Abstract
Background/Aim: Medulloblastoma (MBL), an archetypal primitive neuroectodermal tumor of the cerebellum, is the most common pediatric central nervous system malignancy representing approximately 20% of all childhood brain tumors. Herein, we report on a new xenotransplantable tumor cell line, derived from a 6-year-old female patient with cerebellar medulloblastoma, and the completele proteome molecular characterization of subsequent tumors from MBL xenotrasplanted mice. Materials and Methods: Tumors were grown in nude mice as subcutaneous xenografts (MBLX) composed of small round cells with hyperchromatic nuclei and scant cytoplasm. Tumor specimen were extracted from animals upon their sacrifice and their molecular proteomic content was analyzed by 2-DE coupled to MALDI-TOF MS analysis. Results: Altogether 350 single-gene products were identified through the current approach, reported as the MBLX database. Conclusion: This new xenotransplantable tumor model, offers the scientific community valuable insight on the validity of xenografts altogether, while providing the means for a novel experimental model towards the study of human MBL.
Keywords: Pediatric medulloblastoma, pediatric brain tumors, central nervous system tumors, xenografts, xenotransplantable tumors
Tumors of the central nervous system (CNS), are amongst the most lethal types of neoplasms and have traditionally been the centerfold of scientific and medical interest, especially when affecting the younger population, where disease consequences are devastating (1). Proteomics having unarguably been established as one of the most significant technologies of modern science, are used in research efforts targeting cancer mechanisms (onco-proteomics). Essentially, proteomics during the last two decades have been important in a re-definition of the standards for CNS onco-biology and pediatric cancer research in particular (2).
Medulloblastoma (MBL) an archetypal primitive neuroectodermal tumor of the cerebellum, is the most common pediatric CNS malignancy representing approximately 20% of all childhood brain tumors (3). Despite aggressive multi-modal therapy with surgery, radiation and chemotherapy, 5-year survival rates rarely approach >60% (4). Treatment causes long-term morbidity including endocrine and growth disturbances, as well as neurocognitive dysfunction that are particularly severe in younger children. Prognosis and treatment still remain dismal for infants <3 years due to the tendency of disease to disseminate thoughout the CNS at its’ early stages and due to delay/avoidance of craniospinal radiation therapy for reduction of severe effects on the intellect (5). Consequently, efforts are being made to stratify patients according to their individual risk of tumor relapse or progression.
Mice are among the most extensively-used animal models for studying human disease, in particular cancer, since they represent a controllable experimental and manageable living system, closely-mimicking disease conditions of the human organism. Xenografts, are effective testing models, when it comes to analysis of tumor characteristics, while offering great possibilities for drug testing as well as testing of therapy protocols. These positive attributes are further complemented by the fact that experiments involving xenografts offer ease of laboratory handling and direct sample acquisition amidst experimental sessions.
Our group has previously reported on the establishment and proteomic characterization of two novel clear cell sarcoma (CCS) cell lines (soft-tissue melanoma) from a living patient and their transplantation in mice leading to corresponding xenografts (6). Furthermore, we recently reported on the complete characterization of the proteome of primary pediatric medulloblastoma (MBL) tumors (7).
In the present study, in combination with the two aforementioned experimental embankments, we report on a new xenotransplantable tumor cell line, derived from a 6-year-old female patient with cerebellar medulloblastoma. Tumors were grown in nude mice as serially-transplantable subcutaneous xenografts composed of small round cells with hyperchromatic nuclei and scant cytoplasm. This xenotransplanted tumor (MBLX), is considered to provide an adequate experimental model for the study of human medulloblastoma.
Since only a limited number of studies focusing on the potential power of xenograft models to investigate tumor-associated protein expression exist, the current proteomics approach probes on the signaling pathways and the molecular/functional signatures across the proteome of medulloblastoma-xenotransplanted tumor tissues aiming to further elucidate the human medulloblastoma character.
Materials and Methods
Primary tumor and cell culture. A sample from a tumor specimen was obtained from a 6-year-old patient diagnosed with MBL who underwent resection at the Aghia Sofia Children’s’ Hospital, Athens, Greece. The patient was neither submitted to pre-operative radiotherapy nor neoadjuvant chemotherapy. The excised tumor following hematoxylin and eosin (H&E) staining was examined by light microscopy.
Immediately after excision, the tumor tissue was mechanically-disaggregated and yielded a single-cell suspension which was plated in 100-mm dishes and suspension flasks, and incubated at 37˚C in 5% CO2/air atmosphere. Non-viable cells were removed by density-gradient centrifugation through Ficoll-Hypaque (LSM, Organon Teknika Co., Durham, NC, USA). Tumor cells were immediately processed following surgery completion, with no freeze-thaw cycle interventions. Our Institution’s Ethics committee approved of the current study’s protocol.
Tumor-disseminated cells were grown in RPMI 1640 medium (Invitrogen Ltd., London, UK) supplemented with heat-inactivated FCS (Invitrogen), 2 mM L-glutamine and a mixture of streptomycin-penicillin. After 5 days, a large number of small cells with spindle-to-dendritic shapes were observed to be growing. These cells increased rapidly in number, and were transferred to larger culture vessels within 2 weeks after culture initiation.
Animals. All experiments were performed following the directions of the National and European Convention for the Protection of Animals and the European Directive (2010/63/EU). Male nude mice (n=5), aged 6-8 weeks, (NOD.CB17-Prkdcscid/J were obtained from Jackson labs (Bar Harbor, ME, USA) and maintained under pathogen-free conditions in the animal facility of our Institution until use. The animals were maintained in sterile conditions throughout the experiments (temperature 24±2˚C, relative humidity 55±5% and a 12-h photoperiod) in polycarbonate cages. They were fed common rodent feed and water ad libitum.
Xenografts. Viable cells (4×106) (obtained in previous steps of the experimental procedure) to a total volume of 0.25 ml were s.c. injected into left and right flanks of ether-anaesthetized 5-week-old female nu/nu mice. Tumor sizes were measured weekly and volume proportions were performed following the formula: width2 × length ×0.52. At 8 to 9 weeks, mice were sacrificed and tumors were excised. For xenograft establishment the following were performed: when tumors reached a size of approximately 1.5 cm3, they were excised from animals, cut into 1-2 mm3 fragments and transplanted to another group of mice (P1 and P2) using the method described above. At the time of transplantation and tumor removal animals were anesthetized under isoflurane.
Histological analysis. Tissue from the tumors developed in mice were fixed in 10% phosphate buffered formalin, embedded in paraffin, serially-sectioned at 3-μm and then stained with H&E for histological examination. With regard to tumor analysis, histological type, degree of differentiation, nuclear atypia and extension of invasion, were examined.
Xenograft serum collection. Prior to transplantation serum was collected by retro-orbital punction from control animals. Blood was also collected form tumor-bearing animals upon sacrifice. Blood samples were centrifuged for 15 min at 1,400 × g and plasma was stored at −80˚C, until 2-DE analysis.
Sample preparation for proteomics. Tissues were powderized through grinding in liquid nitrogen. Further homogenization was performed in a glass Wheaton (tight) homogenizer in 8 M urea, 40 mM Tris-HCL (pH 8.5), 2 M thiourea, 4% CHAPS, 1% dithioerythritol (DTE), 0.2% IPG buffer pH 3-10 (Bio-Rad, Hercules, CA, USA). The homogenate was left at room temperature for 1 h and centrifuged at 13,000 × g for 30 min. De-salting was performed with Ultrafree-4 centrifugal filter unit. The protein content of the supernatant was determined using the Bradford quantification method. A similar procedure was followed for preparation of sera obtained from xenotrasplanted mice.
2-Dimentional protein separation and mass spectrometric (MS) identification. Two-dimensional gel electrophoresis was performed as previously reported (8) Samples of 1 mg total protein were applied on 18 cm, PI 3-10 NL, IPG strips (Bio-Rad Lab). IPG strips had been previously prepared for IEF by 20 h rehydration in a buffer of 8 M urea, 4% CHAPS and 1% DTE. After IEF (180,000 Vh) IPG strips were equilibrated in 0.5% (w/v) DTE and then 4% (w/v) iodoacetamide. SDS-PAGE was performed on 12% acrylamide gels with application of 40 mA/gel.
With regard to mass spectrometric (MS) analysis of protein spots, peptide mass fingerprinting was performed as previously described (9). Briefly, all spots on all gels were excised and placed into 96-well microtiter plates. Excised spots treated with 5μl of 20 μg/ml recombinant trypsin (Roche diagnostics, Basel, Swiss) solution. Petide extraction was performed after 16 h by application of 10 μl of 50% acetonitrile containing 0.3% trifluroacetic acid. Sample application to a target plate and MALDI-ToF/MS analysis, as well as peptide matching and protein searching were carried out as described previously (10). Tryptic peptide mixtures (1 μl) were applied on an anchor chip MALDI plate with 1 μL of matrix solution (Sigma), and the internal standard peptides des-Arg-bradykinin (Sigma, 904.4681 Da) and adrenocorticotropic hormone fragment 18-39 (Sigma, 2465.1989 Da). Peptide mixtures were analyzed in a MALDI-ToF mass spectrometer (Ultraflex II, Bruker Daltonics, Bremen, Germany). Laser shots (n=400) of intensity between 40% and 60% were collected and summarized and the peak list was created using the FlexAnalysis v2.2 software (Bruker). Tryptic autodigest as well as commonly-occurring keratin contaminant peaks were filtered-out prior to protein identification process. Peptide matching and protein searches were performed with MASCOT Server 2 (Matrix Science). Peptide masses were compared to the theoretical peptide masses of all available proteins from Homo sapiens in the SWISS-PROT database. Stringent criteria were used for protein identification with a maximum allowed mass error of 10 ppm and a minimum of four matching peptides. A probability score with p<0.05 was used as the criterion for affirmative protein identification.
Functional clustering and classification. All identified MBL xenograft proteins were assigned their gene symbol via the Uniprot Knowledgedbase database (http://www.uniprot.org/). Functional relationships of the identified proteins were determined by submitting protein entry gene symbols to the STRING database (string-db.org). The simplified version of the produced network, involving the most important proteins with regard to tumorigenesis, was adopted.
Results
Patient tumors transplanted to mice create stable MBLX models. A resected primary pediatric MBL with sufficient excess tumor tissue was first disseminated and the obtained extracted cells were implanted into NOD-SCID mice (n=5) and patient-derived tumor xenografts generated stable growth, that could be serially passaged. Models that failed initial engraftment due to absence of tumor cells in the tissue fragment used to implant were excluded from our analysis. All generated MBLX were derived from a single patient who was treatment-free.
Characterisation of MBL xenogratft tumors. To determine protein expression patterns of established xenografts, tumor tissue resections were subjected to 2-DE analysis. To strengthen the quality and reproducibility of the expression profile, the experimental analytical approach consisted of two technical replicates for each examined sample.
The number of protein spots detected in each tissue sample varied, with a mean of 1,250±120 spots per 2-DE gel. More than 5,500 spots were excised from gels and analyzed by MS. The identification process resulted to a list of 3,450 protein entries. Only proteins identified/present in the entity of all samples analyzed were included in the final xanograft medulloblastoma protein/database.
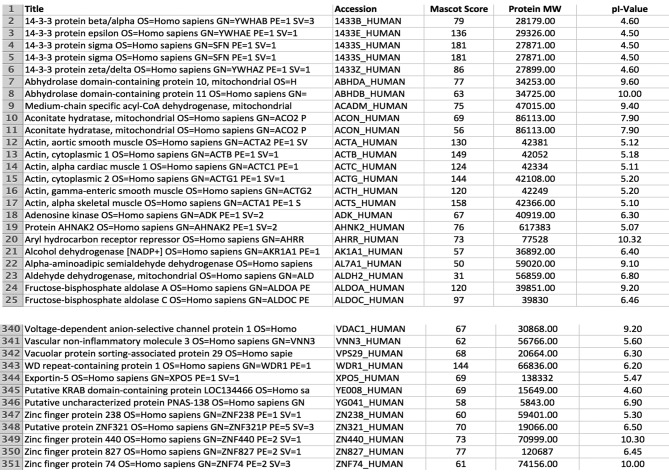
MBLX proteins are mostly present in the acidic region (pI 4-7), although theoretical pI values predict protein accumulation in pI 8-10 as well. Key elements of these entries are summarized in Figure 1. This Figure denotes proteins with their accession number, average Mascot score, average number of matched peptides, MW, theoretical pI and, mean number of spots per gel used for identification. Altogether 350 different gene products (proteins) were expressed in all examined xenograft samples.
Figure 1. Final protein database of a xenotransplanted pediatric medulloblastoma tumor (MBLX). Only proteins identified/present in all samples (n=10) were included in the final reference MBLX, consisting of 350 entries. An Excel file form (an exemplary abstract is shown for reasons of convenience and comprehension), is presented including identified protein accession numbers (according to UniProt), protein name and description, MASCOT score used for identification, molecular weight (MW) and isoelectric points (pI).
A characteristic master gel image revealing the proteome pattern of newly-established MBL xenografts by means of 2-DE, consisting of annotations for the majority of 350 identified gene products, is presented in Figure 2.
Figure 2. Two-dimensional reference map of xenotransplanted pediatric medulloblastoma tumors. Extracted proteins from the tumors were separated on 3-10 non-linear IPG strips, followed by 12% SDS-polyacrylamide gel electrophoresis, as descried in Materials and Methods. 2-DE gels were stained with Coomassie blue and protein spots were excised and further analyzed by MALDI-TOF MS. The proteins that were found present in all samples analyzed are designated with their accession numbers.
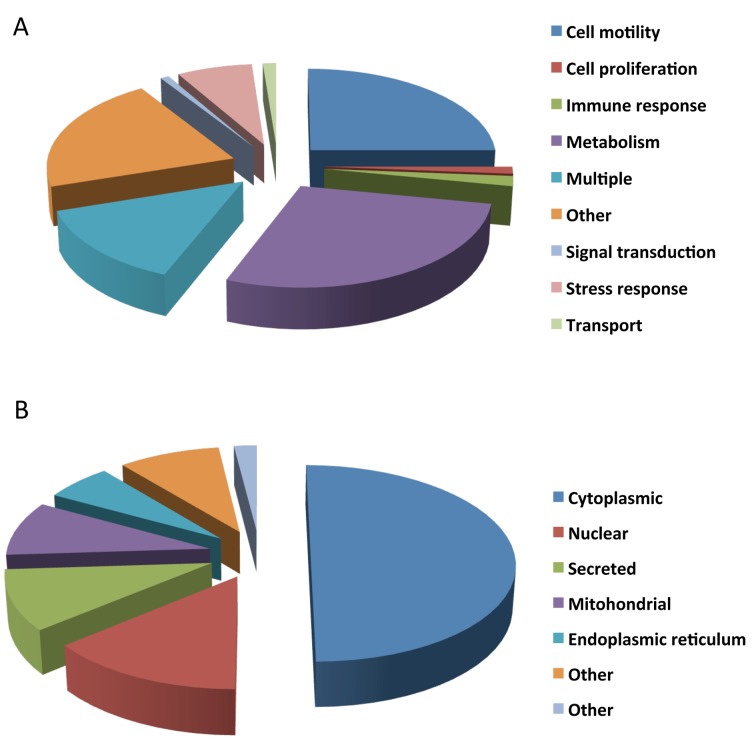
Protein classification. Existing gene ontology information was used to constellate proteins expressed in the MBL xenografts (Figure 1) according to their cellular function and localization (Figure 3A and B).
Figure 3. Classification of xenotransplanted pediatric medulloblastoma tumor specimen according to their biological process (A) and subcellular localization (B). The distribution frequencies with regard to the specified categories within the given charts are indicated in % of the total number of protein entries.
As observed, MBLX proteins are involved in several biological processes that facilitate tumor-relevant adjustments regarding growth, invasion, apoptosis as well as inflammation. The majority of identified proteins (24%) are involved in cell motility, while a significant number (25%) is engaged in cell metabolism, 14% are involved in transport-molecule procedures (carrier molecules), 12% have roles in signal transduction (2%), stress response (6%), or exert multiple functions (19%). Proteins with known, but not further categorized ontology information (15%) are summarized under “other” (Figure 3A).
Regarding their cellular distribution, the majority of proteins are localized in the cytoplasm (50%), a smaller number (20%) are nuclear, 17% of proteins are secreted in the extracellular environment and 8.5% of the MBLX proteins are mitochondrial.
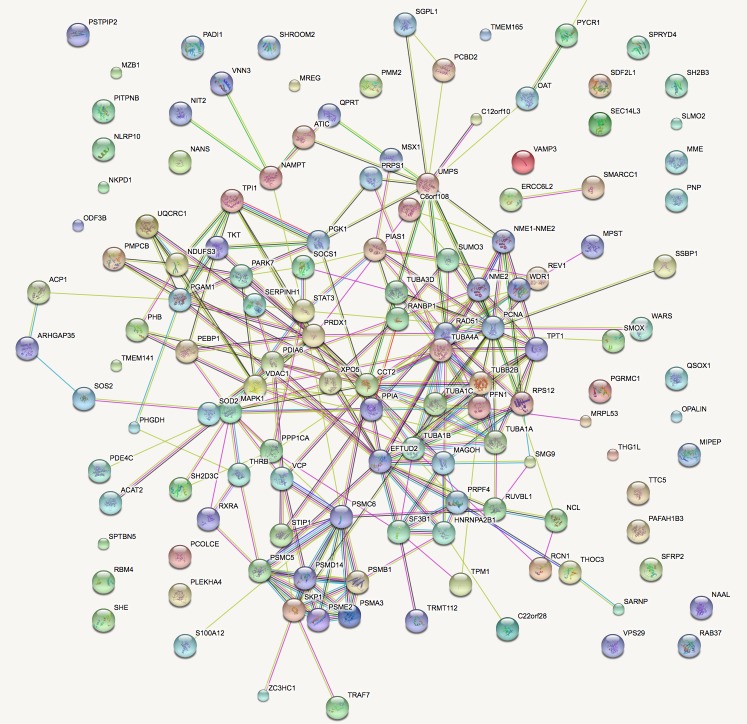
Sub-network analysis. To further interpret our findings from a biological point-of-view, all protein entries implicated in procedures correlated to tumorigenesis were analyzed by the STRING database. Functional analysis identified relationships and biological functions most significant to the dataset of identified proteins in tumor-bearing animals, that are in tight correlation with the MBL phenotype. The rank product list, representing proteins present in MBL-bearing mice is presented in its “extended view” form (Figure 4).
Figure 4. Interaction networks and enriched functional annotations of proteins whose functional analysis identified relationships and biological functions most significant to the cancer development. Thicker network lines demonstrate strong protein relation as well as neighboring positions.
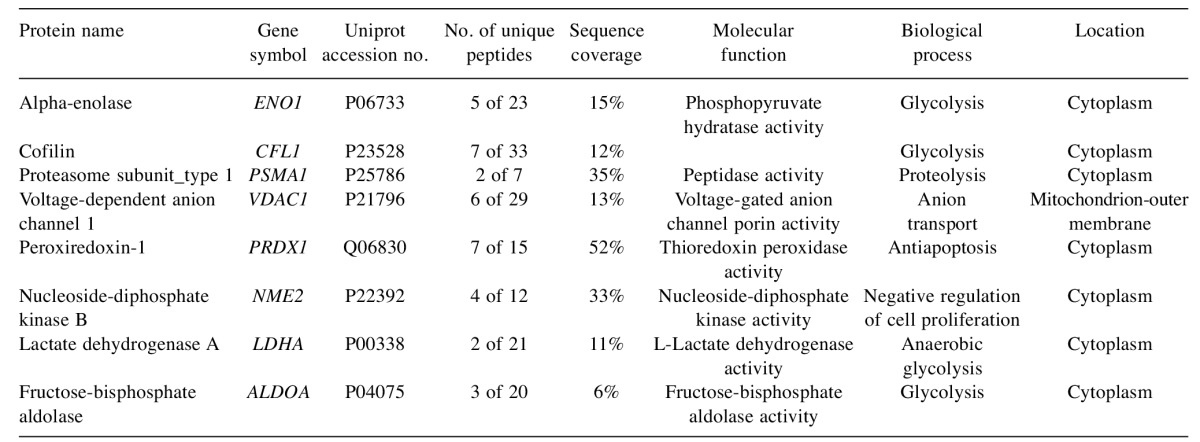
Identification of tumor-derived proteins present in xenografted mice. In order to determine which of the identified proteins were in fact “tumor-derived” in xenotransplanted mice, a selection procedure was followed. Serum was obtained from mice both before (control mice) and after transplantation (xenografted mice), samples were pooled and proteins were separated by 2-DE (see Materials and Methods section). Following tryptic digestion, peptides were subsequently analyzed by MALDI-TOF MS. Data analysis followed, and identified proteins were divided in two groups: human-specific proteins (identified only in the Human Uniprot database) and homologous proteins (present in both the Human and Mouse Uniprot databases). Further comparison between databases was undertaken and homologous proteins were finally annotated as tumor-derived only in case where their expression was higher in sera of xenotransplated mice by at least 4-fold. Annotations were derived from the Gene Ontology database. Overall eight proteins were identified as tumor-derived (Table I). Of those, 7 were annotated as cytoplasmic proteins by the Gene Ontology database.
Table I. Proteins annotated as tumor-derived in xenograft-bearing mice.
Identified proteins were divided in two groups: human-specific proteins (identified only in the Human Uniprot database) and homologous proteins (present in both the Human and Mouse Uniprot databases). Further comparison between databases was undertaken and homologous proteins were finally annotated as tumor-derived only in case where their expression was higher in sera of xenotransplated mice by at least 4-fold.
Discussion
In cancer research, established cell lines have been proven to be potent tools towards understanding of biological and molecular characteristics of malignancies and are widely used in terms of making more accurate diagnoses and developing therapeutic strategies. Even more valuable tools towards this target are xenografts representing models of this disease that are more relevant to the clinical situation.
In the present study, it is safe to say, that we observed a somewhat different protein profile compared to human pediatric primary MBL tumor resection, as was recorded in our previous study (7). The present study aimed at elucidating the molecular character of MBL-xenotransplanted tumors, as well as their ability to grow as xenografts in mice. While having achieved a successful xenotransplantation, the data of MBLX tumors did not indicate as many similarities as one would expect with original primary MBL tumors. However, the illustrated fidelity and diversity is sufficient enough in offering an advantage of MBLX models as preclinical models, over cell lines or other in vivo tumor models, including xenografts formed from established cell lines and genetically- engineered mouse models. While genetically-engineered mouse models provide the advantage of studying the biology of a single or combination of genetic abnormalities, their utility is limited to the models available that represent few specific genetic abnormalities related to histological types of cancer (11,12).
Although to our knowledge no such studies have been previously performed in the field of pediatric oncology, the low recapitulation of the proteomic landscape when MBLX was compared to primary MBL tumors is inconsistent with omics studies comparing xenografts of breast and colon cancers to their corresponding patient tumors (13,14). Our data, are in line with data derived from cell lines, that in previous comparisons to patient tumors have shown low correlation rates between their point-mutation frequencies, and marked differences in their mRNA and copy number profiles (15,16).
One can acknowledge the fact that in MBLX tumors, key elements of the PI3K-mTOR pathway, in contrast to our previous study, were not detected. Whereas it is widely-accepted that the IGF system plays an important role in neuronal development (17) and most MBLs overexpress the IGF-1 receptor (IGF-1R), and more than half of MBLs express the activated phosphorylated form of IGF-1R (18,19); no expression of the above molecules was apparent in our MBLX examined tissues. Neither phosphoinositide 3-kinase catalytic subunit type-3 (PIK3C3, Q8NEB9), a catalytic subunit of class III PI3Ks which forms a heterodimer with p150, a regulatory subunit of class III PI3Ks; nor RAF proto-oncogene serine/threonine-protein kinase (Raf-1, P04049) a member of the serine/threonime (AKT) complex with critical regulatory link functions, determining cell fate decisions e.g. proliferation, differentiation, apoptosis, survival and oncogenic transformation was present. Since Raf-1 activation initiates the mitogen-activated protein kinase (MAPK) cascade that comprises a sequential phosphorylation of the dual-specific MAPK kinases seen downstream as part of the mTOR cascade; different pathways must be activated during the tumorigenesis process in MBLX-bearing animals, distinct form those present in human.
Overall while the total number of identified proteins was not different between the two groups (350 total molecules in MBLX tumors vs. 382 in MBL primary tumors), we must emphasize that this is but an initial study, emphasizing on the capability of producing such MBL xenografts and whilst offering the molecular background/proteomic signature of MBLX tumors. However, one cannot deny the fact that the small sample size of tumors examined herein discretely elongate MBLX tumors form their actual human counterparts whose condition should “theoretically” closely mimic.
On the other hand, several of the detected proteins have been reported to be present in MBL pathophysiology. A series of detected molecules in the MBLX profile contribute to negative regulation of apoptosis: calreticulin (CALR), keratin, type-II cytoskeletal-1 (K22E) keratin, type-I cytoskeletal 18 (KRT18), heat shock 70-kDa protein-1A (HSPA1A), HSP 90-alpha (HS90A), peroxiredoxin-1 (PRDX1). While we observed a notable absence of key tumor elements such as drebrin (Q16643) (a de-stabilizing neuron actin filament associate molecule), programmed cell death 6-interacting protein (PDC6I) (a key regulator of tumor growth and angiogenesis), protein lyric or AEG1 (Q86UE4) (an activator of NF-kB and promoter of anchorage-independent growth of immortalized melanocytes and astrocytes) and secreted protein acidic and rich in cysteine (SPARC) (P09486) (an inducer of neuronal differentiation as a cell defence mechanism against tumor transformation) (20).
Key elements attributing to differentiation of the MBLX profile form that of MBL primary tumors may be associated with the non-orthotopic nature of the xenografts as well as the methodological approach used for protein characterization of MBLX samples. Being a method bearing many drawbacks, 2-DE and subsequent MALDI-TOF MS analysis, limits results to the identification of the most abundant proteins as well as those present in the acidic region of pI. Taking this in consideration future studies are in progress, in which more in-depth analysis of the protein profile of xenotransplanted tumors will be carried out by technologies of far greater penetrating capabilities.
Overall, in the present study, we report on the successful xenotransplantation of a primary MBL tumor and its initial molecular proteomics characterization by means of 2-DE coupled to MALDI-TOF MS analysis and bioinformatics clustering of obtained results. Having observed a distinct detachment of protein components of the MBLX tissues from those of primary MBL tumors many questions remain unanswered regarding the nature of mouse xenografts as well as their utility in anticancer research.
Acknowledgements
The Authors would like to thank the patient and her family without whom advances in pediatric brain tumor research would not be possible.
References
- 1.Izbicka E, Campos D, Marty J, Carrizales G, Mangold G, Tolcher A. Molecular determinants of differential sensitivity to docetaxel and paclitaxel in human pediatric cancer models. Anticancer Res. 2006;26:1983–1988. [PubMed] [Google Scholar]
- 2.Anagnostopoulos AK, Tsangaris GT. The proteomics of pediatric brain tumors. Expert Rev Proteomics. 2014;11:641–648. doi: 10.1586/14789450.2014.939633. [DOI] [PubMed] [Google Scholar]
- 3.Packer RJ, Cogen P, Vezina G, Rorke LB. Medulloblastoma: clinical and biologic aspects. Neuro Oncol. 1999;3:232–250. doi: 10.1215/15228517-1-3-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Packer RJ. Chemotherapy for medulloblastoma/primitive neuroectodermal tumors of the posterior fossa. Ann Neurol. 1990;6:823–828. doi: 10.1002/ana.410280615. [DOI] [PubMed] [Google Scholar]
- 5.Ashley DM, Merchant TE, Strother D, Zhou T, Duffner P, Burger PC, Miller DC, Lyon N, Bonner MJ, Msall M, Buxton A, Geyer R, Kun LE, Coleman L, Pollack IF. Induction Chemotherapy and conformal radiation therapy for very young children with nonmetastatic medulloblastoma: Children’s oncology group study P9934. J Clin Oncol. 2012;30:3181–3186. doi: 10.1200/JCO.2010.34.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimas K, Tsimplouli C, Anagnostopoulos AK, Mahaira L, Iliopoulou E, Perez S, Vougas K, Tsangaris GT. The proteome profile of two cell lines and their xenografts isolated from a patient with clear cell sarcoma (soft tissue melanoma) Cancer Genomics Proteomics. 2008;5:175–237. [PubMed] [Google Scholar]
- 7.Anagnostopoulos AK, Papathanassiou C, Karamolegou K, Anastasiadou E, Dimas KS, Kontos H, Koutsopoulos A, Prodromou N, Tzortzatou-Stathopoulou F, Tsangaris GT. Proteomic studies of pediatric medulloblastoma tumors with 17p deletion. J Proteome Res. 2015;14:1076–1088. doi: 10.1021/pr501219f. [DOI] [PubMed] [Google Scholar]
- 8.Anagnostopoulos AK, Dimas KS, Papathanassiou C, Braoudaki M, Anastasiadou E, Vougas K, Karamolegou K, Kontos H, Prodromou N, Tzortzatou-Stathopoulou F, Tsangaris GT. Proteomics studies of childhood pilocytic astrocytoma. J Proteome Res. 2011;10:2555–2565. doi: 10.1021/pr200024m. [DOI] [PubMed] [Google Scholar]
- 9.Anagnostopoulos AK, Kolialexi A, Mavrou A, Vougas K, Papantoniou N, Antsaklis A, Kanavakis E, Fountoulakis M, Tsangaris GT. Proteomic analysis of amniotic fluid in pregnancies with Klinefelter syndrome foetuses. J Proteomics. 2010;73:943–950. doi: 10.1016/j.jprot.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Anagnostopoulos AK, Vougas K, Kolialexi A, Mavrou A, Foundoulakis M, Tsangaris GT. The protein profile of the human immature T-cell line CCRF-CEM. Cancer Genomics Proteomics. 2005;2:271–300. [PubMed] [Google Scholar]
- 11.Rodriguez E, Mannion L, D'Santos P, Griffiths M, Arends MJ, Brindle KM, Lyons SK. Versatile and enhanced tumour modelling in mice via somatic cell transduction. J Pathol. 2014;232:449–457. doi: 10.1002/path.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trejo CL, Juan J, Vicent S, Sweet-Cordero A, McMahon M. MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res. 2012;72:3048–3059. doi: 10.1158/0008-5472.CAN-11-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D awrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansén S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Jänne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Pham NA, Tong J, Sakashita S, Allo G, Kim L, Yanagawa N, Raghavan V, Wei Y, To C, Trinh QM, Starmans MH, Chan-Seng-Yue MA, Chadwick D, Li L, Zhu CQ, Liu N, Li M, Lee S, Ignatchenko V, Strumpf D, Taylor P, Moghal N, Liu G, Boutros PC, Kislinger T, Pintilie M, Jurisica I, Shepherd FA, McPherson JD, Muthuswamy L, Moran MF, Tsao MS. Molecular heterogeneity of non-small cell lung carcinoma patient-derived xenografts closely reflect their primary tumors. Int J Cancer. 2017;140:662–673. doi: 10.1002/ijc.30472. [DOI] [PubMed] [Google Scholar]
- 15.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nature Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modeling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Yosef T, Eden A, Benvenisty N. Characterization of murine BCAT genes: Bcat1, a c-Myc target, and its homolog, Bcat2. Mamm. Genome. 1998;7:595–597. doi: 10.1007/s003359900825. [DOI] [PubMed] [Google Scholar]
- 18.Hutson SM, Berkich D, Drown P, Xu B, Aschner M, LaNoue KF. Role of branched-chain aminotransferase isoenzymes and gabapentin in neurotransmitter metabolism. J Neurochem. 1998;2:863–874. doi: 10.1046/j.1471-4159.1998.71020863.x. [DOI] [PubMed] [Google Scholar]
- 19.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;5:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 20.Tsangaris GT, Papathanasiou C, Adamopoulos PG, Scorilas A, Vorgias CE, Prodromou N, Stathopoulou FT, Stravopodis DJ, Anagnostopoulos AK. Pediatric Ependymoma: A Proteomics Perspective. Cancer Genomics Proteomics. 2017;14:127–136. doi: 10.21873/cgp.20025. [DOI] [PMC free article] [PubMed] [Google Scholar]