Abstract
Background: Both HPV-positive and -negative cervical cancers are primarily associated with features of cell cycle and cytoskeletal disruption; however, the actual biological processes affected remain elusive. To this end, we systematically characterized the intracellular proteomic profiles of four distinct and informative cervical cell lines. Materials and Methods: Cell extracts from a normal cervical (HCK1T) and three cervical cancer cell lines, one HPV-negative (C33A), and two HPV-positive, SiHa (HPV16+) and HeLa (HPV18+), were analyzed by 2-dimensional electrophoresis and differentially expressed proteins were identified by MALDI-TOF mass spectrometry, while differential expression was confirmed by western blot analysis. Results: In total, 133 proteins were found differentially expressed between the normal and the cervical cancer lines. Bioinformatics analysis revealed the actin cytoskeleton signaling pathway to be significantly affected, while up-regulation of cofilin-1, an actin depolymerizing factor, was documented and further validated by western blotting. Furthermore, two-way comparisons among the four cell lines, revealed a set of 18 informative differentially expressed proteins. Conclusion: These novel identified proteins provide the impetus for further functional studies to dissect the mechanisms operating in the two distinct pathways of cervical carcinogenesis.
Keywords: Cervical cancer, Cervical cell lines, HCK1T cell line, Proteomics, 2-DE, Cofilin-1
Cervical cancer represents the fourth most common and fatal form of cancer among women worldwide (1). More than 90% of cervical cancer cases arise as a consequence of a human papilloma virus (HPV) infection. Currently, 210 different HPV types have been officially recognized (2), and – based on their carcinogenic potential – have been classified either as low-risk HPV types, or as high-risk and potentially carcinogenic (3). Of the high-risk group, the five most common HPV types include HPV 16, 18, 45, 30, and 33, while types 16 and 18 alone, account for about 70% of all cervical cancer cases (4).
In the early phase of cervical carcinogenesis, HPV initially infects the proliferating cells of the basal layer of the cervical stratified epithelium through abrasions of the mucosal epithelium. Following infection, the HPV genome remains in the form of nuclear extrachromosomal episome, and due to the repression of the viral E6 and E7 oncoprotein synthesis, the viral DNA replication occurs at very low levels (5). Following the gradual differentiation of the basal cells and their migration to the upper layers of the epithelium, an increased expression of E6 and E7 oncoproteins occurs, leading to inhibition of apoptosis, while the viral genome is replicated further. At this point, structural proteins and mature viral particles are produced, and the shed virus can then initiate a new infection. These infections resolve within 1-2 years, but they can last up to several decades. However, very few of them are persistent and gradually progress to cancer (6), reflecting a dynamic interplay of viral mechanisms of immune escape and suppression of the negative regulators of the cell growth, combined with defects of cellular response by the host (5). The final events of the neoplastic transformation are associated frequently (45-80%) with the integration of the HPV DNA into the host genome and the ensuing overexpression of E6 and E7 oncoproteins, leading to further proliferation of the transformed cells.
Since both HPV-positive and HPV-negative cervical cancers are primarily associated with features of cell-cycle and cytoskeletal disruption (5), the precise elucidation of the contribution of the putative individual oncogenic drivers induced by the presence or the absence of HPV in cervical cancer is imperative. Therefore, utilization of informative cell lines with or without the HPV genome, can provide valuable insights on these mechanisms. To this end, our group has initiated a comprehensive approach to elucidate the specific oncogenic drivers operating in cervical cancer, both at the transcriptional (7) and the proteomic level (8). In these studies, we have identified for the first time, four novel transcription modules (7), involved in cervical cancer, exhibiting synergy between groups of transcription regulators, while certain modules were annotated to specific biological processes, such as cell cycle, apoptosis, transcription and development. In our recent study (8), by employing proteomic approaches on the secretome of four informative cervical cell lines, we have identified 67 differentially expressed proteins, displaying mainly catalytic, binding or structural molecule activity, while bioinformatics analysis identified the transcription factor NRF2 as an important regulator of differentially expressed proteins in the cancer cell lines. Thus, such comparative proteomic approaches employing cervical cell lines, represent a valuable tool to further explore the precise mechanisms involved in viral infection and protein dysfunction interplay that can lead to cervical carcinogenesis (5,6). Furthermore, a review (6) of recent studies on the proteomics of cervical cancer cell lines, revealed that their major focus relies either on the variable effects of several drugs or on the modulation of a specific gene expression on their proteome composition.
Therefore, based on the above data, in the present study, we further investigated these mechanisms, by employing 2-dimensional electrophoresis (2-DE) and bioinformatics analysis, and systematically characterized the intracellular proteomic profiles of four distinct informative cervical cell lines, either with or without the presence of HPV, and identified specific biochemical similarities and differences, reflecting particular aberrant pathways of carcinogenesis, which can be eventually validated further and assessed as putative biomarkers of cervical pathology.
Materials and Methods
Cell lines culture and sample preparation. HeLa (HPV 18+), SiHa (HPV 16+) and C33A (HPV–negative) cervical cancer cell lines, were purchased from ATCC (Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) (Gibco-Invitrogen, Waltham, MA, USA) at 37˚C, 5% CO2 as previously described (9). ΗCK1T cells were a kind offer of Tohru Kiyono (10) and were cultured as proposed (11) in Defined Keratinocyte Serum-Free Medium (SFM) (Gibco BRL, San Francisco, CA, USA) supplemented with 5 ng/ml Epidermal Growth Factor (EGF) (Gibco BRL) and 50 μg/ml of Bovine Pituitary Extract (BPE) (Gibco BRL). When the cells reached a concentration of 106 cells per ml, they were trypsinized and harvested, and the pellets were washed in Phosphate Buffered Saline (PBS) 3 times.
Pellets were homogenized in 2D-Buffer (7 M Urea, 2 M thiourea, 4% CHAPS, 1% DTE) using mild sonication (water bath sonication). After centrifugation at 16,000 × g for 20 min, the total cell extract was obtained as a supernatant. Protein concentration was measured with the Bradford assay.
2D Electrophoresis. From each cell extract, 80 μg were loaded on 7-cm immobilized pH gradient (IPG) strips of pH 3-10 NL (Bio-Rad, Hercules, CA, USA) in isoelectric focusing (IEF) cell trays, following the addition of 2% Bio-Lyte 3/10 Ampholytes for isoelectric focusing. For the preparative gels only, the amount of protein loaded from each cell line was 400 μg. IEF was performed in Bio-Rad PROTEAN IEF cell for 14,000 VHR as follows. Step 1: 50 V, rapid, 14 h (active rehydration); step 2: 250 V, rapid, 30 min; step 3: 4,000 V, linear, 1 h; step 4: 4000 V, rapid, 11000 VHR (focusing); step 5: 100 V, rapid, 24 h (conservation). For the second dimension electrophoresis, the equilibration of the strips and the reduction and alkylation of the proteins were performed in equilibration buffer (6 M Urea, 1.5 M Tris HCl, pH 8.8, 30% Glycerol, 2% SDS) containing 0.03 M DTE for 20 min and then in equilibration buffer containing 0.136 M Iodoacetamide for 20 min. Strips were then sealed with agarose on top of a 12% SDS-polyacrylamide gel, which was run at 80 V for 10 min and then at 160 V. Following electrophoresis, the gels were fixed in 30% methanol, 10% acetic acid for 30 min, 2 h and then overnight. Visualization of the spots was achieved with silver staining. Each gel was sensitized in 0.8 mM sodium thiosulfate pentahydrate solution for 1 min, washed with ultrapure water for 1 min, stained in 12 mM silver nitrate solution for 20 min and washed again with ultrapure water for 10 sec. Visualization of the spots was performed with 50 ml of development solution (3% w/v potassium carbonate, 12.5 μl formalin-formaldehyde 37%, 5.25 μl 10% w/v sodium thiosulfate pentahydrate). When the development was completed, the gel was washed with ultrapure water for 10 sec and the stop solution (2% acetic acid) was added and left for 10 min. After two washes with ultrapure water for 10 min, the gels were scanned with a GS-800 imaging densitometer (Bio-Rad). Four 2D gels from each cell line were prepared and were used for the analysis.
Comparative analysis. The comparative analysis of the 2D gels images was performed with the PDQuest 2D-Gel analysis software v.8.1.0 (Bio-Rad). The individual protein spot quantity was normalized with the total optical density of the gel. All protein spots with a cancer/normal ratio <0.5 or >2 were considered as differentially expressed and were included for further analysis.
Spot preparation and protein identification. The spots were picked manually from the corresponding preparative 2D gels, which were stained with Coomassie Colloidal Blue overnight, placed in 96-well plates, and 100 μl of destaining solution (40% ACN, 50 mM ammonium bicarbonate) was added in each spot. The plates were shaken for 15 min and this step was repeated three times. Then, the spots were washed with 100 μl ultrapure water for 5 min, reduced with 100 μl of 10 mM DTE dissolved in 100 mM ammonium bicarbonate, pH 8.5 for 10 min and alkylated with 100 μl of 54 mM iodoacetamide, dissolved in 100 mM ammonium bicarbonate, pH 8.5 for 10 min in the dark. The spots were washed with 100 μl of 100 mM ammonium bicarbonate, pH 8.5 for 5 min and then dried in a Savant SpeedVac™ concentrator (Thermo Fisher Scientific, Logan, UT, USA). Finally, 3 μl of 10 ng/μl trypsin in 10 mM ammonium bicarbonate was added, and left overnight. The products of tryptic digestion were extracted from the gel by the addition of 5 μl of extraction solution (50% ACN, 0.1% TFA) for 30 min. Then, 1 μl of peptides from each spot were mixed on a stainless steel MALDI target plate with 1 μl of matrix solution (50% v/v ACN, 0.1% TFA v/v, 0.7% v/v α-cyano-4-hydroxycinnamic acid) containing the peptides des-Arg-bradykinin, 904.4681Da (Sigma-Aldrich Corp., St. Louis, MO, USA), and the adrenocorticotropic hormone fragment 18-39, 2465.1989Da (Sigma-Aldrich), as internal standards. For peptide identification, Matrix Assisted Laser Desorption Ionization-Time of Flight/Time of Flight Mass Spectrometry (MALDI-TOF/TOF MS) was performed in an Ultraflex TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA, USA). Peak list was created with the Flexanalysis v2.2 software (Bruker Daltonics), peptide matching and protein searches were performed automatically by Mascot Server database (Matrix Science, Boston, MA, USA), signal/noise threshold ratio was set at 2.5. For peptide identification, monoisotopic masses were used and a mass tolerance of 0.0025% (25 ppm) was allowed. Cysteine carbamidomethylation and methionine oxidation were set as fixed and variable modifications, respectively. One miscleavage was allowed. The peptide masses were compared with the theoretical peptide masses of all available proteins from Homo sapiens using the Swiss-Prot database. The probability score with p<0.05 identified by the software, was used as the criterion for the affirmative protein identification.
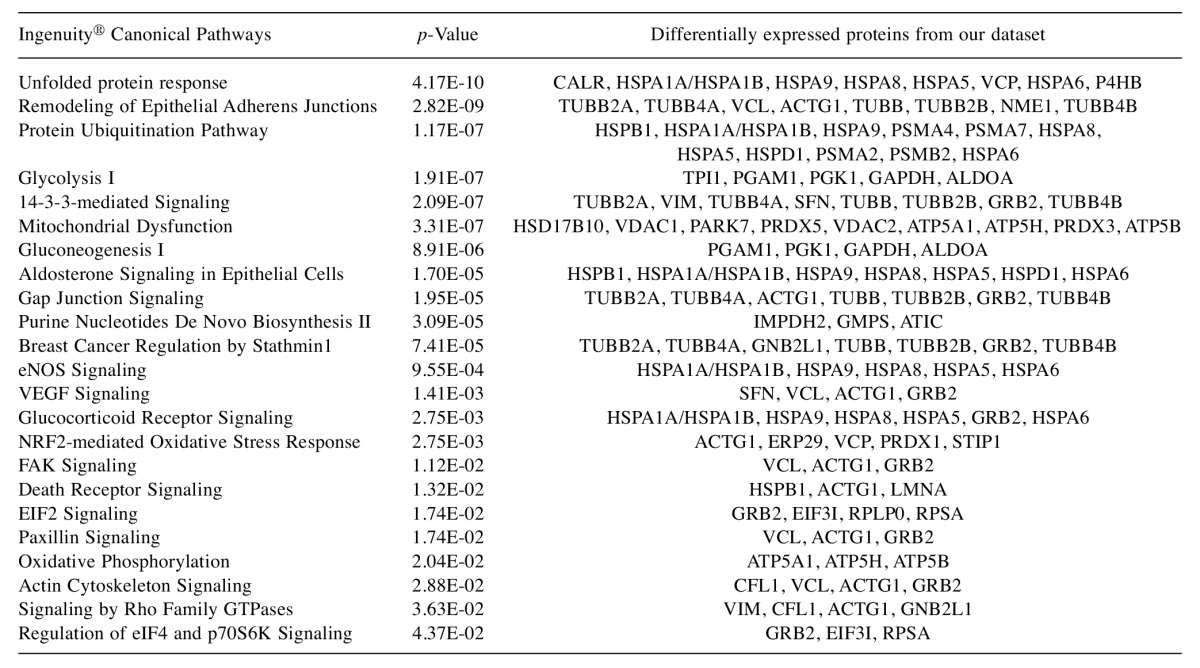
Bioinformatics analysis. Functional annotation of the differentially expressed proteins was performed manually using information from UniProt (http://www.uniprot.org/) and the literature. Pathway analysis was generated by QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN, Redwood City, CA, USA; www.qiagen.com/ ingenuity). Ingenuity® Pathway Analysis output was manually curated in order to remove redundant terms, results unrelated to cancer biology, and pathways with fewer than three differentially expressed proteins from our dataset. Moreover, only statistically significant (p≤0.05, Fisher’s exact test) canonical pathways were selected.
Western blot analysis. Four 50 μg samples of cell extract dissolved in Laemli’s buffer from each cell line were loaded in 15% SDS-polyacrylamide gel after incubation at 90˚C for 10 min. The gel was run at 40 V for 15 min and then at 120 V in transfer buffer (3.03 g Tris, 14.4 g Glycine, 200 ml Methanol for 1 liter total volume). The transfer was performed in transfer buffer for 2 h at 290 mA at 4˚C. Then, the membrane was stained with Ponceau-S stain for 5 min, washed with ultrapure water for 5 min three times, followed by the addition of blocking solution (5% w/v non-fat dried milk in TBS-Tween 0.1% v/v) and incubation for 2 h. The membrane was washed with TBS-Tween 0.1% v/v successively for 15 min, 5 min, and 5 min, and the primary mouse antibody sc-53934 for Cofilin-1 (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was added in a 1:500 dilution and left at 4˚C overnight. The next day, the three washes were repeated and the secondary sc-2005 goat anti-mouse antibody IgG-HRP (Santa Cruz Biotechnology) was added in a 1:2,000 dilution and left at room temperature for 2 h. The three washes were repeated, ECL was added and left for 1 min; its excess was removed, followed by film exposure and development.
Results
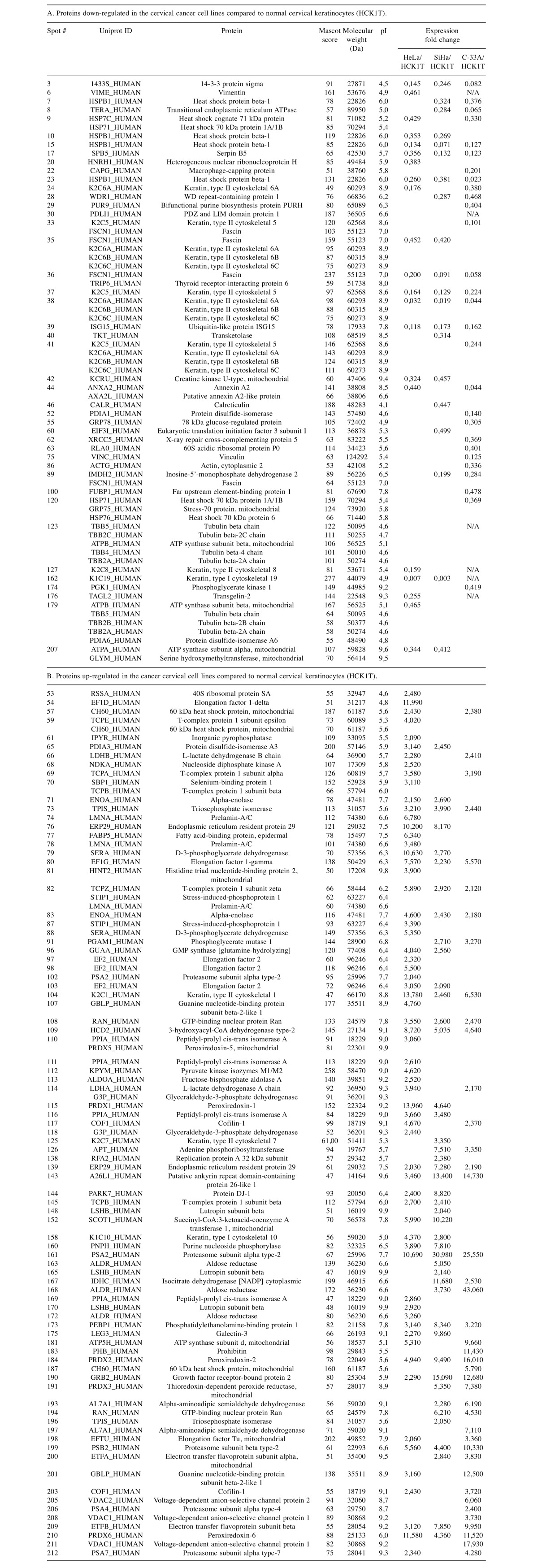
Proteomic analysis. In order to investigate the quantitative differences in protein expression resulting from malignant transformation of the cervical epithelium, the proteomic profiles of four cervical cell lines were analyzed. The four cell lines that were used were the following: HCK1T (Human Cervical Keratinocytes), a normal cervical epithelium cell line; HeLa, a cervical cancer cell line positive for HPV18; SiHa, a cervical cancer cell line positive for HPV16; and C-33A, a cervical cancer cell line negative for HPV. Four 2D gels were run for each cell line total extract. A representative gel is shown in Figure 1 for each cell line. The initial analysis was focused on the comparison between the normal cervical cell line (HCK1T) and the group of the three cervical cancer cell lines (HeLa, SiHa, C-33A). In this comparison, 43 spots, that corresponded to 48 unique proteins, were found down-regulated (cancer/normal ratio <0.5) in the cancer cell lines. Almost half of these proteins (42%), are associated to cytoskeleton and 21% of them are involved in metabolism (Figure 2). Moreover, 85 spots, that corresponded to 65 unique proteins, were found up-regulated (cancer/normal ratio >2) in the cancer cell lines. A significant percentage of these proteins are also associated to cytoskeleton (8%) and metabolism (42%) (Figure 2). Proteins that were found deregulated included cofilin-1; vinculin; vimentin; fascin; annexin A2; transgelin-2; alpha-enolase; triosephosphate isomerase; glyceraldehyde-3-phosphate dehydrogenase; peptidyl-prolyl cis-trans isomerase A; fructose-bisphosphate aldolase A; peroxiredoxins 1, 2, 5 and 6; protein DJ-1; and growth factor receptor-bound protein 2. A complete list of the differentially expressed proteins is presented in Table I.
Figure 1. Annotated 2D gel images of cervical cell lines with differentially expressed proteins between normal and cancer states. A. HCK1T cell extract. The annotated proteins were found up-regulated in HCK1T (normal cervical epithelium) in comparison with the cancer cell lines. B. HeLa cell extract. The annotated proteins were found up-regulated in HeLa in comparison with HCK1T. C. SiHa cell extract. The annotated proteins were found up-regulated in SiHa in comparison to HCK1T. D. C-33A cell extract. The annotated proteins were found up-regulated in C-33A in comparison to HCK1T. The numbers of the spots correspond to proteins listed in Table I.
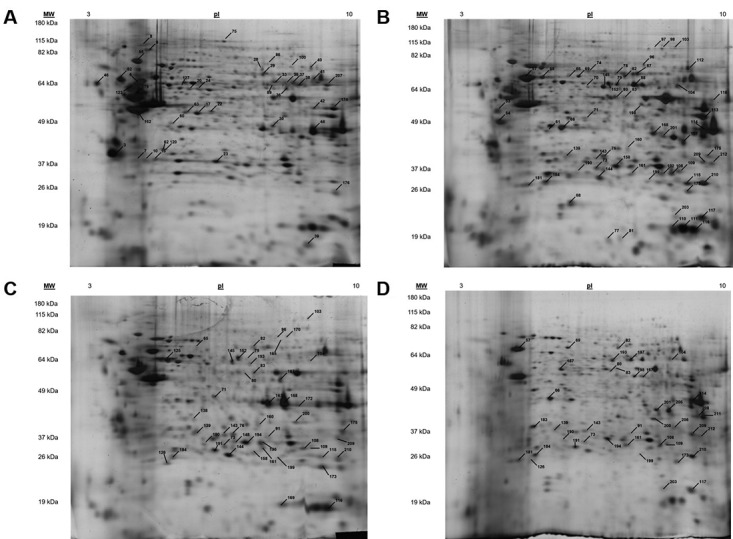
Figure 2. Functional annotation of the differentially expressed proteins from the comparison between normal and cancer cervical cell lines. A. Functional annotation of the proteins found down-regulated in cancer cell lines compared to normal. B. Functional annotation of the proteins found up-regulated in the cancer cell lines compared to normal.
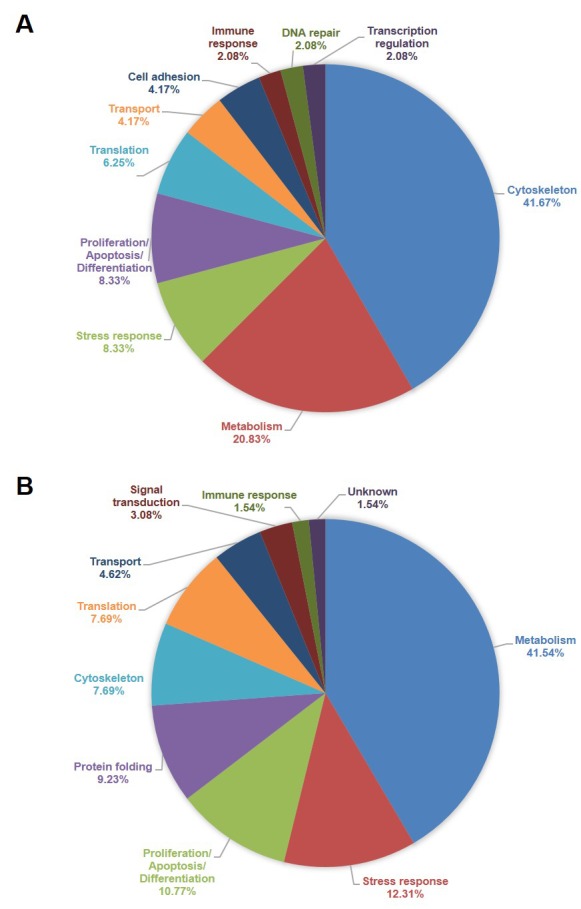
Table I. List of differentially expressed proteins following comparison between normal and cancer cervical cell lines.
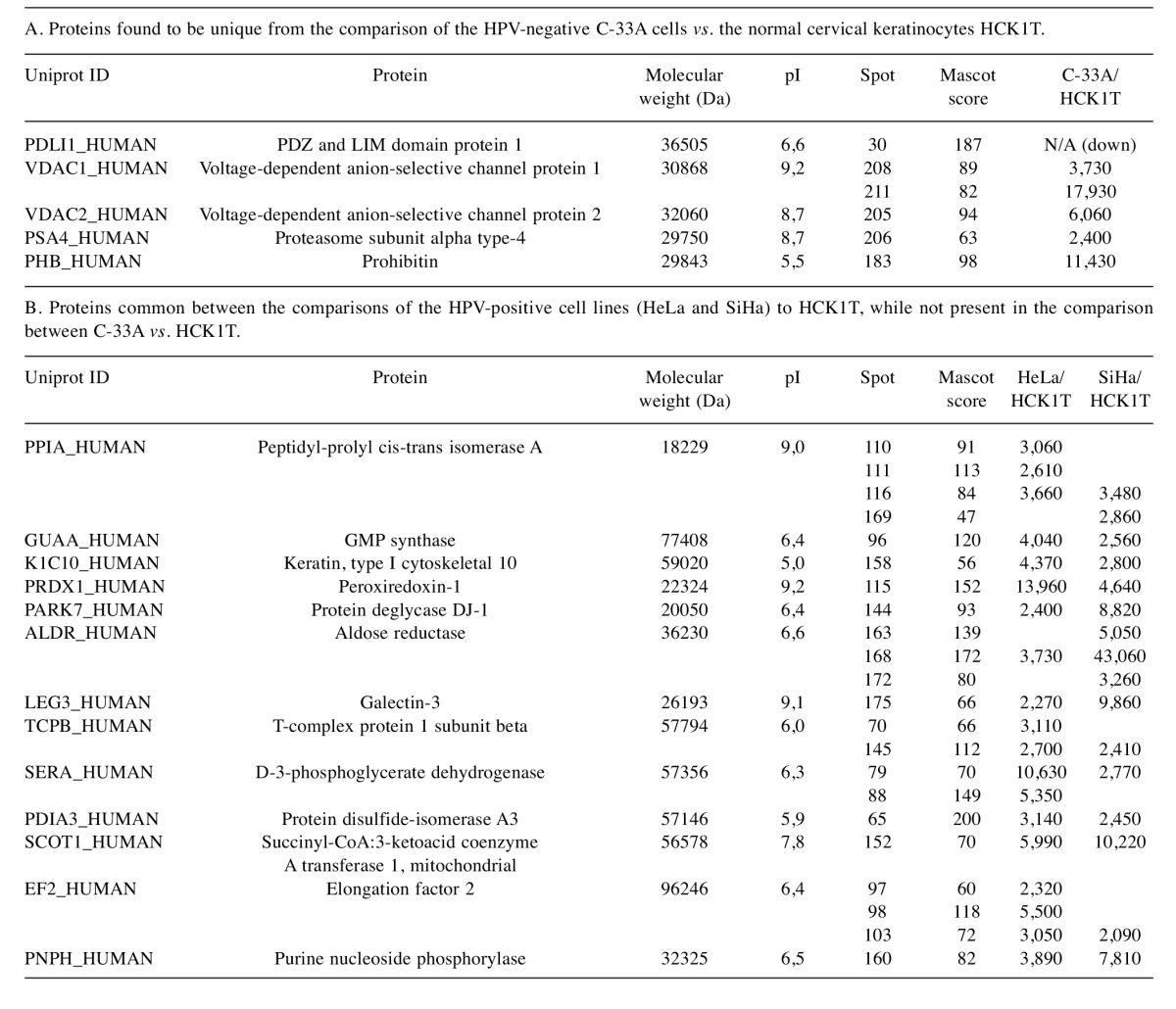
The overlap of differentially expressed proteins among the three comparisons was examined and is presented in a Venn diagram in Figure 3. Five proteins only emerged from the comparison between the HPV-negative cervical cancer cell line (C-33A) and the normal cervical keratinocytes (HCK1T), while 13 proteins composed the HPV-positive “core”, emerging from the individual comparisons between the HPV-positive cervical cancer cell lines (HeLa and SiHa) with the HCK1T (Table II).
Figure 3. Venn diagram of the differentially expressed proteins in the three individual comparisons. Differentially expressed proteins in HeLa vs. HCK1T; SiHa vs. HCK1T; and C-33A vs. HCK1T, are represented with the blue, red and green circles, respectively. Five proteins are unique in the comparison of the HPV-negative cell line (C-33A) to the normal cervical keratinocytes (HCK1T), and 13 proteins are common between the comparisons of the HPV-positive cell lines (HeLa and SiHa) to HCK1T, while not present in the comparison between C-33A vs. HCK1T.
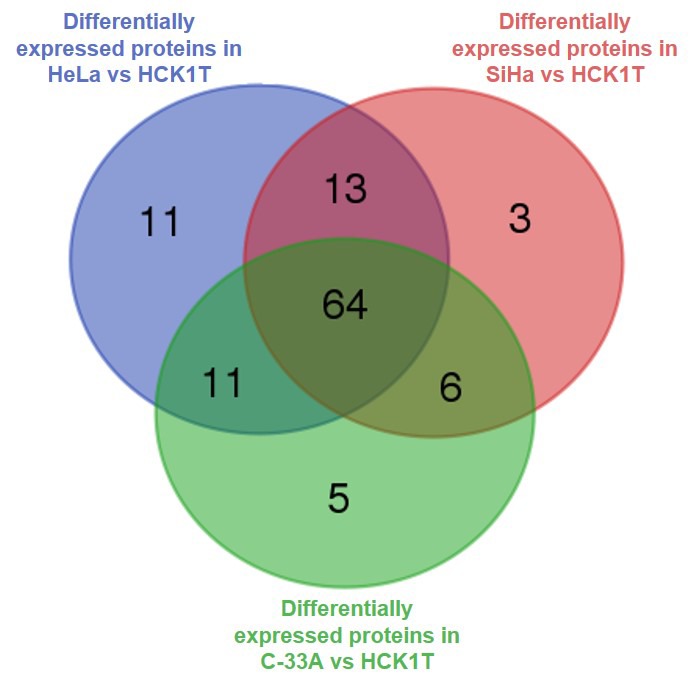
Table II. Differentially expressed proteins that are unique following the comparisons of HPV-negative and HPV-positive cervical cancer cell lines with normal cervical keratinocytes.
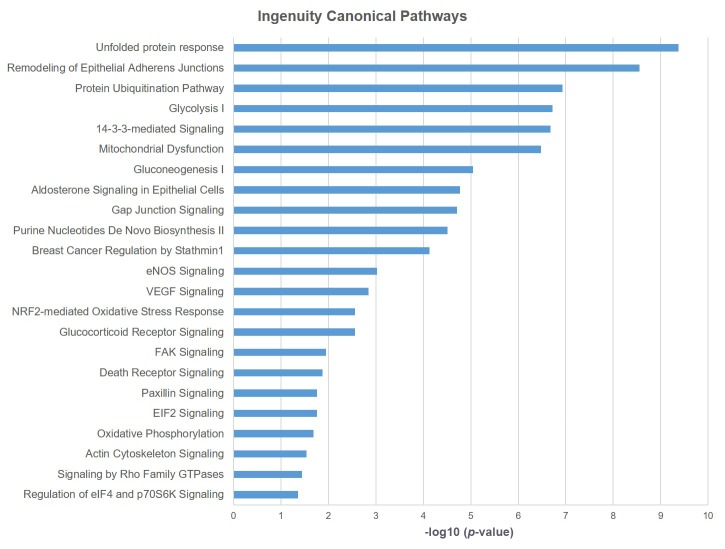
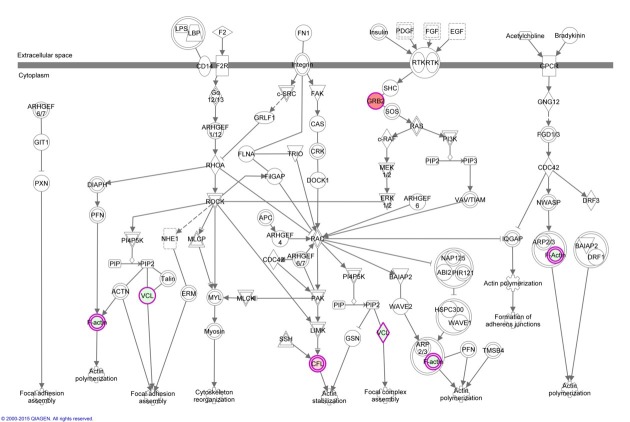
Bioinformatics analysis. Pathway analysis of the differentially expressed proteins between the normal (HCK1T) and the three cancer (HeLa, SiHa, C-33A) cell lines, using the Ingenuity® Pathway Analysis (IPA) software, revealed that the actin cytoskeleton signaling pathway is associated in a statistically significant manner (p=0.029) (Figure 4 and Figure 5, Table III). As mentioned previously, biological function annotation also documented that a large percentage of all the differentially expressed proteins, are associated to cytoskeleton. Based on these indications, and the known biological contribution to cancer pathogenesis, proteins involved in cytoskeletal processes were further investigated. Thus, cofilin-1 (CFL1), an actin depolymerizing factor (ADF) that is implicated in aggressive cancer cell behavior (12-17), was selected for further validation. The main function of cofilin-1 is the depolymerization of F-actin, a biological process crucial for normal mitosis, cytokinesis and cell migration (18). Cofilin-1, was actually up-regulated in all three cancer cell lines (Table I).
Figure 4. Ingenuity® Canonical Pathways deregulated between normal and cancer cervical cell lines. Only statistically significant pathways are shown in a descending order of p-value using Fisher’s exact test.
Figure 5. Actin cytoskeleton signaling is a statistically significant affected pathway in cervical cancer cells, as documented from the Ingenuity® Pathway Analysis of the differentially expressed proteins, between cervical cancer cell lines and normal cervical keratinocytes. Nodes filled with red color represent proteins that were found up-regulated in cervical cancer cell lines and nodes filled with green color represent proteins that were found down-regulated in the cervical cancer cell lines. Double-lined nodes represent groups of proteins or complexes.
Table III. List of Ingenuity® Pathway Analysis results from the comparison between normal and cancer cervical cell lines. Only statistically significant pathways are shown in a descending order of p-value, using Fisher’s exact test.
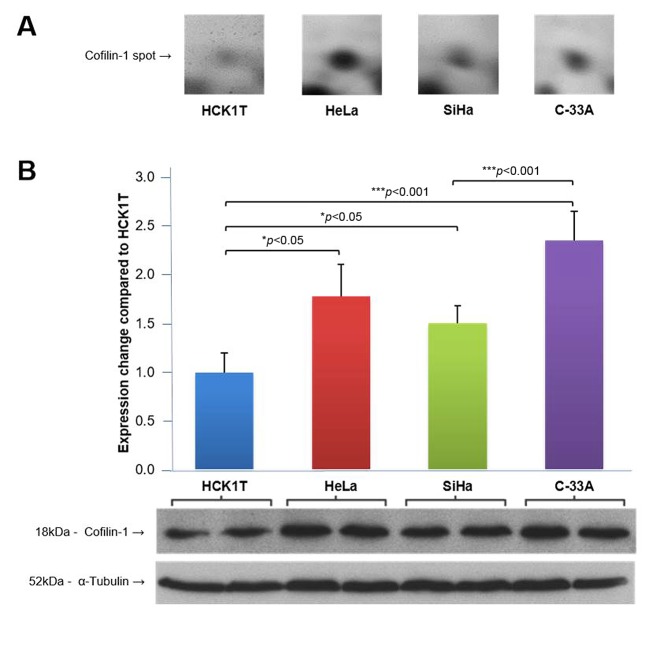
Confirmation of the proteomic analysis results. In the 2D gels, cofilin-1 was found up-regulated in HeLa and C-33A cell lines (HeLa/HCK1T ratio: 3.6, C-33A/HCK1T ratio: 3.0). Its levels were also higher in SiHa compared to the ones in HCK1T, but did not reach the 2-fold threshold (SiHa/HCK1T ratio: 1.3) as shown in Figure 6A. The up-regulation of cofilin-1 in the 2D gels was confirmed by western blot analysis on the four cell lines (Figure 6B). As shown in Figure 6, the levels of cofilin-1 in the western blot analysis follow the same expression pattern as in the 2D gels, thus confirming the proteomic analysis results.
Figure 6. Cofilin-1 levels in cervical cell lines. A. Cofilin-1 levels in proteomics analysis. Fold change is expressed in comparison with HCK1T, as the average from the two identified spots on 2D gels, measured with PDQuest software. B. Cofilin-1 levels in western blot analysis. Fold change is expressed in comparison with HCK1T representing the average from four biological replicates for each cell line, measured with the Quantity One 1-D Analysis software (Bio-Rad). Tubulin levels were used for signal normalization in western blots. Mean values and standard deviation bars are shown for each cell line, and p-value was calculated with Student’s t-test.
Discussion
The comparison of proteomic profiles between the three cervical cancer cell lines (HeLa, SiHa, C-33A) and the normal cervical keratinocytes (HCK1T) total cell extract, revealed a total of 113 differentially expressed proteins (cancer/normal ratio <0.5 or >2). Around 60% of these proteins are associated to cytoskeleton or metabolism. Moreover, bioinformatics analysis of the differentially expressed proteins revealed that actin cytoskeleton signaling represents a statistically significant pathway deregulated in cervical cancer. This finding led us to the investigation of cofilin-1, in the context of cervical cancer and actin cytoskeleton remodeling, and the independent confirmation of its expression trend by western blot analysis.
Functional annotation of the differentially expressed proteins indicated that nearly 63% of all deregulated proteins are involved in metabolism. More specifically, 67% of the proteins involved in metabolism are up-regulated in the cervical cancer cell lines compared to normal cervical keratinocytes. This finding could serve as a validation of our approach, since uncontrolled proliferation occurring during carcinogenesis, generates additional anabolic and energy demands. To sustain survival and proliferation, cancer cells have to activate or enhance metabolic pathways that utilize the available nutrients for production of metabolic precursors for cell anabolism, and maintain the reduction–oxidation balance (19). Hence, molecules and regulators that participate in such metabolic processes are expected to be deregulated in cancer cells when compared to normal ones. Such typical examples of enzyme proteins that emerged from our analysis, included alpha-enolase; triosephosphate isomerase; glyceraldehyde-3-phosphate dehydrogenase; peptidyl-prolyl cis-trans isomerase A; fructose-bisphosphate aldolase A; and peroxiredoxins 1, 2, 5 and 6.
Actin cytoskeleton signaling, a statistically significant pathway that emerged from bioinformatics analysis of the differentially expressed proteins, is known to play a pivotal role in cancer, since actin cytoskeleton remodeling is essential for cell proliferation and migration. The process, during which a cancer cell migrates from the original site of the tumor to a new site, consists of specific steps, known as the ‘metastatic cascade’. To metastasize, a cancer cell has to detach from the primary tumor site, migrate, intravasate, translocate through vessels, extravasate, and finally, attach and grow a secondary tumor at a new site. Due to their plasticity, cancer cells can move utilizing either mesenchymal or amoeboid motility, depending on the physical properties of the extracellular matrix, the degree of extracellular proteolysis and on the soluble signaling factors. This whole procedure requires extensive cell cytoskeleton reorganization to achieve the desired cell movements and shape alterations. Actin cytoskeleton remodeling requires the fine orchestration among actin microfilaments, intermediate filaments and microtubules (20,21).
In our study, cofilin-1, which is involved in the Actin cytoskeleton signaling, was actually up-regulated in HeLa and C-33A (>2-fold expression change), while it was also found in higher levels in SiHa (1.3-fold expression change) compared to HCK1T. This expression trend was confirmed by western blot analysis on the four cell lines total cell extract, employing an antibody previously validated and recommended for use in western blot, according to the guidelines (22).
Cofilin-1 is a small protein of ~19 kD whose name stands for cofilamentous protein. It plays a key role in actin dynamics, cell division, chemotaxis and cancer cell migration (18,23). Cofilin-1 severs and depolymerizes actin filaments, thus increasing the free barbed ends where actin polymerization occurs (24,25). However, different concentrations of cofilin-1 have different effects on actin filament severing and nucleation. Low concentration favors severing, while high concentration favors nucleation (26). Cofilin-1 also has a Nuclear Localization Signal (NLS) and can transfer monomeric actin in the nucleus where rod-like structures of actin are formed in response to heat shock, ATP-depletion and dimethyl sulfoxide (DMSO) treatment, cytochalasin D or high cytosolic G-actin concentration. However, its biological role in the nucleus remains unclear (18).
The function of cofilin-1 does not only depend on its concentration, but also on several environmental factors. Phosphorylated cofilin-1 on ser-3 is considered to be inactive because of its lower affinity for actin. LIM kinase 1 (LIMK1), LIM kinase 2 (LIMK2) and testicular protein kinase 1/2 (TESK1/2) phospohorylate cofilin-1 on ser3, while slingshot-1L (SSH1L) phosphatase and chronophin (CIN) induce its dephosphorylation (18). Upon stimulation by epidermal growth factor (EGF), cofilin-1 becomes activated through dephosporylation and dissociation from phosphatidylinositol 4,5-bisphosphate (PIP2) in order to reorganize the cytoskeleton for chemiotactic migration (27). Changes on the intracellular pH, regulated by Na+-H+ exchanger 1 (NHE1), can also affect cofilin-1 activity. When pH is higher than normal (6.8-7.4), cofilin-1 can be also activated through dissociation from the inhibitory complexes with cortactin and PIP2 (25,28,29).
In the context of cancer metastasis and cell migration, cofilin-1 is active at the leading edge of migrating cells protrusions and its levels have been studied in various types of cancer (25). In non-small cell lung cancer (NSCLC), high cofilin-1 levels were correlated with lower overall survival rate, cellular invasiveness and resistance to drugs, and particularly, to cisplatin (30-33). Coflin-1 was also found up-regulated in breast cancer and its levels correlated with tumor size and stage (34-36). Overexpression of cofilin-1 can predict shorter progression-free survival in advanced ovarian cancer patients, receiving standard therapy (37). Immunohistochemistry studies on prostate tissue sections revealed expression of cofilin-1 in 70% of prostate cancer samples, while benign prostate hyperplasia samples were negative for the protein. In the same study, cofilin-1 levels were significantly associated with the Gleason score and the presence of lymph node metastasis (38). Furthermore, proteomic analysis of saliva from patients with head and neck squamous cell carcinoma, revealed significantly increased levels of cofilin-1 compared to the control group (39). Additionally, high expression levels of cofilin-1 were associated with large tumor size, high TNM stage, lymph node metastasis, and decreased overall survival in immunohistochemical studies of patients with squamous cell and adenosquamous carcinoma, and adenocarcinoma of the gallbladder (40). Finally, high levels of cofilin-1 expression compared to normal tissue have also been documented in pancreatic cancer (41) and oral carcinoma (42).
As mentioned above, in our study, the differentially expressed proteins were obtained from the comparison of normal cervical keratinocytes with the three different cervical cancer cell lines. These cervical cancer cell lines differ primarily by the presence or absence and the type of the HPV. Specifically, HeLa is positive for HPV18, SiHa is positive for HPV16, and C-33A is negative for HPV. These cell lines were carefully chosen for our analysis in order to investigate the differences between the HPV-positive and HPV-negative types of cervical cancers. The availability of the 18 informative differentially expressed proteins (Figure 3 and Table II) revealed from these two-way comparisons among the four cell lines, provide the impetus for further functional studies to dissect the molecular mechanisms that could play a role in the two distinct pathways of cervical carcinogenesis.
Our study is the first to provide an insight into the differences of the total proteome of cervical cancer cell lines in direct comparison to normal cervical keratinocytes. Most of the proteomic studies utilizing cervical cancer cell lines, focus on the differences that occur in the protein expression pattern as an effect of a drug treatment, such as doxorubicin, oxymatrine and cisplatin (43-45), or under stress conditions, like UVB irradiation and hypoxia (46,47), or after the induced and/or inhibited expression of specific genes, as in the cases of HVP16 E6 gene, transgelin-2 and parkin (48-50). Moreover, our group has recently studied the secretome of these cervical cancer cell lines compared to normal cervical keratinocytes (8). Comparative analysis of the secretome revealed 67 differentially expressed proteins, out of which 36 were also identified as differentially expressed in our study. Furthermore, Lin et al. (51) proposed several putative biomarkers for the rare and very aggressive type of neuroendocrine cervical cancer by comparing the proteomic profile of HM-1, a neuroendocrine cervical cancer cell line, to CaSki, ME-180, and HeLa, that exhibit a non-neuroendocrine origin (51). Their study disclosed 82 differentially expressed proteins and further confirmed the differential expression of transgelin, galectin-1 and PGK-1 in all cell lines employing western blotting. Interestingly, transgelin and PGK-1 were also found differentially expressed in our analysis, suggesting a pivotal role of these proteins in cervical pathology.
In conclusion, the proteomic comparison of the three cervical cancer cell lines with normal cervical keratinocytes, revealed novel proteins that are potentially deregulated in cervical cancer and could be further investigated as putative biomarkers and pharmacological targets. Moreover, bioinformatics analysis indicated that these proteins are involved in processes and pathways that are already documented to be active during carcinogenesis, confirming the validity of the proteomics results. The expression trend of cofilin-1, that was found up-regulated in the cancer group, was confirmed by western blot. Although cofilin-1 has been studied thoroughly in the context of various types of cancer, to our knowledge, it has not been studied yet in cervical cancer. Therefore, the up-regulation of the protein in the cancer cell lines we documented, indicates that cofilin-1 could also be overexpressed in cervical cancer biopsies. Nevertheless, confirmation of this hypothesis in a cohort of well-characterized clinical samples of different clinical stages is definitely needed, and could lead to the use of cofilin-1 as a valuable marker of either cancerous and/or precancerous lesions of the cervical epithelium.
Acknowledgements
This study was funded by the Oncology Program of the Central Council of Health of the Ministry of Health, Grant No. 70-3-9209 to Nicholas P. Anagnou, and by the European Union’s European Social Fund (ESF) and Greek National Funds through the Program THALIS, under the Operational Program Education and Lifelong Learning of the National Strategic Reference Framework (NSRF), Grant No. 70-3-11830 to Kalliopi I. Pappa. The authors wish to thank Dr. Tohru Kiyono (National Cancer Centre Research Institute, Tokyo, Japan) for his generous gift of the HCK1T normal cervical cell line.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.International human papillomavirus reference center on. Available from: http://www.hpvcenter.se/index.php. 2017.
- 3.Arbyn M, Tommasino M, Depuydt C, Dillner J. Are 20 human papillomavirus types causing cervical cancer. J Pathol. 2014;234:431–435. doi: 10.1002/path.4424. [DOI] [PubMed] [Google Scholar]
- 4.de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, Vallejos CS, de Ruiz PA, Lima MA, Guimera N, Clavero O, Alejo M, Llombart-Bosch A, Cheng-Yang C, Tatti SA, Kasamatsu E, Iljazovic E, Odida M, Prado R, Seoud M, Grce M, Usubutun A, Jain A, Suarez GA, Lombardi LE, Banjo A, Menendez C, Domingo EJ, Velasco J, Nessa A, Chichareon SC, Qiao YL, Lerma E, Garland SM, Sasagawa T, Ferrera A, Hammouda D, Mariani L, Pelayo A, Steiner I, Oliva E, Meijer CJ, Al-Jassar WF, Cruz E, Wright TC, Puras A, Llave CL, Tzardi M, Agorastos T, Garcia-Barriola V, Clavel C, Ordi J, Andujar M, Castellsague X, Sanchez GI, Nowakowski AM, Bornstein J, Munoz N, Bosch FX, Retrospective International Survey , HPV Time Trends Study Group Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 5.Schiffman M, Doorbar J, Wentzensen N, de Sanjose S, Fakhry C, Monk BJ, Stanley MA, Franceschi S. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers. 2016;2:16086. doi: 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 6.Kontostathi G, Zoidakis J, Anagnou NP, Pappa KI, Vlahou A, Makridakis M. Proteomics approaches in cervical cancer: Focus on the discovery of biomarkers for diagnosis and drug treatment monitoring. Expert Rev Proteomics. 2016;13:731–745. doi: 10.1080/14789450.2016.1210514. [DOI] [PubMed] [Google Scholar]
- 7.Pappa KI, Polyzos A, Jacob-Hirsch J, Amariglio N, Vlachos GD, Loutradis D, Anagnou NP. Profiling of discrete gynecological cancers reveals novel transcriptional modules and common features shared by other cancer types and embryonic stem cells. PLoS One. 2015;10:e0142229. doi: 10.1371/journal.pone.0142229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kontostathi G, Zoidakis J, Makridakis M, Lygirou V, Mermelekas G, Papadopoulos T, Vougas K, Vlamis-Gardikas A, Drakakis P, Loutradis D, Vlahou A, Anagnou NP, Pappa KI. Cervical cancer cell line secretome highlights the roles of transforming growth factor-beta-induced protein ig-h3, peroxiredoxin-2, and nrf2 on cervical carcinogenesis. Biomed Res Int. 2017;2017:4180703. doi: 10.1155/2017/4180703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makridakis M, Gagos S, Petrolekas A, Roubelakis MG, Bitsika V, Stravodimos K, Pavlakis K, Anagnou NP, Coleman J, Vlahou A. Chromosomal and proteome analysis of a new t24-based cell line model for aggressive bladder cancer. Proteomics. 2009;9:287–298. doi: 10.1002/pmic.200800121. [DOI] [PubMed] [Google Scholar]
- 10.Narisawa-Saito M, Handa K, Yugawa T, Ohno S, Fujita M, Kiyono T. Hpv16 e6-mediated stabilization of erbb2 in neoplastic transformation of human cervical keratinocytes. Oncogene. 2007;26:2988–2996. doi: 10.1038/sj.onc.1210118. [DOI] [PubMed] [Google Scholar]
- 11.Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007;27:3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condeelis JS, Wyckoff JB, Bailly M, Pestell R, Lawrence D, Backer J, Segall JE. Lamellipodia in invasion. Semin Cancer Biol. 2001;11:119–128. doi: 10.1006/scbi.2000.0363. [DOI] [PubMed] [Google Scholar]
- 14.Dowling P, Meleady P, Dowd A, Henry M, Glynn S, Clynes M. Proteomic analysis of isolated membrane fractions from superinvasive cancer cells. Biochim Biophys Acta. 2007;1774:93–101. doi: 10.1016/j.bbapap.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Sidani M, Wessels D, Mouneimne G, Ghosh M, Goswami S, Sarmiento C, Wang W, Kuhl S, El-Sibai M, Backer JM, Eddy R, Soll D, Condeelis J. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J Cell Biol. 2007;179:777–791. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rheenen J, Condeelis J, Glogauer M. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J Cell Sci. 2009;122:305–311. doi: 10.1242/jcs.031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tania N, Prosk E, Condeelis J, Edelstein-Keshet L. A temporal model of cofilin regulation and the early peak of actin barbed ends in invasive tumor cells. Biophys J. 2011;100:1883–1892. doi: 10.1016/j.bpj.2011.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CY, Leu J, Lee YJ. The actin depolymerizing factor (adf)/cofilin signaling pathway and DNA damage responses in cancer. Int J Mol Sci. 2015;16:4095–4120. doi: 10.3390/ijms16024095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez A, Kamphorst JJ, Markert EK, Schug ZT, Tardito S, Gottlieb E. Cancer metabolism at a glance. J Cell Sci. 2016;129:3367–3373. doi: 10.1242/jcs.181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrie RJ, Yamada KM. At the leading edge of three-dimensional cell migration. J Cell Sci. 2012;125:5917–5926. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: Cell cytoskeleton in cancer metastasis. Br J Pharmacol. 2014;171:5507–5523. doi: 10.1111/bph.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlen M. Response to: Should we ignore western blots when selecting antibodies for other applications. Nat Meth. 2017;14:215–216. doi: 10.1038/nmeth.4194. [DOI] [PubMed] [Google Scholar]
- 23.Nishida E, Maekawa S, Sakai H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry. 1984;23:5307–5313. doi: 10.1021/bi00317a032. [DOI] [PubMed] [Google Scholar]
- 24.Elam WA, Kang H, De la Cruz EM. Biophysics of actin filament severing by cofilin. FEBS Lett. 2013;587:1215–1219. doi: 10.1016/j.febslet.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of adf/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 27.van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, Desmarais V, Yip SC, Backer JM, Eddy RJ, Condeelis JS. Egf-induced pip2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magalhaes MA, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, Oser M, Chen X, Koleske AJ, Condeelis J. Cortactin phosphorylation regulates cell invasion through a ph-dependent pathway. J Cell Biol. 2011;195:903–920. doi: 10.1083/jcb.201103045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frantz C, Barreiro G, Dominguez L, Chen X, Eddy R, Condeelis J, Kelly MJ, Jacobson MP, Barber DL. Cofilin is a ph sensor for actin free barbed end formation: Role of phosphoinositide binding. J Cell Biol. 2008;183:865–879. doi: 10.1083/jcb.200804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller CB, De Bastiani MA, Becker M, Franca FS, Branco MA, Castro MA, Klamt F. Potential crosstalk between cofilin-1 and egfr pathways in cisplatin resistance of non-small-cell lung cancer. Oncotarget. 2015;6:3531–3539. doi: 10.18632/oncotarget.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker M, De Bastiani MA, Muller CB, Markoski MM, Castro MA, Klamt F. High cofilin-1 levels correlate with cisplatin resistance in lung adenocarcinomas. Tumour Biol. 2014;35:1233–1238. doi: 10.1007/s13277-013-1164-6. [DOI] [PubMed] [Google Scholar]
- 32.Muller CB, de Barros RL, Castro MA, Lopes FM, Meurer RT, Roehe A, Mazzini G, Ulbrich-Kulczynski JM, Dal-Pizzol F, Fernandes MC, Moreira JC, Xavier LL, Klamt F. Validation of cofilin-1 as a biomarker in non-small cell lung cancer: Application of quantitative method in a retrospective cohort. J Cancer Res Clin Oncol. 2011;137:1309–1316. doi: 10.1007/s00432-011-1001-5. [DOI] [PubMed] [Google Scholar]
- 33.Castro MA, Dal-Pizzol F, Zdanov S, Soares M, Muller CB, Lopes FM, Zanotto-Filho A, da Cruz M Fernandes, Moreira JC, Shacter E, Klamt F. Cfl1 expression levels as a prognostic and drug resistance marker in nonsmall cell lung cancer. Cancer. 2010;116:3645–3655. doi: 10.1002/cncr.25125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaheed SU, Rustogi N, Scally A, Wilson J, Thygesen H, Loizidou MA, Hadjisavvas A, Hanby A, Speirs V, Loadman P, Linforth R, Kyriacou K, Sutton CW. Identification of stage-specific breast markers using quantitative proteomics. J Proteome Res. 2013;12:5696–5708. doi: 10.1021/pr400662k. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Tong X. Expression of the actin-binding proteins indicates that cofilin and fascin are related to breast tumour size. J Int Med Res. 2010;38:1042–1048. doi: 10.1177/147323001003800331. [DOI] [PubMed] [Google Scholar]
- 36.Kabbage M, Chahed K, Hamrita B, Guillier CL, Trimeche M, Remadi S, Hoebeke J, Chouchane L. Protein alterations in infiltrating ductal carcinomas of the breast as detected by nonequilibrium ph gradient electrophoresis and mass spectrometry. J Biomed Biotechnol. 2008;2008:564127. doi: 10.1155/2008/564127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishimura S, Tsuda H, Kataoka F, Arao T, Nomura H, Chiyoda T, Susumu N, Nishio K, Aoki D. Overexpression of cofilin 1 can predict progression-free survival in patients with epithelial ovarian cancer receiving standard therapy. Hum Pathol. 2011;42:516–521. doi: 10.1016/j.humpath.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Lu LI, Fu NI, Luo XU, Li XY, Li XP. Overexpression of cofilin 1 in prostate cancer and the corresponding clinical implications. Oncol Lett. 2015;9:2757–2761. doi: 10.3892/ol.2015.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dowling P, Wormald R, Meleady P, Henry M, Curran A, Clynes M. Analysis of the saliva proteome from patients with head and neck squamous cell carcinoma reveals differences in abundance levels of proteins associated with tumour progression and metastasis. J Proteomics. 2008;71:168–175. doi: 10.1016/j.jprot.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Yang ZL, Miao X, Xiong L, Zou Q, Yuan Y, Li J, Liang L, Chen M, Chen S. Cfl1 and arp3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinomas of gallbladder. Cancer Invest. 2013;31:132–139. doi: 10.3109/07357907.2012.756113. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Kuramitsu Y, Ueno T, Suzuki N, Yoshino S, Iizuka N, Zhang X, Oka M, Nakamura K. Differential expression of up-regulated cofilin-1 and down-regulated cofilin-2 characteristic of pancreatic cancer tissues. Oncol Rep. 2011;26:1595–1599. doi: 10.3892/or.2011.1447. [DOI] [PubMed] [Google Scholar]
- 42.Polachini GM, Sobral LM, Mercante AM, Paes-Leme AF, Xavier FC, Henrique T, Guimaraes DM, Vidotto A, Fukuyama EE, Gois-Filho JF, Cury PM, Curioni OA, Michaluart P Jr., Silva AM, Wunsch-Filho V, Nunes FD, Leopoldino AM, Tajara EH. Proteomic approaches identify members of cofilin pathway involved in oral tumorigenesis. PLoS One. 2012;7:e50517. doi: 10.1371/journal.pone.0050517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filippova M, Filippov V, Williams VM, Zhang K, Kokoza A, Bashkirova S, Duerksen-Hughes P. Cellular levels of oxidative stress affect the response of cervical cancer cells to chemotherapeutic agents. Biomed Res Int. 2014;2014:574659. doi: 10.1155/2014/574659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M, Su BS, Chang LH, Gao Q, Chen KL, An P, Huang C, Yang J, Li ZF. Oxymatrine induces apoptosis in human cervical cancer cells through guanine nucleotide depletion. Anticancer Drugs. 2014;25:161–173. doi: 10.1097/CAD.0000000000000012. [DOI] [PubMed] [Google Scholar]
- 45.Singh M, Bhui K, Singh R, Shukla Y. Tea polyphenols enhance cisplatin chemosensitivity in cervical cancer cells via induction of apoptosis. Life Sci. 2013;93:7–16. doi: 10.1016/j.lfs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Perluigi M, Giorgi A, Blarzino C, De Marco F, Foppoli C, Di Domenico F, Butterfield DA, Schinina ME, Cini C, Coccia R. Proteomics analysis of protein expression and specific protein oxidation in human papillomavirus transformed keratinocytes upon UVB irradiation. J Cell Mol Med. 2009;13:1809–1822. doi: 10.1111/j.1582-4934.2008.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sorensen BS, Horsman MR, Vorum H, Honore B, Overgaard J, Alsner J. Proteins upregulated by mild and severe hypoxia in squamous cell carcinomas in vitro identified by proteomics. Radiother Oncol. 2009;92:443–449. doi: 10.1016/j.radonc.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 48.Evans W, Filippova M, Filippov V, Bashkirova S, Zhang G, Reeves ME, Duerksen-Hughes P. Overexpression of HPV16 E6* alters β-integrin and mitochondrial dysfunction pathways in cervical cancer cells. Cancer Genomics Proteomics. 2016;13:259–273. [PubMed] [Google Scholar]
- 49.Yakabe K, Murakami A, Kajimura T, Nishimoto Y, Sueoka K, Sato S, Nawata S, Sugino N. Functional significance of transgelin-2 in uterine cervical squamous cell carcinoma. J Obstet Gynaecol Res. 2016;42:566–572. doi: 10.1111/jog.12935. [DOI] [PubMed] [Google Scholar]
- 50.Song DG, Kim YS, Jung BC, Rhee KJ, Pan CH. Parkin induces up-regulation of 40s ribosomal protein sa and posttranslational modification of cytokeratins 8 and 18 in human cervical cancer cells. Appl Biochem Biotechnol. 2013;171:1630–1638. doi: 10.1007/s12010-013-0443-4. [DOI] [PubMed] [Google Scholar]
- 51.Lin LH, Chang SJ, Hu RY, Lin MW, Lin ST, Huang SH, Lyu PC, Chou HC, Lai ZY, Chuang YJ, Chan HL. Biomarker discovery for neuroendocrine cervical cancer. Electrophoresis. 2014;35:2039–2045. doi: 10.1002/elps.201400014. [DOI] [PubMed] [Google Scholar]