 9.14 | – |
| TcCYP725A2 | non-A | 85 | CYP725 | CYP725A | 485 | 54.49 | 9.04 | S |
| TcCYP725A3 | non-A | 85 | CYP725 | CYP725A | 497 | 55.86 | 8.77 | S |
| TcCYP725A4 | non-A | 85 | CYP725 | CYP725A | 502 | 56.85 | 8.13 | – |
| TcCYP725A5 | non-A | 85 | CYP725 | CYP725A | 493 | 55.32 | 9.41 | S |
| TcCYP725A6 | non-A | 85 | CYP725 | CYP725A | 495 | 55.53 | 9.35 | S |
| TcCYP725A9 | non-A | 85 | CYP725 | CYP725A | 493 | 55.35 | 8.68 | S |
| TcCYP725A10 | non-A | 85 | CYP725 | CYP725A | 484 | 54.56 | 9.1 | S |
| TcCYP725A11 | non-A | 85 | CYP725 | CYP725A | 509 | 58.07 | 9.48 | – |
| TcCYP725A12 | non-A | 85 | CYP725 | CYP725A | 479 | 54.11 | 9.01 | – |
| TcCYP725A13 | non-A | 85 | CYP725 | CYP725A | 484 | 54.50 | 8.98 | S |
| TcCYP725A14 | non-A | 85 | CYP725 | CYP725A | 484 | 55.03 | 8.85 | S |
| TcCYP725A15 | non-A | 85 | CYP725 | CYP725A | 479 | 54.20 | 9.21 | – |
| TcCYP725A16 | non-A | 85 | CYP725 | CYP725A | 478 | 54.12 | 9.17 | – |
| TcCYP725A17 | non-A | 85 | CYP725 | CYP725A | 484 | 55.09 | 9.03 | S |
| TcCYP725A18 | non-A | 85 | CYP725 | CYP725A | 498 | 56.29 | 9.11 | S |
| TcCYP725A19 | non-A | 85 | CYP725 | CYP725A | 497 | 55.82 | 8.89 | S |
| TcCYP725A20 | non-A | 85 | CYP725 | CYP725A | 503 | 56.79 | 8.91 | – |
| TcCYP725A21 | non-A | 85 | CYP725 | CYP725A | 509 | 58.25 | 9.4 | – |
| TcCYP725A22 | non-A | 85 | CYP725 | CYP725A | 499 | 57.24 | 8.77 | – |
| TcCYP725A23 | non-A | 85 | CYP725 | CYP725A | 489 | 55.66 | 9.16 | S |
| TcCYP728Q11 | non-A | 85 | CYP728 | CYP728Q | 490 | 56.10 | 9.28 | – |
| TcCYP728Q12 | non-A | 85 | CYP728 | CYP728Q | 483 | 54.47 | 8.79 | S |
| TcCYP728Q13 | non-A | 85 | CYP728 | CYP728Q | 479 | 54.76 | 9.02 | S |
| TcCYP728S2 | non-A | 85 | CYP728 | CYP728S | 494 | 55.70 | 9.06 | – |
| TcCYP728S3 | non-A | 85 | CYP728 | CYP728S | 482 | 54.23 | 8.98 | S |
| TcCYP729B25 | non-A | 85 | CYP729 | CYP729B | 482 | 55.11 | 9.1 | S |
| TcCYP736C8 | A | 71 | CYP736 | CYP736C | 509 | 57.51 | 6.48 | S |
| TcCYP736E20 | A | 71 | CYP736 | CYP736E | 514 | 58.03 | 6.72 | C |
| TcCYP736E21 | A | 71 | CYP736 | CYP736E | 503 | 56.85 | 6.82 | S |
| TcCYP736E22 | A | 71 | CYP736 | CYP736E | 503 | 56.83 | 6.79 | S |
| TcCYP736E23 | A | 71 | CYP736 | CYP736E | 502 | 56.53 | 6.77 | C |
| TcCYP736E24 | A | 71 | CYP736 | CYP736E | 514 | 58.18 | 6.56 | S |
| TcCYP750B2 | A | 71 | CYP750 | CYP750B | 507 | 57.00 | 6.93 | S |
| TcCYP750C16 | A | 71 | CYP750 | CYP750C | 517 | 59.02 | 6.23 | – |
| TcCYP750C17 | A | 71 | CYP750 | CYP750C | 525 | 59.76 | 7.11 | S |
| TcCYP750C18 | A | 71 | CYP750 | CYP750C | 520 | 58.93 | 7.62 | S |
| TcCYP750C19 | A | 71 | CYP750 | CYP750C | 526 | 60.31 | 6.5 | S |
| TcCYP750C20 | A | 71 | CYP750 | CYP750C | 526 | 60.10 | 6.39 | S |
| TcCYP750C21 | A | 71 | CYP750 | CYP750C | 512 | 57.97 | 6.74 | – |
| TcCYP750C22 | A | 71 | CYP750 | CYP750C | 521 | 59.26 | 7.23 | S |
| TcCYP750C23 | A | 71 | CYP750 | CYP750C | 518 | 58.57 | 8.77 | – |
| TcCYP750C24 | A | 71 | CYP750 | CYP750C | 522 | 59.54 | 7.21 | – |
| TcCYP750C25 | A | 71 | CYP750 | CYP750C | 509 | 57.13 | 7.63 | S |
| TcCYP750C8 | A | 71 | CYP750 | CYP750C | 508 | 57.16 | 8.03 | – |
| TcCYP750C3 | A | 71 | CYP750 | CYP750C | 531 | 59.70 | 8.4 | C |
| TcCYP750C26 | A | 71 | CYP750 | CYP750C | 512 | 57.45 | 6.09 | – |
| TcCYP750C27 | A | 71 | CYP750 | CYP750C | 509 | 57.09 | 8.38 | S |
| TcCYP750C28 | A | 71 | CYP750 | CYP750C | 513 | 57.94 | 7.63 | S |
| TcCYP750C29 | A | 71 | CYP750 | CYP750C | 512 | 57.77 | 7 | S |
| TcCYP750C30 | A | 71 | CYP750 | CYP750C | 484 | 54.39 | 8.37 | S |
| TcCYP782B7 | A | 71 | CYP782 | CYP782B | 508 | 58.10 | 6.96 | S |
| TcCYP864B7 | non-A | 72 | CYP864 | CYP864B | 519 | 58.62 | 8.06 | S |
| TcCYP866A17 | non-A | 72 | CYP866 | CYP866A | 520 | 58.89 | 9.36 | S |
| TcCYP866A18 | non-A | 72 | CYP866 | CYP866A | 515 | 58.36 | 9.24 | S |
| TcCYP866A19 | non-A | 72 | CYP866 | CYP866A | 515 | 58.26 | 9.38 | S |
| TcCYP866A20 | non-A | 72 | CYP866 | CYP866A | 515 | 58.45 | 9.18 | S |
| TcCYP866B7 | non-A | 72 | CYP866 | CYP866B | 530 | 59.96 | 9.44 | S |
| TcCYP866B10 | non-A | 72 | CYP866 | CYP866B | 516 | 58.46 | 9.29 | S |
| TcCYP867B5 | A | 71 | CYP867 | CYP867B | 509 | 57.32 | 8.98 | C |
| TcCYP867E3 | A | 71 | CYP867 | CYP867E | 507 | 57.31 | 9.16 | – |
| TcCYP867F22 | A | 71 | CYP867 | CYP867F | 526 | 59.38 | 6.39 | – |
| TcCYP867F23 | A | 71 | CYP867 | CYP867F | 532 | 60.10 | 7.06 | S |
| TcCYP867G20 | A | 71 | CYP867 | CYP867G | 512 | 58.19 | 7.73 | S |
| TcCYP947A88 | non-A | 85 | CYP947 | CYP947A | 496 | 56.18 | 9.09 | S |
T. chinensis CYP450s were further compared with three other typical plant species, such as angiosperms A. thaliana and Medicago sativa and the gymnosperm P. glauca (Tables 3, 4). The results showed that CYP750 was the largest A-type family in T. chinensis but absent in A. thaliana and M. sativa. Conversely, CYP71, CYP79, CYP81, CYP82, CYP83, and CYP89 families were found in A. thaliana and M. sativa, but not in T. chinensis and P. glauca. For the non-A-type CYP450s, the CYP725, and CYP866 families were found in T. chinensis and P. glauca, but absent in A. thaliana and M. sativa. CYP72, CYP87, CYP96, CYP714, CYP721, and CYP722 families were only found in A. thaliana and M. sativa. Moreover, two new gymnosperm-specific families, named CYP864 and CYP947, were discovered in T. chinensis. The above results showed that distinct genetic differences exist between the gymnosperm and angiosperm, and P. glauca is a good reference for T. chinensis in comparative studies of CYP450 genes.
Table 3.
Comparison of A-type CYP450 families among A. thaliana (At), M. sativas (Ms), P.glauca (Pg) and T.chinensis (Tc).
| Family | At | Ms | Pg | Tc |
|---|---|---|---|---|
| CYP71 | 52 | 37 | 0 | 0 |
| CYP79 | 7 | 3 | 0 | 0 |
| CYP81 | 18 | 5 | 0 | 0 |
| CYP82 | 5 | 10 | 0 | 0 |
| CYP83 | 2 | 9 | 0 | 0 |
| CYP89 | 7 | 9 | 0 | 0 |
| CYP750 | 0 | 0 | 6 | 18 |
| CYP867 | 0 | 0 | 1 | 5 |
| CYP73 | 1 | 1 | 1 | 2 |
| CYP75 | 1 | 0 | 6 | 2 |
| CYP76 | 8 | 6 | 5 | 9 |
| CYP77 | 5 | 2 | 1 | 5 |
| CYP78 | 6 | 1 | 6 | 2 |
| CYP92 | 0 | 1 | 2 | 0 |
| CYP98 | 3 | 1 | 1 | 0 |
| CYP701 | 1 | 1 | 1 | 1 |
| CYP703 | 1 | 1 | 0 | 1 |
| CYP705 | 26 | 0 | 0 | 0 |
| CYP736 | 0 | 1 | 17 | 6 |
| CYP782 | 0 | 0 | 0 | 1 |
Table 4.
Comparison of non-A-type CYP450 families among A. thaliana (At), M. sativas (Ms), P. glauca (Pg), and T. chinensis (Tc).
| Family | At | Ms | Pg | Tc |
|---|---|---|---|---|
| CYP72 | 9 | 7 | 0 | 0 |
| CYP87 | 1 | 2 | 0 | 0 |
| CYP96 | 13 | 5 | 0 | 0 |
| CYP714 | 2 | 3 | 0 | 0 |
| CYP721 | 1 | 1 | 0 | 0 |
| CYP722 | 1 | 1 | 0 | 0 |
| CYP725 | 0 | 0 | 1 | 21 |
| CYP866 | 0 | 0 | 3 | 6 |
| CYP51 | 1 | 1 | 1 | 1 |
| CYP74 | 2 | 4 | 5 | 3 |
| CYP85 | 2 | 1 | 1 | 0 |
| CYP86 | 11 | 3 | 18 | 3 |
| CYP88 | 2 | 3 | 1 | 0 |
| CYP90 | 4 | 4 | 3 | 1 |
| CYP94 | 6 | 4 | 1 | 9 |
| CYP97 | 3 | 4 | 4 | 2 |
| CYP704 | 3 | 14 | 4 | 1 |
| CYP707 | 4 | 3 | 2 | 0 |
| CYP710 | 4 | 1 | 1 | 1 |
| CYP711 | 1 | 2 | 1 | 0 |
| CYP715 | 1 | 1 | 0 | 2 |
| CYP716 | 2 | 3 | 3 | 3 |
| CYP718 | 1 | 0 | 0 | 1 |
| CYP720 | 1 | 1 | 5 | 4 |
| CYP724 | 1 | 0 | 1 | 0 |
| CYP727 | 0 | 0 | 1 | 0 |
| CYP728 | 0 | 0 | 0 | 5 |
| CYP729 | 0 | 1 | 0 | 1 |
| CYP864 | 0 | 0 | 0 | 1 |
| CYP947 | 0 | 0 | 0 | 1 |
Phylogenetic analysis
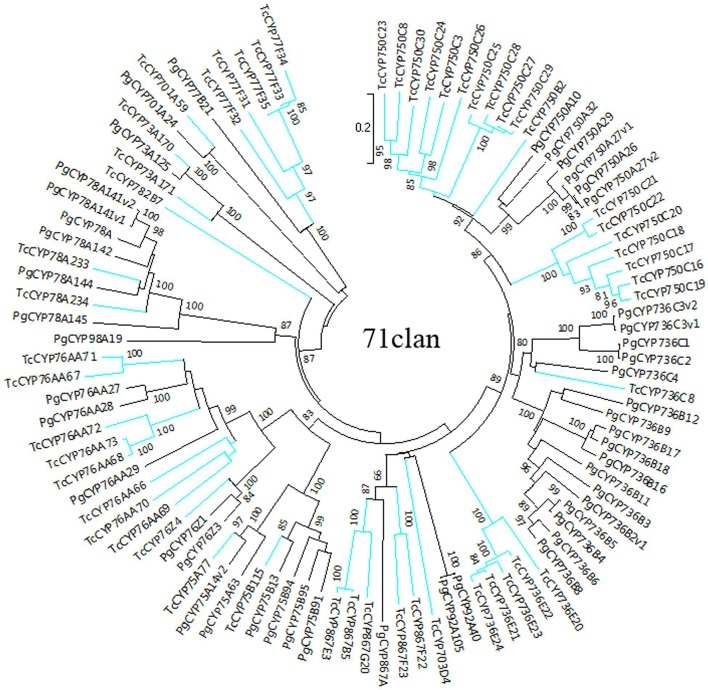
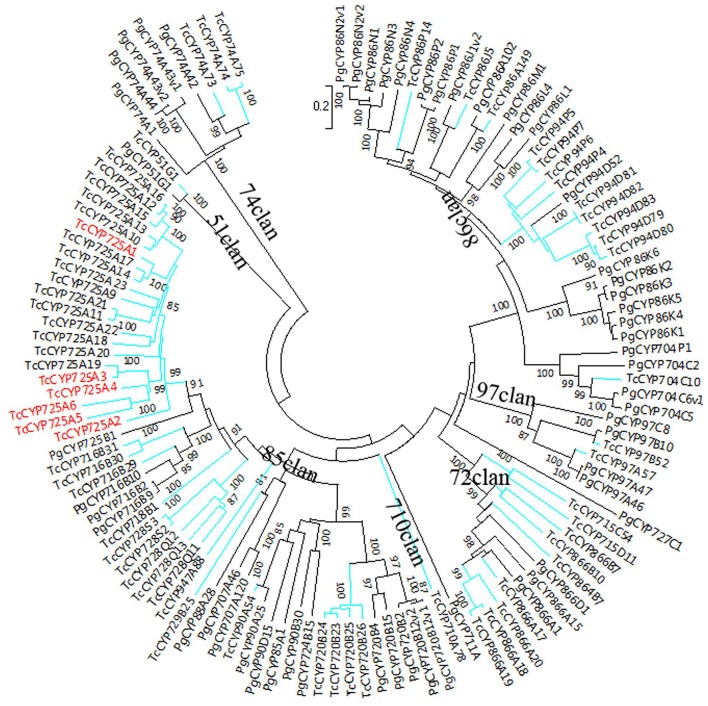
The sequences of P. glauca CYP450 proteins and the 118 full-length T. chinensis CYP450 proteins were used to construct NJ phylogenetic trees for A-type (Figure 1) and non-A-type (Figure 2) CYP450s, separately, using the MEGA7 package. The results showed that P. glauca subfamilies are grouped with T. chinensis CYP450s. Based on phylogenetic trees, 44.1% (52 genes) of the 118 full-length CYP450s are A-type and belong to 11 families. The remaining 55.9% (66 genes) CYP450s are non-A-type and are distributed to 18 families and 7 clans. The A-type CYP450s (71 clan) have been identified to be related to the biosynthesis of secondary compounds. The Figure 2 showed that non-A-type CYP450s include a more diverse group of genes belonging to the remaining 7 clans. These genes involved in the metabolic pathways of primary products (such as carotenoid, oxylipin, etc.), plant hormone and secondary products. 5 CYP450s have been previously identified to be involved in Taxol biosynthesis, such as TcCYP725A1 (taxane 5-alpha-taxadienol-10-beta-hydroxylase, T10βH), TcCYP725A2 (taxane 13-alpha-hydroxylase, T13αH), TcCYP725A4 (taxadiene 5-alpha-hydroxylase, T5αH), TcCYP725A5 (taxoid 7-beta-hydroxylase, T7βH), and TcCYP725A6 (taxoid 2-alpha-hydroxylase, T2αH). In addition, TcCYP725A3, which is highly homologous with taxane 14β-hydroxylase (T14βH) in T. cuspidata. It is suggested that the CYP725A subfamily underwent independent evolution to carry its unique function.
Figure 1.
Phylogenetic tree of A-type CYP450 proteins from T. chinensis (Tc) and P. glauca (Pg). The spokes corresponding to Tc and Pg CYP450s are shown in blue and black, respectively.
Figure 2.
Phylogenetic tree of non-A-type CYP450 proteins from T. chinensis (Tc) and P. glauca (Pg). The spokes corresponding to Tc and Pg CYP450s are shown in blue and black, respectively. The genes marked in red are described in the text.
Physicochemical and structural analyses of T. chinensis CYP450s
The physicochemical parameters of each CYP450 gene were calculated using ExPASy. Most of the members had relative molecular weights close to 55 kDa. Approximately four-fifths of the CYP450 proteins had relatively high isoelectric points (pI > 7); the remaining proteins, particularly those in CYP736 family, had pI < 7. TargetP was used to predict the localizations of 118 T. chinensis CYP450 proteins, most of them were predicted to be anchored on the endoplasmic reticulum. Hitherto, no plant CYP450s have been found to be located in the mitochondria.
All typical conserved structures of CYP450 proteins were present in non-A-type T. chinensis CYP450s (Figure S1), including the cysteine heme-iron ligand signature motif (with PFG element), the PERF motif, K-helix region, and I-helix region. Interestingly, the conserved I-helix region did not exist in A-type T. chinensis CYP450s. For the heme-binding motifs, the A-type CYP450s displayed the signature “PFGxGRRxCxG,” whereas “xFxxGxRxCxG” was found in non-A-type CYP450s. Consistent with previous studies, PERF motifs of A-type and non-A-type CYP450s were different. In T. chinensis, PERF motifs were “PERF” for A-type and “FxPx” for non-A-type. Moreover, the EXXR motif of A-type was consistent with non-A-type CYP450 proteins. These elements ensure structural stability and flexibility, thereby enabling proteins to bind to appropriate substrates.
Annotation of T. chinensis CYP450s
KEGG pathway-based analysis was performed to further understand the functions of the CYP450 genes. In total, the 118 full-length CYP450s were assigned to 16 KEGG pathways (Figure S2). Significantly, only 2 CYP450s were mapped to the diterpenoid biosynthesis pathway, including TcCYP729B25 and TcCYP701A59. CYP701 is related to the biosynthesis of diterpenoid acids, gibberellins.
Information on the specific metabolic pathways in gymnosperms was highly limited, resulting the CYP725s that are related to diterpenoid Taxol biosynthesis were mapped to the carotenoid biosynthesis pathway. The most represented CYP725 family in Taxus (Table 4) plays an important role in the biosynthesis of the diterpenoid anti-cancer drug, Taxol. Moreover, the acquired T2αH, T5αH, T7βH, T10βH, T13αH, and T14βH have high sequence similarity with each other. With an amino acid sequence similarity higher than 70%, the taxoid CYP450 monooxygenases are more conserved than any other known plant of the CYP450 type. Taxoid hydroxylases, with their unique structures and substrate selectivities, form an especially cohesive group. The novel CYP725 proteins identified in this study may be related to Taxol biosynthesis. However, the functional importance of these proteins remains to be determined.
Expression profile analysis of CYP450s
Illumina transcriptome sequencing technology was used to analyze the gene expressions partners of all 118 full-length CYP450s. These data sets were generated from total RNAs isolated from two Taxus cell lines, CA and NA, and MeJA-mediated Taxus cells harvested 16 h after inoculation (Li et al., 2012; Zhang et al., 2015).
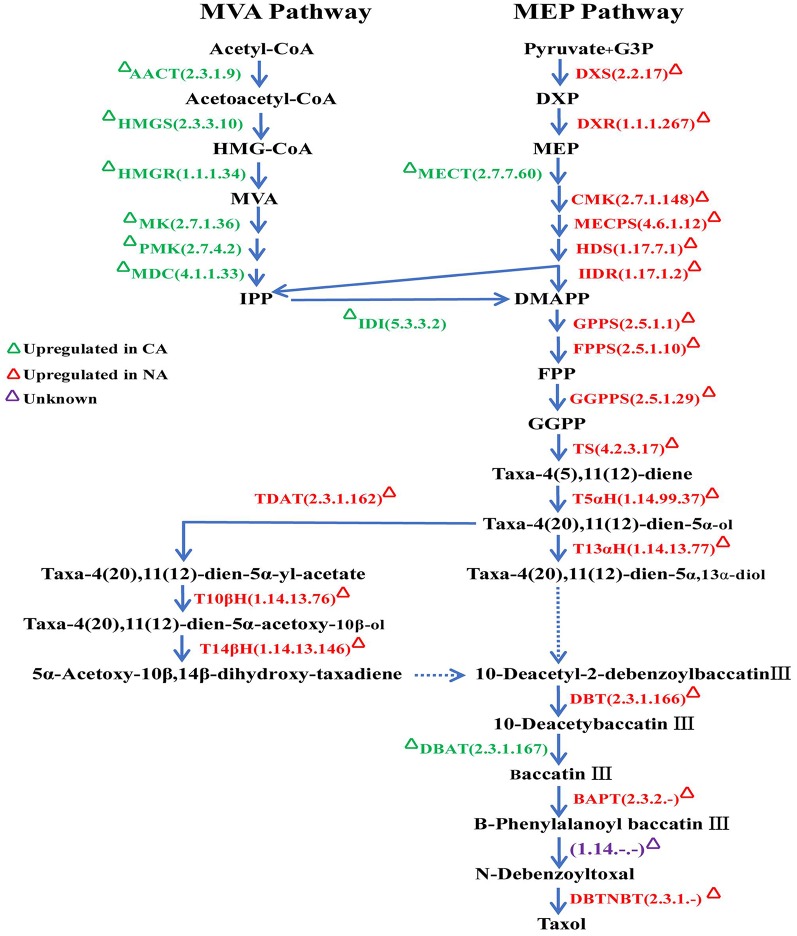
From these Illumina data, all 118 full-length CYP450 genes expressed in different Taxus cell lines, CA and NA, were hierarchically clustered using HemI Heatmap Illustrator v1.0 software (Figure 3, Table S2). According to previous research (Song et al., 2014), contents of secondary metabolites in NA were significantly higher than in CA. The amount of Taxol was 1.88 times higher than that in CA. The downregulation of secondary metabolites in CA may be due to the decreased activity of specific enzymes, including CYP450 monooxygenases. Figure 4 showed highly different expression profiles in NA and CA. TcCYP73A171, TcCYP74A74, TcCYP75A77, TcCYP75B115, TcCYP76AA71, TcCYP76Z4, TcCYP728Q12, TcCYP736E22, and TcCYP750C20 were lowly expressed in CA, but were highly expressed in NA. Moreover, TcCYP76AA67, TcCYP90A54, and TcCYP750B2 members were not detected in CA. CYP450 oxygenases that potentially related to the Taxol biosynthesis were mainly analyzed. The identified transcripts involved in the Taxol biosynthetic pathway and their specific expression levels in CA and NA were shown in Figure 4. Most enzymes of the native methylerythritol phosphate (MEP) pathway were highly expressed in NA (Table S3). Co-expression with a Taxol biosynthesis marker gene, taxadiene synthase (TS), the acquired taxoid hydroxylases were also highly expressed in NA. The expression levels of T2αH and T7βH greatly increased more than 10-fold, T10βH and T13αH increased two-fold, and T5αH increased slightly. In this study, 10 novel CYP725A genes (TcCYP725A9, TcCYP725A10, TcCYP725A11, TcCYP725A16, TcCYP725A18, TcCYP725A19, TcCYP725A20, TcCYP725A21, TcCYP725A22, and TcCYP725A23) showed higher expression levels in NA.
Figure 3.
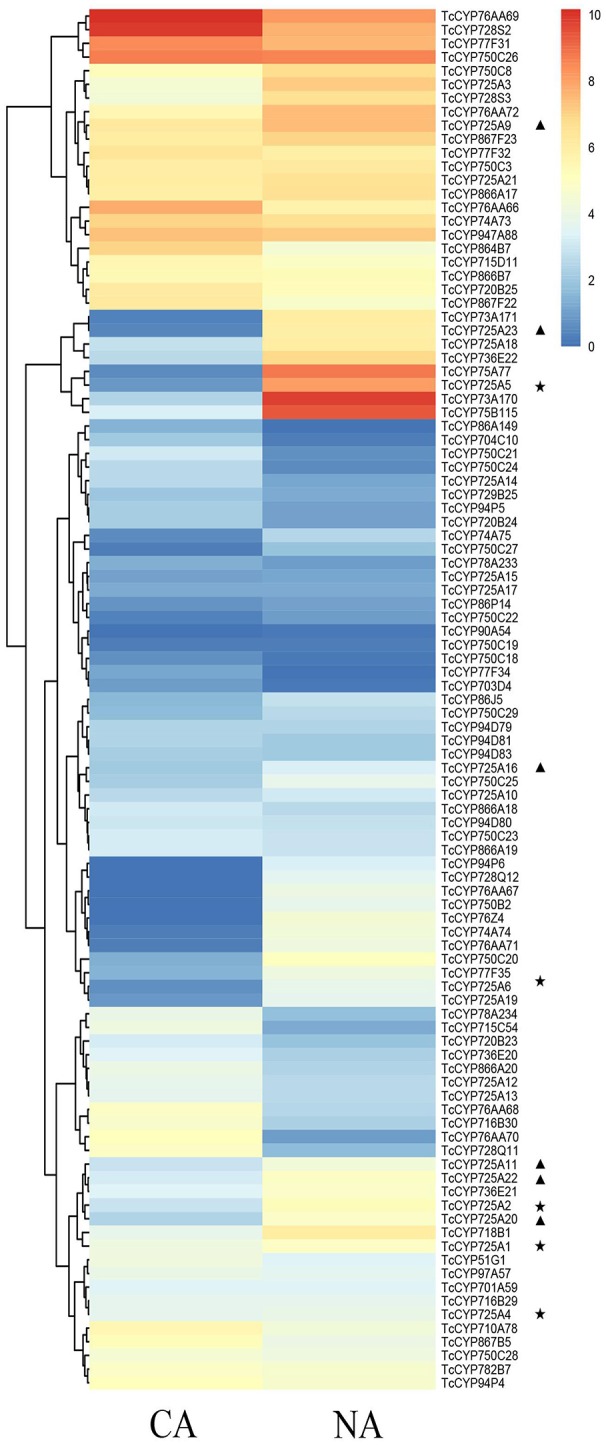
Expression pattern of T. chinensis CYP450s in CA and NA cell lines according to the analysis of RNA-Seq dataset. The color scale shows the expression quantity (red: high expression; blue: low expression). Heat map was created using HemI. Heatmap Illustrator v1.0. “⋆” indicates the known taxoid hydroxylase genes, and “▴” indicates the candidate genes.
Figure 4.
Schematic representation of the taxol biosynthetic pathway. Enzymes were marked according to their specific expression in CA and NA. AACT, acetyl-CoA C-acetyltransferase; HMGS, hydroxymethylglutaryl-CoA synthase; HMGR, hydroxymethylglutaryl-CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MDC, diphosphomevalonate decarboxylase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; MECT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; MECPS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase; IDI, isopentenyl-diphosphate Delta-isomerase; GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; TS, taxadiene synthase; T5αH (CYP725A4), taxadiene 5alpha-hydroxylase; T13αH (CYP725A2), taxane 13-alpha-hydroxylase; TDAT, taxadien-5-alpha-ol O-acetyltransferas; T10βH (CYP725A1), taxane 10-beta-hydroxylase; T14βH (CYP725A3), taxoid 14beta-hydroxylase; DBT, 2-alpha-hydroxytaxane 2-O-benzoyltransferase; DBAT, 10-deacetylbaccatinIII 10-O-acetyltransferase; BAPT, Baccatin III amino phenylpropanoyl-13-O-transferase; DBTNBT, 3′-N-debenzoyl-2′-deoxytaxol N-benzoyl transferase.
Previous research confirmed that taxane biosynthesis is regulated by MeJA elicitation in T. chinensis cells (Li et al., 2012). Transcriptome profiles of T. chinensis cells at 16 h (Tm16) after MeJA treatment and those of mock-treated cells (Tm0) showed that the mRNA levels of most defined hydroxylase genes for taxol biosynthesis increased at 16 h after MeJA elicitation; moreover, genes corresponding to T5αH, T7βH, and T10βH were significantly up-regulated, and genes corresponding to T2αH and T13αH were slightly up-regulated (Table S3). For the 15 novel CYP725 genes, TcCYP725A9, TcCYP725A11, TcCYP725A12, TcCYP725A13, TcCYP725A16, TcCYP725A20, TcCYP725A22, and TcCYP725A23 were up-regulated at 16 h after MeJA elicitation (Figure 5, Table S4).
Figure 5.
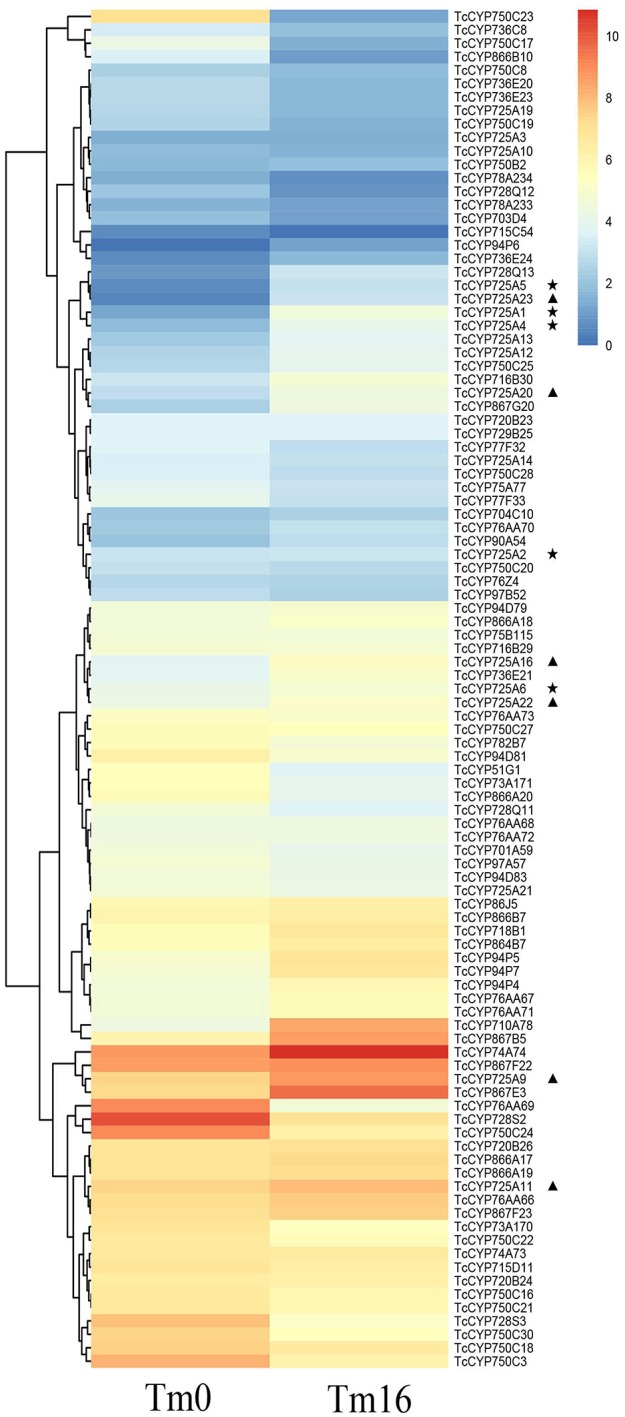
Expression pattern of T. chinensis CYP450s under MeJA elicitationin according to the analysis of RNA-Seq dataset. The color scale shows the expression quantity (red, high expression; blue, low expression). Heat map was created using HemI. Heatmap Illustrator v1.0. “⋆” indicates the known taxoid hydroxylase genes, and “▴” indicates the candidate genes.
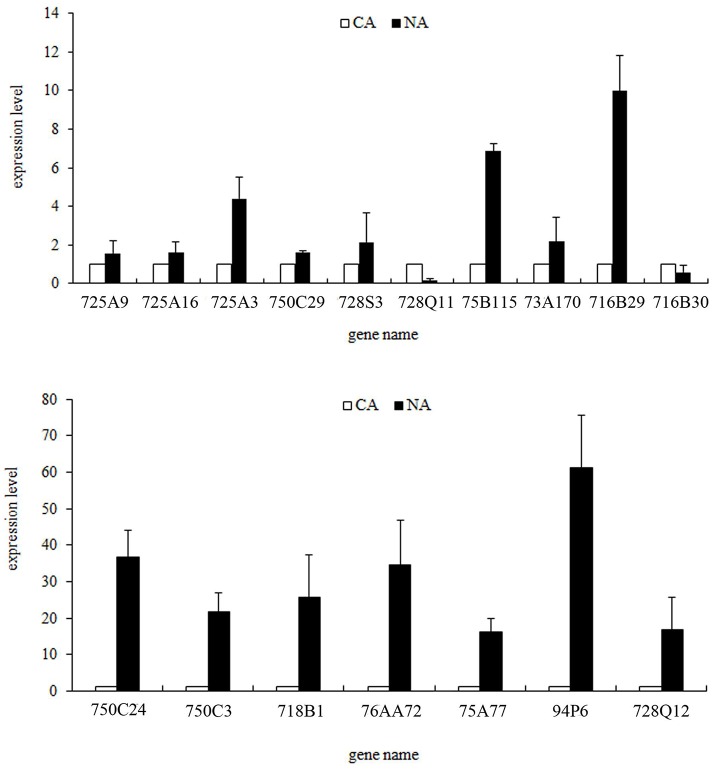
To verify expression profiles obtained from Illumina sequencing, we performed qRT-PCR on 17 randomly selected CYP450 genes (Figure 6, Table S5). Consistent with the Illumina data, most genes showed strong expression levels in NA. The expression fold changes of some genes, such as TcCYP73A170, TcCYP716B29, TcCYP750C3, and TcCYP716B were higher than RNA-seq results. qRT-PCR results validated that the RNA-seq data is reliable.
Figure 6.
qRT-PCR confirmation of the expression profiles of some randomly selected CYP450 genes. Fold changes of transcript levels in CA and NA are shown. Error bars indicate the mean (SEM).
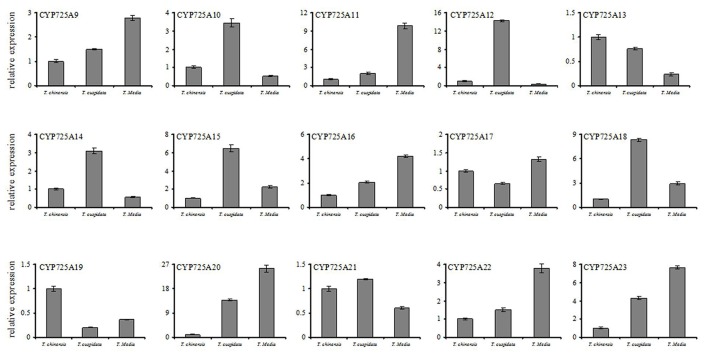
The different expression levels of 15 novel CYP725 genes among three different species such as T. chinensis, T. cuspidate, and T. media were also investigated (Figure 7). The qRT-PCR profiles showed that CYP725 genes had different expression profiles in different Taxus species. All genes but CYP725A13 and CYP725A19 showed a low expression level in T. chinensis. Six genes (TcCYP725A9, TcCYP725A11, TcCYP725A16, TcCYP725A20, TcCYP725A22, and TcCYP725A23) showed the highest expression level in T. media, followed by T. cuspidata, and little amount in T. chinensis. The taxol content in T. media (0.0186%) was higher than T. cuspidata (0.0138%) and T. chinensis (0.0109%). The expression rule of these six genes were coincident with Taxol content.
Figure 7.
Comparisons of the expression profiles of 15 novel CYP725 genes in different 5-year-old Taxus species, including T. chinensis, T. cuspidate, and T. Media. The bars represent the standard deviation (n = 3).
Identification of candidates CYP450s involved in taxol biosynthesis
CYP450s that were potentially related to Taxol biosynthesis were further identified based on the three following criteria: (1) belonging to CYP725 family. To date, the CYP450 enzymes identified in Taxol biosynthesis are primarily CYP725s, (2) expression level was corresponding with the known Taxol biosynthesis hydroxylases CYP725A1, CYP725A2, CYP725A4, CYP725A5, and CYP725A6, (3) expression profile was consistent with Taxol content. Among the 118 full-length CYP450s, 15 novel genes belong to the CYP725 family (from CYP725A9 to CYP725A23). More importantly, TcCYP725A9, TcCYP725A11, TcCYP725A16, TcCYP725A20, TcCYP725A22, and TcCYP725A23 were highly expressed in NA and up-regulated after MeJA elicitation. The expression pattern of these six CYP725s were similar to the known Taxol biosynthesis hydroxylases. Interestingly, all these six CYP725 genes were expressed at the highest level in T. Media, and the lowest in T. chinensis. Their expression rule were coincident with Taxol content in Taxus organs. Therefore, they are likely to involved in Taxol biosynthesis, and their specific functions require further study.
Cis-regulatory elements in the promoters of TCCYP725s
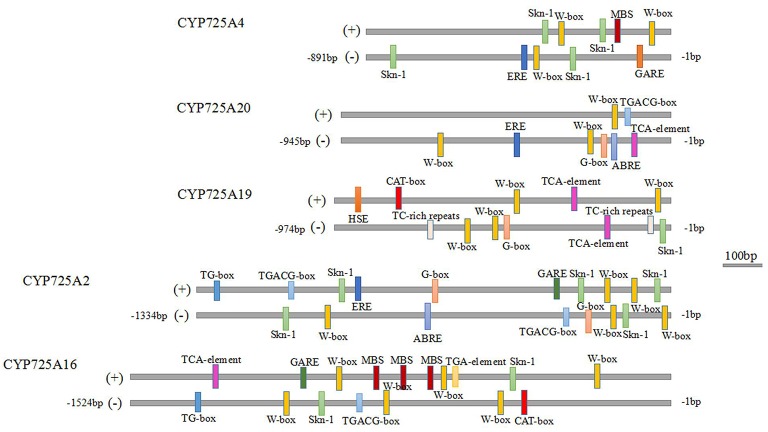
As Taxus genome information was not complete, only the promoter fragments of 5 TcCYP725s were identified (Accession Numbers: MF598831-MF598835). In addition to the common cis elements CAAT-box and TATA-box, 14 types of cis-acting elements in the TcCYP725s were discovered (Figure 8). The Skn-1 motif contributed to gene expression in the endosperm, and the CAT-box was required for gene expression in the meristem. The common cis-acting elements G-box and TG-box were responsive to light. All the other cis-regulatory elements are related to stresses and hormones, such as TC-rich repeats (required for defense and stresses), the TCA element (salicylic acid response), MBS (drought inducibility), HSE (heat response), W-box (responsive to fungal elicitors and plant hormones), the GARE motif (gibberellin response), the TGACG motif (MeJA response), ABRE (abscisic acid response), TGA (auxin response), and ERE (ethylene response). The Skn-1 motif is present in most of CYP725 genes, indicating that CYP725s play important roles in immature tissues. In addition, the W-box was widely found in CYP725 genes. Previous study showed that TcWRKY1 protein regulated Taxol biosynthesis in T. chinensis cells by specifically interacting with the two W-box of 10-deacetylbaccatin III-10β-O-acetyl transferase (DBAT; Li S. et al., 2013). These putative cis-acting elements would contribute to future researches of the transcriptional regulation of Taxol biosynthesis.
Figure 8.
Cis-acting elements in the promoter regions of CYP725 genes. The scale bar represents 100 bp.
Discussion
Taxol, a complex diterpenoid, is a highly effective antimitotic drug with excellent activity against many types of cancer. The lack of detailed information on Taxus CYP450s, such as centralized resource, systematic nomenclature, and biological functions, has significantly hampered research efforts to elucidate biosynthetic pathways for the medicinal ingredients in Taxus. To our knowledge, this study was the first to overcome these limitations by (1) identifying a large set of CYP450s; (2) establishing a systematic nomenclature for these CYP450s; (3) mining the candidate CYP450s that are likely related to Taxol biosynthesis; and (4) analyzing the cis-regulatory elements in the promoters of CYP725 genes to provides useful information to the transcriptional regulation of Taxol biosynthesis.
T. chinensis CYP450s identified in this study
Totally, 118 full-length and 175 partial CYP450 genes were identified in T. chinensis. The number of CYP450 genes in T. chinensis is of the same order of magnitude as the number of CYP450s found in other gymnosperm and angiosperm plants, that is, 307 in P. glauca, 272 in A. thaliana, 332 in soybean, 334 in flax, and 455 in rice (Guttikonda et al., 2010; Babu et al., 2013; Warren et al., 2015). The previously identified five genes involved in Taxol biosynthesis, such as TcCYP725A1 (T10βH, AAN52360.1), TcCYP725A2 (T13αH, AAX59903.1), TcCYP725A4 (T5αH, AAU93341.1), TcCYP725A5 (T7βH, AAR21106.1), and TcCYP725A6 (T2αH, AAV54171.1), were included in the 118 full-length CYP450s. Moreover, TcCYP725A3, which is highly homologous with T14βH (Accession No. Q84KI1) in T. cuspidata, was explored in this study (Jennewein et al., 2003). This inferred that the CYP450s data in our research were representatives, and the remaining 112 novel full-length CYP450 sequences were of great value for the advanced research and applications of CYP450s in Taxus.
Gymnosperm-specific CYP450s in T. chinensis
Some known gymnosperm-specific CYP450 subfamilies were discovered in T. chinensis. In the CYP71 clan, 8 CYP76AA and 18 CYP750 gymnosperm-specific members were identified among the 118 full-length CYP450s. Previous research revealed that the gymnosperms CYP76AA25 and CYP750B1 can catalyze the hydroxylation of sabinene to trans-sabin-3-ol from Thuja plicata (Bohlmann et al., 2015). The CYP85 clan, which includes many gymnosperm CYP450s, is involved in the biosynthesis of plant metabolites (Hamberger and Bohlmann, 2006; Zerbe et al., 2013): the CYP720B is a conifer-specific subfamily with four members in T. chinensis, and the CYP720B4 has been characterized in the biosynthesis of dehydroabietic acid, an ingredient related to the insect resistance of Picea sitchensis (Hamberger et al., 2011); the CYP716B is a gymnosperm-specific subfamily, with three members found in T. chinensis, and the sole functionally determined CYP716B gene is a taxoid 9α-hydroxylase in Ginkgo biloba (Zhang et al., 2014). Furthermore, two new gymnosperm-specific families were discovered in this study, namely, CYP864 in the 72 clan and CYP947 in the 85 clan (Communicated with Professor David Nelson, unpublished work. The gymnosperm CYP450 names have just been expanded by naming transcriptome data from the 1KP project). Two new gymnosperm-specific families that were recently discovered in P. glauca, including 71 clan family CYP867 and 72 clan family CYP866, were also found in T. chinensis (Warren et al., 2015). The exact functions of these new gymnosperm-specific families require further investigation.
Potential candidate CYP450s involved in terpenoid biosynthesis
Terpenoids are one of the most widespread classes of secondary metabolites in higher plants, which are biosynthesized from basic isoprene units (C5H8) and further modified by various oxidoreductases, acyltransferases, dehydrogenases, and glucosyltransferases. CYP450-dependent oxidative modification is essential for the terpenoid biosynthesis. Hitherto, more than 50 CYP450 genes, which belong to CYP51, CYP71, CYP72, CYP76, CYP88, CYP93, CYP97, CYP701, CYP705, CYP706, CYP707, CYP714, CYP716, CYP720, CYP725, CYP735, and some unassigned families related to the biosynthesis of terpenoids in medicinal plants have been identified (Zhao et al., 2014). The structural diversity of terpenoid compounds depends on the rearrangement modifications of their skeletal structures and extensive oxidative modification (Zerbe et al., 2013). Therefore, it shouldn't be surprising that so many CYP450s families have been found related to their biosynthesis.
Taxus spp. specifically employs up to eight CYP450-mediated oxidatives to create the diterpene, Taxol (Jennewein and Croteau, 2001; Kaspera and Croteau, 2006). So far, the C-2, C-5, C-7, C-10, and C-13 hydroxylases have been successfully obtained (Jennewein et al., 2001, 2004; Schoendorf et al., 2001; Chau and Croteau, 2004; Chau et al., 2004). Unfortunately, the genes responsible for C-1 hydroxylation, oxetane formation, C-9 oxidation, and C-2′ hydroxylation remain unknown. In this study, an intriguing result was that 6 CYP725s (TcCYP725A9, TcCYP725A11, TcCYP725A16, TcCYP725A20, TcCYP725A22, and TcCYP725A23) were found to belong to the candidates that were involved in Taxol biosynthesis. By a blast search, TcCYP725A9, TcCYP725A11, TcCYP725A16, TcCYP725A22, and TcCYP725A23 were found to show high sequence similarity (>66%) to the T10βH (Schoendorf et al., 2001). The T10βH transformed taxadien-5α-yl acetate to taxadien-5α-acetoxy-10β-ol, which was an important intermediate in the biosynthesis of Taxol. In addition, the BLAST analysis of TcCYP725A20 revealed that the most homology (63%) found in public databases was with T13αH, an enzyme capable of hydrolyzing the taxadien-5α-ol at its C-13 position. The above information may also suggested that these 6 CYP725 genes were likely candidate contribute to the formation of Taxol.
Future plans
Because the whole genome information of T. chinensis is unavailable, 5′ Race and 3′ Race can amplify numerous full-length CYP450 genes based on the current study. Functional predictions of the candidate CYP725s will be performed by heterologous expression in yeast. Linking in vivo feeding studies to cell-free enzyme systems, with available taxanes or suspension-cultured cells as ingredients, will enable better understanding of the Taxol biosynthetic pathway. Ultimately, we can reconstruct this secondary metabolite pathway in a microbial system to establish the engineered production of Taxol. Moreover, the further researches of the transcriptional regulation of Taxol biosynthesis will be done, based on the putative cis-acting elements in the promoters of taxoid hydroxylase.
Author contributions
Conceived and designed the experiments: CF, LY, WL. Performed the experiments: WL, SZ, KD. Analyzed the data: MZ, KD. Contributed reagents/materials/analysis tools: SZ, YC. Wrote the paper: WL.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. David Nelson at University of Tennessee Health Science Center for the systematic classification of T. chinensis CYP450s. This project is supported by The Specialized Research Fund for the Doctoral Program of Higher Education (No. 2012142130009) and Independent innovation fund project of Huazhong University of Science and Technology (No. 2013TS079).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01468/full#supplementary-material
References
- Ajikumar P. K., Xiao W. H., Tyo K. E., Wang Y., Simeon F., Leonard E., et al. (2010). Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330, 70–74. 10.1126/science.1191652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu P. R., Rao K. V., Reddy V. D. (2013). Structural organization and classification of cytochrome P450 genes in flax (Linum usitatissimum L.). Gene 513, 156–162. 10.1016/j.gene.2012.10.040 [DOI] [PubMed] [Google Scholar]
- Bohlmann J., Gesell A., Blaukopf M., Madilao L., Macaire M. S., Stephen G. W., et al. (2015). The gymnosperm cytochrome P450 CYP750B1 catalyzes stereospecific monoterpene hydroxylation of (+)-sabinene in thujone biosynthesis in Thuja plicata. Plant Physiol. 168, 94–106. 10.1104/pp.15.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple C. (1998). Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Biol. 49, 311–343. 10.1146/annurev.arplant.49.1.311 [DOI] [PubMed] [Google Scholar]
- Chau M., Croteau R. (2004). Molecular cloning and characterization of a cytochrome P450 taxoid 2α-hydroxylase involved in Taxol biosynthesis. Arch. Biochem. Biophys. 427, 48–57. 10.1016/j.abb.2004.04.016 [DOI] [PubMed] [Google Scholar]
- Chau M., Jennewein S., Walker K., Croteau R. (2004). Taxol biosynthesis: molecular cloning and characterization of a cytochrome P450 taxoid 7β-hydroxylase. Chem Biol. 11, 663–672. 10.1016/j.chembiol.2004.02.025 [DOI] [PubMed] [Google Scholar]
- Chen H., Wu B., Nelson D. R., Wu K., Liu C. (2014). Computational identification and systematic classification of novel cytochrome P450 genes in Salvia miltiorrhiza. PLoS ONE 9:e115149. 10.1371/journal.pone.0115149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg G. M., Schepartz S. A., Suffness M., Grever M. R. (1993). The taxol supply crisis. New NCI policies for handling the large-scale production of novel natural product anticancer and anti-HIV agents. J. Nat. Prod. 56, 1657–1668. 10.1021/np50100a001 [DOI] [PubMed] [Google Scholar]
- Croteau R., Ketchum R. E., Long R. M., Kaspera R., Wildung M. R. (2006). Taxol biosynthesis and molecular genetics. Phytochem. Rev. 5, 75–97. 10.1007/s11101-005-3748-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson P. B. (2002). The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr. Drug Metab. 3, 561–597. 10.2174/1389200023337054 [DOI] [PubMed] [Google Scholar]
- de Jong P., Catanese J. J., Osoegawa K., Shizuya H., Choi S., Chen Y. J. (2001). Initial sequencing and analysis of the human genome. Nature 412, 565–566. 10.1038/35087627 [DOI] [Google Scholar]
- Durst F., Nelson D. R. (1995). Diversity and evolution of plant P450 and P450-reductases. Drug Metabol. Drug Interact. 12, 189–206. 10.1515/DMDI.1995.12.3-4.189 [DOI] [PubMed] [Google Scholar]
- Guo J., Zhou Y. J., Hillwig M. L., Shen Y., Yang L., Wang Y., et al. (2013). CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. U.S.A. 110, 12108–12113. 10.1073/pnas.1218061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttikonda S. K., Trupti J., Bisht N. C., Chen H., An Y. Q. C., Pandey S., et al. (2010). Whole genome co-expression analysis of soybean cytochrome P450 genes identifies nodulation-specific P450 monooxygenases. BMC Plant Biol. 10:243. 10.1186/1471-2229-10-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B., Bohlmann J. (2006). Cytochrome P450 mono-oxygenases in conifer genomes: discovery of members of the terpenoid oxygenase superfamily in spruce and pine. Biochem. Soc. Trans. 34, 1209–1214. 10.1042/BST0341209 [DOI] [PubMed] [Google Scholar]
- Hamberger B., Ohnishi T., Hamberger B., Séguin A., Bohlmann J. (2011). Evolution of diterpene metabolism: sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol. 157, 1677–1695. 10.1104/pp.111.185843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasemann C. A., Kurumbail R. G., Boddupalli S. S., Peterson J. A., Deisenhofer J. (1995). Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure 3, 41–62. 10.1016/S0969-2126(01)00134-4 [DOI] [PubMed] [Google Scholar]
- Jennewein S., Croteau R. (2001). Taxol: biosynthesis, molecular genetics, and biotechnological applications. Appl. Microbiol. Biotechnol. 57, 13–19. 10.1007/s002530100757 [DOI] [PubMed] [Google Scholar]
- Jennewein S., Long R. M., Williams R. M., Croteau R. (2004). Cytochrome P450 taxadiene 5α-hydroxylase, a mechanistically unusual monooxygenase catalyzing the first oxygenation step of taxol biosynthesis. Chem. Biol. 11, 379–387. 10.1016/j.chembiol.2004.02.022 [DOI] [PubMed] [Google Scholar]
- Jennewein S., Rithner C. D., Williams R. M., Croteau R. (2003). Taxoid metabolism: taxoid 14β-hydroxylase is a cytochrome P450-dependent monooxygenase. Arch. Biochem. Biophys. 413, 262–270. 10.1016/S0003-9861(03)00090-0 [DOI] [PubMed] [Google Scholar]
- Jennewein S., Rithner C. D., Williams R. M., Croteau R. B. (2001). Taxol biosynthesis: taxane 13α-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 98, 13595–13600. 10.1073/pnas.251539398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspera R., Croteau R. (2006). Cytochrome P450 oxygenases of Taxol biosynthesis. Phytochem. Rev. 5, 433–444. 10.1007/s11101-006-9006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhu Y., Guo X., Sun C., Luo H., Song J., et al. (2013). Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng CA Meyer. BMC Genomics 14:245 10.1186/1471-2164-14-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. T., Zhang P., Zhang M., Fu C. H., Zhao C. F., Dong Y. S., et al. (2012). Transcriptional profile of Taxus chinensis cells in response to methyl jasmonate. BMC Genomics 13:295. 10.1186/1471-2164-13-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang P., Zhang M., Fu C., Yu L. (2013). Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol. 15, 19–26. 10.1111/j.1438-8677.2012.00611.x [DOI] [PubMed] [Google Scholar]
- Li Y., Gou J., Chen F., Li C., Zhang Y. (2016). Comparative transcriptome analysis identifies putative genes involved in the biosynthesis of Xanthanolides in Xanthium strumarium L. Front Plant. Sci. 7:1317. 10.3389/fpls.2016.01317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W. K., Fossella F. V., Winn R. J., Shin D. M., Hynes H. E., Gross H. M., et al. (1993). Phase II study of taxol in patients with untreated advanced non-small-cell lung cancer. J. Natl. Cancer Inst. 85, 384–388. 10.1093/jnci/85.5.384 [DOI] [PubMed] [Google Scholar]
- Nelson D. R. (2009). The cytochrome p450 homepage. Hum. Genomics. 4, 59–65. 10.1186/1479-7364-4-1-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B., Street N. R., Wetterbom A., Zuccolo A., Lin Y. C., Scofield D. G., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584. 10.1038/nature12211 [DOI] [PubMed] [Google Scholar]
- Paquette S. M., Bak S., Feyereisen R. (2000). Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 19, 307–317. 10.1089/10445490050021221 [DOI] [PubMed] [Google Scholar]
- Rasool S., Mohamed R. (2016). Plant cytochrome P450s: nomenclature and involvement in natural product biosynthesis. Protoplasma 253, 1197–1209. 10.1007/s00709-015-0884-4 [DOI] [PubMed] [Google Scholar]
- Ro D. K., Paradise E. M., Ouellet M., Fisher K. J., Newman K. L., Ndungu J. M., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. 10.1038/nature04640 [DOI] [PubMed] [Google Scholar]
- Schoendorf A., Rithner C. D., Williams R. M., Croteau R. B. (2001). Molecular cloning of a cytochrome P450 taxane 10β-hydroxylase cDNA from Taxus and functional expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 98, 1501–1506. 10.1073/pnas.98.4.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G. H., Zhao C. F., Zhang M., Fu C. H., Zhang H., Yu L. J. (2014). Correlation analysis of the taxane core functional group modification, enzyme expression, and metabolite accumulation profiles under methyl jasmonate treatment. Biotechnol Prog. 30, 269–280. 10.1002/btpr.1864 [DOI] [PubMed] [Google Scholar]
- Sun G., Yang Y., Xie F., Wen J. F., Wu J., Wilson I. W., et al. (2013). Deep sequencing reveals transcriptome re-programming of Taxus × media cells to the elicitation with methyl jasmonate. PLoS ONE 8:e62865. 10.1371/journal.pone.0062865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. J., Zhang W., Zhang J. W., Guo M. J., Zhuang Y. P. (2016). Optimization of a broth conductivity controlling strategy directed by an online viable biomass sensor for enhancing Taxus cell growth rate and Taxol productivity. RSC Adv. 6, 40631–40640. 10.1039/C5RA26540A [DOI] [Google Scholar]
- Warren R. L., Keeling C. I., Yuen M. M. S., Raymond A., Taylor G. A., Vandervalk B. P., et al. (2015). Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Plant J. 83, 189–212. 10.1111/tpj.12886 [DOI] [PubMed] [Google Scholar]
- Wheeler A. L., Long R. M., Ketchum R. E. B., Rithner C. D., Williams R. M., Croteau R. (2001). Taxol biosynthesis: differential transformation of taxadien-5α-ol and its acetate ester by cytochrome P450 hydroxylases from Taxus suspension cells. Arch. Biochem. Biophys. 390, 265–278. 10.1006/abbi.2001.2377 [DOI] [PubMed] [Google Scholar]
- Williams P. A., Cosme J., Sridhar V., Johnson E. F., McRee D. E. (2000). Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell. 5, 121–131. 10.1016/S1097-2765(00)80408-6 [DOI] [PubMed] [Google Scholar]
- Wu Q., Sun C., Luo H., Li Y., Niu Y., Sun Y., et al. (2011). Transcriptome analysis of Taxus cuspidata needles based on 454 pyrosequencing. Planta Med. 77, 394–400. 10.1055/s-0030-1250331 [DOI] [PubMed] [Google Scholar]
- Yan C., Duan W., Lyu S., Li Y., Hou X. (2017). Genome-wide identification, evolution, and expression analysis of the ATP-binding cassette transporter gene family in Brassica rapa. Front Plant. Sci. 8:349. 10.3389/fpls.2017.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbe P., Hamberger B., Yuen M. M., Chiang A., Sandhu H. K., Madilao L. L., et al. (2013). Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 162, 1073–1091. 10.1104/pp.113.218347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Dong Y., Nie L., Lu M., Fu C., Yu L. (2015). High-throughput sequencing reveals miRNA effects on the primary and secondary production properties in long-term subcultured Taxus cells. Front Plant. Sci. 6:604. 10.3389/fpls.2015.00604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Han Z., Sun G., Hoffman A., Wilson I. W., Yang Y., et al. (2014). Molecular cloning and characterization of a cytochrome P450 taxoid 9a-hydroxylase in Ginkgo biloba cells. Biochem Biophys. Res. Commun. 443, 938–943. 10.1016/j.bbrc.2013.12.104 [DOI] [PubMed] [Google Scholar]
- Zhao Y. J., Cheng Q. Q., Su P., Chen X., Wang X. J., Gao W., et al. (2014). Research progress relating to the role of cytochrome P450 in the biosynthesis of terpenoids in medicinal plants. Appl. Microbiol. Biotechnol. 98, 2371–2383. 10.1007/s00253-013-5496-3 [DOI] [PubMed] [Google Scholar]
- Zhou K., Qiao K., Edgar S., Stephanopoulos G. (2015). Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 33, 377–383. 10.1038/nbt.3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.