Abstract
The genetic modifications through breeding of crop plants have long been used to improve the yield and quality. However, precise genome editing (GE) could be a very useful supplementary tool for improvement of crop plants by targeted genome modifications. Various GE techniques including ZFNs (zinc finger nucleases), TALENs (transcription activator-like effector nucleases), and most recently clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 (CRISPR-associated protein 9)-based approaches have been successfully employed for various crop plants including fruit trees. CRISPR/Cas9-based approaches hold great potential in GE due to their simplicity, competency, and versatility over other GE techniques. However, to the best of our knowledge no such genetic improvement has ever been developed in date palm—an important fruit crop in Oasis agriculture. The applications of CRISPR/Cas9 can be a challenging task in date palm GE due to its large and complex genome, high rate of heterozygosity and outcrossing, in vitro regeneration and screening of mutants, high frequency of single-nucleotide polymorphism in the genome and ultimately genetic instability. In this review, we addressed the potential application of CRISPR/Cas9-based approaches in date palm GE to improve the sustainable date palm production. The availability of the date palm whole genome sequence has made it feasible to use CRISPR/Cas9 GE approach for genetic improvement in this species. Moreover, the future prospects of GE application in date palm are also addressed in this review.
Keywords: CRISPR/Cas9, date palm, genome editing, multiplexing, loss of and gain-of-functions
Introduction
The erosion of plant genetic resources and the global climate changes confront us with enormous challenges including biotic and abiotic stresses (Arzani and Ashraf, 2016). Add to these limitations the fact that date palm (Phoenix dactylifera L.) is composed of genetically discrete clones representing thousands of cultivars without the benefits of a dynamic mutation-recombination system (Al-Khayri et al., 2015). The generation of an explosion of knowledge and technology related to genomics and genetics over the last few decades is promising in providing powerful tools for future development of higher-yielding cultivars (Arzani and Ashraf, 2016). The genome editing (GE) tools like zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the contemporary clustered regularly interspaced short palindromic repeats (CRISPR) along with CRISPR-associated protein 9 (Cas9) have established their hierarchy in editing plant genomes. By using these GE tools, genome modifications have been accomplished in various plants by ZFNs (Shukla et al., 2009; Townsend et al., 2009; Zhang and Voytas, 2011; Curtin et al., 2013; Pater et al., 2013; Qi et al., 2013) and TALENs (Christian et al., 2013; Sun et al., 2013; Wendt et al., 2013; Zhang et al., 2013). Recently developed CRISPR/Cas9 has gained popularity among the scientific community in a short period of time. In comparison to all these three GE tools, CRISPR/Cas9 offers several advantages over TALENs and ZFNs which include target design simplicity, improved efficacy and precision, multiplexing, least off-targets, ability to target multiple alleles, cost effective, easy delivery and execution, and availability of in silico techniques to design and evaluate the designed single-guide RNA (sgRNA). Additionally, in ZFNs and TALENs sequence specificity is conferred by DNA-binding domain of protein while in CRISPR/Cas9 system sgRNA mediate this, no protein engineering is involved in CRISPR system. Unlike to predecessor GE—CRISPR system can cleave methylated target sequence (reviewed by Bortesi and Fischer, 2015). Moreover, the protein domain engineering of the target DNA is a pre-requisite for multiplexing using TALEN and ZFNs, which makes these techniques less-suitable and limited for multiplexing (Lowder et al., 2017). Whereas, multiplexing in CRISPR/Cas9 system requires only multiple sgRNAs jointly expressed with the Cas9 (detailed discussion in Section “Verifications of the Genome Editing Events in Date Palm”). CRISPR/Cas9 system has been modified into a two-component system; Cas9 and an sgRNA. The Cas9 protein cleaves the target DNA by a synthetic sgRNA, which comprised of crRNA and tracrRNA. CRISPR/Cas9 has been successfully used as a GE tool to confer resistance against citrus canker (Jia et al., 2016; Peng et al., 2017), rice blast (Wang et al., 2016; Kanda et al., 2017), powdery mildew (Nekrasov et al., 2017; Zhang et al., 2017), phytophthora (Fang and Tyler, 2016), and multiple plant viruses (Iqbal et al., 2016; Khatodia et al., 2017) in the model plants as well as in the commercial crops. Moreover, it has also been successfully tested, either transiently or through stable transformation, for precise GE and knockout mutations in the woody plants citrus (Jia and Wang, 2014), populous (Fan et al., 2015; Zhou et al., 2015), and apple (Nishitani et al., 2016). Thus, programmable GE in woody fruit trees holds a substantial potential elucidating off-screen molecular mechanisms in governing flowers, fruits, and whole plant developments.
Nevertheless, no GE tool has been exploited in date palm genome engineering. Date palm (family Palmae or Arecaceae) is a high ranked diploid (2n = 36), monocotyledonous, dioecious, perennial woody fruit tree and has high socio-economic significance in Oasis agriculture (Mahmoudi et al., 2008). The average date palm genome is about 670 Mb in size (Al-Dous et al., 2011; Al-Mssallem et al., 2013) and comprises of 18 chromosomes. It is native to Arabian Peninsula, possibly originated from southern Iraq and has vast diversity from Mauritania to Pakistan (Pintaud et al., 2011). Other regimes where date palm is being cultivated are sub-Saharan African countries, Australia, United States (California), Peru, and some other warmer parts of the world. Currently, 450 date palm varieties are grown in Kingdom of Saudi Arabia and more than 2000 cultivars in the world. This majestic plant is known as “Tree of life” since the settlements of ancient human in the hot and barren parts of the world. Besides a nutritious source of human diet, it is a continuous source of raw material for housing, sheltering, and handicrafts in the harsh dry environments of Southwest Asia and North Africa (Jain et al., 2011). Nevertheless, over the time date palm agronomy has been shifted to monoculture instead of traditional cultivation. Such a situation brought severe genetic erosion of many productive cultivars and trembled the date palm agro-biodiversity in many areas (Jain et al., 2011). Additionally, several biotic and abiotic stresses further worsen the situation.
Previously, biotechnological approaches, such as plant tissue culture, marker-assisted breeding and DNA finger printing, have been used in date palm genomics but failed to bring a significant improvement. Additionally, conventional agronomic practices and breeding approaches in date palm are not cost effective as performing three backcrosses it usually takes 30 years of breeding. For the sustainability of date palm, employment of new techniques in date palm breeding programs is highly needed to develop tolerant varieties and enrich the existing germplasm. This can be achieved by modifying the date palm genome against various biotic and abiotic stresses by overexpressing or downregulating the key genes involved in biochemical pathways, or by engineering resistance against various pests and diseases. Additionally, the dissection of genetic information in date palm would help in understanding the role of various genes involved in sex determination, enzymatic reactions controlling fruit ripening, fruit sweetness, and fruit quality. In order to determine the universal efficacy of CRISPR/Cas9, extensive investigation in date palm is also necessary.
Major Abiotic and Biotic Constraints In Date Palm Cultivation
During the long life span, high salinity, extreme drought regimes, and blazing heat are major abiotic stresses affecting date palm. Our understanding about the molecular mechanisms regarding abiotic stresses is still very limited, however, a proteomic analysis recently identified 47 differentially expressed proteins in salt and drought affected date palm plants (El Rabey et al., 2015). Among various biotic stresses, the bayoud disease infection (caused by Fusarium oxysporum) in North Africa (Sedra, 2007), red palm weevil (Rhynchophorus ferrugineus) in Middle East Asia and Mediterranean regions (Ferry and Gomez, 2002) are the most important (Supplementary Table 1). Furthermore, 34 diverse fungal and Oomyces species have been found associated with date palm root diseases (Al-Sadi et al., 2012).
The mounting climatic and population scenario, long date palm breeding time along with biotic factors have left no choice for the scientists except development of genetic resistance for sustainability of date palm. However, developing a high yielding, resistant and good fruit quality cultivar demands a stringent, comprehensive and reliable methodology. The CRISPR/Cas9-based approaches can be harnessed to target the genomes of date palm’s pathogens directly or indirectly. The date palm insect/pests can be targeted by CRISPR/Cas9-based approaches through “gene drives” to circumvent their population (Figure 1).
FIGURE 1.
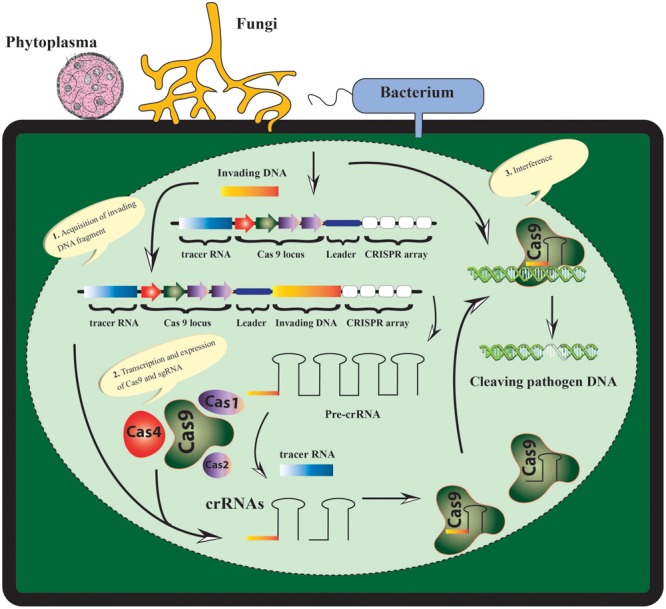
The CRISPR/Cas9-based resistance model in date palm depicting the recognition and disruption of the pathogen genetic material in three steps: acquisition, expression, and interference). During acquisition the invading DNA is integrated and duplicated into the CRISPR-locus at the leader side. The expression step involves the active transcription and expression of pre-CRISPR RNA (Pre-crRNA), which is further processed into mature crRNAs specifically with the help of different Cas proteins. During the third step of interference, any complementary target region of the foreign genetic material is recognized and cleaved as guided by crRNA and Cas9 protein.
Limitations In Date Palm Functional Genomics
Despite of a rich agricultural history and economic importance of date palm, the application of high-throughput technologies started during last decade. The palm family has been an ignored group to understand their developmental potential for genetic improvement (Bekheet and Hanafy, 2011). The first genetic map of the date palm “cultivar Khalas” has been constructed quite recently (Mathew et al., 2015), soon after sequencing of chloroplast (Khan et al., 2005; Yang et al., 2010; Sabir et al., 2014), mitochondria (Fang et al., 2012), and the whole nuclear genome (Al-Dous et al., 2011; Al-Mssallem et al., 2013). Furthermore, the molecular variations in the date palm genome have also been cataloged recently (Hazzouri et al., 2015). The whole genome sequencing of date palm also facilitated other relative studies including transcriptomic analysis (Yin et al., 2012), miRNAs expression profiling (Xiao et al., 2013; Xin et al., 2015; Yaish et al., 2015), comparative analysis of P. dactylifera and oil palm fruits (Bourgis et al., 2011), construction of genetic models (Zhang et al., 2011), and genetic diversity using single-nucleotide polymorphism (SNPs) data (Sabir et al., 2014; Mathew et al., 2015). Very recently, the applications of -omics coupled with bioinformatics were employed to determine and characterize simple sequence repeats (SSRs) for the endowment of SSR database in date palm genome (Zhao et al., 2012; Mokhtar et al., 2016). Moreover, same approaches have also been employed recently for in silico characterization and molecular structuring of DnMRE11, a gene involved in double stranded DNA-breaks repair, in date palm cultivar “Deglet Noor” (Stracker and Petrini, 2011; Rekik et al., 2015). A miRNA profiling of the date palm cultivar “Khalas” revealed 153 conserved homologs, 89 variants, whereas 180 novel miRNAs directly involved in the salt adaptation (Yaish et al., 2015). An in silico analysis by the same researchers unraveled that these miRNAs could directly regulate many salt tolerance related genes in leaves and roots, respectively. Similarly, computational studies of miRNA in date palm revealed their involvement in fruit development (Xin et al., 2015) and evolution (Xiao et al., 2013). Besides, the recent gene annotation in date palm no experimental validations are still available for their expression and biological functions in this species (Bourgis et al., 2011; Zhang et al., 2012). Therefore, predicting the expression of a relevant gene in date palm is not as authenticated as its ortholog in other plant species. Likewise, most of the available information regarding date palm genome is from a single cultivar “Khalas,” which may or may not match with the cultivar used in other studies. Thus, exploring the intrinsic molecular mechanisms governing certain gene functions and transcriptional regulation in date palm is vital for its sustainable production across the globe. Although, genetic diversity in different date palm cultivar can be assessed through NGS and SNPs techniques (Yang et al., 2010; Fang et al., 2012; Khan et al., 2012; Sabir et al., 2014) but studying functional genomics in date palm is very tedious due to longer vegetative regimes, low efficacy of genetic transformation, and limited survival of mutants. The evasion of genetic barriers in the genetic improvement of such tree plants is possible through the application of genetic transformation and GE tools.
A Generalized Stepwise and Basic Strategy for CRISPR/Cas9 Implications In Date Palm
A stepwise methodology for successful execution of CRISPR/Cas9 system in date palm is outlined in the succeeding section and the Figure 2.
FIGURE 2.
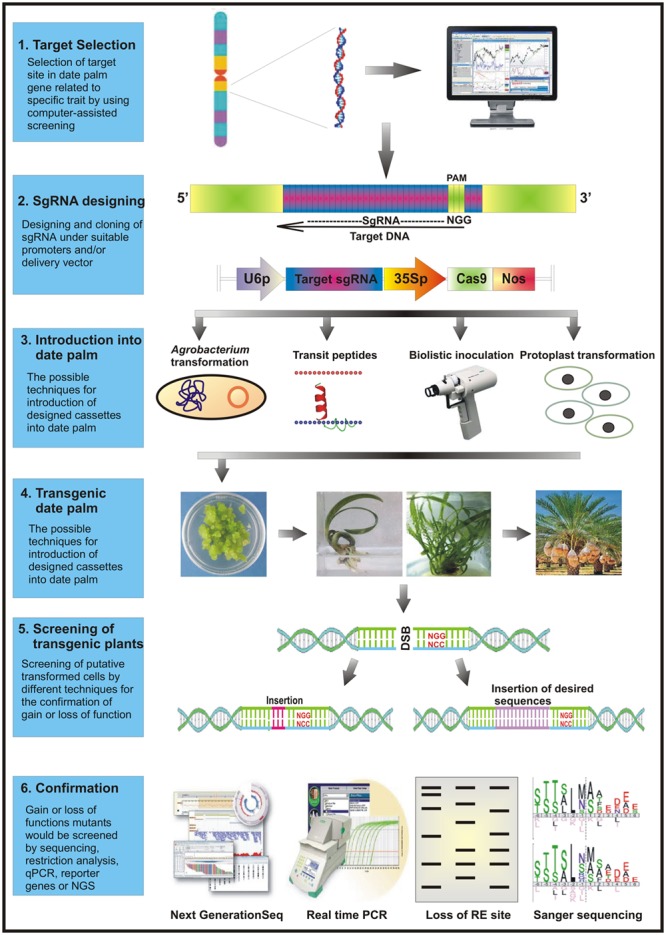
A schematic diagram representing the execution of CRISPR/Cas9 based system in date palm genome editing. The most vulnerable target sites in the desired gene(s) are selected specifically using online available web sourcing to design primers for complementary 20-nucleotides. The target specific sgRNA and Cas9 cassettes are constructed either in a single binary vector or separate expression vectors. These cassettes are then co-transformed in vivo into the plant cells employing a suitable transformation method. Following the putative transformation, the mutated cells are screened an analyzed for target-specific mutations using reporter genes, endonucleases, polyacrylamide gel electrophoreses, or high throughput sequencing techniques. The successfully transformed cells are then selected for further downstream applications and analysis.
Data Mining and Target Selection (sgRNA Design)
The primary and the most important step in GE is the selection of a target region in the genome. The major challenges during the selection of a target region in date palm may include genome polymorphism, off-targets (discussed in section “Countering off-targets in date palm genome editing”), presence of introns (that may get further aggravated by alternative splicing) and presence of SNPs. However, many tools are available to deal with such problems and generally categorized into three major steps: (i) selection of target region or sites/sgRNA designing, (ii) verification of the designed sgRNA for possible off-targets, and (iii) evaluation of on- and off-target cleavage rates (Lee et al., 2016). To ascertain successful date palm genome engineering, several available web-based bioinformatics and data mining tools (like CCTop, ATUM, MIT CRISPR design, Alt-RTM CRISPR-Cas9 System, CHOPCHOP, CROP-IT, GT-Scan, sgRNA Designer, Cas-OFFinder, etc.) can impart very promising role.
Selection of CRISPR/Cas9 System
Successful execution of CRISPR/Cas9 system in date palm requires expression of bacterial Cas9 protein into date palm cells. Codon optimization and selection of suitable promoter(s) is a pre-requisite to accomplish CRISPR/Cas9 system in plants (Belhaj et al., 2013). Several plant promoters (CaMV 35s, CMV, EF1A, LTR, and UBO) have been implicated to drive Cas9 expression. In many studies, plant RNAIII promoters, like U3 and U6, have also been used successfully (Belhaj et al., 2013).
Another important consideration is the selection of efficient Cas9. Generally, eukaryotic optimized SpCas9 have efficient GE ability and used vastly in eukaryotes. Nevertheless, many studies have used plant codon-optimized version of Cas9 (Jiang et al., 2013; Li et al., 2013; Miao et al., 2013; Shan et al., 2013) with improved efficacy. Beside these versions of Cas9, a dCas9 version lacking nuclease ability has been developed to employ this tool as gene silencing [CRISPR interference (CRISPRi)] and gene activation [CRISPR activation (CRISPRa)] rather than GE tool (Gilbert et al., 2013; Lei et al., 2013). Such modifications will further widen the applications of CRISPR/Cas9 system and can potentially be opted for date palm genome modifications and regulations.
CRISPR/Cas9 Cassette Delivery into Date Palm
Next step is the assembly of whole cassette (carrying sgRNA, CRISPR, Cas9 and, if needed, a nuclear localization signal) into any suitable plant based expression vector to deliver the assembled construct into date palm genome. Successful delivery of the system into date palm with high precision will be a challenging task. To deliver CRISPR/Cas9 constructs into date palm tissues various methods such as protoplast transformation, polyethylene glycol-mediated transformation, biolistic inoculations, transit peptides (Lee et al., 2008), or some plant virus-based vectors can be used (Butler et al., 2016). However, stable genetic transformation in date palm through particle bombardment (Mousavi et al., 2014) and/or Agrobacterium-mediated genetic transformation of callus, embryos and immature tissues is the most common and successful method for effective applications of CRISPR/Cas9 in plants (Ma et al., 2016). Direct transformation of date palm genome can be tricky therefore, adapting a DNA-free strategy to transfect date palm protoplast by the preassembled cassettes would be a good choice. Such strategy is highly useful for the vegetatively propagated perennial plant species (Woo et al., 2015).
Regeneration and Screening of Targeted Mutations in Date Palm
After successful transfection, the date palm plantlets can be regenerated on selection media and subjected to screening of gene editing events by employing different strategies (Figure 2). The phenotypic and genotypic screening requires detection and confirmation of the targeted mutation (Hua et al., 2017).
Verifications of the Genome Editing Events in Date Palm
Screening and confirmation of specific gene edited mutants induced by CRISPR system is not only crucial but important too for downstream processing.
A quick way to verify the efficacy of GE is the use of reporter genes (such as GFP, RFP, YFP, and GUS) as a marker of editing events. To use this system, the reporter gene should bear a frame shifting at target site or alternatively contain duplicated region that could be corrected by CRISPR/Cas9 system after GE (Siebert and Puchta, 2002; Jiang et al., 2013; Mao et al., 2013; Feng et al., 2014). Alternatively, an internal or introduced endonuclease site can be targeted during Cas9/sgRNA cleavage (Jiang et al., 2013; Shan et al., 2013; Xie and Yang, 2013). The perturbed endonuclease site can be confirmed through PCR amplification and ultimately a successful GE will be ensured.
The successful GE events through CRISPR/Cas9 system can also be confirmed by using polyacrylamide gel electrophoresis (PAGE). Single-stranded DNAs with nucleotide variations can exhibit different migration rate due to change in the DNA conformations. This technique referred as single-stranded conformation polymorphism and could be used to detect targeted mutations induced by CRISPR/Cas9 (Zhang Y. et al., 2016). High-resolution melting assay offers another powerful tool for GE verifications, where mutation is determined in PCR amplicons on the basis of difference in their melting temperature. Though, sensitivity is low and sequence of mutated target gene cannot be determined but can be practiced for a preliminary screening of mutations induced by CRISPR/Cas9 system (Dahlem et al., 2012; Fauser et al., 2014).
Next generation or high-throughput sequencing of the PCR amplicons or whole genome is highly sensitive and efficacious method of detecting the mutation in the target region. Although, this method is expensive and time consuming but is highly reliable and robust to detect low frequency mutations and off-target mutations in the whole genome (Fauser et al., 2014; Feng et al., 2014). Using old fashioned PCR amplification, subsequent cloning and sequencing of the targeted gene through Sanger sequencing platform can provide relatively inexpensive and efficient method for the confirmation of mutations at the target site. This approach is convenient for determining either simple mutations or complicated chimeric mutations (Zhou et al., 2014; Ma et al., 2015). A real-time quantitative PCR can enable the measurement of transcript levels in both coding and non-coding targets. These tools greatly facilitate the analysis of targeted sites.
Gene Stacking Using Multiplex CRISPR/Cas9 Model in Date Palm
Precise modification of multiple genes, involved in controlling a particular trait, in one go have been a long standing interest of genome scientists. Recently, a powerful system for multiplex genome engineering has been employed in rice, where hijacking of endogenous tRNA-processing mechanism was achieved to generate multiple sgRNAs from a single construct (Xie et al., 2015). In this system, constructs are designed in polycistronic tRNA–sgRNA (PTG) form with repeating units of target specific spacer and sgRNA scaffolds, which can be separated by conserved tRNA for multiplexing (Figure 3). During transcription, the endogenous RNases will cleave the PTG leaving multiple sgRNAs that direct the Cas9 to respective target sites. The tRNAs will also lead to sgRNA over expression by increasing the Pol III transcriptions. By using this method, 3–31 times increased level of GE with 15–19% higher mutation have been achieved when compared to other CRISPR/Cas9-based multiplexing approaches (Xie et al., 2015).
FIGURE 3.
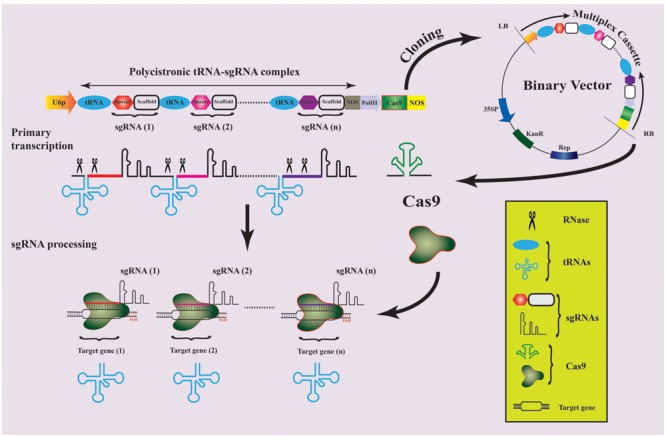
Schematic presentation of multiplex genome editing strategy in date palm. A preferred assemblage of multiplex cassette is shown for different sgRNAs to target different genes simultaneously. The spacers and sgRNA scaffold can be inserted between adjacent tRNAs followed by a NOS terminator at the end. The second cassette can be expressed from Pol-III promotor and NOS terminator sequences sharing the same binary vector with sgRNA cassette. The whole multiplex cassette can then be transcribed and expressed separately in the date palm genome to carry out genome editing.
PTG-based multiplex CRISPR/Cas9 system can be used for date palm GE because of its certain advantages over other multiplexing techniques, which include use of short 5′ spacer sequences prior to sgRNAs, editing/deletion of non-coding regions (Xie et al., 2015) and efficient production of multiple sgRNAs from one construct using endogenous RNA-processing machinery with improved multiplex GE abilities (Xie et al., 2015; Ding et al., 2016).
The successful implications of PTG-based method include GE of allopolyploid wheat (Wang et al., 2016) (Table 1) and Zea mays where enhanced mutagenesis efficiency was observed (Qi et al., 2016). These implications are not limited to plants but quite recently, three human genes related to histone deacetylase (HDAC) have been targeted successfully (Dong et al., 2017). By utilizing the universal tRNA-based approach, successful manipulation of multiple genomic loci in date palm can potentially be achieved against different biotic and abiotic challenges (Supplementary Table 1 and Figure 3) particularly red palm weevil and bayoud disease. Comparative genomic studies of date palm deciphered that more than 50% date palm predicted ORFs matches with rice ORFs (Al-Dous et al., 2011) while, proteome comparison showed that ∼8000 gene families of date palm are common to both monocot (rice and sorghum) and dicot plants (Arabidopsis thaliana and grapevine) (Al-Mssallem et al., 2013). Thus, different date palm genes can be targeted by designing the sgRNAs referring to the successful multiplex GE in various crops (Table 1).
Table 1.
Examples of different plant species (genes) multiplexed using CRISPR/Cas9 GE approach.
| Organism | Target(s)/gene(s) | Description of gene(s) | Cas9 promoter | sg Promoter | Binary vector/backbone | Reference |
|---|---|---|---|---|---|---|
| Dicotyledonous plants | ||||||
| Arabidopsis thaliana | GLV family | GOLVEN gene family regulating root stem cells | Arabidopsis ubiquitin 10 promoter | Arabidopsis U6 promoter | pCUT binary vector | Peterson et al., 2016 |
| PYR1, PYL1, PYL2, PYL4, PYL5, PYL8 | PYR/PYL gene family | pAtUBQ1 promoter | AtU6-26, AtU3b, and At7SL-2 promoters | pEx-6XsgR-PYL114285-Cas9 binary vector | Zhang Z. et al., 2016 | |
| AtPDS3 | Phytoene desaturase; photobleached phenotype | Constitutive 35SPPDK | Arabidopsis U6 polymerase III promoter | pFGC-RCS binary vector | Li et al., 2013 | |
| AtRACK1b, AtRACK1c | Receptor for activated C kinase 1 (RACK1) family | |||||
| CHLI1, CHL12 | Magnesium-chelatase subunit I (CHLI); pale green to albino plants | AtUBQ1 promoter | AtU6 promoter | pCAMBIA1300 | Mao et al., 2013 | |
| TT4 | TRANSPARENT TESTA 4 | |||||
| RTEL1 | Regulator of telomere length 1, AT1G79950 | Ubiquitin 4-2 promoter from Petroselinum crispum (PcUbi4-2) | Arabidopsis U6-26 promoter (AtU6-26) | Binary vectors was derived from pPZP201; pDe-Cas9-D10A | Schiml et al., 2014 | |
| At5g55580 | Mitochondrial transcription termination factor (mTERF) | Maize ubiquitin promoter (Pubi) or the cauliflower mosaic virus 35S promoter (P35S) | U3 and U6 small nuclear RNA promoters from Arabidopsis; AtU3b, AtU3d, AtU6-1, AtU6-29 | pYLCRISPR/Cas9 binary vectors based on the pCAMBIA1300 backbone | Ma et al., 2015 | |
| Tomato (Solanum lycopersicum) | SlyGABA-TP1, SlyGABA-TP2, SlyGABA-TP3, SlyCAT9, and SlySSADH | γ-Aminobutyric acid (GABA) metabolic pathway | Ubiquitin promotor | LacZ-AtU3d, AtU3d, AtU3b, AtU3b, AtU6-1 or AtU6-29 | pYLCRISPR/Cas9 | Li et al., 2017 |
| SlAGO7 | ARGONAUTE7 (SlAGO7) for post-transcriptional silencing of AUXIN RESPONSE FACTOR | CaMV 35S promoter | A. thaliana U6 promoter | pAGM4723 | Brooks et al., 2014 | |
| Solyc08g041770, Solyc07g021170, Solyc12g044760 | Three homologs of Solyc11g064850 control tomato reproductive development | |||||
| Nicotiana benthamiana | PDS | Phytoene desaturase gene related with albino leaf phenotype | CaMV 35S promoter | CaMV 35S | pBI121 | Upadhyay et al., 2013 |
| PDS | Albino leaf phenotype | CaMV 35S promoter | Pea early browning virus (PEBV) promoter | pK2GW7, TRV2 RNA2 vector | Ali et al., 2015 | |
| PCNA | Proliferating cell nuclear antigen gene related with DNA replication | |||||
| Nicotiana tabacum | XylTA, XylTB | β(1,2)-Xylosyltransferase (XylT) | 35SPPDK | U6 promoter | pFGC-pcoCas9 | Mercx et al., 2017 |
| FucTA, FucTB, FucTC, FucTD | α(1,3)-Fucosyltransferase (FucT) plant specific glycans | |||||
| NtPDS | Albino leaf phenotype | 2 × CaMV 35S promoter | Arabidopsis U6-26 promoter | pORE O4 | Gao et al., 2015 | |
| NtPDR6 | Multiple branches | |||||
| Soybean (Glycine max) | Glyma01g38150; Glyma11g07220; Glyma04g36150; and Glyma06g18790 | A. thaliana DDM1 and MET1 orthologs | 2 × CaMV 35S promoter | Medicago truncatula U6.6 polymerase III promoter | p201N Cas9 | Jacobs et al., 2015 |
| Monocotyledonous plants | ||||||
| Rice (Oryza sativa) | DEP1, EP3, Gn1a, GS3, GW2 | Panicle architecture and yield related genes | 2× 35S promoter | U6 promoter | pC1300-Cas9 binary vector | Shen et al., 2017 |
| BADH2 | Betaine aldehyde dehydrogenase 2 | |||||
| QTL | Major quantitative trait loci | |||||
| Hd1 | Heading date 1 | |||||
| LPA1 | Loose plant architecture 1 | |||||
| MPK1, MPK2, MPK5, MPK6 | Mitogen-activated protein kinase genes | Rice ubiquitin promoter | Rice U3 promoter | pRGEB32 binary vector | Minkenberg et al., 2017 | |
| OsEPSPS | 5-Enolpyruvylshikimate 3-Phosphate synthase | ZmUbi promoter | Oryza sativa U6 (OsU6) promoter | pCambia binary vector | Wang et al., 2017 | |
| OsBEL | Bentazon sensitive lethal | |||||
| OsPDS | Phytoene desaturase | |||||
| SWEET genes | Sugar efflux transporter genes related with disease susceptibility | Maize ubiquitin 1 promoter | Rice small nuclear RNA U6 | pCAMBIA1300-based destination vector pUbi-Cas9 | Zhou et al., 2014 | |
| OsCPS4, CYP99A2 | For production of labdane-related diterpenoids, a group of phytoalexins | |||||
| OsKO1, OsKOL5 | Diterpenoid synthetic genes | |||||
| CYP76M5, CYP76M6 | Cytochrome P450 gene | |||||
| OsPDS | Mutation resulted in albino phenotype | CaMV 35S promoter | OsU3 promoter | pCAMBIA1300 | Zhang et al., 2014 | |
| OsPMS3 | Non-coding RNA | |||||
| OsDERF1 | AP2 domain containing protein for drought-resistant | |||||
| OsMSH1 | DNA mismatch repair protein; pleiotropic phenotype | |||||
| Rice MAPKs (MPK1/2/5/6) | Mitogen-activated protein kinase (MAPK) involved in biotic and abiotic signaling pathways | Rice ubiquitin promoter plus the complete 5′ untranslated region (UBIp) | Rice U3 snoRNA promoter (U3p) | pRGEB32 | Xie et al., 2015 | |
| FT-like (FTL) genes | Premature leaf senescence | Maize ubiquitin promoter (Pubi) or the cauliflower mosaic virus 35S promoter (P35S) | U3 and U6 small nuclear RNA promoters from rice: OsU3, OsU6b, OsU6c, OsU6a | pYLCRISPR/Cas9 binary vectors based on the pCAMBIA1300 backbone | Ma et al., 2015 | |
| OsGSTU, OsMRP15, OsAnP | Genes for anthocyanin accumulation | |||||
| OsWaxy | Decrease amylose content | |||||
| Triticum aestivum (wheat) | PDS | Phytoene desaturase; albino leaf phenotype | CaMV 35S promoter | CaMV 35S promoter | pBI121 | Upadhyay et al., 2013 |
| INOX | Inositol oxygenase enzyme | |||||
| TaGW2 | Grain weight | Ubiquitin gene promoters | Wheat U3 and U6 promoters | pBUN421-GLM | Wang et al., 2016 | |
| TaLpx-1 | lipoxygenase 1 related with disease resistance | |||||
| TaMLO | Mildew Resistance Locus | |||||
| Maize (Zea mays) | ZmAgo18a, ZmAgo18b | Argonaute 18 | Maize ubiquitin 1 gene promoter | Rice U6 small nuclear RNA gene promoters | pMCG1005 binary vector | Char et al., 2017 |
| a1, a4 | Dihydroflavonol 4-reductase or anthocyaninless | |||||
| LIG | Liguleless1 | Ubiquitin promoter | Maize U6 polymerase III promoter | pSB11-Ubi-Cas9; U6:sgRNA | Svitashev et al., 2015 | |
| Ms26, Ms45 | Male fertility genes | |||||
| RPL, PPR | For plant development | Maize U6 promoter | Maize U6 promoter | pCAMBIA3301 | Qi et al., 2016 | |
| lncRNAs | Two reverse overlapping long non-coding RNAs | |||||
Countering Off-Targets In Date Palm Genome Editing
The robustness of CRISPR/Cas9 has become unprecedented though, yet off targets are major limiting factor in the applications of this system. However, in larger eukaryotic genomes avoiding off-targets effects by choosing minimum sequences of sgRNA albeit with least similarity to unrelated sequences is nearly impossible. To dealt such problems in date palm GE can be challenging and some of the strategies have been proposed in this section. Several studies have revealed that Cas9 has the ability to cleave the target region even if several mismatches are present between sgRNA and the target sequence, thus capable to perform off-target activity. Unlike previous studies, which showed that 20 bp long sgRNA endow high level of specificity (Fu et al., 2013; Hsu et al., 2013; Pattanayak et al., 2013), new findings showed improved GE specificity by using less than 20 bp sgRNA (Fu et al., 2014). It has been empirically proven that generally up to three nucleotide mismatches, especially at 5′ end, between sgRNA and target sequence can be tolerated by CRISPR/Cas9 system (Ran et al., 2013). However, the crucial role in determining specificity is being imparted by sgRNA seed sequence—a region of 12 nucleotides following the PAM—and two nucleotides of the PAM sequence (Larson et al., 2013). Off-targets can also be substantially minimized by adding few guanidine residues at 5′ end (Cho et al., 2014) and/or by reducing the length of the sgRNA up to 17 nucleotides (Fu et al., 2014). Such reduction in size may render the RNA–DNA complex more sensitive to mismatches, perhaps by reducing binding energy at the sgRNA–DNA interface.
Another substantial way of eliminating off-targets is the use of modified Cas9 nuclease. The use of different mutant variants of Cas9 (SpCas9 VQR and VRER) (Kleinstiver et al., 2015) or Cas9 orthologs StlCas9 (derived from Streptococcus thermophilus) could also be used for highly efficient GE with modified PAM recognition specificity (Kleinstiver et al., 2015). A promising study showed no detectable off-targets effects in rabbits accompanying with up to 50-fold improved specificity, as compared to conventional Cas9, where single strand of target sequences was nicked by inactivating one of the two conserved nuclease domains of Cas9 (Honda et al., 2015). Such single-stranded nicks are not capable of causing mutation because of base excision repair mechanism. Cleaving the target DNA sequence by pairing the two nickases can generate double stranded sticky ends break (Mali et al., 2013; Ran et al., 2013). The obvious reason in enhanced specificity is expansion of target sequences from 20 to 40 bp by the use of two sgRNAs. However, in Arabidopsis and rice plants, the use of paired Cas9 nickases failed to outperform the nuclease in promoting NHEJ or HDR repair, but may reduce off-targets effects (Fauser et al., 2014; Mikami et al., 2016). Over the time, the Cas9 specificity has been improved by adopting certain strategies. In a recent study, a structure-guided approach was employed to modify alanine groove (with positively charged amino acid) that potentially disrupted the interaction of Cas9 with non-target DNA strand thus, leading to increase specificity by enhancing Cas9 reliance on the sgRNA base pairing to the target sequence (Slaymaker et al., 2016). Such Cas9 mutants (Cas9s) demonstrated enhanced, but variable, editing efficiency across multiple genomic targets with significant reduction in the off-targets (Crosetto et al., 2013). As an alternate approach, the Cas9 structure was explored to identify those amino acids, which are essential in stabilizing the target DNA strand and the 5′ end of the sgRNA (Kleinstiver et al., 2016). By mutating such amino acids, a modified Cas9 (SpCas9-HF1) was designed that not only efficiently cleave target sequence but also showed higher sensitivity to mismatches between the sgRNA and target sequences. In order to obtain desired changes free of off-targets through CRISPR/Cas9, opting such techniques could be a valuable tool in date palm GE.
The Limitations Of Date Palm Genome Editing
Besides all the fascination, GE in date palm can have certain bottle-necks. The genomes of outcrossing species have high heterozygosity and thus, occurrence of SNPs is quite frequent and can affect the efficiency of GE in these species. Outcrossing phenomena brings high allelic heterozygosity, polymorphism, and genetic instability in the date palm genome (Jubrael et al., 2005). The genome draft of date palm cultivars Agwa, Fahal, Khalas, and Sukary has identified a SNP range of 3.85–6.63/kb in the nuclear genome (Al-Mssallem et al., 2013). Moreover, in the cultivar Khalas about 0.9 million new SNPs have also been reported. A high frequency of SNPs in the coding region is also reported from other woody perennial trees such as Populus trichocarpa, Populus tremula, and Eucalyptus (Tsai and Xue, 2015). Because, most of the plant models used for GE through CRISPR/Cas9 are highly homozygous, therefore few studies are available, which addressed sequence polymorphisms. However, the occurrence of allelic heterozygosity and sequence polymorphism may affect the GE in outcrossing, woody perennial plants (Fan et al., 2015; Tsai and Xue, 2015). The GE of 4-coumarate:CoA ligase (4CL) genes was accomplished by successfully targeting 4CL1, 4CL2 genes with 100% biallelic mutation in transgenic popular plants. Despite of 89% sequence identity with 4CL1, the third target gene of 4CL family (4CL5) could not be mutated due to the presence of SNP near PAM sequence (Zhou et al., 2015). Different web-based programs for sgRNA design in plants have limited utility to harness outcrossing plant species. This may be due to lack of multiploidy coverage of plant genomes on the available resources. The identification of SNPs requires the genome sequence information of the respective cultivar to be used for GE. However, the cultivar chosen for whole genome sequencing may have unique biological features different from other cultivars. Thus, it is easy to speculate that the whole genome of one date palm cultivar may or may not explicitly represent the exact features of similar genes between two cultivars. The whole genome sequencing of different date palm cultivars revealed the existence of intra- and inter-varietal SNPs not only in the intergenic region but also in the coding regions (Yang et al., 2010; Khan et al., 2012; Al-Mssallem et al., 2013; Sabir et al., 2014). Under such circumstances, it is highly tricky to choose and design sgRNAs for date palm GE and other related applications using CRISPR/Cas9.
Improving the plant traits through conventional breeding is time consuming phenomena as it relies on extensive back-crossing and introgression of naturally existing genetic variation. The mounting scenario of increasing population could not be paced with conventional breeding. Nonetheless, plant breeding can be accelerated through modern GE tools especially in date palm with long breeding cycles and vegetative propagation. CRISPR/Cas9 system can be executed to eliminate genes that negatively regulate date palm quality traits and stacking molecular traits at desired locus. In gene stacking, genes can be introduced in close proximity to a desired locus into a crop with a low risk of segregation. However, such executions are tedious to achieve through classical breeding (Ainley et al., 2013). Thus, establishing the generic recipient lines through CRISPR/Cas9-based approaches would accelerate the date palm breeding program by reducing the time span.
Delivering CRISPR/Cas9 components into the date palm genome via genetic transformation of somatic embryos may possibly introduce random integration of bacterial plasmid sequences and thus, may arise current GMO interceptions. One possibility to override any transgene is a continuous back-crossing for several generations, which is not workable in date palm. It requires around 30 years accomplishing three backcrosses in date palm. The application of next-generation DNA-free CRISPR/Cas9 approaches [such as CRISPR/Cas9 ribonucleoproteins (RNPs)] may offer a plausible solution to address current GMO-regulations in such plants (Kanchiswamy, 2016; Lu et al., 2017). The CRISPR/Cas9 RNPs approach has successfully been used for grapevine and apple protoplast transformation to cope with Erysiphe necator and Erwinia amylovora, respectively by delivering CRISPR/Cas9 RNPs directly into the protoplast (Malnoy et al., 2016). The date palm protoplasts transfection may constitute dynamic and versatile CRISPR/Cas9 GE to tackle long procedure of functional analysis for the concerned traits. This method has already been deployed in human, animals, and some plants in terms of high efficiency, reduced off-targets and rapid GE (Liu et al., 2015; Woo et al., 2015; Kanchiswamy, 2016).
Crispr-Based Future Applications In Date Palm
The contemporary CRISPR versions are revolutionizing the functional genomics and molecular biology especially in characterization of the genes functions through gain and/or loss-of functions. In date palm, random mutation induced through irradiation and magnetic field has been employed to create genetic variability and selection against abiotic stresses and bayoud disease (Jain et al., 2011). However, such randomly induced mutagenesis has a severe drawback of enormous background mutation load (Braatz et al., 2017). Instead of using conventional mutagenic systems, the use of CRISPR/Cas9 system in date palm GE will definitely open up new horizons in date palm biological research. The classical mutagenic approaches are not necessarily suitable for inactivation of every gene under study due to variable nature of gene integration, which could end up in appearance of transgenes (Khatodia et al., 2016). However, site-specific mutagenesis through CRISPR/Cas9 system will enable the scientists to study gene expression of such inaccessible genes and targeting multiple loci in large date palm genome. It can also be employed to target various pathogen effectors, explore secondary metabolites in date palm fruit and improving quantitative and qualitative traits (Figure 4).
FIGURE 4.
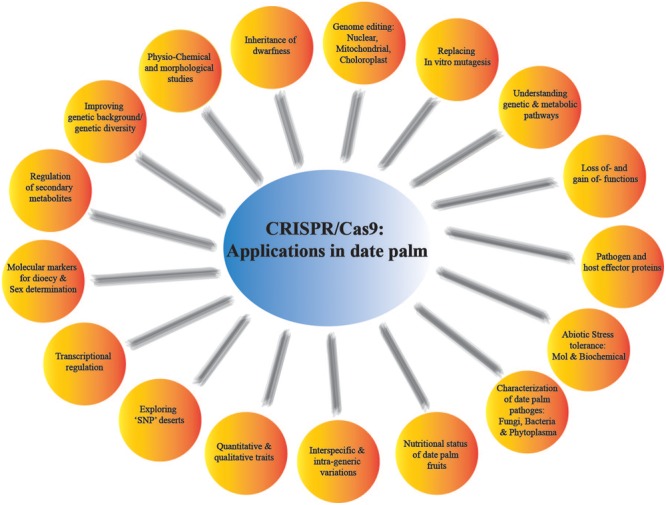
Depicted future applications of CRISPR/Cas9 in date palm genome editing and beyond.
Moreover, sex determination in the new borne date palm progeny is not possible until the plants are matured. There are several marker-assisted approaches available to determine dioecious status of the date palm sprouts, but the exact sex determinants can be explored by CRISPR/Cas9-based approaches. By employing the gene knocking ability, CRISPR system can be used to identify genetic markers for sex determination in date palm. The targeted mutation of many negatively regulated resistance genes have been knockout or knockdown for example, OsERF922 against rice blast (Liu et al., 2012; Wang et al., 2016) and TMS5 for temperature sensitivity (Zhou et al., 2014), in rice. CRISPR/Cas9-based GE approaches provide a throughput tool for functional genomics and genetic improvement in crop plants. It can help in promoting yield components, overall plant architecture, nutrient uptake efficiency, and promoting natural plant adaptation to various stresses. Additionally, CRISPR/Cas9 system can be explored to curtail phytoplasma diseases in date palm. The additional applications may help in regulating secondary metabolites production from date palm fruits and production of biofuel from the date palm wastes (Figure 4).
Many agronomically important loci are located in the “SNP deserts” in the plant genome (Xu et al., 2012). SNP deserts in date palm genome recruit high density of abiotic and biotic resistance genes as compared to the whole genome (Al-Mssallem et al., 2013). These SNP deserts can be targeted to devise resistance strategies against major abiotic and biotic stresses in date palm through CRISPR/Cas9 system. Many such characters such as up-regulation of antioxidants, enhanced flavor, pests and disease resistance can be promoted through GE in date palm.
Concluding Remarks
In future, the genetic improvement in date palm would accelerate progress against abiotic and biotic challenges for sustainable production using GE technologies. The drawbacks of conventional breeding in date palm could be circumvented by genetic manipulation and transcriptional control of various physiological and metabolic processes. The site-specific gene insertion, specificity in targeted mutation and controlled genetic manipulations can make CRISPR/Cas9 a novel tool in the date palm breeder’s kit. The availability of the date palm full genome sequence opened up new territories for yield improvement through GE and genetic manipulation as well as the applications of high-throughput genome sequencing may revolutionize date palm biotechnology by introducing targeted genetic modifications and new functionalities. It will also pave the way to improve flowering, sex determination, juvenility control and manipulation of fruit ripening, fruit quality and nutritional value in date palm. The futuristic outcomes of the application of CRISPR/Cas9 in date palm GE will not only address the basic biological questions but will definitely reduce the concerns of common people due to its non-GMO nature.
Author Contributions
MS conceived the idea, produced the first draft of the manuscript, and helped to draw the figures. ZI collected the data, helped to draw the figures, and participated to produce the first draft of the manuscript. MT helped to formulate all the tables. MS participated to retrieve the data and helped to prepare first draft of the manuscript. MK helped to write first draft of the manuscript. AA-K and SA-K helped to finalize the final draft of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01469/full#supplementary-material
References
- Ainley W. M., Sastry-Dent L., Welter M. E., Murray M. G., Zeitler B., Amora R., et al. (2013). Trait stacking via targeted genome editing. Plant Biotechnol. J. 11 1126–1134. 10.1111/pbi.12107 [DOI] [PubMed] [Google Scholar]
- Al-Dous E. K., George B., Al-Mahmoud M. E., Al-Jaber M. Y., Wang H., Salameh Y. M., et al. (2011). De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat. Biotechnol. 29 521–527. 10.1038/nbt.1860 [DOI] [PubMed] [Google Scholar]
- Ali Z., Abul-faraj A., Li L., Ghosh N., Piatek M., Mahjoub A., et al. (2015). Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 8 1288–1291. 10.1016/j.molp.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Al-Khayri J. M., Jain S. M., Johnson D. V. (2015). Date Palm Genetic Resourse and Utilization. Berlin: Springer. [Google Scholar]
- Al-Mssallem I. S., Hu S., Zhang X., Lin Q., Liu W., Tan J., et al. (2013). Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 4 2274 10.1038/ncomms3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sadi A. M., Al-Jabri A. H., Al-Mazroui S. S., Al-Mahmooli I. H. (2012). Characterization and pathogenicity of fungi and oomycetes associated with root diseases of date palms in Oman. Crop Prot. 37 1–6. 10.1016/j.cropro.2012.02.011 [DOI] [Google Scholar]
- Arzani A., Ashraf M. (2016). Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit. Rev. Plant Sci. 35 146–189. 10.1080/07352689.2016.1245056 [DOI] [Google Scholar]
- Bekheet S. A., Hanafy M. S. (2011). “Date palm biotechnology,” in Towards Sex Determination of Date Palm eds Jain S. M., Al-Khayri J. M., Johnson D. V. (Dordrecht: Springer; ) 551–566. 10.1007/978-94-007-1318-5_26 [DOI] [Google Scholar]
- Belhaj K., Chaparro-Garcia A., Kamoun S., Nekrasov V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9 39–47. 10.1186/1746-4811-9-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi L., Fischer R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33 41–52. 10.1016/j.biotechadv.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Bourgis F., Kilaru A., Cao X., Ngando-Ebongue G. F., Drira N., Ohlrogge J. B., et al. (2011). Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. U.S.A. 108 12527–12532. 10.1073/pnas.1106502108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braatz J., Harloff H.-J., Mascher M., Stein N., Himmelbach A., Jung C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174 935–942. 10.1104/pp.17.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C., Nekrasov V., Lippman Z. B., Van Eck J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 System. Plant Physiol. 166 1292–1297. 10.1104/pp.114.247577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler N. M., Baltes N. J., Voytas D. F., Douches D. S. (2016). Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front. Plant Sci. 7:1045 10.3389/fpls.2016.01045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char S. N., Neelakandan A. K., Nahampun H., Frame B., Main M., Spalding M. H., et al. (2017). An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15 257–268. 10.1111/pbi.12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., Bae S., et al. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24 132–141. 10.1101/gr.162339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Qi Y., Zhang Y., Voytas D. F. (2013). Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 (Bethesda) 3 1697–1705. 10.1534/g3.113.007104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N., Mitra A., Silva M. J., Bienko M., Dojer N., Wang Q., et al. (2013). Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10 361–365. 10.1038/nmeth.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S. J., Anderson J. E., Starker C. G., Baltes N. J., Mani D., Voytas D. F., et al. (2013). “Targeted mutagenesis for functional analysis of gene duplication in legumes,” in Legume Genomics: Methods and Protocols ed. Rose R. J. (Totowa, NJ: Humana Press; ) 25–42. [DOI] [PubMed] [Google Scholar]
- Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., Locke A. S., et al. (2012). Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8:e1002861 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Li H., Chen L. L., Xie K. (2016). Recent advances in genome editing using CRISPR/Cas9. Front. Plant Sci. 7:703 10.3389/fpls.2016.00703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., Xie K., Chen Y., Yang Y., Mao Y. (2017). Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem. Biophys. Res. Commun. 482 889–895. 10.1016/j.bbrc.2016.11.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Rabey H. A., Al-Malki A. L., Abulnaja K. O., Rohde W. (2015). Proteome analysis for understanding abiotic stress (salinity and drought) tolerance in date palm (Phoenix dactylifera L.). Int. J. Genomics 2015 1–11. 10.1155/2015/407165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D., Liu T., Li C., Jiao B., Li S., Hou Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in populus in the first generation. Sci. Rep. 5:12217 10.1038/srep12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Tyler B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 17 127–139. 10.1111/mpp.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Wu H., Zhang T., Yang M., Yin Y., Pan L., et al. (2012). A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome. PLoS ONE 7:e37164 10.1371/journal.pone.0037164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F., Schiml S., Puchta H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79 348–359. 10.1111/tpj.12554 [DOI] [PubMed] [Google Scholar]
- Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 4632–4637. 10.1073/pnas.1400822111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry M., Gomez S. (2002). The red palm weevil in the Mediterranean area. J. Int. Palm Soc. 46 172–178. [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., et al. (2013). High frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31 822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32 279–284. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang G., Ma S., Xie X., Wu X., Zhang X., et al. (2015). CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 87 99–110. 10.1007/s11103-014-0263-0 [DOI] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri K. M., Flowers J. M., Visser H. J., Khierallah H. S. M., Rosas U., Pham G. M., et al. (2015). Whole genome re-sequencing of date palms yields insights into diversification of a fruit tree crop. Nat. Commun. 6 8824 10.1038/ncomms9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A., Hirose M., Sankai T., Yasmin L., Yuzawa K., Honsho K., et al. (2015). Single-step generation of rabbits carrying a targeted allele of the tyrosinase gene using CRISPR/Cas9. Exp. Anim. 64 31–37. 10.1538/expanim.14-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Wang C., Huang J., Wang K. (2017). A simple and efficient method for CRISPR/Cas9-induced mutant screening. J. Genet. Gen. 44 207–213. 10.1016/j.jgg.2017.03.005 [DOI] [PubMed] [Google Scholar]
- Iqbal Z., Sattar M. N., Shafiq M. (2016). CRISPR/Cas9: a tool to circumscribe cotton leaf curl disease. Front. Plant Sci 7:475 10.3389/fpls.2016.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T. B., LaFayette P. R., Schmitz R. J., Parrott W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16 10.1186/s12896-015-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S. M., Al-Khayri J., Johnson D. V. (2011). Date Palm Biotechnology. New York, NY: Springer Science+Business Media B.V. 10.1007/978-94-007-1318-5 [DOI] [Google Scholar]
- Jia H., Orbovic V., Jones J. B., Wang N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14 1291–1301. 10.1111/pbi.12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Wang N. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:e93806 10.1371/journal.pone.0093806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41:e188 10.1093/nar/gkt780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubrael J. M. S., Udupa S. M., Baum M. (2005). Assessment of AFLP-based genetic relationships among date palm (Phoenix dactylifera L.) varieties of Iraq. J. Am. Soc. Hortic. Sci. 130 442–447. [Google Scholar]
- Kanchiswamy C. N. (2016). DNA-free genome editing methods for targeted crop improvement. Plant Cell Rep. 35 1469–1474. 10.1007/s00299-016-1982-2 [DOI] [PubMed] [Google Scholar]
- Kanda Y., Yokotani N., Maeda S., Nishizawa Y., Kamakura T., Mori M. (2017). The receptor-like cytoplasmic kinase BSR1 mediates chitin-induced defense signaling in rice cells. Biosci. Biotechnol. Biochem. 81 1497–1502. 10.1080/09168451.2017.1325710 [DOI] [PubMed] [Google Scholar]
- Khan A., Khan I. A., Heinze B., Azim M. K. (2012). The chloroplast genome sequence of date palm (Phoenix dactylifera L. cv. ‘Aseel’). Plant Mol. Biol. Rep. 30 666–678. 10.1007/s11105-011-0373-7 [DOI] [Google Scholar]
- Khan M. S., Khalid A. M., Malik K. A. (2005). Phage phiC31 integrase: a new tool in plastid genome engineering. Trends Plant Sci. 10 1–3. 10.1016/j.tplants.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Khatodia S., Bhatotia K., Passricha N., Khurana S. M. P., Tuteja N. (2016). The CRISPR/Cas genome-editing tool: application in improvement of crops. Front. Plant Sci. 7:506 10.3389/fpls.2016.00506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatodia S., Bhatotia K., Tuteja N. (2017). Development of CRISPR/Cas9 mediated virus resistance in agriculturally important crops. Bioengineered 8 274–279. 10.1080/21655979.2017.1297347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., Pattanayak V., Prew M. S., Tsai S. Q., Nguyen N. T., Zheng Z., et al. (2016). High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529 490–495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., Prew M. S., Tsai S. Q., Topkar V. V., Nguyen N. T., Zheng Z., et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523 481–485. 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M. H., Gilbert L. A., Wang X., Lim W. A., Weissman J. S., Qi L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8 2180–2196. 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Cradick T. J., Fine E. J., Bao G. (2016). Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing. Mol. Ther. 24 475–487. 10.1038/mt.2016.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Kim J. K., Lee S., Choi S., Kim S., Hwang I. (2008). Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20 1603–1622. 10.1105/tpc.108.060541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S. Q., Larson Matthew H., Gilbert, Luke A., Doudna, Jennifer A., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. F., Aach J., Norville J. E., McCormack M., Zhang D., Bush J., et al. (2013). Multiplex and homologous recombination-mediated plant genome editing via guide RNA/Cas9. Nat. Biotechnol. 31 688–691. 10.1038/nbt.2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li R., Li X., Fu D., Zhu B., Tian H., et al. (2017). Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 10.1111/pbi.12781 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Chen X., Liu J., Ye J., Guo Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63 3899–3911. 10.1093/jxb/ers079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Gaj T., Yang Y., Wang N., Shui S., Kim S., et al. (2015). Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat. Protoc. 10 1842–1859. 10.1038/nprot.2015.117 [DOI] [PubMed] [Google Scholar]
- Lowder L. G., Paul J. W., Qi Y. (2017). “Multiplexed transcriptional activation or repression in plants using CRISPR-dCas9-based systems,” in Plant Gene Regulatory Networks: Methods and Protocols eds Kaufmann K., Mueller-Roeber B. (New York, NY: Springer; ) 167–184. [DOI] [PubMed] [Google Scholar]
- Lu H. P., Liu S. M., Xu S. L., Chen W. Y., Zhou X., Tan Y. Y., et al. (2017). CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol. J. 10.1111/pbi.12788 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8 1274–1284. 10.1016/j.molp.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Ma X., Zhu Q., Chen Y., Liu Y. G. (2016). CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol. Plant 9 961–974. 10.1016/j.molp.2016.04.009 [DOI] [PubMed] [Google Scholar]
- Mahmoudi H., Hosseininia G., Azadi H., Fatemi M. (2008). Enhancing date palm processing, marketing and pest control through organic culture. J. Organ. Syst. 3 29–39. [Google Scholar]
- Mali P., Aach J., Stranges P. B., Esvelt K. M., Moosburner M., Kosuri S., et al. (2013). Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31 833–838. 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoy M., Viola R., Jung M. H., Koo O. J., Kim S., Kim J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7:1904 10.3389/fpls.2016.01904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang H., Xu N., Zhang B., Gao F., Zhu J. K. (2013). Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6 2008–2011. 10.1093/mp/sst121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew L. S., Seidel M. A., George B., Mathew S., Spannagl M., Haberer G., et al. (2015). A genome-wide survey of date palm cultivars supports two major subpopulations in Phoenix dactylifera. G3 (Bethesda) 5 1429–1438. 10.1534/g3.115.018341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercx S., Smargiasso N., Chaumont F., De Pauw E., Boutry M., Navarre C. (2017). Inactivation of the β(1,2)-xylosyltransferase and the α(1,3)-fucosyltransferase genes in Nicotiana tabacum BY-2 cells by a multiplex CRISPR/Cas9 strategy results in glycoproteins without plant-specific glycans. Front. Plant Sci. 8:403 10.3389/fpls.2017.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23 1233–1236. 10.1038/cr.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami M., Toki S., Endo M. (2016). Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. 57 1058–1068. 10.1093/pcp/pcw049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkenberg B., Xie K., Yang Y. (2017). Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J. 89 636–648. 10.1111/tpj.13399 [DOI] [PubMed] [Google Scholar]
- Mokhtar M. M., Adawy S. S., El-Assal S. E.-D. S., Hussein E. H. A. (2016). Genic and intergenic SSR database generation, SNPs determination and pathway annotations, in date palm (Phoenix dactylifera L.). PLoS ONE 11:e0159268 10.1371/journal.pone.0159268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi M., Mousavi A., Habashi A. A., Dehsara B. (2014). Genetic transformation of date palm (Phoenix dactylifera L. cv. ‘Estamaran’) via particle bombardment. Mol. Biol. Rep. 41 8185–8194. 10.1007/s11033-014-3720-6 [DOI] [PubMed] [Google Scholar]
- Nekrasov V., Wang C., Win J., Lanz C., Weigel D., Kamoun S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7 482 10.1038/s41598-017-00578-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani C., Hirai N., Komori S., Wada M., Okada K., Osakabe K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6 1–8. 10.1038/srep31481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pater S., Pinas J. E., Hooykaas P. J. J., Zaal B. J. (2013). ZFN-mediated gene targeting of the Arabidopsis protoporphyrinogen oxidase gene through Agrobacterium-mediated floral dip transformation. Plant Biotechnol. J. 11 510–515. 10.1111/pbi.12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., Liu D. R. (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31 839–843. 10.1038/nbt.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A., Chen S., Lei T., Xu L., He Y., Wu L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 10.1111/pbi.12733 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. A., Haak D. C., Nishimura M. T., Teixeira P. J. P. L., James S. R., Dangl J. L., et al. (2016). Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS ONE 11:e0162169 10.1371/journal.pone.0162169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintaud J. C., Ludeña B., Aberlenc Bertossi F., Zehdi S., Gros Balthazard M., Ivorra S., et al. (2011). “Biogeography of the date palm (Phoenix dactylifera L., Arecaceae): insights on the origin and on the structure of modern diversity,” in Proceedings of the Ist International Symposium on Date Palm Algeria: 19–38. [Google Scholar]
- Qi W., Zhu T., Tian Z., Li C., Zhang W., Song R. (2016). High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 16:58 10.1186/s12896-016-0289-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Li X., Zhang Y., Starker C. G., Baltes N. J., Zhang F., et al. (2013). Targeted deletion and inversion of tandemly arrayed genes in Arabidopsis thaliana using zinc finger nucleases. G3 (Bethesda) 3 1707–1715. 10.1534/g3.113.006270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu Patrick D., Lin C. Y., Gootenberg, Jonathan S., Konermann S.et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154 1380–1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekik I., Chaabene Z., Grubb C. D., Drira N., Cheour F., Elleuch A. (2015). In silico characterization and molecular modeling of double-strand break repair protein MRE11 from Phoenix dactylifera vs deglet nour. Theor. Biol. Med. Model. 12 23 10.1186/s12976-015-0013-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir J. S. M., Arasappan D., Bahieldin A., Abo-Aba S., Bafeel S., Zari T. A., et al. (2014). Whole mitochondrial and plastid genome SNP analysis of nine date palm cultivars reveals plastid heteroplasmy and close phylogenetic relationships among cultivars. PLoS ONE 9:e94158 10.1371/journal.pone.0094158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiml S., Fauser F., Puchta H. (2014). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80 1139–1150. 10.1111/tpj.12704 [DOI] [PubMed] [Google Scholar]
- Sedra M. (2007). “Bayoud disease of date palm in North Africa: recent distribution and remarks about its characterization, diagnosis and origin,” in Proceedings of the Fourth Symposium Date Palm Hofuf: King Faisal University. [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31 686–688. 10.1038/nbt.2650 [DOI] [PubMed] [Google Scholar]
- Shen L., Hua Y., Fu Y., Li J., Liu Q., Jiao X., et al. (2017). Rapid generation of genetic diversity by multiplex CRISPR/Cas9 genome editing in rice. Sci. China Life Sci. 60 506–515. 10.1007/s11427-017-9008-8 [DOI] [PubMed] [Google Scholar]
- Shukla V. K., Doyon Y., Miller J. C., DeKelver R. C., Moehle E. A., Worden S. E., et al. (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459 437–441. 10.1038/nature07992 [DOI] [PubMed] [Google Scholar]
- Siebert R., Puchta H. (2002). Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 14 1121–1131. 10.1105/tpc.001727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I. M., Gao L., Zetsche B., Scott D. A., Yan W. X., Zhang F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351 84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker T. H., Petrini J. H. J. (2011). The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12 90–103. 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Li N., Huang G., Xu J., Pan Y., Wang Z., et al. (2013). Site-specific gene targeting using transcription activator-like effector (TALE)-based nuclease in Brassica oleracea. J. Integrat. Plant Biol. 55 1092–1103. 10.1111/jipb.12091 [DOI] [PubMed] [Google Scholar]
- Svitashev S., Young J. K., Schwartz C., Gao H., Falco S. C., Cigan A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169 931–945. 10.1104/pp.15.00793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend J. A., Wright D. A., Winfrey R. J., Fu F., Maeder M. L., Joung J. K., et al. (2009). High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459 442–445. 10.1038/nature07845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. J., Xue L. J. (2015). CRISPRing into the woods. GM Crops Food 6 206–215. 10.1080/21645698.2015.1091553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S. K., Kumar J., Alok A., Tuli R. (2013). RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 3 2233–2238. 10.1534/g3.113.008847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang C., Liu P., Lei C., Hao W., Gao Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11:e0154027 10.1371/journal.pone.0154027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Mao Y., Lu Y., Tao X., Zhu J.-K. (2017). Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol. Plant 10 1011–1013. 10.1016/j.molp.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Wendt T., Holm P. B., Starker C. G., Christian M., Voytas D. F., Brinch-Pedersen H., et al. (2013). TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol. Biol. 83 279–285. 10.1007/s11103-013-0078-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J. W., Kim J., Kwon S. I., Corvalan C., Cho S. W., Kim H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33 1162–1164. 10.1038/nbt.3389 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Xia W., Yang Y., Mason A. S., Lei X., Ma Z. (2013). Characterization and evolution of conserved microRNA through duplication events in date palm (Phoenix dactylifera). PLoS ONE 8:e71435 10.1371/journal.pone.0071435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Minkenberg B., Yang Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 112 3570–3575. 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Yang Y. (2013). RNA-guided genome editing in plants using A CRISPR-Cas system. Mol. Plant 6 1975–1983. 10.1093/mp/sst119 [DOI] [PubMed] [Google Scholar]
- Xin C., Liu W., Lin Q., Zhang X., Cui P., Li F., et al. (2015). Profiling microRNA expression during multi-staged date palm (Phoenix dactylifera L.) fruit development. Genomics 105 242–251. 10.1016/j.ygeno.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Xu X., Liu X., Ge S., Jensen J. D., Hu F., Li X., et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30 105–111. 10.1038/nbt.2050 [DOI] [PubMed] [Google Scholar]
- Yaish M. W., Sunkar R., Zheng Y., Ji B., Al-Yahyai R., Farooq S. A. (2015). A genome-wide identification of the miRNAome in response to salinity stress in date palm (Phoenix dactylifera L.). Front. Plant Sci. 6:946 10.3389/fpls.2015.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Zhang X., Liu G., Yin Y., Chen K., Yun Q., et al. (2010). The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS ONE 5:e12762 10.1371/journal.pone.0012762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Zhang X., Fang Y., Pan L., Sun G., Xin C., et al. (2012). High-throughput sequencing-based gene profiling on multi-staged fruit development of date palm (Phoenix dactylifera L.). Plant Mol. Biol. 78 617–626. 10.1007/s11103-012-9890-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Voytas D. F. (2011). “Targeted mutagenesis in Arabidopsis using zinc-finger nucleases,” in Plant Chromosome Engineering: Methods and Protocols ed. Birchler J. A. (Totowa, NJ: Humana Press; ) 167–177. [DOI] [PubMed] [Google Scholar]
- Zhang G., Pan L., Yin Y., Liu W., Huang D., Zhang T., et al. (2012). Large-scale collection and annotation of gene models for date palm (Phoenix dactylifera L.). Plant Mol. Biol. 79 521–536. 10.1007/s11103-012-9924-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P., Zhang B., Gou F., Feng Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12 797–807. 10.1111/pbi.12200 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Bai Y., Wu G., Zou S., Chen Y., Gao C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91 714–724. 10.1111/tpj.13599 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liang Z., Zong Y., Wang Y., Liu J., Chen K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7 1–8. 10.1038/ncomms12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7 30–38. 10.1186/1746-4811-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Williams R., Prakash C. S., He G. (2012). Identification and characterization of gene-based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol. 12:237 10.1186/1471-2229-12-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang F., Li X., Baller J. A., Qi Y., Starker C. G., et al. (2013). Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 161 20–27. 10.1104/pp.112.205179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Mao Y., Ha S., Liu W., Botella J. R., Zhu J. K. (2016). A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 35 1519–1533. 10.1007/s00299-015-1900-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Liu B., Weeks D. P., Spalding M. H., Yang B. (2014). Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 12 10903–10914. 10.1093/nar/gku806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jacobs T. B., Xue L. J., Harding S. A., Tsai C. J. (2015). Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol. 208 298–301. 10.1111/nph.13470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


