Abstract
Background/Aim: T-cadherin is a tumor suppressor gene, its predictive value in colorectal cancer (CRC) still remains controversial. In this study, we aimed to evaluate the association between T-cadherin promoter methylation and CRC by performing a meta-analysis. Materials and Methods: The relevant literature was searched using the PubMed, Cochrane Library, Web of Science and Google Scholar databases for articles published until December 2016. The effect sizes were estimated by measuring an odds ratio (OR) with a 95% confidence interval (CI). Sensitivity analysis was performed to examine the heterogeneity and funnel plots were constructed to evaluate publication bias. Results: Nine studies, including 488 samples were included in this meta-analysis. The pooled OR of T-cadherin promoter methylation in cancer tissues was 16.73 (95%CI=6.24-44.87), 19.48 (95%CI=5.64-67.31) and 2.23 (95%CI=1.05-4.75) compared to normal tissues, adjacent tissues and premalignant tissues, respectively. The relationship between T-cadherin promoter methylation and clinicopathological features were also analyzed. However, a significant association was not observed between T-cadherin promoter methylation status and gender, tumor stage, and lymph node status (p>0.05). Conclusion: The methylation status of T-cadherin promoter was strongly associated with CRC risk. However, T-cadherin promoter methylation may have a limited prognostic value for CRC patients.
Keywords: T-cadherin, CRC, methylation, premalignant lesions, progression
At present, colorectal cancer (CRC) is among the most common malignant diseases in the western world. Many Asian countries have experienced a two- to four-fold increase in the frequency of CRC during the past few decades (1,2). Despite the recent and main improvements in diagnostic and therapeutic opportunities, overall survival is poor for CRC.
DNA methylation is one of the key epigenetic modifications in eukaryotes, regulating genes microRNAs expression (3), and gene alternative splicing (4). In addition, epigenetic alterations can change gene expression levels including direct hyper-methylation that influence gene expression. Furthermore, abnormal DNA methylation plays a role in the development of colorectal cancer (5). T-cadherin, also known as cadherin 13 (CDH13), is a tumor-suppressor gene (TSG), its loss in tumor cells is associated with tumor malignancy, invasiveness and metastasis. The mechanism for T-cadherin suppression is associated with hypermethylation of the T-cadherin gene promoter region. T-cadherin promoter hypermethylation has been detected in breast (6) and lung cancer (7), in pituitary adenoma (8), diffuse large B cell lymphoma (9), and nasopharyngeal carcinoma (10). Moreover, T-cadherin gene has been suggested as a promising early detecting marker for CRC. Herein, we conducted a meta-analysis to evaluate the diagnostic ability of the T-cadherin promoter methylation test in CRC.
Materials and Methods
Literature search strategy. A systemic literature search for studies published prior to November, 2016 was conducted in the PubMed, Cochrane Library, Web of Science and Google Scholar databases without any language restrictions. The following keywords and search terms were used: (CDH13 OR cadherin 13 OR H-cadherin OR T-cadherin) and (colorectal cancer OR colorectal tumor OR colorectal carcinoma OR colorectal neoplasm) and (‘methylation or hypermethylation or epigenetic). The retrieved studies were carefully examined to exclude potential duplicates or overlapping data. Titles and abstracts of articles selected from the initial search were first scanned, and then full papers of potential eligible studies were reviewed.
Selection of studies. Eligibility of studies for inclusion was independently assessed by two investigators. Studies were eligible for inclusion if all the following criteria were fulfilled: (i) The studies were case–control or cohort studies; (ii) diagnosis of CRC was proven by histopathological analysis; (iii) the methylation frequency of the T-cadherin gene was sufficient for the case-control or cohort studies; (iv) the data provided should have been sufficient to estimate T-cadherin promoter methylation in cancer versus premalignant, adjacent, or normal tissues; (v) the studies showed an association between T-cadherin promoter methylation and clinicopathological parameters, including gender, tumor differentiation, tumor stage, and lymph node status. If the data sets overlapped or were duplicated, we only extracted the most detailed or recent information.
Data extraction. Data were extracted by two of the authors independently using the same standardized form. The fields extracted included first author, year of publication, the original country of patients, ethnicity, sample size, the number of T-cadherin promoter methylations in individual cases and controls in individuals, detection method of methylation, and clinicopathological Parameters. For the articles with the same population resources or overlapping data sets, the paper that included the largest population or contained more useful information was included.
Statistical analysis. All statistical tests were two-sided, and all statistical analyses were carried out with SPSS 16.0 and Stata Statistical Software 13.0. A random-effects model was used to estimate pooled ORs in order to take into account the heterogeneity of the risk estimates and to provide more conservative estimates compared with the fixed effects model. Statistical heterogeneity between studies was assessed with the chi-square statistic and quantified by I2, a statistic that represents the percentage of total variation contributed by between-study variation. A significant heterogeneity was defined as a p-value <0.10. To investigate potential sources of between studies heterogeneity, subgroup analyses was conducted. Bias was assessed using the tests by Egger, and Begg, and the contour enhanced funnel plots.
Sensitivity analysis. Sensitivity analysis is the study of how the uncertainty in the output of a mathematical model or system can be apportioned to different sources of uncertainty in its inputs. Then sensitivity analyses were carried out to assess whether the summary estimates are robust to inclusion of studies by omitting one study from the pooled analysis each time.
Results
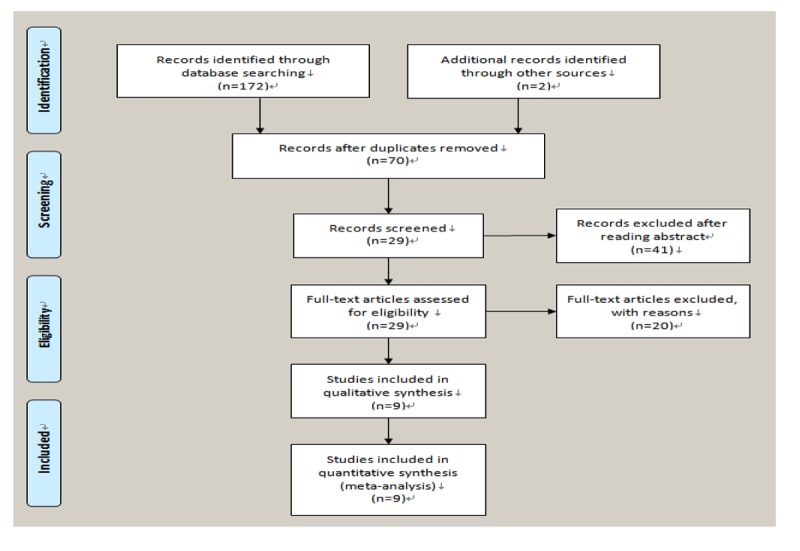
Study selection and characteristics. The characteristics of eligible studies are summarized in Table I. A total of 9 studies (11-19), including 488 cases with CRC, 48 premalignant tissues, 298 adjacent tissues, and 144 normal, met the inclusion criteria for this meta-analysis (Figure 1). The total number of patients was 998. All studies were based on data of retrospective analyses. Among the studies, 4 focused on Asian subjects, and 5 examined Caucasians subjects. Six studies reported the association between T-cadherin promoter methylation and CRC in CRC versus normal tissues; 5 studies in CRC versus adjacent tissues; 3 studies in CRC versus premalignant lesions; and, 4 studies assessed the association of T-cadherin promoter methylation with clinicopathological features.
Table I. Characteristics of the included studies.
M, Methylation; MS-MLPA, methylation-specific multiplex ligation-dependent probe assay; MSP, methylation-specific polymerase chain reaction; N, the number of samples.
Figure 1. Flow chart showing the process articles inclusion.
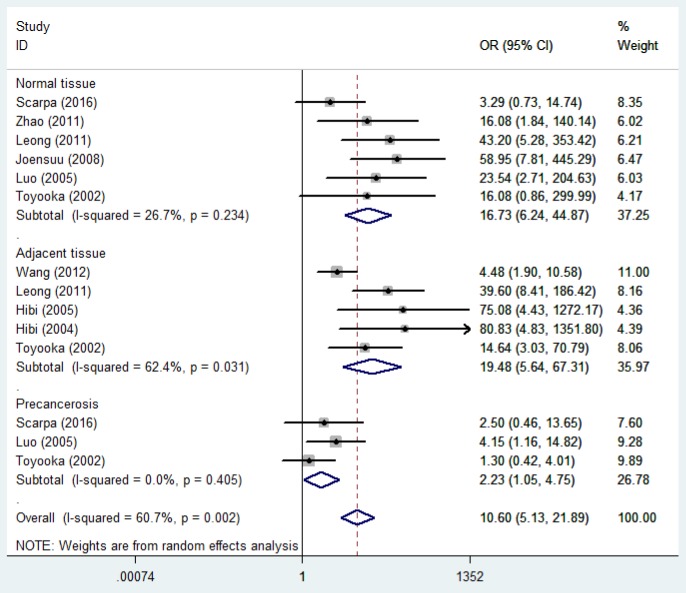
T-cadherin promoter methylation and risk of CRC. The heterogeneity among studies was significant (p<0.05 and I2=60.7%), and therefore, the random-effects model was used. Our findings demonstrated that the frequency of T-cadherin promoter methylation in cancer was significantly higher in cancer tissues compared to normal, adjacent, and premalignant tissues (cancer tissues vs. normal tissues: OR=16.73, 95%CI=6.24-44.87; cancer tissues vs. adjacent tissues: OR=19.48, 95% CI=5.65-67.31; cancer tissues vs. premalignant tissues: OR=2.23, 95% CI=1.05-4.75), which suggested a statistically significant increase in the likelihood of T-cadherin promoter methylation in CRC compared to the controls (Figure 2).
Figure 2. Forest plot of the association between T-cadherin promoter methylation and CRC among different groups.
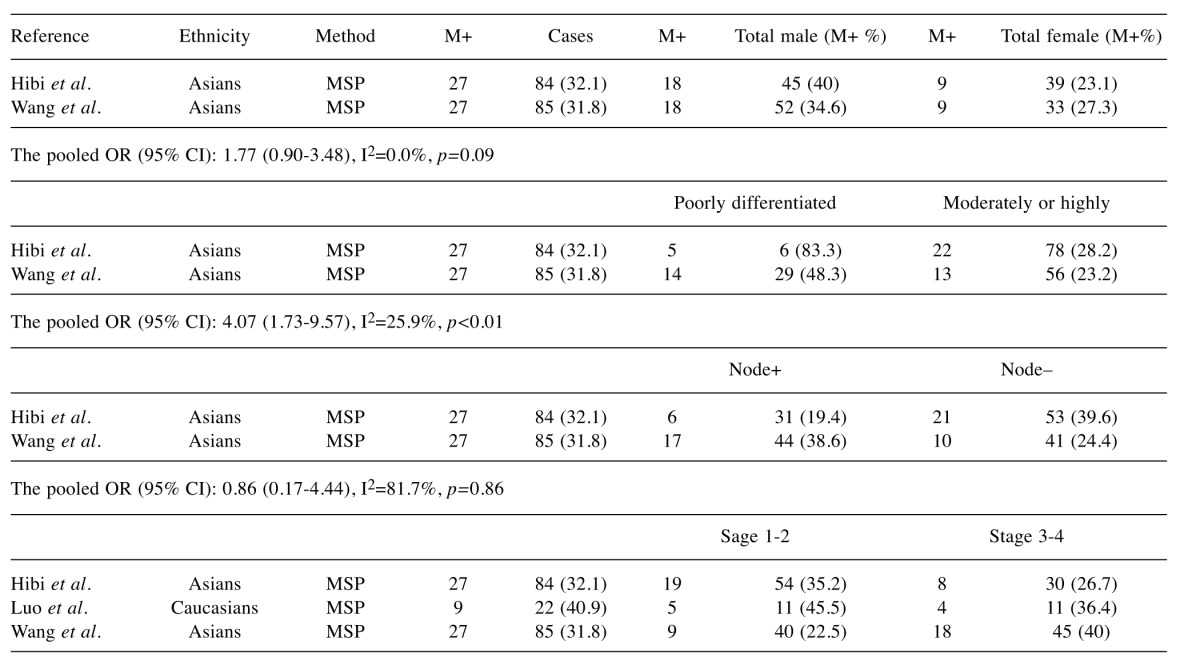
The analyses of T-cadherin promoter methylation and gender, tumor grade and lymph node status used the random effects model; we found that the OR was 1.77 (95%CI=0.90-3.48), 1.17 (95%CI=0.63-2.15), and 0.86 (95%CI=0.17-4.44), respectively, as assessed by the random effect model. This result indicated that a significant association was not found. The pooled OR (OR=4.07, 95%CI=1.73-9.57) from 2 studies indicated that T-cadherin promoter methylation was significantly higher in poorly-differentiated CRC than in moderately- or highly-differentiated CRC (Table II). Hence, when T-cadherin used as a potential biomarker for CRC diagnosis should be cautious.
Table II. T-cadherin promoter methylation and clinicopathological features.

95% CI, 95% Confidence interval; M, methylation; MSP, methylation-specific polymerase chain reaction; OR,odds ratio; total, the number of samples.
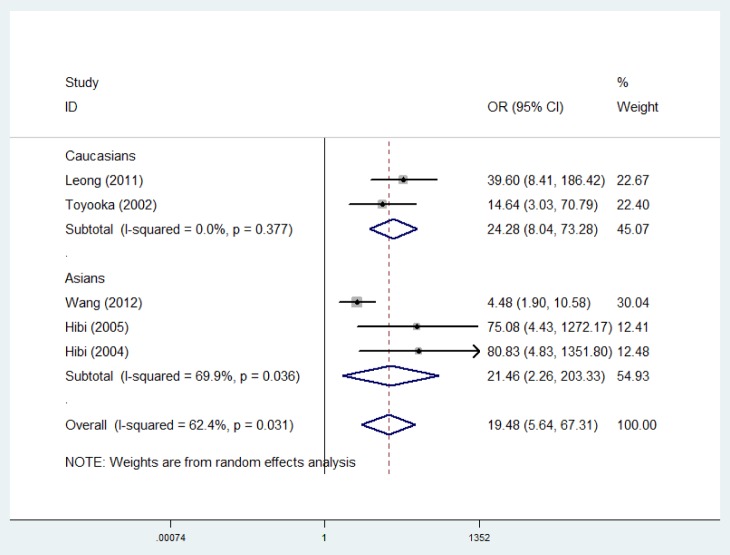
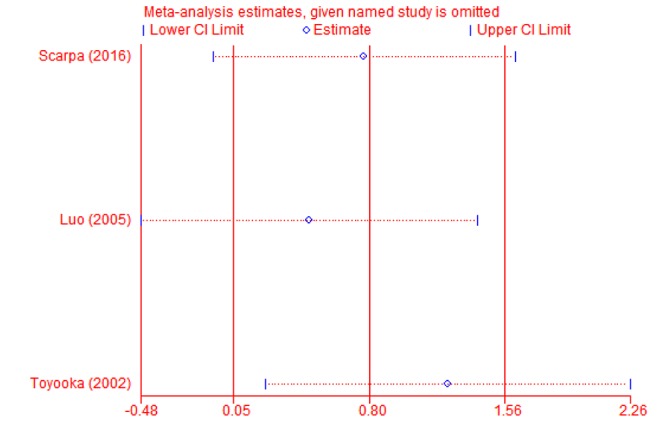
Sub-group and sensitivity analyses. Sub-group analysis was carried-out to confirm the stability of the relationship. When cancer tissues were compared to adjacent tissues, the result demonstrated that T-cadherin promoter methylation had significantly increased the risk of CRC in Asians and Caucasians (OR=21.46, 95% CI=2.26-203.33; OR=24.28, 95% CI=8.04-73.28, respectively) (Figure 3). Then, sensitivity analysis was subsequently performed to detect the influence of individual study on the pooled estimate by omitting one study from the pooled analysis each time (CRC vs. normal tissues; CRC versus adjacent tissues; CRC vs. precancerosis tissues). The exclusion of each single study did not significantly change the pooled OR, suggesting that the results of the meta-analysis were robust (Figure 4, Figure 5 and Figure 6).
Figure 3. Forest plot of subgroup analysis on ethnicity for CDH13 promoter methylation in CRC versus adjacent tissues.
Figure 4. Sensitivity analysis of T-cadherin in CRC versus normal tissues.
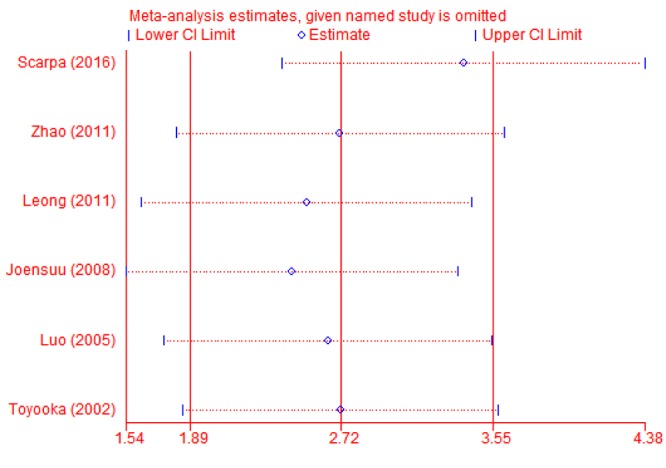
Figure 5. Sensitivity analysis of T-cadherin in CRC versus adjacent tissues.
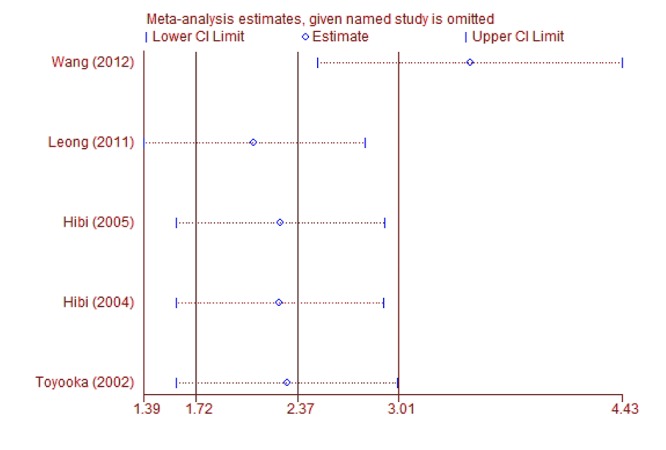
Figure 6. Sensitivity analysis of T-cadherin in CRC versus precancerosis tissues.
Discussion
Epigenetic alterations are much more frequent in CRC than genetic alterations. In addition to epigenetic alteration of expression of miRNAs, other common types of epigenetic alterations in cancers that change gene expression levels include direct hypermethylation or hypomethylation of CpG islands of protein-encoding genes and alterations in histones and chromosomal architecture that influence gene expression. As an example, 147 hypermethylations and 27 hypomethylations of protein-coding genes were frequently associated with colorectal cancers. Of the hypermethylated genes, 10 were hypermethylated in 100% of colon cancers, and many others were hypermethylated in more than 50% of colon cancers (20). In addition, 11 hypermethylations and 96 hypomethylations of miRNAs were also associated with colorectal cancer.
However, the potential of T-cadherin promoter methylation to be a biomarker for CRC has not yet been evaluated. The findings of the current study showed that T-cadherin promoter methylation was significantly higher in CRC patients than in normal, adjacent, and premalignant tissues, suggesting that the methylation of T-cadherin may be involved in the development of CRC. And the result denoted that T-cadherin promoter methylation had a significantly increased risk of CRC in Asians and Caucasians patients, which indicated that these populations were susceptible to the promoter methylation of T-cadherin. Furthermore, the OR value in Caucasians (OR=24.28) was significantly higher than in Asians (OR=21.46), which revealed that T-cadherin promoter methylation may be a relatively more important risk factor among Caucasian populations. In addition, we conducted meta-analyses to determine the correlations between T-cadherin promoter methylation and clinicopathological characteristics. The results showed that the T-cadherin promoter methylation was not associated with gender, tumor stage, and lymph node status.
T-cadherin could play a navigating role in the growing tumor vessels, which in the absence of contact inhibition from the stromal cells, grow into the surrounding tumor tissue. And T-cadherin loss in tumor cells is associated with tumor malignancy, invasiveness and metastasis. Thus, tumor progression in colorectal cancer correlates with downregulation of Т-cadherin expression. In vitro, Т-cadherin is implicated in regulation of cell growth, survival and proliferation. In cultured VSМС and primary astrocytes, the expression of Т-cadherin depends оn proliferation status with maximum at confluency suggesting its regulation of cell growth by contact inhibition. Known mitogens such as platelet-derived growth factor (PDGF)-BB, epidermal growth factor (EGF) or insulin-like growth factor (IGF) elicit а reversible dose- and time-dependent decrease in Т-cadherin expression in cultured VSMCs. In nude mice, expression of T-cadherin leads to complete inhibition of subcutaneous tumor growth. Seeding T-cadherin-expressing cells on plastic coated with recombinant aminoterminal fragments of T-cadherin resulted in suppression of cell growth and was found to be associated with increased expression of p21. In T-cadherin-deficient С6 glioma сеll lines, its overexpression results in growth suppression involving p21CIP1/WAF1 production and G2 arrest.
Certain limitations of the current meta-analysis should be considered. First, the numbers of studies are relatively small, and may not consider all confounding factors though they reported adjusted estimates. And the inclusion of articles published only in English and Chinese might lead to a selection bias. Second, all the studies are based on Asians and Caucasians, other ethnicities, such as Africans, were limited. Further studies are needed to investigate the role of T-cadherin in on African countries. As we know, there are significant differences such as etiology, biology features, clinical types, and prognosis in the risk of CRC in different ethnic groups within a given geographical area. Third, blood or feces samples were insufficient. More studies based on urine and blood samples are very essential to evaluate whether T-cadherin promoter methylation can become a non-invasive biomarker for the detection and diagnosis of CRC in the future. Finally, no attempt was made to identify unpublished work and grey literature, for example university theses or conference proceedings. As a result, publication bias may have influenced the results.
Conclusion
This integrated analysis of pooled data provides strong evidence that the methylation status of the T-cadherin promoter is significantly associated with CRC. However, T-cadherin promoter methylation was not correlated with gender, tumor stage, and lymph node status in cancer. Large-scale, prospective clinical trials with advanced methodologies are still required to verify the findings and provide a higher level of evidence.
Funding
The project is sponsored by The Ph.D Start-up Fund of Liaoning Province, China (No. 201601413) and Liaoning BaiQianWan Talents Program, type C project, No.13.
References
- 1.Sung JJ, Lau JY, Goh KL, Leung WK, Asia Pacific Working Group on Colorectal Cancer. Increasing incidence of colorectal cancer in Asia: Implications for screening. Lancet Oncol. 2005;6:871–876. doi: 10.1016/S1470-2045(05)70422-8. [DOI] [PubMed] [Google Scholar]
- 2.Coleman MP, Quaresma M, Berrino F, Lutz JM, De Angelis R, Capocaccia R, Baili P, Rachet B, Gatta G, Hakulinen T, Micheli A, Sant M, Weir HK, Elwood JM, Tsukuma H, Koifman S, E Silva GA, Francisci S, Santaquilani M, Verdecchia A, Storm HH, Young JL, CONCORD Working Group Cancer survival in five continents: A worldwide population based study. Lancet Oncol. 2008;9:730–756. doi: 10.1016/S1470-2045(08)70179-7. [DOI] [PubMed] [Google Scholar]
- 3.He Y, Cui Y, Wang W, Gu J, Guo S, Ma K, Luo X. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia. 2011;13:841–53. doi: 10.1593/neo.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores K, Wolschin F, Corneveaux JJ, Allen AN, Huentelman MJ, Amdam GV. Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genomics. 2012;13:480. doi: 10.1186/1471-2164-13-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuebel KE, Chen W, Cope L, Glöckner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, van Engeland M, Ting AH, Jair K, Yu W, Toyota M, Imai K, Ahuja N, Herman JG, Baylin SB. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3(9):1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riener MO, Nikolopoulos E, Herr A, Wild PJ, Hausmann M, Wiech T, Orlowska-Volk M, Lassmann S, Walch A, Werner M. Microarray comparative genomic hybridization analysis of tubular breast carcinoma shows recurrent loss of the CDH13 locus on 16q. Human Pathol. 2008;39:1621–1629. doi: 10.1016/j.humpath.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Toyooka KO, Toyooka S, Virmani AK, Sathyanarayana UG, Euhus DM, Gilcrease M, Minna JD, Gazdar AF. Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Cancer Res. 2001;61:4556–4560. [PubMed] [Google Scholar]
- 8.Qian ZR, Sano T, Yoshimoto K, Asa SL, Yamada S, Mizusawa N, Kudo E. Tumor-specific downregulation and methylation of the CDH13 (H-cadherin) and CDH1 (E-cadherin) genes correlate with aggressiveness of human pituitary adenomas. Modern Pathol. 2007;20:1269–1277. doi: 10.1038/modpathol.3800965. [DOI] [PubMed] [Google Scholar]
- 9.Ogama Y, Ouchida M, Yoshino T, Ito S, Takimoto H, Shiote Y, Ishimaru F, Harada M, Tanimoto M, Shimizu K. Prevalent hyper-methylation of the CDH13 gene promoter in malignant B cell lymphomas. Int J Oncol. 2004;25:685–691. [PubMed] [Google Scholar]
- 10.Sun D, Zhang Z, Van DN, Huang G, Ernberg I, Hu L. Aberrant methylation of CDH13 gene in nasopharyngeal carcinoma could serve as a potential diagnostic biomarker. Oral Oncol. 2007;43:82–87. doi: 10.1016/j.oraloncology.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Leong KJ, Wei W, Tannahill LA, Caldwell GM, Jones CE, Morton DG, Matthews GM, Bach SP. Methylation profiling of rectal cancer identifies novel markers of early-stage disease. Br J Surg. 2011;98:724–734. doi: 10.1002/bjs.7422. [DOI] [PubMed] [Google Scholar]
- 12.Joensuu EI, Abdel-Rahman WM, Ollikainen M, Ruosaari S, Knuutila S, Peltomäki P. Epigenetic signatures of familial cancer are characteristic of tumor type and family category. Cancer Res. 2008;68:4597–4605. doi: 10.1158/0008-5472.CAN-07-6645. [DOI] [PubMed] [Google Scholar]
- 13.Hibi K, Kodera Y, Ito K, Akiyama S, Nakao A. Aberrant methylation of HLTF, SOCS-1, and CDH13 genes is shown in colorectal cancers without lymph node metastasis. Dis Colon Rectum. 2005;48:1282–1286. doi: 10.1007/s10350-004-0947-7. [DOI] [PubMed] [Google Scholar]
- 14.Luo L, Chen WD, Pretlow TP. CpG island methylation in aberrant crypt foci and cancers from the same patients. Int J Cancer. 2005;115:747–751. doi: 10.1002/ijc.20936. [DOI] [PubMed] [Google Scholar]
- 15.Hibi K, Nakayama H, Kodera Y, Ito K, Akiyama S, Nakao A. CDH13 promoter region is specifically methylated in poorly differentiated colorectal cancer. Br J Cancer. 2004;90:1030–1033. doi: 10.1038/sj.bjc.6601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyooka S, Toyooka KO, Harada K, Miyajima K, Makarla P, Sathyanarayana UG, Yin J, Sato F, Shivapurkar N, Meltzer SJ, Gazdar AF. Aberrant methylation of the CDH13 (H-cadherin) promoter region in colorectal cancers and adenomas. Cancer Res. 2002;62:3382–3386. [PubMed] [Google Scholar]
- 17.Zhao L, Yu JP. Methylation status of CDH13 gene promoter in colon cancer. Chin J Postgrad Med. 2011;34:23–24. [Google Scholar]
- 18.Scarpa M, Scarpa M, Castagliuolo I, Erroi F, Kotsafti A, Basato S, Brun P, D’Incà R, Rugge M, Angriman I, Castoro C. Aberrant gene methylation in non-neoplastic mucosa as a predictive marker of ulcerative colitis-associated CRC. Oncotarget. 2016;7:10322–10331. doi: 10.18632/oncotarget.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Yuan X, Jiao N, Zhu H, Zhang Y, Tong J. CDH13 and FLBN3 genemethylation are associated with poor prognosis in colorectal cancer. Pathol Oncol Res. 2012;18:263–270. doi: 10.1007/s12253-011-9437-0. [DOI] [PubMed] [Google Scholar]
- 20.Schnekenburger M, Diederich M. Epigenetics Offer New Horizons for Colorectal Cancer Prevention. Curr Colorectal Cancer Rep. 2012;8(1):66–81. doi: 10.1007/s11888-011-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]