Abstract
Orally ingested bacteria interact with intestinal mucosa and may impact immunity. However, insights in mechanisms involved are limited. In this randomized placebo-controlled cross-over trial, healthy human subjects were given Lactobacillus plantarum supplementation (strain TIFN101, CIP104448, or WCFS1) or placebo for 7 days. To determine whether L. plantarum can enhance immune response, we compared the effects of three stains on systemic and gut mucosal immunity, by among others assessing memory responses against tetanus toxoid (TT)-antigen, and mucosal gene transcription, in human volunteers during induction of mild immune stressor in the intestine, by giving a commonly used enteropathic drug, indomethacin [non-steroidal anti-inflammatory drug (NSAID)]. Systemic effects of the interventions were studies in peripheral blood samples. NSAID was found to induce a reduction in serum CD4+/Foxp3 regulatory cells, which was prevented by L. plantarum TIFN101. T-cell polarization experiments showed L. plantarum TIFN101 to enhance responses against TT-antigen, which indicates stimulation of memory responses by this strain. Cell extracts of the specific L. plantarum strains provoked responses after WCFS1 and TIFN101 consumption, indicating stimulation of immune responses against the specific bacteria. Mucosal immunomodulatory effects were studied in duodenal biopsies. In small intestinal mucosa, TIFN101 upregulated genes associated with maintenance of T- and B-cell function and antigen presentation. Furthermore, L. plantarum TIFN101 and WCFS1 downregulated immunological pathways involved in antigen presentation and shared downregulation of snoRNAs, which may suggest cellular destabilization, but may also be an indicator of tissue repair. Full sequencing of the L. plantarum strains revealed possible gene clusters that might be responsible for the differential biological effects of the bacteria on host immunity. In conclusion, the impact of oral consumption L. plantarum on host immunity is strain dependent and involves responses against bacterial cell components. Some strains may enhance specific responses against pathogens by enhancing antigen presentation and leukocyte maintenance in mucosa. In future studies and clinical settings, caution should be taken in selecting beneficial bacteria as closely related strains can have different effects. Our data show that specific bacterial strains can prevent immune stress induced by commonly consumed painkillers such as NSAID and can have enhancing beneficial effects on immunity of consumers by stimulating antigen presentation and memory responses.
Keywords: intestinal mucosal immunity, adaptive immunity, Lactobacillus plantarum, indomethacin, non-steroidal anti-inflammatory drug, probiotics
Introduction
Commensal Lactobacilli species may play an active role in intestinal immune homeostasis (1–9). Lactobacilli likely regulate immune cells via the interaction of bacterial cell-wall components or secreted bacterial products with immune or epithelial cells in human gut mucosa (8, 10, 11). These interactions seem to contribute to more than just tolerance to the beneficial microbes. As demonstrated in several vaccination studies Lactobacilli, such as plantarum strains derived from WCFS1, have a positive impact on immune responses (12–18). Furthermore, these bacteria can activate tolerogenic cellular pathways in human intestinal mucosal cells (3, 5, 10).
In previous studies, we demonstrated that Lactobacillus plantarum, a member of lactic acid bacteria with a “generally recognized as safe” status (3, 19), had different effects on human dendritic and peripheral blood mononuclear cells (10, 11). Using comparative genomic hybridization, we identified a number of bacteriocins and cell-wall components involved in the glycosylation of cell wall teichoic acids associated with these differential effects (8, 10). The differential expression of L. plantarum genes may contribute to the observed differences in activation of toll-like receptor (TLR) 2–4 and CD14 antigens in the host (10). As a consequence of differences in TLR-binding dendritic cells L. plantarum strains induce alterations in production of the pro-inflammatory cytokine IL-12 and the regulatory cytokine IL-10 (10). The immunomodulatory effects of L. plantarum may benefit human gut homeostasis, in particular in case of pathogenic or pharmacological induced mucosal stress. Non-steroidal anti-inflammatory drugs (NSAIDs), which are commonly used painkillers, are well known for their negative side effects on gut mucosal integrity and immunity, which are mediated through inhibition of cyclooxygenase and subsequent prostaglandin deficiency. But these effects have also been shown to be modulated by gut microbiota via among others TLR4 signaling (20).
The current clinical trial has been conducted to study the effects of three L. plantarum strains on immunity and intestinal barrier function. The findings regarding immune responses are presented in the current report, while the results on gut barrier function have been published previously (2). We have shown that small intestinal permeability of healthy volunteers increased, indicating gut barrier dysfunction, after administration of indomethacin (a NSAID) which could not be reversed by intake of L. plantarum strains. However, one of the strains, i.e., L. plantarum TIFN101, did demonstrate profound effects on mucosal gene transcription related to mucosal structure, while the findings regarding the modulation of these processes by L. plantarum WCFS1 and CIP4448 were more moderate. This indicates strain-dependent modulation of gut barrier function by the tested L. plantarum strains (2).
To investigate mechanism by which L. plantarum may influence in vivo human mucosal and systemic immune activity under mild mucosal stress conditions such as intake of NSAIDs, we conducted a randomized double-blind placebo-controlled cross-over human trial. Three L. plantarum strains were selected for their different immunomodulating effect in vitro (3, 10, 11). The three strains have different effects on TLRs signaling and differently stimulate cytokine production in vitro in immune cells (3, 10, 11). Healthy volunteers received the NSAID indomethacin, which induces mild and reversible damage to the gastrointestinal lining (21–24). Study participants subsequently consumed one of the three L. plantarum strains or placebo for a week. Polarized T-cell responses in the peripheral circulation, as well as the responses after re-stimulation with cell extract of L. plantarum strains, superantigen, or tetanus were studied to determine whether these bacteria could enhance systemic immunity against one of the stimuli. Duodenal biopsies were taken and used to study the effect of the strains on mucosal gene regulation. Furthermore, the bacterial strains were sequenced and annotated to identify putative gene clusters associated with the differential responses induced by the probiotics.
Materials and Methods
This study was approved by the University Hospital Maastricht Ethical Committee and has been registered in the US National Library of Medicine (NCT01456767).1 A more detailed description of the trial design has been published previously (2).
Study Design and Study Participants
Ten healthy volunteers, seven females and three males (26.3 ± 10.1 years, BMI of 21.8 ± 2.40 kg/m2), without a history of gastrointestinal symptoms and free of medication, were tested on four separate occasions in a randomized cross-over design. Four intervention periods included the 7-day intake of L. plantarum strains WCFS1 (WCFS), CIP104448 (CIP48), or TIFN101, or placebo. Before and after the intervention, blood samples were taken to study the effect of L. plantarum consumption on T-cell polarization. Furthermore, at day 7, duodenal biopsies were taken for whole-genome expression microarrays. No side effects or complications were reported.
All included subjects gave their written informed consent. The healthy volunteers consumed their habitual diet and kept a gastrointestinal symptoms diary. The night before the start day, the volunteers ingested 75 mg of the NSAID indomethacin. On the start day, the volunteers ingested another dosage of 50 mg indomethacin to conform to a previously established protocol to establish mild gastrointestinal mucosal stress (25, 26). Subsequently, the volunteers consumed the bacterium or placebo supplements for a period of 7 days during breakfast and during dinner. The vials containing bacteria or placebo were non-transparent. On the seventh day, blood samples were taken again and tissue samples were obtained from the horizontal part of the duodenum by standard flexible gastroduodenoscopy at approximately 15 cm distal to the pylorus. The duodenal mucosa was chosen as the duodenum is readily accessible for sampling. The interventions were performed with an interval of 4 weeks to allow a wash-out period and also healing of the biopsy-sampling region.
Bacterial Strains and Growth Conditions
Three L. plantarum strains form the NIZO strain-collection were selected: WCFS1, CIP104448, and TIFN101. In previous studies TIFN101 has been referred to as CIP104450 (11, 27). The bacteria were cultured at 37°C in man, rogosa, and sharpe medium (Merck). Detailed protocols for culturing, harvesting, freeze drying, storing, and viability determination of Lactobacillus species have been published (1). Maltodextrin and glucose were added to a final concentration of 20 and 2% (wt/vol), respectively, to obtain bacterial preparations (WCFS1, 2.6 × 109 CFU; CIP104448, 2.4 × 109 CFU; and TIFN101, 5.6 × 109 CFU); placebo controls contained the two sugars.
Cell Staining
Blood of study participants was collected in EDTA-containing tubes and processed for fluorescence-activated cell sorting (FACS) analysis (Table S1 in Supplementary Material). Isotype controls were used at the same dilution as the antibody.
To study T-cell polarization, 200 µl of blood was diluted with 200 µl of RPMI1640 supplemented with heat-inactivated 10% fetal calf serum (FCS) and incubated with either phorbol-myristate-acetate (PMA; 80 nM Sigma-Aldrich, Steinheim, Germany) and 2 nM calcium ionophore (Ca-Io; Sigma-Aldrich) (4 h), Staphylococcus aureus-enterotoxin B (SEB) (5 µg/ml Sigma, Deisenhofen, Germany) (24 h), tetanus toxoid (TT; 1.5 Lf/ml, RIVM, Bilthoven, The Netherlands) (24 h), or bacterial lysates (30 µg/ml) (24 h). Stimulation with bacterial lysates was performed 1 week after treatment with one of the three strains. After stimulation, red blood cells were lysed with ammonium chloride, washed (PBS with 2% FCS), and incubated with different antibody cocktails.
To stain for T-cells and T-cell subsets, cells were incubated with an antibody cocktail consisting of anti-CD3, anti-CD8, and anti-CD45RO for 30 min in the dark on ice. After washing with buffer, cells were incubated with streptavidin-Pacific Orange (1:100 Invitrogen) for 15 min on ice. After washing and centrifugation, pelleted cells were resuspended in Fix/Perm solution (eBioscience) for 45 min on ice. After washing in Perm-solution cells were incubated in mouse-serum for 15 min, followed by incubation with the cytokine antibody mix (anti-IL-4, anti-IFNγ, anti-IL-17, and anti-IL-21) or an isotype control mix for 30 min on ice. Cells were then washed with Perm solution (three times), resuspended in wash-buffer and measured by flow-cytometry.
For staining NK-cells, cells were incubated with an antibody cocktail consisting of anti-CD3, anti-CD16, anti-CD56, anti-CD335, and anti-CD161 (NK-cell staining), or with an isotype control cocktail for NK-cells consisting of anti-CD3, anti-CD16, anti-CD56, and isotype controls for anti-CD335 and CD161 for 30 min in the dark on ice. After washing with washing buffer, cells were fixed in FACS-lysis solution (BD Biosciences) for 30 min on ice, washed and analyzed by flow-cytometry.
RNA Isolation and Microarray
Total RNA was isolated from duodenal biopsy samples of the study participants using Trizol reagent (1 ml) (Invitrogen, Breda, The Netherlands). RNA was purified using the Qiagen RNeasy Micro kit (Qiagen, The Netherlands) and quantified on a NanoDrop ND-1000 spectrophotometer (Isogen Life Science, The Netherlands). RNA quality was confirmed using an Agilent 2100 bioanalyzer (Agilent Technologies, The Netherlands). RNA was judged suitable for array-hybridization only if samples exhibited intact bands corresponding to 18S and 28S ribosomal subunits and displayed no chromosomal peaks or RNA-degradation products.
Total RNA (100 ng) was used for whole transcript cDNA synthesis using the Ambion WT expression kit (Life Technologies, The Netherlands) and was subsequently labeled using the Affymetrix GeneChip WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA, USA). Samples were hybridized to human whole genome Affymetrix GeneChip Human Gene 1.1 ST arrays, washed, stained, and scanned on an Affymetrix GeneTitan instrument. Details on array handling can be found in the Affymetrix GeneTitan Instrument User Guide for Expression Array Plates (P/N 702933 Rev.2).
Duodenal Mucosa Microarray Data Analysis
Microarray analysis was performed utilizing MADMAX for statistical analysis (28). All arrays met the quality criteria. The probes on the Human Gene 1.1 ST arrays were redefined according to Dai et al. (29) based on the NCBI Entrez database (CDF version 15.1). In this way, the Human Gene 1.1 ST array targets 19,682 unique genes. Normalized expression values were obtained from the raw intensity values by using the robust multi-array analysis preprocessing algorithm available in the AffyPLM library using default settings (30). Microarray data were filtered and probe sets with at least five probes and expression values higher than 20 on at least five arrays with an interquartile range (IQR) >0.2 (log2 scale) across all samples were selected for further statistical analysis. In addition, an IQR cut-off of 0.2 was used to filter out genes that showed no variation between the conditions. Differentially expressed genes were identified using linear models, applying moderated t-statistics that implemented empirical Bayes regularization of SEs in the library limma (31). The moderated t-statistic was extended by a Bayesian hierarchical model to define an intensity-based moderated t-statistic to adjust for independence of variances relative to the degree of identity and relation between variance and signal intensity (32). Genes were defined as significantly changed when the p-value was <0.05 for pairwise comparisons. The datasets generated from microarray profiling experiments have been deposited to the publicly accessible database repository Gene Expression Omnibus (GSE74988).
Pathway Analysis
Gene set enrichment analysis (GSEA2) was performed using MADMAX and gene sets with a false discovery rate <0.2 were considered significantly enriched. GSEA takes into account the broader context in which gene products function, namely in physically interacting networks such as biochemical, metabolic, or signal transduction routes, and has the advantage that it is unbiased because no gene selection step is used. Possible transcription factors were identified using upstream regulator analysis in ingenuity pathway analysis (IPA; Ingenuity Systems, Redwood City, CA, USA).
Bacterial Genome Sequencing and Annotation
For DNA preparation, bacteria were pelleted, washed, and resuspended in TES buffer (N-[tris(hydroxymethyl)methyl]-2-aminoet hanesulfonic acid). Cells were lysed with lysozyme (360 mg/ml) and mutanolysin (140 U/ml) by incubation for 2 h at 37°C. Subsequently, 300 µl water and 80 µl of a 20% SDS solution were added. The DNA extraction was done using phenol/chloroform (3×). DNA was precipitated with isopropanol and washed with 70% ethanol. Samples were treated with 100 µg/ml RNAse (Sigma) for 1 h at 37°C. DNA paired-end libraries with barcoding were made and sequenced using Illumina technology (Baseclear Leiden). The genome sequences of L. plantarum strains CIP104448 and TIFN 101 have been deposited in NCBI/GenBank under accession numbers JSUW00000000 and JSUX00000000, respectively. The contig sequences were submitted to the RAST automatic annotation server,3 which provided ORF calling and automatic annotation. The annotated contigs of CIP48 and TIFN101 were ordered by comparing them to the circular template genome of L. plantarum WCFS1 (33), and comparing them to each other. Contigs/genes that did not match to the WCFS1 genome were annotated in more detail using BLASTP4 and InterProscan.5 Ortholog groups (OGs) in the three genomes were determined using OrthoMCL.6
Statistics
Flow-cytometry results are expressed as the mean ± SEM. Normal distribution was confirmed by the Kolmogorov–Smirnov test. The two-sided Student’s t-test was used for changes in immune-cell populations after L. plantarum treatment. Gene expression data are depicted as the median (range). The two-sided Mann–Whitney U-test was used to determine changes in expression profiles after L. plantarum treatment in vivo. A p-value <0.05 was considered statistically significant.
Results
Cell Frequencies and T-Cell Polarization after Treatment with L. plantarum Strains
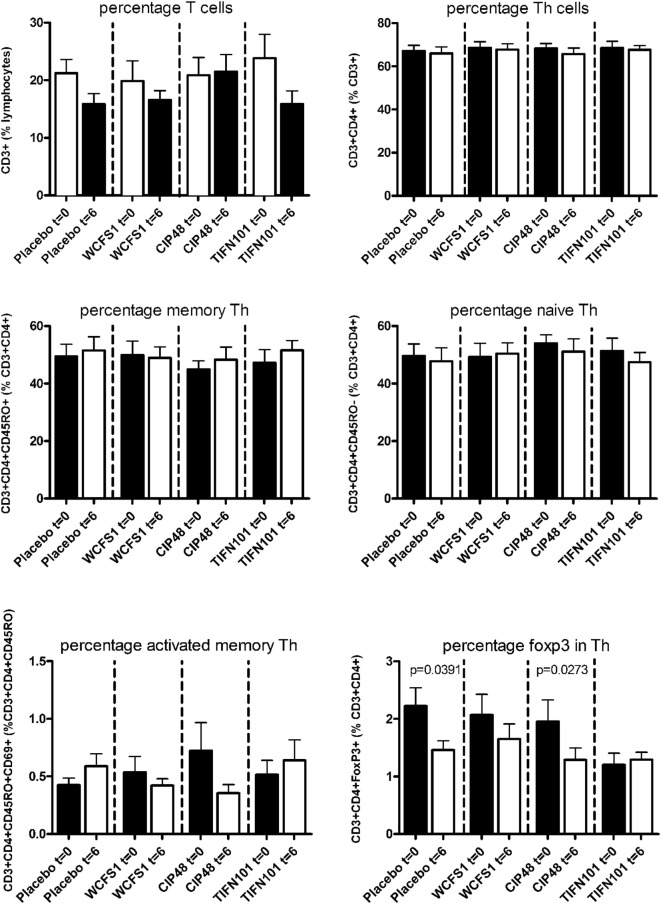
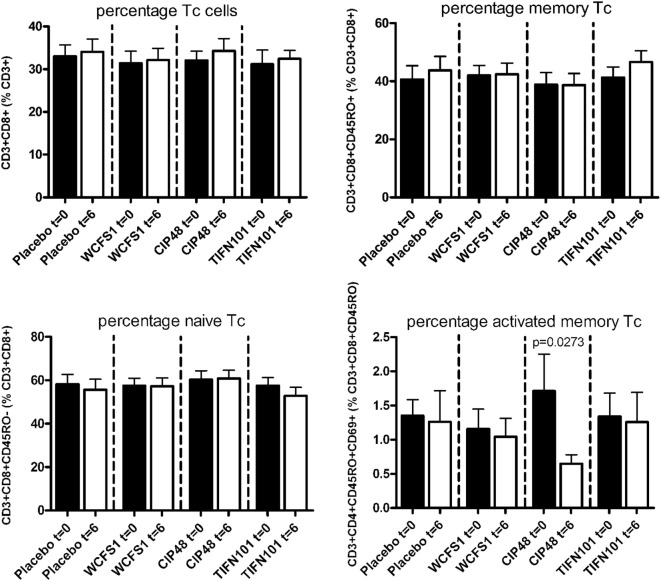
In peripheral blood samples, we did not observe differences in the frequencies of total CD3+ cells, CD3+CD4+ cells (naïve or memory), or the CD3+CD4+ activated memory cells after treatment with any of the L. plantarum strains. However, the percentage of CD4+Foxp3+ cells was significantly decreased following placebo and CIP48 treatment, but not after TIFN101 treatment (Figure 1). Moreover, although we did not observe any effect with indomethacin treatment on CD3+CD8+ (naïve and memory) cells, activated memory-cells exhibited a statistically significantly decrease after CIP48 treatment (p < 0.05) (Figure 2).
Figure 1.
Effects of three Lactobacillus plantarum strains on the frequency of CD4+ T-cell populations in systemic circulation. Data are expressed as mean ± SEM. Statistical significant differences were determined by using two-sided Student’s t-tests. A p-value <0.05 was considered statistically significant. A p-value <0.1 was considered a statistical trend.
Figure 2.
Effects of three Lactobacillus plantarum strains on the frequency of CD8+ T-cell populations in systemic circulation. Data are expressed as mean ± SEM. Statistical significant differences were determined by using two-sided Student’s t-tests. A p-value <0.05 was considered statistically significant. A p-value <0.1 was considered a statistical trend.
Treatment did not influence percentages of NK-cells or NKT-cells. There was also no change in the percentages of the NK-cell subtypes (i.e., CD56high and CD56dim); and the expression of CD161 (KLRB1), mediating cytotoxicity (33, 34), was not affected either by the L. plantarum treatments (results not shown).
T-cell polarization was studied after three types of T-cell stimulation: (i) non-specific polyclonal stimulation with PMA/Ca2+ or superantigen Staphylococcus aureus-enterotoxin B (SEB) to study whether the total responsiveness was influenced by L. plantarum treatment, (ii) stimulation with a previously administered vaccine-antigen (TT) to study stimulation of specific memory responses, and (iii) stimulation with cell extracts of the specific L. plantarum strains in order to investigate whether specific immune responses against the L. plantarum were stimulated.
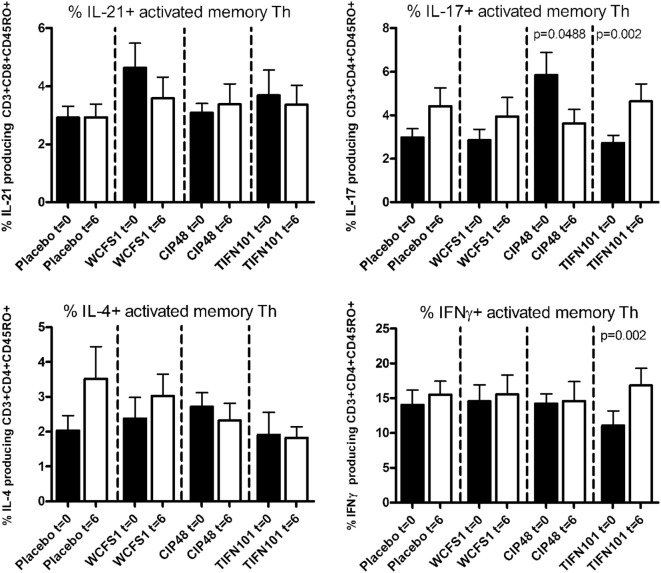
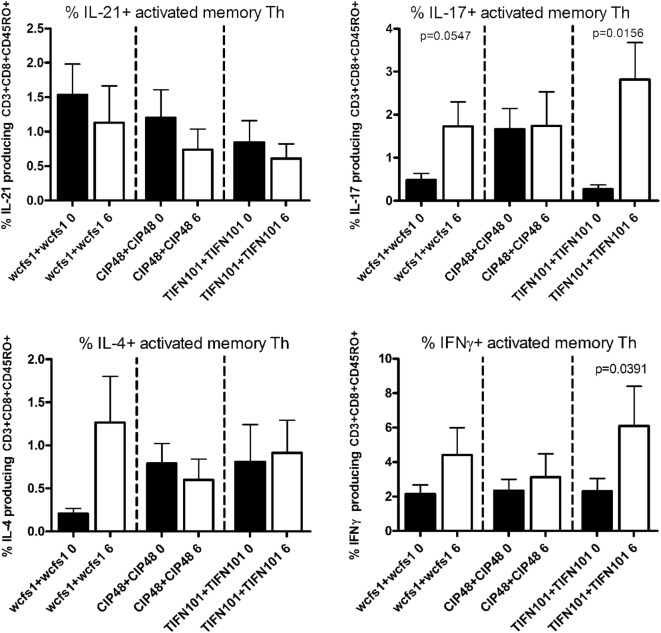
After non-specific stimulation with PMA/Ca-ionophore or SEB, we quantified the percentage of IFNγ, IL-4, IL-17, or IL-21 positive Th cells and memory Th cells. Treatment with placebo or the administered L. plantarum strains did not influence cytokine production of the total population of Th cells or of Th memory cells after non-specific stimulation with PMA/Ca-ionophore. Although no differences were found after SEB-stimulation in cytokine production by the total Th cell population after the three L. plantarum treatments (results not shown), we did observe differences in cytokine production of the Th memory cells after L. plantarum treatment. After stimulation with SEB, we observed a decreased percentage of IL-17-producing activated memory Th cells following treatment with CIP48 and an increased percentage of IL-17-producing activated memory Th cells after treatment with TIFN101 (Figure 3). Moreover, the percentage of IFNγ-producing activated memory Th cells was also increased after TIFN101 treatment (Figure 3).
Figure 3.
Effects of three Lactobacillus plantarum strains on the frequency of IL-21, IL-17, IL-4, and IFN-γ-producing Staphylococcus aureus enterotoxin B superantigen (SEB)-stimulated memory-CD45RO+ Th cells. Data are expressed as mean ± SEM. Statistical significant differences were determined by using two-sided Student’s t-tests. A p-value <0.05 was considered statistically significant. A p-value <0.1 was considered a statistical trend.
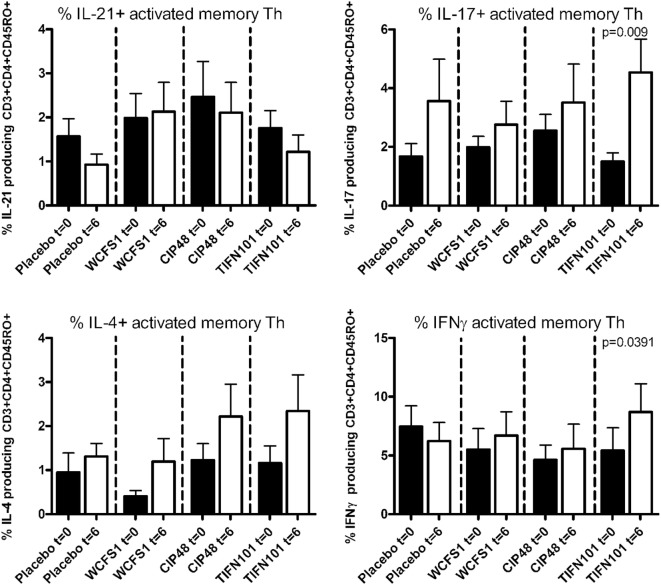
Treatment with L. plantarum strains also modulated cytokine production following a more specific stimulation by TT (Figure 4). After TIFN101 treatment, the percentage IL-17 and the percentage IFN-γ-producing memory Th cells were significantly increased, while no effect on cytokine production by memory Th cells after TT-stimulation was observed with the other L. plantarum strains.
Figure 4.
Effects of three Lactobacillus plantarum strains on the frequency of IL-21, IL-17, IL-4, and IFN-γ-producing tetanus toxoid-stimulated memory-CD45RO+ Th cells. Data are expressed as mean ± SEM. Statistical significant differences were determined by using two-sided Student’s t-tests. A p-value <0.05 was considered statistically significant. A p-value <0.1 was considered a statistical trend.
Finally, we stimulated human blood samples with cell extracts of the L. plantarum strain that the volunteers had consumed in the study (Figure 5). We observed that subjects, who were treated with WCFS, showed a trend toward an increased IL-17 response after stimulation with WCFS cell extracts. Other cytokines were not affected after this treatment. There were no differences in cytokine production in subjects treated with CIP48, when their blood samples were stimulated with CIP48 cell extract. When subjects were treated with TIFN101, their activated memory cells exhibited increased IL-17 and IFN-γ-production following stimulation with TIFN101 cell extract.
Figure 5.
Effects of three Lactobacillus plantarum strains on the frequency of IL-21, IL-17, IL-4, and IFN-γ-producing memory-CD45RO+ Th cells stimulated with bacterial cell extracts, matched to the strain consumed. Data are expressed as mean ± SEM. Statistical significant differences were determined by using two-sided Student’s t-tests. A p-value <0.05 was considered statistically significant. A p-value <0.1 was considered a statistical trend.
Transcriptional Response in Duodenal Mucosa upon Exposure to L. plantarum Strains
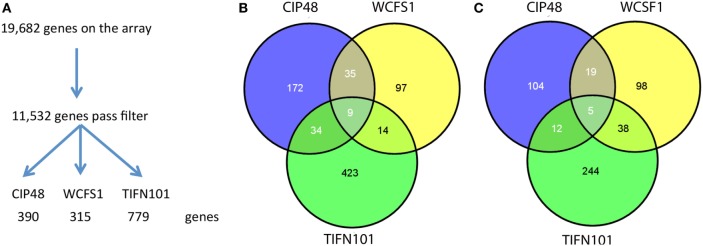
Differential gene expression profiles were found in the stressed intestinal mucosa of the subjects consuming each bacterial strain; 315 genes were differentially regulated with L. plantarum WCFS, 390 with CIP48, and 779 with TIFN101, as compared to the placebo intervention (Figure 6A). Of these genes, the different bacterial strains shared only relatively small numbers of upregulated and downregulated genes (Figures 6B,C, respectively). Shared genes were mainly involved in general cellular functions and metabolism.
Figure 6.
Flowchart of the microarray analysis (A) and the number of unique genes regulated in human intestinal biopsies after consumption of three different Lactobacillus plantarum strains [L. plantarum WCFS1 (WCFS), L. plantarum CIP104448 (CIP48), L. plantarum TIFN101 (TIFN101)]. Intensity >20 on at least five arrays, interquartile range >0.2, at least seven probes per gene. Venn diagrams of the number of upregulated (B) and downregulated (C) genes in the intestinal biopsies after consumption of L. plantarum and indomethacin.
The 10 genes that were most highly up- or downregulated are listed in Tables 1–3. Of the most highly induced genes after TIFN101 consumption 80% are related to immunity: immunoglobulin lambda variable 6–57, putative V-set and immunoglobulin domain-containing protein 6-like, immunoglobulin lambda variable 7–46, interferon regulatory factor 4, GDNF family, CD27, CD79a, and plasminogen activator. WCFS and TIFN101 shared the downregulation of six small nucleolar RNAs, i.e., snoRNA (H)C/D(ACA) box 6, 14b, 53, 57, 60, 388, while CIP48 had a complete different profile of up and downregulation.
Table 1.
Lactobacillus plantarum WCFS1 (WCSF): 10 most upregulated and downregulated genes.
| Gene name | IBMT p-value | Mean fold versus placebo | |
|---|---|---|---|
| Top 10: upregulated genes placebo versus WCFS | |||
| Kinesin family member 20B | KIF20B | 0.01 | 1.34 |
| microRNA 186 | MIR186 | 0.03 | 1.31 |
| Guanylate cyclase activator 2A (guanylin) | GUCA2A | 0.04 | 1.31 |
| Integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | ITGA4 | 0.01 | 1.31 |
| Centromere protein E, 312 kDa | CENPE | 0.05 | 1.30 |
| Putative homeodomain transcription factor 1 | PHTF1 | 0.00 | 1.30 |
| Spindle and kinetochore-associated complex subunit 2 | SKA2 | 0.01 | 1.29 |
| Killer cell lectin-like receptor subfamily D, member 1 | KLRD1 | 0.00 | 1.29 |
| Gamma-aminobutyric acid A receptor, alpha 2 | GABRA2 | 0.02 | 1.27 |
| Retinitis pigmentosa GTPase regulator | RPGR | 0.01 | 1.27 |
| Top 10: downregulated genes placebo versus WCFS | |||
| Small nucleolar RNA, H/ACA box 16A | SNORA16A | 0.02 | −1.35 |
| Small nucleolar RNA, C/D box 53 | SNORD53 | 0.04 | −1.36 |
| Contactin 3 (plasmacytoma associated) | CNTN3 | 0.00 | −1.38 |
| Small nucleolar RNA, C/D box 6 | SNORD6 | 0.04 | −1.39 |
| Small nucleolar RNA, H/ACA box 57 | SNORA57 | 0.00 | −1.44 |
| Small nucleolar RNA, H/ACA box 60 | SNORA60 | 0.00 | −1.44 |
| Small nucleolar RNA, H/ACA box 14A | SNORA14A | 0.03 | −1.49 |
| Small nucleolar RNA, H/ACA box 38B (retrotransposed) | SNORA38B | 0.01 | −1.58 |
| Small nucleolar RNA, H/ACA box 14B | SNORA14B | 0.01 | −1.58 |
| Small Cajal body-specific RNA 4 | SNORA16A | 0.01 | −1.72 |
Table 3.
Lactobacillus plantarum TIFN101 (TIFN101): 10 most upregulated and downregulated genes.
| Gene name | IBMT p-value | Mean fold versus placebo | |
|---|---|---|---|
| Top 10: upregulated genes placebo versus TIFN101 | |||
| Immunoglobulin lambda variable 6–57 | IGLV6-57 | 0.01 | 1.63 |
| Putative V-set and immunoglobulin domain-containing protein 6-like | LOC642131 | 0.00 | 1.55 |
| Immunoglobulin lambda variable 7–46 (gene/pseudogene) | IGLV7-46 | 0.04 | 1.48 |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 | HS3ST3B1 | 0.00 | 1.41 |
| Interferon regulatory factor 4 | IRF4 | 0.00 | 1.40 |
| GDNF family receptor alpha 2 | GFRA2 | 0.00 | 1.40 |
| CD27 molecule | CD27 | 0.01 | 1.40 |
| CD79a molecule, immunoglobulin-associated alpha | CD79A | 0.03 | 1.38 |
| Plasminogen activator, tissue | PLAT | 0.00 | 1.37 |
| Der1-like domain family, member 3 | DERL3 | 0.00 | 1.37 |
| Top 10: downregulated genes placebo versus TIFN101 | |||
| Small nucleolar RNA, H/ACA box 38B (retrotransposed) | SNORA38B | 0.04 | −1.40 |
| Small nucleolar RNA, H/ACA box 21 | SNORA21 | 0.03 | −1.40 |
| Small nucleolar RNA, H/ACA box 60 | SNORA60 | 0.01 | −1.41 |
| Small nucleolar RNA, C/D box 53 | SNORD53 | 0.01 | −1.43 |
| Ephrin-A1 | EFNA1 | 0.00 | −1.45 |
| Small nucleolar RNA, C/D box 6 | SNORD6 | 0.02 | −1.46 |
| Small nucleolar RNA, H/ACA box 57 | SNORA57 | 0.00 | −1.47 |
| Small nucleolar RNA, H/ACA box 14B | SNORA14B | 0.00 | −1.73 |
| Family with sequence similarity 5, member C | FAM5C | 0.03 | −1.97 |
| Small Cajal body-specific RNA 4 | SCARNA4 | 0.00 | −2.00 |
Table 2.
Lactobacillus plantarum CIP104448 (CIP48): 10 most upregulated and downregulated genes.
| Gene name | IBMT p-value | Mean fold versus placebo | |
|---|---|---|---|
| Top 10: upregulated genes placebo versus CIP48 | |||
| Coiled-coil domain containing 59 | CCDC59 | 0.01 | 1.33 |
| Aldehyde dehydrogenase 1 family, member L2 | ALDH1L2 | 0.03 | 1.32 |
| KIAA0125 | KIAA0125 | 0.00 | 1.31 |
| Phospholipase C, beta 4 | PLCB4 | 0.01 | 1.31 |
| Coiled-coil domain containing 102B | CCDC102B | 0.04 | 1.29 |
| RAS guanyl releasing protein 3 (calcium and DAG-regulated) | RASGRP3 | 0.03 | 1.28 |
| Peptidase domain containing associated with muscle regeneration 1 | PAMR1 | 0.00 | 1.28 |
| DEP domain containing 1 | DEPDC1 | 0.01 | 1.28 |
| Phospholipase A2, group IIA (platelets, synovial fluid) | PLA2G2A | 0.02 | 1.28 |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 | HS3ST3B1 | 0.04 | 1.28 |
| Top 10: downregulated genes placebo versus CIP48 | |||
| Potassium channel, subfamily K, member 15 | KCNK15 | 0.05 | −1.33 |
| Transient receptor potential cation channel, subfamily V, member 6 | TRPV6 | 0.01 | −1.33 |
| Long intergenic non-protein coding RNA 282 | LINC00282 | 0.03 | −1.35 |
| Ephrin-A1 | EFNA1 | 0.02 | −1.38 |
| Matrix metallopeptidase 10 (stromelysin 2) | MMP10 | 0.05 | −1.41 |
| Angiopoietin-like 4 | ANGPTL4 | 0.03 | −1.47 |
| Heme oxygenase (decycling) 1 | HMOX1 | 0.00 | −1.50 |
| Nuclear factor, interleukin 3 regulated | NFIL3 | 0.01 | −1.52 |
| Major facilitator superfamily domain containing 2A | MFSD2A | 0.02 | −1.59 |
| Glucose-6-phosphatase, catalytic subunit | G6PC | 0.02 | −1.65 |
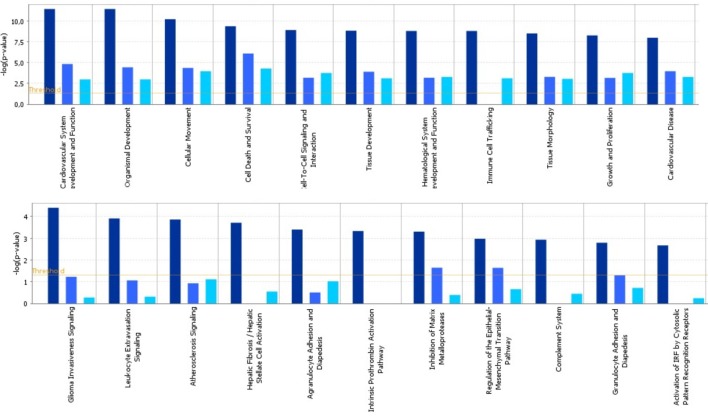
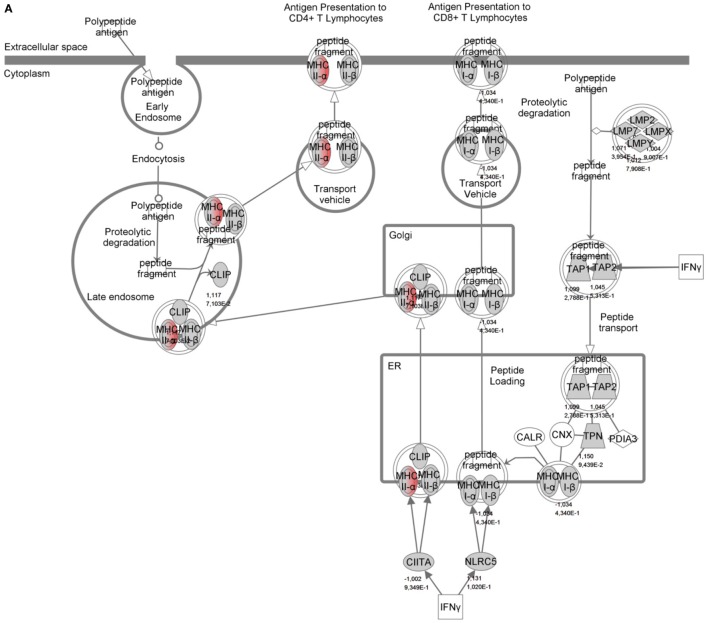
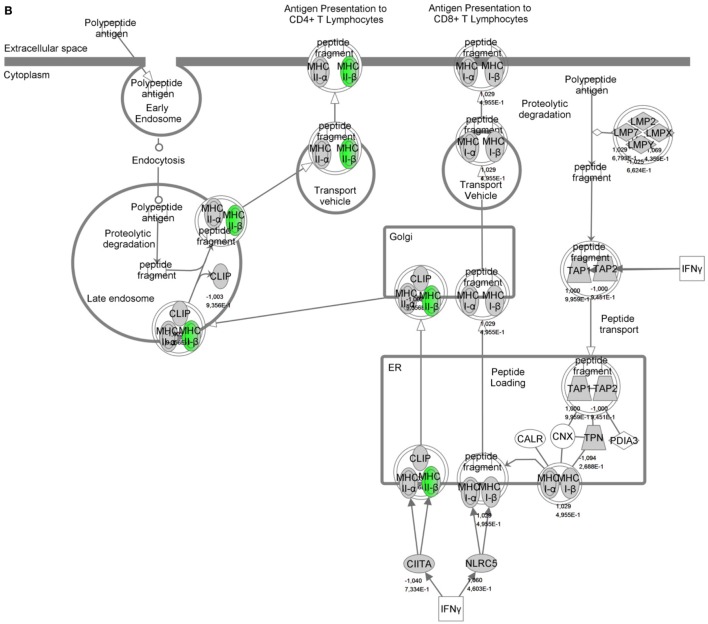
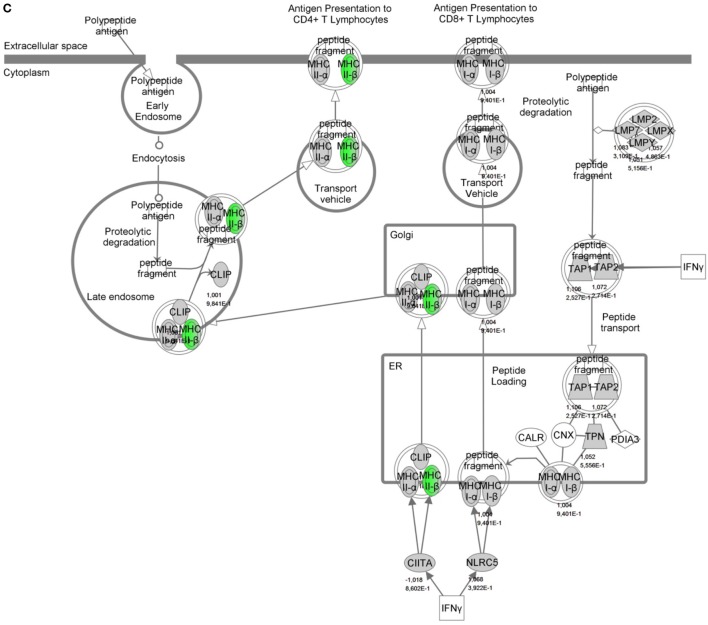
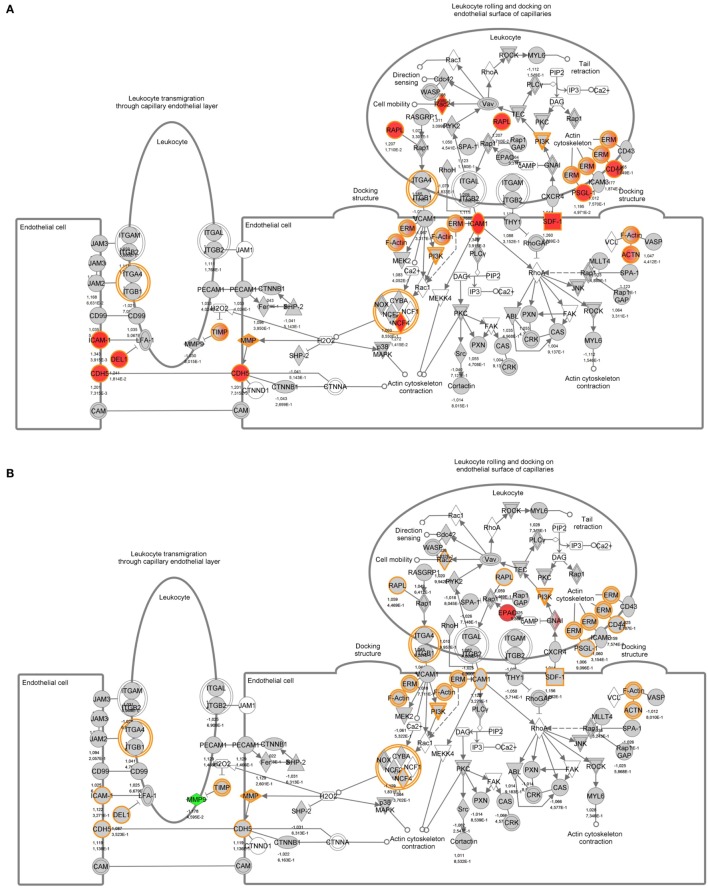
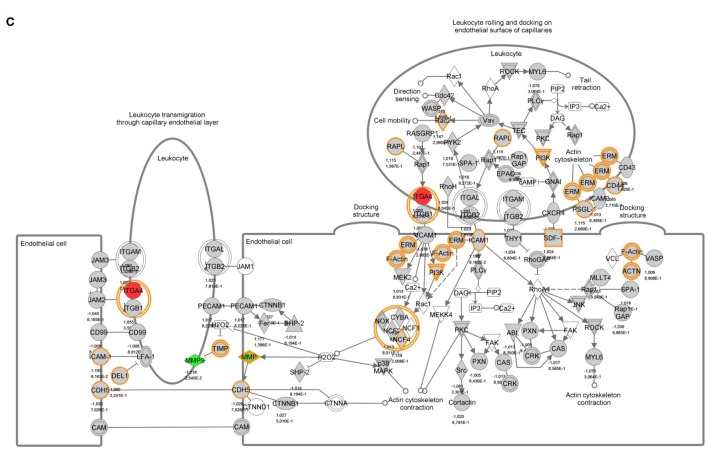
To identify specific transcription factors and to identify pathways regulated by the strains, IPA was performed (Figure 7). TIFN101 induced more changes than CIP48 and WCSF1 in the NSAID-stressed intestine. The most significant set of target-genes in the TIFN101 group were immune response-related genes. TIFN101 upregulated MHC-IIα, while CIP48 and WCFS downregulated MHC-IIβ (Figure 8). Another pathway that might contribute to the enhanced responses in TIFN101 is the upregulation of genes involved in leukocyte extravasation (Figure 9). TIFN101 enhanced RAPL expression, which is a GTPase involved in regulating integrin affinity. Concomitantly, an upregulation of essential adhesion molecules such as ICAM-1 and Cadherin 5 was seen, illustrating the upregulation of immune-cell migration pathways by TIFN101. Also, some regulation of leukocyte extravasation was observed with CIP48 and WCFS. However, it was much less pronounced than for the TIFN101 intervention.
Figure 7.
Cellular pathways are significantly more modulated after consumption of either Lactobacillus plantarum TIFN101 (dark blue, left column) than after consumption of L. plantarum CIP104448 (light blue, middle column) or L. plantarum WCFS1 (cyan, right column). Statistical significance of pathway modulation was calculated via a right-tailed Fisher’s Exact test in ingenuity pathway analysis and represented as −log (p-value); −log values exceeding the depicted threshold were significant (p < 0.05).
Figure 8.
Regulation of genes involved in antigen presentation in non-steroidal anti-inflammatory drug-stressed intestine after consumption of either Lactobacillus plantarum TIFN101 (A), L. plantarum CIP104448 (B), or L. plantarum WCFS1 (C). Red indicates upregulation while green depicts downregulation of the specific gene. TIFN101 upregulated MHC-IIα while CIP48 and WCFS1 downregulated MHC-IIβ.
Figure 9.
Regulation of genes involved in leukocyte extravasation pathways in non-steroidal anti-inflammatory drug-stressed intestine after consumption of either Lactobacillus plantarum TIFN101 (A), L. plantarum CIP104448 (B), or L. plantarum WCFS1 (C). Red indicates upregulation while green depicts downregulation of the specific gene.
Differential Gene Content Profiles between the Three L. plantarum Strains
Lactobacillus plantarum strains CIP48 and TIFN101 were sequenced, annotated, and compared with the genome (chromosome and plasmids) of L. plantarum WCFS1 (35, 36). A total of 3,010 OGs were assigned to the chromosome, based on the ordering of contigs to the template WCFS genome. The three genomes shared 2,455 of the 3,010 chromosomal OGs (=81.5%), which is defined as the core genome for this study. Figure 10 presents the shared and genes and contigs of the L. plantarum strains. When the contigs and OGs/genes are included that do not match to the WCFS chromosome, higher numbers of unique genes were found for the CIP48 and TIFN101 genomes. Many of these extra unique genes are on plasmids (Table S2 in Supplementary Material). All unique genes for L. plantarum TIFN101 and L. plantarum CIP104448 compared to WCFS1 are listed in Tables S3 and S4 in Supplementary Material, and all absent genes in the two strains are listed in Tables S5 and S6 in Supplementary Material, respectively, as these are potential candidate genes for the biological effects of the two strains. L. plantarum CIP48 lacks the complete plantaricin biosynthesis gene cluster, a large set of genes adjacent to this cluster (i.e., OGs 334–348), and the entire gene cluster for EPS biosynthesis. L. plantarum TIFN101 is missing some genes associated with plantaricin biosynthesis as well as genes for exopolysaccharide biosynthesis, many sugar utilization cassettes, and two large LPXTG-anchored mucus-binding proteins (Tables S7A,B in Supplementary Material).
Figure 10.
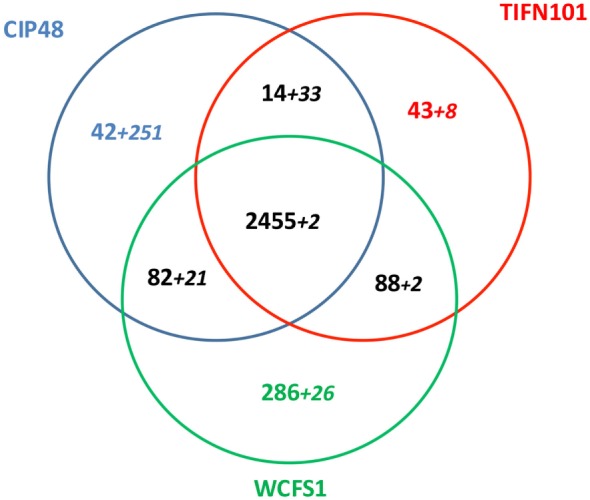
Venn diagram of shared and unique genes in all contigs of Lactobacillus plantarum strains [L. plantarum WCFS1 (WCFS), L. plantarum CIP104448 (CIP48), L. plantarum TIFN101]. The smaller numbers in italics represent ortholog groups that do not match to the chromosome of strain WCFS. These numbers do not include the putative prophage genes in each genome.
Discussion
The current experimental human study was undertaken to investigate the immunomodulatory and potentially beneficial effects of three L. plantarum strains, which were selected in vitro for their differential immune stimulating capacity. Indicators of responses of mucosal and systemic immunity were assessed in healthy individuals undergoing a mild, commonly encountered stressor (25, 26) of the intestine and immune system, i.e., the intake of NSAIDs. We show that three L. plantarum strains had a highly strain-dependent effect on immunity in vivo, which were different to those predicted in vitro.
Indicators of systemic immunomodulatory effects were assessed in peripheral blood samples of the study participants. Consumption of indomethacin induced a reduction in CD4+/Foxp3 regulatory cell frequencies, which was prevented by WCFS and TIFN101 administration. This can be considered a beneficial regulatory effect. CIP48, not preventing the reduction of CD4+Foxp3 and reducing the number of memory cells, had effects that suggest a pro-inflammatory effect of consumption of this bacterium. Furthermore, T-cell polarization was studied after different stimuli. The hypothesis was that bacterial cell-wall components might induce immune responses (1) and enhance systemic immunity as a bystander effect. This happened with WCFS and TIFN101 (Figure 5). However, the most pronounced stimulator of immunity, i.e., TIFN101, also showed enhanced responses against specific pathogenic antigens such as TT and had an overall enhanced response to the SEB challenge. It is unlikely that the enhanced response against TT in the TIFN101 consumers is caused solely by the response against TIFN101 cell components. This is supported by the analysis of the intestinal biopsies that suggest that TIFN101 stimulate specific processes in the intestinal mucosa. TIFN101, in contrast to the other strains, upregulates mucosal transcription processes associated with T and B cell function and antigen presentation, when compared to placebo. It also had a pronounced effect on CD27 upregulation, which is required for generation and long-term maintenance of T cell immunity (37). Furthermore, TIFN101 enhanced expression of MHC-IIα in intestinal mucosa as well as key regulatory molecules such as RAPL. RAPL enhances integrin affinity and the adhesion of T cells (38, 39). These observations in the mucosa may explain the enhanced memory T cell responses observed with TIFN101 consumption. Furthermore, B cell immunity in the mucosa was enhanced as illustrated by upregulation of immunoglobulin regulatory genes and by CD79a. CD79a is also known as B cell antigen receptor complex-associated protein alpha-chain forming, together with CD79b protein, the B cell antigen receptor complex-associated protein (40). CIP48 and WCFS did not have these effects; in fact, they had a tendency to downregulate processes such as antigen presentation in the mucosa.
As a consequence of differences in TLR-binding, L. plantarum strains induce upon incubation with monocytes or dendritic cells different quantities of the pro-inflammatory cytokine IL-12 and the regulatory cytokine IL-10 (10). On the basis of these findings, three strains were selected, L. plantarum WCFS1 (WCFS), L. plantarum CIP104448 (CIP48), and L. plantarum TIFN101 (TIFN101), as their induced IL-10/IL-12 ratio in vitro were classified as pro-inflammatory, neutral, or anti-inflammatory, respectively. However, the immune responses to these strains in vivo were very different to that predicted in vitro. Consumption of NSAID induced a reduction in CD4+/Foxp3 regulatory cell frequencies, which was prevented by WCFS1 and TIFN101 administration. This should be considered to be a beneficial regulatory effect. CIP48 did not prevent NSAID-induced reduction of CD4+Foxp3 T cells and had more negative effects. CIP48 reduced the number of memory cells suggesting a pro-inflammatory, worsening effect of consumption of this bacterium. Our data suggest that solely studying the effects of bacteria on a single cell type in vitro, such as dendritic cells, has limited value, as it does not provide representative insight into the complex interplay between immune and other mucosal cells in vivo.
To our knowledge, this is the first report that bacteria can downregulate snoRNAs in the stressed intestine. SnoRNA are metabolically stable, non-coding RNAs that associate with a set of proteins to form small nucleolar ribonucleoproteins. The majority of snoRNAs function to guide RNAs in the post-transcriptional synthesis of 2′-O-methylated nucleotides and pseudo-uridines in ribosomal RNAs (rRNAs), small nuclear RNAs, and other cellular RNAs, including messenger RNAs (41–43). The relative reduction of several snoRNAs by TIFN101 and WCFS suggest a downregulation of methylation of rRNA (44) and downregulation of 14b diminished pseudo-uridinilation of RNA (45). Usually, this is indicative of a destabilization of cellular processes, but it may also be an indicator of tissue repair (46).
It should be noted that, in the present trial, we used a cross-over design to eliminate potential influences of inter-subject differences. Previous interventions may influence subsequent tests in such a study design. However, a long wash-out period of 4 weeks was chosen and the order of interventions was randomly assigned to each study subject, which should have minimized potential influences between interventions. Furthermore, the impact of L. plantarum consumption observed in this study may be caused by direct effects on host immunity; however, indirect effects via modulation of the resident microbiota may contribute as well.
Finally, full genome sequencing of L. plantarum CIP48 and L. plantarum TIFN101 was applied to identify possible gene clusters that might be responsible for the differential biological effects of the three L. plantarum strains. Several 100 novel L. plantarum genes were found in strain CIP48 and TIFN101 as compared to the known genome of strain WCFS (35, 36), of which only a small number are shared by both CIP48 and TIFN101. The majority of the novel genes appear to be located on plasmids. CIP48 appears to have several plasmids that are not present in TIFN101 or WCFS. Strain CIP48 has specific genes, such as for lantibiotic biosynthesis and several cell-surface proteins, that might explain its differential effect. Notably, however, there are also numerous genes/gene clusters in L. plantarum CIP48 and TIFN101 that are not present in the WCFS genome. Strikingly, CIP48 completely and TIFN101 partially lack the plantaricin biosynthesis clusters. These genes have been linked to strain differences in cytokine production (10, 47), but were shown here not to be associated with immune effects in vivo. Also, both strains lack very large regions important for sugar metabolism. These differences have been attributed to adaptations to environmental factors which, in our opinion, are interesting targets genes and possibly associated with probiotic effects (48). Not only the presence but also the absence of genes may enhance immune effects of bacteria (8).
Conclusion
The current randomized double-blind placebo-controlled cross-over human trial demonstrated immune modulatory effects by orally consumed L. plantarum strains via specific responses against bacterial components as well as direct stimulation of specific immunity in the intestine. Specific bacteria can influence and possibly prevent undesired immune responses during commonly encountered stressors of the intestine such as intake of NSAIDs. Strong strain-dependent effects were found that showed L. plantarum TIFN101 to have a positive effect on host immunity, while CIP48 and WCFS1 had effects that may to be considered as less beneficial. Caution should be taken in selecting beneficial bacteria as even closely related strains can have very different effects. The observation that some bacteria can enhance specific memory cell populations can be helpful in identifying putative immune active human applicable bacterial strains. Furthermore, the current comparative genomics study provides many leads for follow-up experimental work to identify genes that are responsible for or involved in the observed differences in immune effects in humans.
Ethics Statement
This study was carried out in accordance with the recommendations of the University Hospital Maastricht Ethical Committee with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the University Hospital Maastricht Ethical Committee. The study has been registered in the US National Library of Medicine (http://www.clinicaltrials.gov, NCT01456767).
Author Contributions
PV co-supervised and co-designed the study and wrote the manuscript; ZM conducted the study and was involved in manuscript writing; BH and RS performed most analyses and reviewed the manuscript; PB provided the bacterial supplements and placebo; MM, JW, and AM co-supervised the study and interpretation of data; MB conducted the transcriptome analyses; and MF and FT co-supervised and co-designed the study.
Conflict of Interest Statement
PB is employee of NIZO food research, a private commercial company. All other authors declare no conflict of interest.
Acknowledgments
We would like to acknowledge Harm-Jan Pieters for his assistance at the laboratory.
Funding. This work was supported by a project from the Top Institute Food and Nutrition, Wageningen, The Netherlands. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.01000/full#supplementary-material.
References
- 1.Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM, et al. L. plantarum, L. salivarius, and L. lactis attenuate Th2 responses and increase Treg frequencies in healthy mice in a strain dependent manner. PLoS One (2012) 7(10):e47244. 10.1371/journal.pone.0047244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mujagic Z, de Vos P, Boekschoten MV, Govers C, Pieters HH, de Wit NJ, et al. The effects of Lactobacillus plantarum on small intestinal barrier function and mucosal gene transcription; a randomized double-blind placebo controlled trial. Sci Rep (2017) 7:40128. 10.1038/srep40128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, et al. Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci U S A (2009) 106(7):2371–6. 10.1073/pnas.0809919106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Troost FJ, van Baarlen P, Lindsey P, Kodde A, de Vos WM, Kleerebezem M, et al. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genomics (2008) 9:374. 10.1186/1471-2164-9-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Baarlen P, Troost F, van der Meer C, Hooiveld G, Boekschoten M, Brummer RJ, et al. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc Natl Acad Sci U S A (2011) 108(Suppl 1):4562–9. 10.1073/pnas.1000079107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HJ, Kim YJ, Lee SH, Yu J, Jeong SK, Hong SJ. Effects of Lactobacillus rhamnosus on allergic march model by suppressing Th2, Th17, and TSLP responses via CD4(+)CD25(+)Foxp3(+) Tregs. Clin Immunol (2014) 153(1):178–86. 10.1016/j.clim.2014.04.008 [DOI] [PubMed] [Google Scholar]
- 7.Steinberg RS, Lima M, Gomes de Oliveira NL, Miyoshi A, Nicoli JR, Neumann E, et al. Effect of intestinal colonisation by two Lactobacillus strains on the immune response of gnotobiotic mice. Benef Microbes (2014) 5:409–19. 10.3920/BM2013.0075 [DOI] [PubMed] [Google Scholar]
- 8.Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM, et al. The impact of Lactobacillus plantarum WCFS1 teichoic acid D-alanylation on the generation of effector and regulatory T-cells in healthy mice. PLoS One (2013) 8(4):e63099. 10.1371/journal.pone.0063099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM, et al. Probiotics can generate FoxP3 T-cell responses in the small intestine and simultaneously inducing CD4 and CD8 T cell activation in the large intestine. PLoS One (2013) 8(7):e68952. 10.1371/journal.pone.0068952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meijerink M, van Hemert S, Taverne N, Wels M, de Vos P, Bron PA, et al. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One (2010) 5(5):e10632. 10.1371/journal.pone.0010632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Hemert S, Meijerink M, Molenaar D, Bron PA, de Vos P, Kleerebezem M, et al. Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol (2010) 10:293. 10.1186/1471-2180-10-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akatsu H, Arakawa K, Yamamoto T, Kanematsu T, Matsukawa N, Ohara H, et al. Lactobacillus in jelly enhances the effect of influenza vaccination in elderly individuals. J Am Geriatr Soc (2013) 61(10):1828–30. 10.1111/jgs.12474 [DOI] [PubMed] [Google Scholar]
- 13.Bosch M, Mendez M, Perez M, Farran A, Fuentes MC, Cune J. Lactobacillus plantarum CECT7315 and CECT7316 stimulate immunoglobulin production after influenza vaccination in elderly. Nutr Hosp (2012) 27(2):504–9. 10.1590/S0212-16112012000200023 [DOI] [PubMed] [Google Scholar]
- 14.de Azevedo MS, Innocentin S, Dorella FA, Rocha CS, Mariat D, Pontes DS, et al. Immunotherapy of allergic diseases using probiotics or recombinant probiotics. J Appl Microbiol (2013) 115(2):319–33. 10.1111/jam.12174 [DOI] [PubMed] [Google Scholar]
- 15.Licciardi PV, Tang ML. Vaccine adjuvant properties of probiotic bacteria. Discov Med (2011) 12(67):525–33. [PubMed] [Google Scholar]
- 16.Van Puyenbroeck K, Hens N, Coenen S, Michiels B, Beunckens C, Molenberghs G, et al. Efficacy of daily intake of Lactobacillus casei Shirota on respiratory symptoms and influenza vaccination immune response: a randomized, double-blind, placebo-controlled trial in healthy elderly nursing home residents. Am J Clin Nutr (2012) 95(5):1165–71. 10.3945/ajcn.111.026831 [DOI] [PubMed] [Google Scholar]
- 17.West NP, Cripps AW. Are vaccination models suitable to determine whether probiotics have beneficial health effects in the general population? Hum Vaccin Immunother (2013) 9(3):621–4. 10.4161/hv.23254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaqoob P. Ageing, immunity and influenza: a role for probiotics? Proc Nutr Soc (2014) 73(2):309–17. 10.1017/S0029665113003777 [DOI] [PubMed] [Google Scholar]
- 19.Seddik HA, Bendali F, Gancel F, Fliss I, Spano G, Drider D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob Proteins (2017) 9:111–22. 10.1007/s12602-017-9264-z [DOI] [PubMed] [Google Scholar]
- 20.Otani K, Tanigawa T, Watanabe T, Shimada S, Nadatani Y, Nagami Y, et al. Microbiota plays a key role in non-steroidal anti-inflammatory drug-induced small intestinal damage. Digestion (2017) 95(1):22–8. 10.1159/000452356 [DOI] [PubMed] [Google Scholar]
- 21.Lee OY, Kang DH, Lee DH, Chung IK, Jang JY, Kim JI, et al. A comparative study of DA-9601 and misoprostol for prevention of NSAID-associated gastroduodenal injury in patients undergoing chronic NSAID treatment.Arch Pharm Res (2014) 37(10):1308–16. 10.1007/s12272-014-0408-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mokhtare M, Valizadeh SM, Emadian O. Lower gastrointestinal bleeding due to non-steroid anti-inflammatory drug-induced colopathy case report and literature review. Middle East J Dig Dis (2013) 5(2):107–11. [PMC free article] [PubMed] [Google Scholar]
- 23.Tomita T, Sadakata H, Tamura M, Matsui H. Indomethacin-induced generation of reactive oxygen species leads to epithelial cell injury before the formation of intestinal lesions in mice. J Physiol Pharmacol (2014) 65(3):435–40. [PubMed] [Google Scholar]
- 24.Fornai M, Antonioli L, Colucci R, Pellegrini C, Giustarini G, Testai L, et al. NSAID-induced enteropathy: are the currently available selective COX-2 inhibitors all the same? J Pharmacol Exp Ther (2014) 348(1):86–95. 10.1124/jpet.113.207118 [DOI] [PubMed] [Google Scholar]
- 25.Bjarnason I, Hayllar J, MacPherson AJ, Russell AS. Side effects of nonsteroidal anti-inflammatory drugs on the small and large intestine in humans. Gastroenterology (1993) 104(6):1832–47. 10.1016/0016-5085(93)90667-2 [DOI] [PubMed] [Google Scholar]
- 26.Troost FJ, Saris WH, Brummer RJ. Recombinant human lactoferrin ingestion attenuates indomethacin-induced enteropathy in vivo in healthy volunteers. Eur J Clin Nutr (2003) 57(12):1579–85. 10.1038/sj.ejcn.1601727 [DOI] [PubMed] [Google Scholar]
- 27.van Bokhorst-van de Veen H, van Swam I, Wels M, Bron PA, Kleerebezem M. Congruent strain specific intestinal persistence of Lactobacillus plantarum in an intestine-mimicking in vitro system and in human volunteers. PLoS One (2012) 7(9):e44588. 10.1371/journal.pone.0044588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin K, Kools H, de Groot PJ, Gavai AK, Basnet RK, Cheng F, et al. MADMAX – management and analysis database for multiple ~omics experiments. J Integr Bioinform (2011) 8(2):160. 10.2390/biecoll-jib-2011-160 [DOI] [PubMed] [Google Scholar]
- 29.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res (2005) 33(20):e175. 10.1093/nar/gni179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (2003) 4(2):249–64. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol (2004) 3:Article3. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 32.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics (2006) 7:538. 10.1186/1471-2105-7-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs R, Hintzen G, Kemper A, Beul K, Kempf S, Behrens G, et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol (2001) 31(10):3121–7. [DOI] [PubMed] [Google Scholar]
- 34.Tarazona R, Casado JG, Delarosa O, Torre-Cisneros J, Villanueva JL, Sanchez B, et al. Selective depletion of CD56(dim) NK cell subsets and maintenance of CD56(bright) NK cells in treatment-naive HIV-1-seropositive individuals. J Clin Immunol (2002) 22(3):176–83. 10.1023/A:1015476114409 [DOI] [PubMed] [Google Scholar]
- 35.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A (2003) 100(4):1990–5. 10.1073/pnas.0337704100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siezen RJ, Francke C, Renckens B, Boekhorst J, Wels M, Kleerebezem M, et al. Complete resequencing and reannotation of the Lactobacillus plantarum WCFS1 genome. J Bacteriol (2012) 194(1):195–6. 10.1128/JB.06275-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Jochems C, Anderson AM, Talaie T, Jales A, Madan RA, et al. Soluble CD27-pool in humans may contribute to T cell activation and tumor immunity. J Immunol (2013) 190(12):6250–8. 10.4049/jimmunol.1300022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raab M, Wang H, Lu Y, Smith X, Wu Z, Strebhardt K, et al. T cell receptor “inside-out” pathway via signaling module SKAP1-RapL regulates T cell motility and interactions in lymph nodes. Immunity (2010) 32(4):541–56. 10.1016/j.immuni.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Wang H. Integrin signalling and function in immune cells. Immunology (2012) 135(4):268–75. 10.1111/j.1365-2567.2011.03549.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Liu L, Yang Y, He Y, Zhang Y, Wang M, et al. A school-based study of irritable bowel syndrome in medical students in Beijing, china: prevalence and some related factors. Gastroenterol Res Pract (2014) 2014:124261. 10.1155/2014/124261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bratkovic T, Rogelj B. Biology and applications of small nucleolar RNAs.Cell Mol Life Sci (2011) 68(23):3843–51. 10.1007/s00018-011-0762-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet (2011) 12(12):861–74. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 43.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer (2012) 12(2):84–8. 10.1038/nrc3195 [DOI] [PubMed] [Google Scholar]
- 44.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell (1996) 85(7):1077–88. 10.1016/S0092-8674(00)81308-2 [DOI] [PubMed] [Google Scholar]
- 45.Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol (2004) 24(13):5797–807. 10.1128/MCB.24.13.5797-5807.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su H, Xu T, Ganapathy S, Shadfan M, Long M, Huang TH, et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene (2014) 33(11):1348–58. 10.1038/onc.2013.89 [DOI] [PubMed] [Google Scholar]
- 47.Wells JM, Rossi O, Meijerink M, van BP. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A (2011) 108(Suppl 1):4607–14. 10.1073/pnas.1000092107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molenaar D, Bringel F, Schuren FH, de Vos WM, Siezen RJ, Kleerebezem M. Exploring Lactobacillus plantarum genome diversity by using microarrays. J Bacteriol (2005) 187(17):6119–27. 10.1128/JB.187.17.6119-6127.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.