Abstract
In Liberibacter asiaticus, PrbP is an important transcriptional accessory protein that was found to regulate gene expression through interactions with the RNA polymerase β-subunit and a specific sequence on the promoter region. It was found that inactivation of PrbP, using the inhibitor tolfenamic acid, resulted in a significant decrease in the overall transcriptional activity of L. asiaticus, and the suppression of L. asiaticus infection in HLB symptomatic citrus seedlings. The molecular interactions between PrbP and tolfenamic acid, however, were yet to be elucidated. In this study, we modeled the structure of PrbP and identified a ligand binding pocket, TaP, located at the interface of the predicted RNA polymerase interaction domain (N-terminus) and the DNA binding domain (C-terminus). The molecular interactions of PrbP with tolfenamic acid were predicted using in silico docking. Site-directed mutagenesis of specific amino acids was followed by electrophoresis mobility shift assays and in vitro transcription assays, where residues N107, G109, and E148 were identified as the primary amino acids involved in interactions with tolfenamic acid. These results provide insight into the binding mechanism of PrbP to a small inhibitory molecule, and a starting scaffold for the identification and development of therapeutics targeting PrbP and other homologs in the CarD_CdnL_TRCF family.
Keywords: transcriptional accessory protein, Liberibacter asiaticus, tolfenamic acid, antimicrobial, citrus, binding pocket
Introduction
Huanglongbing (HLB) or citrus greening, is a devastating disease threatening the citrus industry in Asia, Africa, and America (Graça, 1991; da Graça and Korsten, 2004; Bové, 2006; Gottwald, 2010; Wang and Trivedi, 2013). The causative agent is the phloem-limited bacteria Liberibacter asiaticus, L. africanus, and L. americanus (Bové, 2006; Gottwald, 2010; Wang and Trivedi, 2013). Although, it was first documented in Asia over a century ago (Graça, 1991; da Graça and Korsten, 2004; Bové, 2006), HLB was not widely recognized until its emergence in the United States between 2004 and 2005 (Bové, 2006). In Florida, the causing agent of HLB has been identified as L. asiaticus (Sagaram et al., 2009; Tyler et al., 2009). Despite efforts of the research community, the laboratory culture conditions of L. asiaticus remain elusive (Davis et al., 2008; Sechler et al., 2009; Parker et al., 2014), which has severely impeded the understanding of the physiology of L. asiaticus, and the development of an effective treatment for HLB.
In 2009, the first complete genome sequence of L. asiaticus was published (Duan et al., 2009). L. asiaticus has a relatively small genome (1.23 Mbps) that lacks genes encoding traditional pathogenicity determinants (Pagliai et al., 2014). Another salient feature revealed by genomic analysis is that only 2% of the L. asiaticus genome is predicted to encode transcription factors (Pagliai et al., 2014), significantly lower than many other common model organisms (e.g., 6.4% in Escherichia coli, 9.4% in Pseudomonas aeruginosa, 6% in Sinorhizobium meliloti; Stover et al., 2000; Pagliai et al., 2014). Interestingly, half of the predicted transcription factors in L. asiaticus were up-regulated in planta when compared to their expression in the psyllid vector (Yan et al., 2013). These data suggest that L. asiaticus may be adapted to each of its hosts (psyllid and citrus) by sensing specific host-derived signals which are sensed by specific transcription factors. These observations led to our hypothesis that the inactivation of critical transcriptional regulators in L. asiaticus may cause pleiotropic effects that result in compromised viability or reduced persistence of the pathogen during citrus infection.
PrbP is a member of CarD_CdnL_TRCF family, a distinct group of proteins that regulate the transcription machinery through interactions with the RNA polymerase (Padmanabhan, 2001; Penalver-Mellado et al., 2006; Stallings et al., 2009; Garcia-Moreno et al., 2010; Weiss et al., 2012; Gallego-García et al., 2014; Bernal-Bernal et al., 2015; Gardner et al., 2016). Myxococcus xanthus encodes for two members of this family, carD and cdnL. Myxococcus CarD, in complex with the zinc-binding protein CarG, was found to regulate various processes including oxidative stress response (induced by blue light) and vegetative growth under starvation conditions (Nicolas et al., 1994; Galbis-Martinez, 2004; Penalver-Mellado et al., 2006; Galbis-Martinez et al., 2008; Elías-Arnanz et al., 2010; Abellón-Ruiz et al., 2014). CdnL is a smaller protein (164 aa) that is essential for M. xanthus viability, and is homologous to the Myxococcus CarD N-terminal domain (Garcia-Moreno et al., 2010; Gallego-García et al., 2014). In Mycobacterium, CarD (a sequence homolog of M. xanthus CdnL) was also found to be essential for viability (Stallings et al., 2009), as well as a critical component for antibiotic resistance and multiple stress responses (Stallings et al., 2009; Weiss et al., 2012; Garner et al., 2014). CarD is also a critical component in pathogenesis and persistence during M. tuberculosis infection in mice (Stallings et al., 2009; Weiss et al., 2012; Garner et al., 2014). Furthermore, Mycobacterium CarD plays a general role in global regulation by binding non-specifically to the promoter region of hundreds of genes, and stabilizing the RNA polymerase open complex (Stallings et al., 2009; Gulten and Sacchettini, 2013; Landick et al., 2014; Rammohan et al., 2015).
Contrary to other PrbP homologs, in L. asiaticus, PrbP binds specifically to the promoter regions of the rplK gene, and the 16S rRNA gene (Gulten and Sacchettini, 2013; Landick et al., 2014; Gardner et al., 2016). Using high-throughput screening of chemical libraries, we identified a small molecule, tolfenamic acid, that binds to PrbP (Figure 1A). Tolfenamic acid is a generic NSAID drug that is used to treat migraines in the UK, Latin America, and European countries (Hakkarainen et al., 1979; Hansen, 1994). In vitro, tolfenamic acid can effectively reduce PrbP interactions with the promoter region of the rplK gene (Gardner et al., 2016). In vivo tests of this potential PrbP inhibitor, using the surrogate strain L. crescens, suggest PrbP interactions with DNA may be physiologically important, as the addition of tolfenamic acid in culture was able to inhibit L. crescens growth (Gardner et al., 2016). Foliar application of tolfenamic acid on HLB-symptomatic leaves and sweet orange (Citrus sinensis) seedlings significantly decreased the overall transcriptional activity of L. asiaticus, and inhibited the infection progression (Gardner et al., 2016). These findings support that tolfenamic acid could be a promising antimicrobial targeting PrbP for therapeutic treatment of HLB, however, the molecular interactions between PrbP and tolfenamic acid are yet to be elucidated.
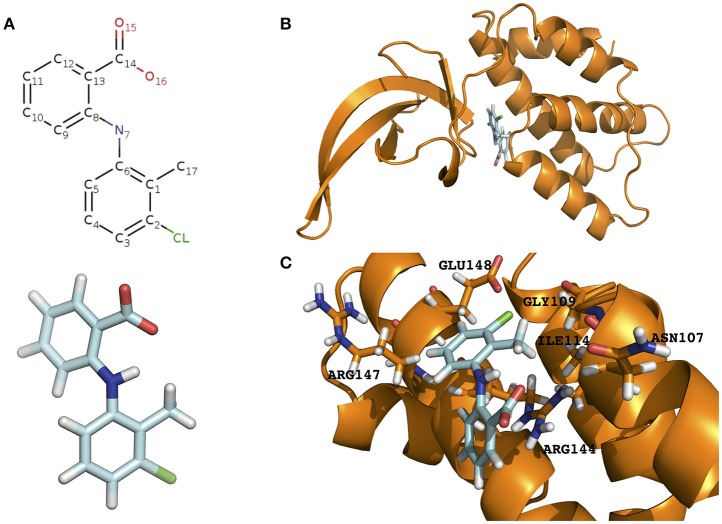
Figure 1.
Prediction of the tolfenamic acid binding pocket in the model PrbP-MI and the amino acids involved in PrbP/ligand interactions. (A) Structure of tolfenamic acid (PDB#TLF). (B) The PrbP-MI model using T. thermophilus CarD co-crystallized with RNA polymerase β-1 lobe (PDB# 4XAX, Bae et al., 2015) as template. Tolfenamic acid is shown docked in TaP located at the interface of RID and CTD. (C) Close-up view of the predicted interacting amino acids in PrbP-MI model. Orange, cartoon representation of the PrbP-MI; blue sticks, structure of tolfenamic acid; orange sticks, predicted interacting amino acids in PrbP.
In this study, we focused on the identification and characterization of the tolfenamic acid binding pocket (TaP, Tolfenamic acid Pocket) in PrbP. A structural model of PrbP was constructed based on crystalized homologs, and in silico docking was used to predict the amino acids that mediate interactions with tolfenamic acid. We then used electrophoresis mobility shift assays (EMSA) and in vitro transcription assays to validate the in silico predictions. In summary, this study provides insights into the inhibition mechanism for PrbP function that could be used to develop therapeutics against members of the CarD_CdnL_TRCF protein family.
Materials and methods
Bioinformatics
The PrbP sequence was retrieved from the NCBI protein database (ACT56890). The structural modeling was performed under automated mode using SWISS-MODEL (Grosdidier et al., 2011). The results of the modeling are provided on Supplementary Table 1. The 3D structure of tolfenamic acid was extracted from ZINC database (ZINC00002188). The in silico docking was performed using SwissDock (Grosdidier et al., 2011) with the following parameters: PASSIVEFLEXIBILITYDISTANCE: 0.0, WANTEDCONFS: 5,000, NBFACTSEVAL: 5,000, NBSEEDS: 250, SDSTEPS: 100, ABNRSTEPS: 250, CLUSTERINGRADIUS: 2.0, MAXCLUSTERSIZE: 8. Prediction of the ligand interactions with PrbP was done by the Molecular Docking server (Bikadi and Hazai, 2009) using default parameters. The docking hits obtained from SwissDock were manually examined and the ligand orientation that matched to the most favorable prediction of ligand/protein interactions predicted in Molecular Docking server was chosen.
To perform multiple sequence alignments, PrbP homolog sequences were retrieved from the NCBI protein database, and alignments were performed using Clustal Omega (Sievers et al., 2011) with default parameters. To perform structural alignments with PrbP homologs that lack 3D-structures, the amino acid sequences were obtained from NCBI and modeled as described above. All available 3D-structures were obtained from Protein Data Bank (Berman et al., 2000). PyMol was used to visualize data and perform structural alignments.
Strains and DNA manipulation
Escherichia coli DH5α and E. coli JM110 cells were used to maintain and replicate all plasmids. E. coli BL21 (DE3) (Novagen) was used for over-expression and further purification of recombinant proteins. The cells were grown in Luria-Bertani (LB) medium, at 37°C, in a rotary shaker (250 rpm). The culture media was supplemented with ampicillin (100 μg/ml), kanamycin (30 μg/ml) and/or chloramphenicol (34 μg/ml) when necessary. All antibiotics and chemicals were purchased from Sigma-Aldrich.
Chromosomal DNA was isolated with Qiagen DNAeasy Kit and plasmid extractions were performed with QIAprep Spin Miniprep Kit (Qiagen). Standard molecular protocols described in Molecular Cloning (Sambrook et al., 2001) were used to perform polymerase chain reactions (PCR), restrictive enzyme digestion, construction of recombinant DNA molecules, and cell transformations. Q5 high fidelity DNA Polymerase (NEB) was used for PCR reactions. PrbP site-directed mutagenesis was performed using the QuikChange Site-directed Mutagenesis kit (Agilent Technologies) following the manufacturer's protocol. All predicted residues were mutated to alanine using the plasmid p15TV-L carrying prbP as the template.
L. asiaticus RNA polymerase subunits β and β′ were cloned into plasmid pACYCDuet-1. Subunits α and ω were cloned into plasmid pRSFDuet-1. Sigma factor 70 was cloned into plasmid p15TV-L. The N-terminal 6X His-tag was fused to subunits β′, ω and sigma factor 70. Reproduction and manipulation of plasmids was done using E. coli DH5α or, in the case of plasmid pACYCDuet-1 and its derivatives, in E. coli JM110. All of the plasmids containing subunits of the RNA polymerase were co-transformed into E. coli BL21 (DE3), and the proteins were simultaneously over-expressed with IPTG for in vivo reconstruction of the L. asiaticus RNA polymerase holoenzyme.
All cloned DNA fragments described herein were verified by DNA sequencing. All of the strains, plasmids and primers are listed in Tables 1, 2.
Table 1.
Strains and Plasmids used in this study.
| Name | Genotype/Description | Origin/Reference |
|---|---|---|
| BACTERIAL STRAINS | ||
| Escherichia coli DH5α | F−Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (, ) phoA supE44 λ– thi-1 gyrA96 relA1 | Invitrogen |
| E. coli BL21 (DE3) | F−ompT hsdSB() gal dcm (DE3) pRARE | Novagen |
| E. coli JM110 | F− traD36 lacIqΔ(lacZ)M15proA+B+/rpsL(StrR) thr leu thi laY galK galT ara fhuA dam dcm glnV44 Δ(lac-proAB) | ATCC |
| EEPrbP | BL21 (DE3) carrying p15TV-prbP. Ampr | Gardner et al., 2016 |
| EEPrbP-N107A | BL21 (DE3) carrying p15TV-prbP(N107A). Ampr | This work |
| EEPrbP-G109A | BL21 (DE3) carrying p15TV-prbP(G109A). Ampr | This work |
| EEPrbP-I114A | BL21 (DE3) carrying p15TV-prbP(I114A). Ampr | This work |
| EEPrbP-R144A | BL21 (DE3) carrying p15TV-prbP(R144A). Ampr | This work |
| EEPrbP-R147A | BL21 (DE3) carrying p15TV-prbP(R147A). Ampr | This work |
| EEPrbP-E148A | BL21 (DE3) carrying p15TV-prbP(E148A). Ampr | This work |
| EERNAPD | BL21 (DE3) carrying p15TV-rpoD, pACYCDuet-rpoB-rpoC, pRSFDuet-rpoZ-rpoA. Ampr. Cmr. Kanr | This work |
| ECPRPLK | DH5α carrying pMiniT-PrplK. Ampr | This work |
| PLASMIDS | ||
| p15TV-L | Expression vector for purification of proteins by nickel affinity chromatography. Ampr | Cheryl Arrowsmith, (Addgene plasmid # 26093) |
| p15TV-prbP | prbP gene from L. asiaticus cloned in the BseRI site of p15TV-L. Ampr | Gardner et al., 2016 |
| p15TV-prbP(N107A) | p15TV-prbP with mutation for N107A. Ampr | This work |
| p15TV-prbP(G109A) | p15TV-prbP with mutation for G109A. Ampr | This work |
| p15TV-prbP(I114A) | p15TV-prbP with mutation for I114A. Ampr | This work |
| p15TV-prbP(R144A) | p15TV-prbP with mutation for R144A. Ampr | This work |
| p15TV-prbP(R147A) | p15TV-prbP with mutation for R147A. Ampr | This work |
| p15TV-prbP(E148A) | p15TV-prbP with mutation for E148A. Ampr | This work |
| p15TV-rpoD | rpoD gene from L. asiaticus cloned in the NdeI-BamHI site of p15TV-L. Ampr | This work |
| pACYCDuet-1 | Expression vector for co-purification of protein complexes by nickel affinity chromatography. Cmr | Novagen |
| pRSFDuet-1 | Expression vector for co-purification of protein complexes by nickel affinity chromatography. Kanr | Novagen |
| pACYCDuet-rpoB | rpoB gene from L. asiaticus cloned in the FseI-PvuI site of pACYCDuet-1 plasmid. Cmr | This work |
| pRSFDuet-rpoZ | rpoZ gene from L. asiaticus cloned in the SalI-NotI site of pRSFDuet-1 plasmid. Kanr | This work |
| pACYCDuet-rpoB-rpoC | rpoC and rpoB genes from L. asiaticus cloned in the PstI-NotI and FseI-PvuI sites of pACYCDuet-1 plasmid respectively. Cmr | This work |
| pRSFDuet-rpoZ-rpoA | rpoA and rpoZ genes from L. asiaticus cloned in the EcoRV-KpnI and SalI-NotI sites of pRSFDuet-1 plasmid respectively. Kanr | This work |
| pMiniT | Cloning vector for PCR amplicon | NEB |
| pMiniT-PrplK | Promoter region (−230 to +54 bp) of rplK gene cloned into insertion site of pMiniT. Ampr | This work |
Table 2.
Primers used in this study.
| Primer | Sequence (5′-3′) |
|---|---|
| EMSA | |
| EMSA_CLIB_00130_Ext_Fw | CTGTTTTCTTCGAGGTTGGTG |
| EMSA_CLIB_00130_Ext_Rv | CCGCATTAAACGCCTTACAA |
| EMSA_CLIB_00130_Fw | CTGATGGTCCGTTTGCTTCT |
| EMSA_CLIB_00130_Rv_Bio | TGCAGAACCCGACTCTATCTG |
| SITE-DIRECTED MUTAGENESIS AND PROTEIN | |
| CLIBASIA_01510_LIC-Fw | TTGTATTTCCAGGGCATGACATTCCAACAGAAAAGAGATG |
| CLIBASIA_01510_LIC-Rv | CAAGCTTCGTCATCA CTATGCGGCTTTATCTTGATTTTC |
| CLIBASIA_01510_Ext-Fw | GAGTGTGCGTTTGTTTGAAAAG |
| CLIBASIA_01510_Ext-Rv | CCCACGCGATCTTATCTGAC |
| CLIB_01510_N107A_FP | CAAGAATACGATGCCAAGATTGCCTCTGGAGACCTAATTGCTATA |
| CLIB_01510_N107A_RP | TATAGCAATTAGGTCTCCAGAGGCAATCTTGGCATCGTATTCTTG |
| CLIB_01510_G109A_FP | TACGATGCCAAGATTAACTCTGCAGACCTAATTGCTATAGCGGAA |
| CLIB_01510_G109A_RP | TTCCGCTATAGCAATTAGGTCTGCAGAGTTAATCTTGGCATCGTA |
| CLIB_01510_I114A_FP | AACTCTGGAGACCTAATTGCTGCAGCGGAAGTTGTCCGTGACTTA |
| CLIB_01510_I114A_RP | TAAGTCACGGACAACTTCCGCTGCAGCAATTAGGTCTCCAGAGTT |
| CLIB_01510_R144A_FP | GCTATATGAATCCGCTCTCAATGCCATGGTCAGAGAAATCGCTGCT |
| CLIB_01510_R144A_RP | AGCAGCGATTTCTCTGACCATGGCATTGAGAGCGGATTCATATAGC |
| CLIB_01510_R147A_FP | ATCCGCTCTCAATCGCATGGTCGCAGAAATCGCTGCTGTAAATAGT |
| CLIB_01510_R147A_RP | ACTATTTACAGCAGCGATTTCTGCGACCATGCGATTGAGAGCGGAT |
| CLIB_01510_E148A_FP | CGCTCTCAATCGCATGGTCAGAGCAATCGCTGCTGTAAATAGTATC |
| CLIB_01510_E148A_RP | GATACTATTTACAGCAGCGATTGCTCTGACCATGCGATTGAGAGCG |
| CLIB_rpoC_Fw_ext | AGCATAACTATTCTGCCAAGACA |
| CLIB_rpoC_Rv_ext | CGGTGAACCATTTGATCGGC |
| CLIB_rpoB_Fw_ext | CAAGGATGAGCAACGGGAGAAGGA |
| CLIB_rpoB_Rv_ext | AGGAATGGGGTGTTTCTGCT |
| CLIB_rpoA_Fw_ext | ACAATCGGACGCAACTCCTT |
| CLIB_rpoA_Rv_ext | GCGACGGATGATTGTTCAGC |
| CLIB_rpoZ_Fw_ext | CATAGAGGTTGGCACCGTCA |
| CLIB_rpoZ_Rv_ext | TCAACATACATCGAATGCCCCA |
| CLIB_rpoC_PstI_Fw | TCGCTGCAGATGCAACAAGAGGTCATGAG |
| CLIB_rpoC_NotI_Rv | ACTGCGGCCGCTTACTCCGCTATACTCCC |
| CLIB_rpoB_FseI_Fw | GCTGGCCGGCCATGGCAAAAGGCGTTGTGTT |
| CLIB_rpoB_PvuI_Rv | TGGCGATCGTTACTTTAATTCACATTTAT |
| CLIB_rpoA_EcoRV_Fw | ATGCTGATATCATGATCCAAAAAAATTGGCAAG |
| CLIB_rpoA_KpnI_Rv | TCGGGTACCTTAGCACTTATCTTCATATT |
| CLIB_rpoZ_SalI_Fw | TGTGTCGACATGGCACGTACTACTGTAGA |
| CLIB_rpoZ_NotI_Rv | ACTGCGGCCGCTCAATCATCTCTCTTATCAG |
| CLIBASIA_00870_NdeI_Fw | GGCCATATGATGACAATAGGAAA |
| CLIBASIA_00870_BamHI_Rv | GGATCCCTAACCATCTAAAAAACTC |
| SEQUENCING | |
| T7 | TAATACGACTCACTATAGGG |
| T7 term | GCTAGTTATTGCTCAGCGG |
| pMiniT_Fw | ACCTGCCAACCAAAGCGAGAAC |
| pMiniT_Rv | TCAGGGTTATTGTCTCATGAGCG |
| ACYCDuetUP1 | GGATCTCGACGCTCTCCCT |
| DuetUP2 | TTGTACACGGCCGCATAAT |
| DuetDOWN1 | GATTATGCGGCCGTGTACAA |
Protein purification
Purification of PrbP and its mutants was performed as previously described (Gardner et al., 2016). The 6X His-tagged fusion proteins were over-expressed in E. coli BL21 (DE3). Cells were grown in LB broth at 37°C, to OD600 = 0.6 and gene expression was induced with 0.5 mM isopropyl-thio-β-D-galactopyranoside (IPTG). Upon induction, the cells were incubated at 17°C for 16 h. The cells were harvested by centrifugation and re-suspended in binding buffer (500 mM NaCl, 5% glycerol, 50 mM HEPES, 5 mM imidazole, pH 7.5). The buffer was amended with Pierce EDTA-free protease inhibitor (Thermo Fisher Scientific). Tris (2-carboxyethyl) phosphine hydrochloride (0.5 mM) was added to the cells immediately before lysing. Cells were lysed by passing through a French pressure cell. The lysates were centrifuged 30 min at 17,000 × g at 4°C and the supernatant was applied to a metal chelate affinity column charged with nickel (His60 Ni Superflow Resin, Clontech). The column was washed extensively with binding buffer containing 25 mM imidazole, and the proteins were eluted from the column in elution buffer (binding buffer with 250 mM imidazole). The purified proteins were dialyzed against 10 mM HEPES (pH 7.5), 500 mM NaCl, 2.5% glycerol, 0.5 mM TCEP, then aliquoted and stored at −80°C. The 6X His-tag of PrbP and its variants was not removed for further experiments.
Purification of the L. asiaticus RNAP holoenzyme was performed using a similar protocol, however, the buffers were modified as follow: The binding, wash and elution buffers contained 500 mM NaCl, 5% glycerol, 50 mM Tris-HCl (pH 8) with 5, 15, and 250 mM imidazole, respectively. The purified protein complex was dialyzed against 10 mM Tris-HCl (pH 8), 250 mM NaCl, 50% glycerol, 0.1 mM EDTA, 0.5 mM TCEP, then aliquoted and stored at −80°C. The fused 6X His-tag was not removed for further experiments.
Electrophoresis mobility shift assays
Electrophoresis mobility shift assays (EMSA) were carried out using the PrbP wild type protein (WT) and PrbP mutants. A fragment of the rplK promoter region was amplified by PCR using pre-labeled 5′-biotin primers, and the biotin labeled product was used for the binding target as previously described (Gardner et al., 2016). The electrophoresis mobility shift assay reaction contained 1 ng of 5′-Biotin labeled DNA probe, 10 mM HEPES (pH 7.5), 250 mM NaCl, 5% glycerol, 12.5 ng/μl of non-specific competitor DNA Poly(dI-dC), purified PrbP WT or mutant PrbP protein (0–7 μM), and tolfenamic acid (0–0.5 mM), as indicated. Following incubation at 37°C for 20 min, the samples were analyzed by electrophoresis using 6% acrylamide-bisacrylamide non-denaturing gels, in ice-cold 0.5x Tris-borate EDTA buffer (TBE), pH 8.3. The DNA was then transferred from the polyacrylamide gel to a Hybond-N+ membrane (GE Healthcare) by electro-blotting at 250 mA for 45 min, using a semi-dry transfer blot (Fisher Scientific). The transferred DNA was UV-crosslinked and the biotin-labeled DNA was detected using the Phototope-Star Detection Kit (NEB). Membranes were exposed to Kodak X-ray films. Vehicle controls were included in all assays.
In vitro transcription run-off assays
The PrbP proteins used for in vitro transcription assays were purified and stored as described above. The recombinant plasmid pMiniT-PrplK was linearized using the restriction enzyme NdeI. All proteins were diluted to the working concentration with transcription buffer (40 mM Tris-HCl, pH 8.0, 50 mM KCl, 10 mM MgCl2,0.1 mM EDTA). Each 20 μl reaction contained template DNA (5 nM), purified PrbP protein (0–5 μM), and tolfenamic acid (0–0.5 mM), in 1x transcription buffer. The mixtures were pre-incubated for 10 min at 37°C prior to adding the partially purified RNA polymerase. A second incubation was then performed at 37°C for 5 min. The transcription reaction was then initiated by the addition of NTPs (2 mM each of ATP, GTP, and CTP, 1.5 mM of UTP, and 0.5 mM biotin-11-UTP). The reactions were terminated by adding 10 mM EDTA after 30 min of incubation at 37°C. The transcripts were purified, concentrated by ethanol precipitation, and analyzed using 6% acrylamide-bisacrylamide, 7M urea gels, in ice-cold 0.5x TBE buffer, at 100 V for 2.5 h. Transcripts were transferred from the gel to a Hybond-N+ membrane (GE Healthcare) by electro-blotting at 380 mA for 40 min in a semi-dry transfer blot (Fisher Scientific). The transferred transcripts were UV-crosslinked and detected using the Phototope-Star Detection Kit (New England Biolabs). The membranes were exposed to Kodak X-ray films to visualize the transcription products. A biotinylated sRNA Ladder (Kerafast) was used as a molecular weight marker.
Results
Model of tolfenamic acid binding in PrbP
The structural model of PrbP was generated using SWISS-MODEL server in automated mode. Several templates were identified with coverages >80%, however, all models had low quality scores (GQME <0.6 and negative Z-scores). Although the models were not highly reliable, we have found in other transcription factors that these models can still be utilized as the starting point for in silico predictions if followed by biochemical validations (Pagliai et al., 2010; Blancato et al., 2016). Using size exclusion chromatography, it was determined that L. asiaticus PrbP is a monomer in solution, with an estimated molecular weight of 24.6 kDa (Supplementary Figure 1A). Since several of the templates retrieved were dimers, those predictions were excluded from further analyses. Among the remaining templates, three models based on the structures of CarD from Thermus (PDB# 4L5G, 4XAX, and 4XLR) were found. These CarD structures showed different conformations of the RNA polymerase interacting domain (RID) and DNA binding domain (CTD) (Srivastava et al., 2013; Bae et al., 2015). The distribution of the two functional domains vary from relaxed in the apo structure (PDB#4L5G), to intermediate when in complex with Taq RNAP β-1 lobe (PDB#4XAX), and to constrained in the transcription initiation complex (PDB#4XLR). Since each conformation has been linked to mechanisms of interaction of Thermus CarD with DNA and the RNA polymerase (Bae et al., 2015), the models derived from these structures were selected for further studies. The models were named PrbP-MR (using the 4L5G template), PrbP-MI (using the 4XAX template), and PrbP-MC (using the 4XLR template; Figure 1B, Supplementary Figure 2). The modeling input sequence and statistics of the models are listed in Supplementary Table 1.
The docking of tolfenamic acid into PrbP-MI was performed using SwissDock in automated mode, as described in Section Materials and Methods. The best scores were obtained with a pocket formed in the interface of the RID and CTD domains (Supplementary Table 2). In the identified pocket, tolfenamic acid was predicted to interact with residues N107, G109, I114, R144, R147, and E148 (Figure 1C), with several possible orientations. Given the large number of possible docking orientations with no significant difference in energy (<1 kcal/mol difference), the Molecular Docking Server interface was used to narrow down and help determine the docking orientation of tolfenamic acid in the pocket. The most favorable orientation (Figure 1C) had a Gibbs free energy (ΔG) of −6.79 kcal/mol. Residue I114 was predicted to interact with tolfenamic acid atom C1 via hydrophobic interactions (3.4 Å). Residue G109 was predicted to interact with atom Cl1 via halogen bond (3.6 Å). Residue E148 was also predicted to interact with atom Cl1 by halogen bond (3.7 Å) as well as with atoms C6 (2.9 Å). Residue R144 was predicted to interact with atoms C1-C2-C3-C7-C8-C9-C10-C12 (2.1–3.8 Å, respectively). The measurements of distances between the atoms in tolfenamic acid and the amino acids in PrbP are shown in Table 3 and Supplementary Figure 2.
Table 3.
In silico prediction of the interactions between tolfenamic acid and residues within the TaP pocket in PrbP.
| Amino acid | Predicted interacting atom(s) on TA | Predicted interaction type(s) | Measurements of distance [Å] |
|---|---|---|---|
| N107 | NP* | NP* | 3.3 |
| G109 | Cl1 | Halogen | 3.6 |
| I114 | C1 | Hydrophobic | 3.4 |
| R144 | C1-C2-C3-C7-C8-C9-C10-C12 | Other | 2.1-3.8 |
| R147 | NP* | NP* | 2.8 |
| E148 | Cl1/C6 | Halogen/other | 3.7/3.9 |
Amino acids within 4 Å radius but the type of interaction has not predicted.
To verify which amino acids in the predicted TaP pocket are required for interactions with tolfenamic acid, six PrbP mutants were constructed by site-directed mutagenesis, where each of the predicted residues was replaced with alanine. The PrbP WT and mutant constructs were purified with up to 90% purity (Supplementary Figure 1B).
Mutations in the TaP decrease the inhibitory effect of tolfenamic acid on PrbP/DNA interactions
The effect of mutations in the TaP pocket, on the ability for PrbP to bind DNA, were first assessed by EMSA, using the known PrbP binding site in the promoter region of rplK (PrplK) as a probe (Gardner et al., 2016). Of the six mutants tested, no significant difference in DNA binding activity was observed, when compared to the PrbP WT (Supplementary Figure 3). The effect of tolfenamic acid was then examined with each PrbP mutant/PrplK complex and compared to the PrbP WT. Complete disruption of the PrbP WT/PrplK complex was consistently observed with 350 μM tolfenamic acid, which corresponds to a protein to ligand ratio of 1:100 (Figure 2). Mutations in I114, R144, and R147 showed a similar pattern to the WT, with disruption of the complex also occurring in presence of 350 μM tolfenamic acid (Supplementary Figure 4). Conversely, the presence of 350 μM tolfenamic acid did not completely disrupt the Protein/DNA complex for mutants N107A, G109A, and E148A. The complete disruption of the complex formed with these mutants was only observed at higher concentrations (500 μM) of tolfenamic acid, which corresponds to a protein to ligand ratio of 1:143 (Figure 2). These results indicate that residues N107, G109, and E148 of the predicted TaP pocket, are directly involved in the binding of tolfenamic acid to PrbP.
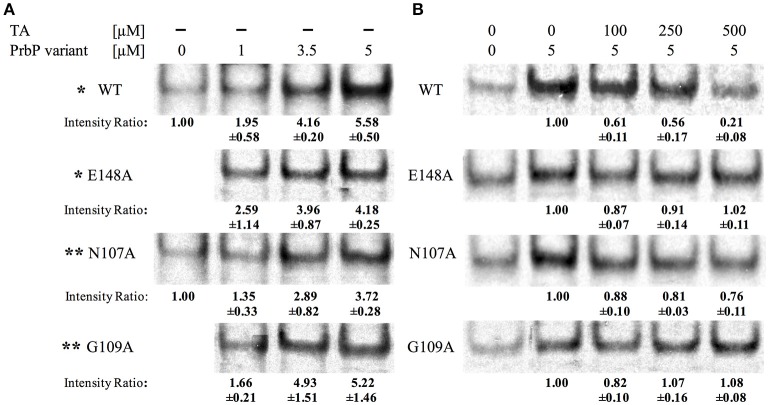
Figure 2.
Mutations N107A, G109A, and E148A in TaP decreases the inhibitory effect of tolfenamic acid on PrbP/DNA interactions. The PrplK probe and 3.5 μM of each of the PrbP mutants (N107A, G109A, and E148A, as indicated in the top of each panel) were incubated with increasing concentrations (0–500 μM) of tolfenamic acid as indicated. TA, tolfenamic acid; First lane of each panel has no protein added. Ratio was calculated by the band intensity of protein: DNA complex and total DNA. Quantification was based on observations from at least 2 replicates.
Effect of mutations in TaP on PrbP interactions with the RNA polymerase
In our previous report, we showed that PrbP binds specifically to the promoter region of rplK and interacts with the β-subunit of the RNA polymerase. Interestingly, the addition of tolfenamic acid significantly decreased expression of rplK and 16S ribosomal genes in vivo, suggesting a role as a transcriptional activator (Gardner et al., 2016). However, when tested in a two-hybrid system, tolfenamic acid only slightly decreased the interaction between PrbP and RpoB. These results suggest that tolfenamic acid, may inhibit PrbP by disrupting or destabilizing DNA binding.
An in vitro transcription assay was used to examine the effect of tolfenamic acid on PrbP, and its interactions with the RNA polymerase during transcription initiation. All five components of the L. asiaticus RNA polymerase (αββ′ω), and the sigma 70 factor, were cloned into a E. coli co-expression system, and purified by affinity chromatography (Supplementary Figure 5). The partially purified RNA polymerase was used for in vitro transcription run-off assays, with plasmid DNA carrying PrplK as a template. When added to the assay reaction, PrbP significantly increased the transcript concentration in a dose dependent manner, with the highest increase in transcription observed with 5 μM PrbP (Figure 3A). These results confirmed that PrbP WT acts as a transcriptional activator for rplK. These findings are consistent with previous studies on CarD from Mycobacterium, where CarD was found to increase transcriptional activity of the Mycobacterium RNAP, while it had no effect on transcription when added to the E. coli RNAP (Davis et al., 2015; Rammohan et al., 2015). The transcription of PrplK was then examined in presence of the TaP mutants, where mutants E148A, N107A, and G109A were all found to behave similar to the WT (Figure 3A).
Figure 3.
Tolfenamic acid affect PrbP interactions with the RNA polymerase. (A) PrbP WT and TaP mutations N107A, G109A, and E148A act as transcriptional activators of rplK. Increasing concentrations (0–5 μM) of PrbP WT and mutants were added as indicated; * and ** indicate that reactions ran on the same gel and share the same control. Ratio was calculated by band intensity normalized to negative control without PrbP or mutants. (B) Effect of tolfenamic acid on PrbP WT or mutants (N107A, G109A, and E148A) interactions with the RNA polymerase. PrbP WT, mutants and increasing concentrations of tolfenamic acid (0–500 μM) were added as indicated. Ratio was calculated by band intensity normalized to the control without tolfenamic acid. Quantification was based on observations from at least 2 replicates.
In vitro transcription was also used to examine the effect of increasing concentrations of tolfenamic acid, on the transcription of rplK. The addition of 250 μM tolfenamic acid was found to decrease rplK transcription by 44% with the PrbP WT, and at higher concentrations (500 μM tolfenamic acid) a 79% decrease in transcription was observed (Figure 3B). No significant decrease in rplK transcription was observed with mutants G109A and E148A. With the N107A mutant, the addition of tolfenamic acid was also found to decrease the transcription of rplK, but to a much lesser extent (19 and 24% with 250 and 500 μM tolfenamic acid, respectively; Figure 3B). The reduced sensitivity to tolfenamic acid observed with PrbP mutants N107A, G109A, and E148A supports an important role of these residues for a ligand inhibition of the PrbP activity. These results also suggest that residues G109 and E148 are more significant for interactions with tolfenamic acid than residue N107.
Residues in the TaP are conserved in the CarD_CdnL_TRCF family
To explore the potential significance of TaP in the CarD_CdnL_TRCF family, multiple linear sequence alignments and structural alignments were performed. In this analysis, we included the sequence of each PrbP homolog with structural data available (Thermus, Mycobacterium, and Myxococcus) as well as members of the Rhizobiaceae family. It was found that residues G109 and E148 are conserved in all members of the CarD_CdnL_TRCF family that were analyzed, while N107 is conserved in the Rhizobiaceae family (Figures 4A,B). The conservation of these residues in the ligand binding pocket suggests that they may be functionally relevant for members of the CarD_CdnL_TRCF family, and that interactions with these proteins may be modulated by an unknown native ligand.
Figure 4.
Sequence and structural alignments of PrbP and CarD_CdnL_TRCF family members. (A) Multiple sequence alignment of PrbP and PrbP homologs of the CarD_CdnL_TRCF family. Sequences were extracted from NCBI database and multiple alignments were performed using Clustal Omega. Black dots, 60% and up conserved; asterisk, completely conserved. (B) Structural alignment of PrbP-MI and PrbP homologs. The homologs from S. meliloti (pink), M. smegmatis (green), M. xanthus (yellow), were modeled using PDB# 4XAX as the template. The crystal structures used are from T. thermophilus (PDB# 4L5G, red, Srivastava et al., 2013), M. tuberculosis (PDB#4KBM, blue, Gulten and Sacchettini, 2013).
Discussion
CarD_CdnL_TRCF is an expanding family of proteins with an atypical DNA binding domain. Members of this family are widely distributed and have been identified in several human and plant pathogens, including Mycobacterium, Clostridium, Bacillus, and Liberibacter species. Initial studies in Myxococcus xanthus identified the role of this family as transcriptional regulators. Additional studies in Myxococcus, Mycobacterium, and Thermus species later revealed that members of this family are important RNA polymerase interacting proteins, that regulate transcription initiation (Stallings et al., 2009; Garcia-Moreno et al., 2010; Bae et al., 2015; Rammohan et al., 2015). In some genera, they are essential proteins (Stallings et al., 2009; Garcia-Moreno et al., 2010), suggesting their potential use as therapeutic targets for new antimicrobial compounds.
PrbP is a newly characterized member of the CarD_CdnL_TRCF family from L. asiaticus (Gardner et al., 2016). In our previous study, we found that PrbP interacts with RNAP, however, contrary to previous reports of homologs, L. asiaticus PrbP binds specifically to the rplK promoter region, and the 16S ribosomal gene (Gulten and Sacchettini, 2013; Landick et al., 2014; Gardner et al., 2016). Tolfenamic acid is a small molecule that was found to disrupt PrbP/DNA interactions in vitro. It was also found to be an effective inhibitor of PrbP in vivo (using HLB-infected citrus seedlings), where the overall transcriptional activity of L. asiaticus was observed to decrease after treatment with tolfenamic acid.
To further examine the effect of tolfenamic acid on PrbP/RNAP interactions, as well as to identify the tolfenamic acid binding pocket in PrbP, we performed in vitro transcription assays. Purification of a bacterial RNA polymerase is usually accomplished by tagging one subunit of the polymerase, followed by affinity chromatography to pull-down the entire complex in the target bacteria (Murakami et al., 1997; China and Nagaraja, 2010; Svetlov and Artsimovitch, 2015). Due to the inability to culture L. asiaticus under standard laboratory conditions, an alternative purification strategy was employed. To this end, all five components of the L. asiaticus RNA polymerase (αββ′ω), and the sigma 70 factor, were cloned into a E. coli co-expression system, and purified (Supplementary Figure 5). In Mycobacterium, the RNAP/open promoter complexes are highly unstable, however, previous studies have shown that PrbP homologs can effectively stabilize mycobacterial RNAP/open promoter complexes, resulting in increased transcriptional activity. Conversely, the open promoter complexes formed by E. coli RNAP are highly stable, and transcriptional activity is unaffected by the presence of PrbP homologs (Davis et al., 2015; Rammohan et al., 2015). Based on these findings, the partially purified L. asiaticus RNAP was used to perform in vitro transcription assays, as any potential E. coli RNAP contaminants should not significantly affect the results of the assay. Similar to the results reported in Mycobacterium for CarD, the addition of PrbP increased the transcription of the rplK promoter by the L. asiaticus RNA polymerase. One observation of the inhibitory effect of tolfenamic acid is that higher concentrations (500 μM) did not completely abolish the activity of PrbP in vitro, however, lower concentrations (70 μM) of tolfenamic acid were previously found to inhibit L. crescens growth and affect the transcriptional activity of L. asiaticus in vivo (Gardner et al., 2016). We hypothesize that even a small decrease in PrbP activity could significantly affect cell viability due to the slow duplication rate of L. crescens (24 h, Fagen et al., 2014). It is also possible that the inhibitory effect of tolfenamic acid on PrbP would affect the expression of other genes that may be essential for growth.
To investigate the inhibition mechanism of PrbP, we tested the possibility that tolfenamic acid could block PrbP binding to RNAP using a E. coli two-hybrid system, and only observed a 15% decrease in the protein-protein interactions (Gardner et al., 2016). Since the low level of inhibition was not sufficient to explain the significant decrease in transcriptional activity observed in vivo (Gardner et al., 2016) and in vitro, the results suggest the mechanism of tolfenamic acid inhibition is unlikely to be the direct binding to the PrbP/RNAP interface. In this report, we used in silico structure modeling and docking to identify TaP as the tolfenamic acid binding pocket in PrbP. Interestingly, TaP is located at the interface of the RNAP and DNA binding domains. Recent literature on CarD from T. thermophiles, where it was suggested that the RID and CTD of CarD are flexible in solution, and that CarD only assumes its active conformation when the CTD is positioned to interact with the promoter DNA (Bae et al., 2015). Based on our findings, and the structural data available, we hypothesize that preventing the formation of the PrbP active conformation is the basis for the inhibition mechanism of tolfenamic acid.
Thermus CarD structures PDB# 4L5G (apo structure) (Srivastava et al., 2013), PDB# 4XAX (in complex with RNAP β-1 lobe) and PDB# 4XLR (with the transcription initiation complex) (Bae et al., 2015) showed that the CTD and RID functional domains of CarD assume different conformations depending on the environment (i.e., presence of RNAP components or transcription initiation complex). These conformations seem to be relaxed (PDB# 4L5G), intermediate (PDB# 4XAX) or constrained (PDB# 4XLR). Using these structures as templates, the structural modeling of PrbP showed that the TaP is only formed in the intermediate conformation (PrbP-MI). Using in silico docking, we tested if tolfenamic acid would bind to the same location if PrbP was modeled using the CarD apo structure from T. thermophilus (PDB# 4L5G) or the structure obtained from CarD crystallized with the transcription initiation complex (PDB# 4XLR). When PrbP was modeled to 4L5G, tolfenamic acid was positioned further away from the interacting amino acids N107, G109, and E148 (average distance 4.3–5.6 Å, Supplementary Figure 2C) when compared to the docking model based on PDB# 4XAX (average distance 3.3–3.9 Å, Supplementary Figure 2A), indicating that the relaxed distribution may not aid in tolfenamic acid binding. Structural alignment of the PrbP-MC model and the tolfenamic acid/PrbP-MI docking prediction, revealed that the tolfenamic acid molecule would clash with amino acids R144 and R147 (Supplementary Figure 2D). These in silico predictions indicate that PrbP may be unable to assume a constrained (“active”) conformation when tolfenamic acid binds to the predicted pocket, hence, inhibiting the transcription activation activity of PrbP.
All together, these in silico analyses suggest that the formation of TaP in PrbP may be dependent on the RID and CTD domain conformation. The conserved nature of the critical amino acids in TaP suggests that the identified binding pocket may also be a critical component for regulating the activity of members of the CarD_CdnL_TRCF family, through interactions with native small molecules. Previous reports on members of the CarD_CdnL_TRCF family have focused on the identification of residues that mediate binding to the promoter DNA, as well as those that mediate interactions with the β1-lobe of the RNAP. The effect of small molecules that modulate these interactions, however, has not been addressed. In L. asiaticus, the expression of prbP was found to remain stable following chemical inactivation of PrbP/DNA interactions, indicating that prbP expression is not auto-regulated at the level of transcription (Gardner et al., 2016). Based on the constitutive nature of PrbP in L. asiaticus, and the finding that tolfenamic acid binding inhibits the activity of PrbP, we hypothesize that the activity of PrbP is modulated by the binding of an unknown signaling molecule which may maintain PrbP in an “inactive” conformation until its activity is required, however, further studies are required to identify the native molecule.
In mycobacteria, the expression of carD is highly induced in response to DNA damage, oxidative stress, and starvation conditions (Stallings et al., 2009). Conversely, the rRNA levels in wild-type M. smegmatis are repressed during starvation and oxidative stress, however, the repression was abolished when CarD was depleted in vivo. Microarray and qRT-PCR experiments confirmed that CarD depletion resulted in transcriptional upregulation of genes encoding ribosomal proteins and translation factors (Stallings et al., 2009). These observations indicate that in mycobacteria, CarD is involved in stringent response. One well-known bacterial stringent control model is the DksA-(p)ppGpp in E. coli (Paul et al., 2004, 2005; Perederina et al., 2004; Potrykus and Cashel, 2008; Atkinson et al., 2011; Lennon et al., 2012; Ross et al., 2013, 2016), where ppGpp is a signaling molecule synthesized during amino acid starvation, that works in conjunction with DksA to regulate transcription. It is proposed that DksA forms a synergetic binding with ppGpp at the RNA polymerase secondary channel rim, while stabilizing ppGpp binding to a second site 60 Å away, resulting in a modified transcription initiation complex (Ross et al., 2013, 2016). In M. tuberculosis and M. smegmatis, which lack DksA, CarD is required for ppGpp to elicit a stringent response (Stallings et al., 2009). Although, investigation of CarD-ppGpp interaction is yet to be done, the fact that mycobacteria CarD can functionally complement an E. coli ΔdksA strain (Stallings et al., 2009) suggests that ppGpp may serve as a native ligand for some members of the CarD_CdnL_TRCF family. Similar to M. tuberculosis and M. smegmatis, L. asiaticus also lacks DksA, suggesting PrbP may be involved in stringent response as well. In L. asiaticus, the signaling molecule is unlikely to be ppGpp, as the enzymes required for its synthesis are absent in the genome, however, a similar regulatory mechanism may exist.
In summary, we identified a conserved ligand binding pocket (TaP), that is involved in modulating the DNA binding activity of PrbP in vitro, and may also serve as the binding site for a native signaling molecule to regulate PrbP activity in vivo. Taken together, these results indicate that members of the CarD_CdnL_TRCF family might be regulated through interactions with small molecules during stress response or during interactions with the host.
Author contributions
LP, FP, and GL designed and analyzed the experiments. LP, CLG, and FP performed experiments. CLG and CFG provided assistance in experiments, contributed in the analysis of results and preparation of the manuscript. LP and GL wrote the paper. GL conceived and coordinated the study.
Conflict of interest statement
A patent application has been submitted for the use of tolfenamic acid for the treatment of HLB. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture http://nifa.usda.gov/ (award number 2015-70016-23029). The content is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
Glossary
Abbreviations
- PrbP
Predicted RNA polymerase binding protein
- HLB
Huanglongbing
- RNAP
RNA polymerase
- EMSA
electrophoretic mobility shift assay
- TaP
tolfenamic acid pocket
- PrplK
rplK promoter region.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01591/full#supplementary-material
References
- Abellón-Ruiz J., Bernal-Bernal D., Abellán M., Fontes M., Padmanabhan S., Murillo F. J., et al. (2014). The CarD/CarG regulatory complex is required for the action of several members of the large set of Myxococcus xanthus extracytoplasmic function σ factors. Environ. Microbiol. 16, 2475–2490. 10.1111/1462-2920.12386 [DOI] [PubMed] [Google Scholar]
- Atkinson G. C., Tenson T., Hauryliuk V. (2011). The RelA/SpoT Homolog (RSH) superfamily: distribution and functional evolution of ppgpp synthetases and hydrolases across the tree of life. PLoS ONE 6:e23479. 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae B., Chen J., Davis E., Leon K., Darst S. A., Campbell E. A. (2015). CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. Elife 4, 1–19. 10.7554/eLife.08505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., et al. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal-Bernal D., Gallego-García A., García-Martínez G., García-Heras F., Jiménez M. A., Padmanabhan S., et al. (2015). Structure-function dissection of Myxococcus xanthus CarD N-terminal domain, a defining member of the CarD-CdnL-TRCF family of RNA polymerase interacting proteins. PLoS ONE 10:e0121322. 10.1371/journal.pone.0121322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikadi Z., Hazai E. (2009). Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminform. 1:15. 10.1186/1758-2946-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancato V. S., Pagliai F. A., Magni C., Gonzalez C. F., Lorca G. L. (2016). Functional analysis of the citrate activator CitO from Enterococcus faecalis implicates a divalent metal in ligand binding. Front. Microbiol. 7:101. 10.3389/fmicb.2016.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bové J. M. (2006). Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37. 10.4454/jpp.v88i1.828 [DOI] [Google Scholar]
- China A., Nagaraja V. (2010). Purification of RNA polymerase from mycobacteria for optimized promoter-polymerase interactions. Protein Expr. Purif. 69, 235–242. 10.1016/j.pep.2009.09.022 [DOI] [PubMed] [Google Scholar]
- da Graça J. V., Korsten L. (2004). Citrus huanglongbing: review, present status and future strategies, in Diseases of Fruits and Vegetables, Vol. 1, ed Naqvi S. A. M. H. (Dordrecht: Springer; ), 229–245. [Google Scholar]
- Davis E., Chen J., Leon K., Darst S. A., Campbell E. A. (2015). Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res. 43, 433–445. 10.1093/nar/gku1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. J., Mondal S. N., Chen H., Rogers M. E., Brlansky R. H. (2008). Co-cultivation of ‘Candidatus Liberibacter asiaticus’ with Actinobacteria from citrus with Huanglongbing. Plant Dis. 92, 1547–1550. 10.1094/PDIS-92-11-1547 [DOI] [PubMed] [Google Scholar]
- Duan Y., Zhou L., Hall D. G., Li W., Doddapaneni H., Lin H., et al. (2009). Complete genome sequence of citrus Huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 22, 1011–1020. 10.1094/MPMI-22-8-1011 [DOI] [PubMed] [Google Scholar]
- Elías-Arnanz M., Padmanabhan S., Murillo F. J. (2010). The regulatory action of the myxobacterial CarD/CarG complex: a bacterial enhanceosome? FEMS Microbiol. Rev. 34, 764–778. 10.1111/j.1574-6976.2010.00235.x [DOI] [PubMed] [Google Scholar]
- Fagen J. R., Leonard M. T., McCullough C. M., Edirisinghe J. N., Henry C. S., Davis M. J., et al. (2014). Comparative genomics of cultured and uncultured strains suggests genes essential for free-living growth of liberibacter. PLoS ONE 9:e84469. 10.1371/journal.pone.0084469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbis-Martinez L., Galbis-Martinez M., Murillo F. J., Fontes M. (2008). An anti-antisigma factor in the response of the bacterium Myxococcus xanthus to blue light. Microbiology 154, 895–904. 10.1099/mic.0.2007/013359-0 [DOI] [PubMed] [Google Scholar]
- Galbis-Martinez M. (2004). The high-mobility group A-type protein CarD of the bacterium Myxococcus xanthus as a transcription factor for several distinct vegetative genes. Genetics 167, 1585–1595. 10.1534/genetics.104.029207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-García A., Mirassou Y., García-Moreno D., Elías-Arnanz M., Jiménez M. A., Padmanabhan S. (2014). Structural Insights into RNA polymerase recognition and essential function of Myxococcus xanthus CdnL. PLoS ONE 9:e108946. 10.1371/journal.pone.0108946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno D., Abellon-Ruiz J., Garcia-Heras F., Murillo F. J., Padmanabhan S., Elias-Arnanz M. (2010). CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucleic Acids Res. 38, 4586–4598. 10.1093/nar/gkq214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner C. L., Pagliai F. A., Pan L., Bojilova L., Torino M. I., Lorca G. L., et al. (2016). Drug repurposing: tolfenamic acid inactivates PrbP, a transcriptional accessory protein in Liberibacter asiaticus. Front. Microbiol. 7:1630. 10.3389/fmicb.2016.01630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner A. L., Weiss L. A., Manzano A. R., Galburt E. A., Stallings C. L. (2014). CarD integrates three functional modules to promote efficient transcription, antibiotic tolerance, and pathogenesis in mycobacteria. Mol. Microbiol. 93, 682–697. 10.1111/mmi.12681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald T. R. (2010). Current epidemiological understanding on citrus Huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. 10.1146/annurev-phyto-073009-114418 [DOI] [PubMed] [Google Scholar]
- Graça J. V. (1991). Citrus greening disease. Annu. Rev. Phytopathol. 29, 109–136. 10.1146/annurev.py.29.090191.000545 [DOI] [Google Scholar]
- Grosdidier A., Zoete V., Michielin O. (2011). SwissDock, a protein-small molecule docking web service based on EADock, DSS. Nucleic Acids Res. 39, W270–W277. 10.1093/nar/gkr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulten G., Sacchettini J. C. (2013). Structure of the Mtb CarD/RNAP β-lobes complex reveals the molecular basis of interaction and presents a distinct DNA-binding domain for Mtb CarD. Structure 21, 1859–1869. 10.1016/j.str.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkarainen H., Vapaatalo H., Gothoni G., Parantainen J. (1979). Tolfenamic acid is as effective as ergotamine during migraine attacks. Lancet 314, 326–328. 10.1016/S0140-6736(79)90343-X [DOI] [PubMed] [Google Scholar]
- Hansen P. E. (1994). Tolfenamic acid in acute and prophylactic treatment of migraine: a review. Pharmacol. Toxicol. 75, 81–82. 10.1111/j.1600-0773.1994.tb02005.x [DOI] [PubMed] [Google Scholar]
- Landick R., Krek A., Glickman M. S., Socci N. D., Stallings C. L. (2014). Genome-wide mapping of the distribution of CarD, RNAP σA, and RNAP β on the Mycobacterium smegmatis chromosome using chromatin immunoprecipitation sequencing. Genomics Data 2, 110–113. 10.1016/j.gdata.2014.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon C. W., Ross W., Martin-Tumasz S., Toulokhonov I., Vrentas C. E., Rutherford S. T., et al. (2012). Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 26, 2634–2646. 10.1101/gad.204693.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Kimura M., Owens J. T., Meares C. F., Ishihama A. (1997). The two alpha subunits of Escherichia coli RNA polymerase are asymmetrically arranged and contact different halves of the DNA upstream element. Proc. Natl. Acad. Sci. U.S.A. 94, 1709–1714. 10.1073/pnas.94.5.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas F. J., Ruiz-Vazquez R. M., Murillo F. J. (1994). A genetic link between light response and multicellular development in the bacterium Myxococcus xanthus. Genes Dev. 8, 2375–2387. 10.1101/gad.8.19.2375 [DOI] [PubMed] [Google Scholar]
- Padmanabhan S. (2001). Domain architecture of a high mobility group A-type bacterial transcriptional factor. J. Biol. Chem. 276, 41566–41575. 10.1074/jbc.M106352200 [DOI] [PubMed] [Google Scholar]
- Pagliai F. A., Gardner C. L., Bojilova L., Sarnegrim A., Tamayo C., Potts A. H., et al. (2014). The transcriptional activator LdtR from ‘Candidatus Liberibacter asiaticus’ mediates osmotic stress tolerance. PLoS Pathog. 10:e1004101. 10.1371/journal.ppat.1004101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliai F. A., Gardner C. L., Pande S. G., Lorca G. L. (2010). LVIS553 transcriptional regulator specifically recognizes novobiocin as an effector molecule. J. Biol. Chem. 285, 16921–16930. 10.1074/jbc.M110.111138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. K., Wisotsky S. R., Johnson E. G., Hijaz F. M., Killiny N., Hilf M. E., et al. (2014). Viability of ‘Candidatus Liberibacter asiaticus’ prolonged by addition of citrus juice to culture medium. Phytopathology 104, 15–26. 10.1094/PHYTO-05-13-0119-R [DOI] [PubMed] [Google Scholar]
- Paul B. J., Barker M. M., Ross W., Schneider D. A., Webb C., Foster J. W., et al. (2004). DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322. 10.1016/j.cell.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Paul B. J., Berkmen M. B., Gourse R. L. (2005). DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. U.S.A. 102, 7823–7828. 10.1073/pnas.0501170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalver-Mellado M., Garcia-Heras F., Padmanabhan S., Garcia-Moreno D., Murillo F. J., Elias-Arnanz M. (2006). Recruitment of a novel zinc-bound transcriptional factor by a bacterial HMGA-type protein is required for regulating multiple processes in Myxococcus xanthus. Mol. Microbiol. 61, 910–926. 10.1111/j.1365-2958.2006.05289.x [DOI] [PubMed] [Google Scholar]
- Perederina A., Svetlov V., Vassylyeva M. N., Tahirov T. H., Yokoyama S., Artsimovitch I., et al. (2004). Regulation through the secondary channel - structural framework for ppGpp-DksA synergism during transcription. Cell 118, 297–309. 10.1016/j.cell.2004.06.030 [DOI] [PubMed] [Google Scholar]
- Potrykus K., Cashel M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- Rammohan J., Ruiz Manzano A., Garner A. L., Stallings C. L., Galburt E. A. (2015). CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res. 43, 3272–3285. 10.1093/nar/gkv078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Sanchez-Vazquez P., Chen A. Y., Lee J.-H., Burgos H. L., Gourse R. L. (2016). ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell 62, 811–823. 10.1016/j.molcel.2016.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Vrentas C. E., Sanchez-Vazquez P., Gaal T., Gourse R. L. (2013). The Magic Spot: a ppGpp Binding Site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50, 420–429. 10.1016/j.molcel.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaram U. S., DeAngelis K. M., Trivedi P., Andersen G. L., Lu S.-E., Wang N. (2009). Bacterial diversity analysis of Huanglongbing pathogen-infected citrus, using phylochip arrays and 16S rRNA gene clone library sequencing. Appl. Environ. Microbiol. 75, 1566–1574. 10.1128/AEM.02404-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Sechler A., Schuenzel E. L., Cooke P., Donnua S., Thaveechai N., Postnikova E., et al. (2009). Cultivation of ‘Candidatus Liberibacter asiaticus’, ‘Ca. L. africanus’, and ‘Ca. L. americanus’ Associated with Huanglongbing. Phytopathology 99, 480–486. 10.1094/PHYTO-99-5-0480 [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. B., Leon K., Osmundson J., Garner A. L., Weiss L. A., Westblade L. F., et al. (2013). Structure and function of CarD, an essential mycobacterial transcription factor. Proc. Natl. Acad. Sci. U.S.A. 110, 12619–12624. 10.1073/pnas.1308270110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings C. L., Stephanou N. C., Chu L., Hochschild A., Nickels B. E., Glickman M. S. (2009). CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138, 146–159. 10.1016/j.cell.2009.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- Svetlov V., Artsimovitch I. (2015). Purification of bacterial RNA polymerase: tools and protocols, in Bacterial Transcriptional Control: Methods in Molecular Biology, eds Artsimovitch I., Santangelo T. J. (New York, NY: Springer; ), 13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler H. L., Roesch L. F. W., Gowda S., Dawson W. O., Triplett E. W. (2009). Confirmation of the Sequence of ‘Candidatus Liberibacter asiaticus’ and assessment of microbial diversity in Huanglongbing-infected citrus phloem using a metagenomic approach. Mol. Plant Microbe Interact. 22, 1624–1634. 10.1094/MPMI-22-12-1624 [DOI] [PubMed] [Google Scholar]
- Wang N., Trivedi P. (2013). Citrus Huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 103, 652–665. 10.1094/PHYTO-12-12-0331-RVW [DOI] [PubMed] [Google Scholar]
- Weiss L. A., Harrison P. G., Nickels B. E., Glickman M. S., Campbell E. A., Darst S. A., et al. (2012). Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J. Bacteriol. 194, 5621–5631. 10.1128/JB.00879-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Sreedharan A., Wei S., Wang J., Pelz-Stelinski K., Folimonova S., et al. (2013). Global gene expression changes in Candidatus Liberibacter asiaticus during the transmission in distinct hosts between plant and insect. Mol. Plant Pathol. 14, 391–404. 10.1111/mpp.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.