Abstract
Background: Fat-enriched diets produce metabolic changes in skeletal muscle, which in turn can mediate changes in gene regulation.
Objective: We examined the high-fat-diet–induced changes in skeletal muscle gene expression by characterizing variations in pre-mRNA alternative splicing.
Methods: Affymetrix Exon Array analysis was performed on the transcriptome of the gastrocnemius/plantaris complex of male obesity-prone Sprague-Dawley rats fed a 10% or 60% fat (lard) diet for 2 or 8 wk. The validation of exon array results was focused on troponin T (Tnnt3). Tnnt3 splice form analyses were extended in studies of rats fed 10% or 30% fat diets across 1- to 8-wk treatment periods and rats fed 10% or 45% fat diets with fat sources from lard or mono- or polyunsaturated fats for 2 wk. Nuclear magnetic resonance (NMR) was used to measure body composition.
Results: Consumption of a 60% fat diet for 2 or 8 wk resulted in alternative splicing of 668 and 726 pre-mRNAs, respectively, compared with rats fed a 10% fat diet. Tnnt3 transcripts were alternatively spliced in rats fed a 60% fat diet for either 2 or 8 wk. The high-fat-diet–induced changes in Tnnt3 alternative splicing were observed in rats fed a 30% fat diet across 1- to 8-wk treatment periods. Moreover, this effect depended on fat type, because Tnnt3 alternative splicing occurred in response to 45% fat diets enriched with lard but not in response to diets enriched with mono- or polyunsaturated fatty acids. Fat mass (a proxy for obesity as measured by NMR) did not differ between groups in any study.
Conclusions: Rat skeletal muscle responds to overconsumption of dietary fat by modifying gene expression through pre-mRNA alternative splicing. Variations in Tnnt3 alternative splicing occur independently of obesity and are dependent on dietary fat quantity and suggest a role for saturated fatty acids in the high-fat-diet–induced modifications in Tnnt3 alternative splicing.
Keywords: high-fat diet, saturated fat, MUFA, PUFA, alternative splicing, troponin T
Introduction
Skeletal muscle comprises ~40% of body weight in nonobese people and accounts for almost 80% of glucose disposal under euglycemic and hyperinsulinemic conditions (1). However, in individuals with type 2 diabetes, the sensitivity of muscle to insulin-stimulated glucose uptake is dramatically impaired. Consequently, insulin-stimulated glucose disposal by muscle is significantly reduced compared with that of nondiabetic subjects (2). The development of insulin resistance in skeletal muscle is associated with lipid accumulation and dysfunctional mitochondria (2–4), and a recent study shows that such changes are in part due to an altered pattern of skeletal muscle gene expression (5). Similarly, in rodent models of type 2 diabetes induced by feeding a high-fat diet, development of insulin resistance is associated with a large number of changes in skeletal muscle gene expression (6). Although many of the alterations in gene expression involve proteins that function in carbohydrate and lipid metabolism, expression of a number of genes that have structural roles or are involved in muscle contraction are also altered, and this is observed in both humans and rodents (7–9). For example, soleus muscle fast skeletal muscle troponin T (Tnnt3) gene expression is significantly lower in the muscle of high-fat-diet–fed female mice compared with those fed a control diet (6). Moreover, our group previously showed that the alternative splicing of the Tnnt3 pre-mRNA responds to both naturally and experimentally induced changes in body mass and that this process is impaired in Zucker rats, a genetic model of obesity (10), suggesting that excessive calorie consumption may impair normal regulation of alternative splicing. Indeed, dietary intake affects Tnnt3 alternative splicing in insect flight muscle (11), but this has yet to be demonstrated in mammals.
Alternative splicing of pre-mRNA is a critically important process for increasing protein functional diversity by generating multiple mature mRNAs from a single gene. Indeed, 90–95% of all human protein coding genes undergo alternative splicing (12, 13). Of the tissues examined, skeletal muscle has one of the highest tissue-specific proportions of alternative splicing (13), and alternative splicing of pre-mRNA is critical in regulating physiological processes in skeletal muscle. For example, a recent study demonstrated that alternatively spliced forms of genes involved in cellular trafficking are necessary for proper functioning of adult skeletal muscle (14). Also, disruption of the alternative splicing of skeletal muscle genes controlling myofibril structure and calcium handling impairs muscle function (15). These findings highlight the importance of understanding factors that affect alternative splicing in skeletal muscle. Unfortunately, although alterations in skeletal muscle gene expression in response to nutrient overconsumption have been studied extensively (16–20), few studies to our knowledge have investigated variations in alternative splicing of pre-mRNA under such conditions. In the present study we used Affymetrix Rat Exon 1.0 ST Arrays to survey transcriptome-wide changes in the pattern of pre-mRNA alternative splicing in the skeletal muscle of rats fed a 10% fat or a 60% fat diet for 2 or 8 wk. Tnnt3 was among the top 10 alternatively spliced genes affected by the high-fat diet. We confirmed the Tnnt3 alternative splicing data from the exon array by PCR and extended them by performing a time course study using a 30% fat diet. In addition, because previous studies have shown differential effects of SFAs compared with unsaturated FAs on alternative splicing of pre-mRNAs in C2C12 myotubes (21), we also compared the effect of feeding diets enriched with MUFAs and PUFAs with one enriched with lard.
Methods
Animal protocol.
The animal protocols for the studies described herein were reviewed and approved by the Penn State College of Medicine Institutional Animal Care and Use Committee. Male obesity-prone Sprague-Dawley rats (OP-CD/463) were obtained from Charles River Laboratories, housed under a reverse 12-h light/12-h dark cycle, and given ad libitum access to water throughout the course of the studies. Diets containing 10% fat (D12450J or D12450H), 30% fat (D08091811), 45% fat (D12451), and 60% fat from lard (D12492) were purchased from Research Diets. For diets with varying FA quantity and type, the lard present in D12450H and D12451 was replaced with MUFAs from high-oleic sunflower oil (D16010601 and D07062503, respectively), or PUFAs from safflower oil (D16010602 and D02062102, respectively). The exact nutrient composition and FA profiles for each of the diets used are provided in Supplemental Tables 1–3. The diets were designed to have the same nutrient content per calorie. No statistically significant differences were observed for daily food intake (grams per day) between groups (data not shown). On the day of harvesting muscle, rats were placed under isoflurane anesthesia (EZ Anesthesia), and the gastrocnemius and plantaris complex (hereafter referred to as muscle) was removed from anesthetized rats, frozen between aluminum blocks precooled in liquid nitrogen, and stored at −80°C until further analysis. Rats were killed after the removal of muscle and while under anesthesia. Rats were placed into a Bruker MiniSpec whole-body composition analyzer to measure lean and fat tissue by NMR the day of or 2 d before tissue harvest.
Affymetrix rat exon 1.0 ST array.
Total RNA (2 μg; n = 4/diet group from individual rats) was treated with DNase I per the manufacturer’s protocol (Invitrogen). RNA quality was assessed by using the Agilent 2100 Bioanalyzer and RNA Nano 6000 Chip (Agilent Technologies), ensuring that all samples had RNA integrity number values >8. Sense-stranded cDNA was prepared from 100 ng total RNA by using a Whole Transcript Expression Kit (Life Technologies, Thermo Fisher). The sense-stranded cDNA was fragmented and labeled according to the manufacturer’s protocol by using a GeneChip Whole Transcript Terminal Expression Kit (Affymetrix) at the Penn State Genomics Core Facility, University Park, Pennsylvania. Fragmentation and labeling was confirmed by using an RNA Nano Chip on the Agilent Bioanalyzer. The GeneChip Hybridization Wash and Stain Kit (Affymetrix) was used to complete subsequent hybridization, washing, and staining of the arrays. Samples were hybridized to the GeneChip Rat Exon 1.0 ST Array (Affymetrix) in the GeneChip Hybridization Oven 640 (Affymetrix) according to the manufacturer’s protocol. After hybridization, the arrays were washed and stained on the Affymetrix GeneChip Fluidics Station 450. Stained arrays were scanned by using the GeneChip Scanner 3000 7G (Affymetrix).
Quantification of Tnnt3 pre-mRNA splice form abundance.
Total RNA was extracted from frozen, powdered muscle by using TRIzol Reagent (Invitrogen, Thermo Fisher) according to the manufacturer’s protocol. Total RNA (1 μg) was reverse transcribed to cDNA by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher). The Tnnt3 splice form characterization and quantification were performed as described previously (10).
Data analysis and statistics.
Raw Affymetrix .CEL files were uploaded to AltAnalyze v2.1.0 containing probeset annotations for Rattus norvegicus from EnsMart65. Affymetrix Power Tools with the use of the robust multi-array average as implemented by AltAnalyze (22–24) was used to perform data normalization. Gene-level expression was determined by using the core meta-probeset annotation list (known reference sequence (RefSeq) and full-length mRNA transcripts) (25). Probesets were filtered to include those that had a detection above a background P value of <0.05, a nonlog expression value >70, and were present in each experimental condition. Differentially expressed probeset (i.e., alternatively spliced pre-mRNA) statistics were determined by using the finding isoforms by using the robust multichip analysis (FIRMA) method within AltAnalyze. Briefly, the FIRMA model calculates a score based on how much a probeset’s expression value deviates from the expected gene expression value (i.e., residuals) (26). A probeset’s FIRMA score is used to calculate the fold change and summary statistics between the control and experimental groups. In addition, AltAnalyze uses Affymetrix Power Tools to calculate a microarray detection of alternative splicing (MiDAS) P value for each probeset. Briefly, each probeset expression value within a gene is normalized to the gene-level expression value. The MiDAS method is based on the ratio of gene-level corrected probeset expression values between experimental groups (27). Both the FIRMA and MiDAS methods used the extended meta-probeset annotation list (core meta-probeset and cDNA based annotations) (25) for alternative splicing analysis. Probesets were considered differentially expressed if they had FIRMA fold changes of >∣1.5∣, a FIRMA P value <0.05, and a MiDAS P value <0.05. An exon array workflow diagram is shown in Supplemental Figure 1. Gene ontology (GO) Elite and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Mapping analyses were performed on the genes containing differentially expressed probesets. Fisher’s exact P values for GO and KEGG analyses were calculated by using AltAnalyze.
The log2 signal intensities from differentially expressed probesets at 2 and 8 wk were subjected to a 3-way principal component (PC) analysis by using the singular-value decomposition algorithm (28). Heatmaps were generated with the log2 signal intensities from differentially expressed probesets at 2 or 8 wk by using the hierarchical ordered partitioning and collapsing hybrid (HOPACH) clustering algorithm (29). Briefly, samples and probesets are grouped into columns and rows, respectively. The HOPACH clustering algorithm builds a hierarchical tree of data (i.e., clusters of probesets) by grouping them based on similarity of their features.
Two-factor ANOVA was used to determine the effect of time, fat quantity, or fat type on the Tnnt3 splice form expression. The Holm-Sidak test was used post hoc to determine specific differences if main effects or the interaction was significant. GraphPad Prism v7.0 was used to perform ANOVA tests and to generate figures. The mean Tnnt3 relative abundances are displayed in the figures. Statistical analysis was performed on arcsine-transformed relative abundance to achieve normality.
Results
Visualization of high-fat-diet–induced changes in the alternative splicing of pre-mRNA.
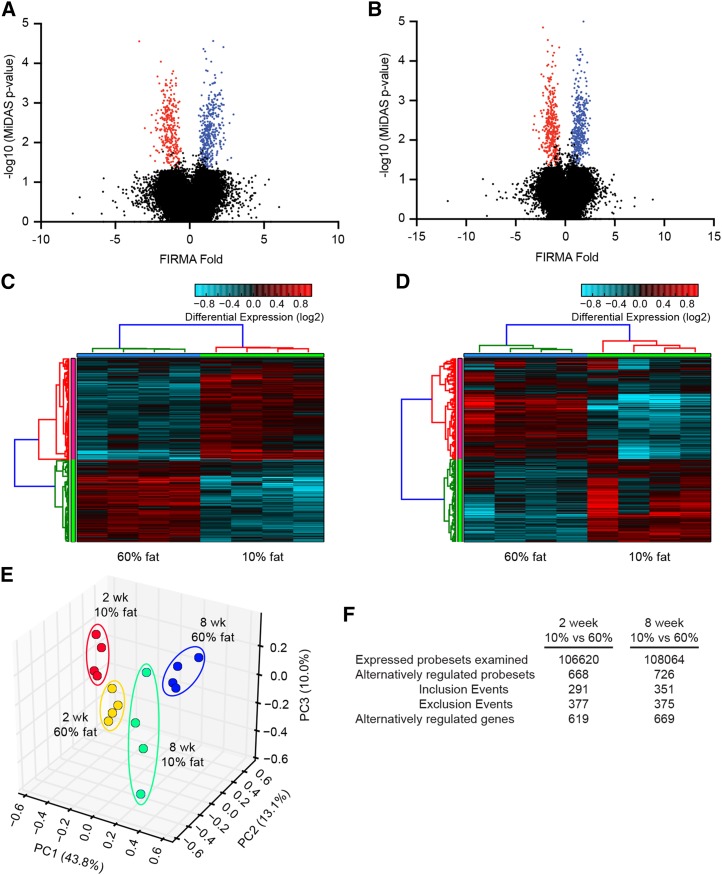
After data normalization and filtering, 106,620 and 108,064 of 366,685 probesets from rats fed experimental diets for 2 or 8 wk, respectively, met the criteria for subsequent alternative splicing analysis. Within this probeset subset, 668 and 726 differentially expressed probesets were detected in the muscle of rats fed a 60% fat diet for 2 or 8 wk, respectively, compared with rats fed a 10% fat diet (Figure 1F). Differentially expressed probesets are visualized in Figure 1A, B as volcano plots (probeset’s FIRMA fold change value against the −log10 of the MiDAS P values) comparing the muscle from rats fed experimental diets for 2 or 8 wk. Consumption of a 60% fat diet for 2 wk caused the differential inclusion or exclusion of 291 and 377 probesets, respectively, compared with rats fed the 10% fat diet. Moreover, 351 and 375 probesets were differentially included or excluded, respectively, from the muscle of rats fed the 60% fat diet for 8 wk compared with the rats fed the 10% fat diet. The differential expression of probesets affected 619 and 669 distinct genes from rats fed a 60% fat diets for 2 or 8 wk, respectively. (Figure 1F). HOPACH clustering heatmaps show an overall uniform log2 probeset expression pattern from the muscle of rats fed a 10% or 60% fat diet for 2 (Figure 1C) or 8 wk (Figure 1D). PC analysis of the differentially expressed probesets indicated that the first 3 PCs explain 66.9% of the variation in the data with PC1 responsible for 43.8% of the variation, whereas PC2 and PC3 account for 13.1% and 10.0% of the variation, respectively (Figure 1E). These data demonstrate that consumption of a high-fat diet causes changes in the pattern of alternative splicing in skeletal muscle.
FIGURE 1.
Visualization of differentially expressed probesets from the Rat Exon 1.0 ST Array from rats fed a 10% or 60% fat diet for 2 or 8 wk. FIRMA fold change values plotted against the −log10 of the MiDAS P value from rats fed a 10% or 60% fat diet for 2 (A) or 8 wk (B). Probeset’s with a FIRMA fold change of >∣1.5∣, a FIRMA fold change P value <0.05, and a MiDAS P value <0.05 were deemed to be differentially expressed compared with rats fed a 10 kcal% fat diet. Positive and negative FIRMA fold change values are reflective of excluded (blue) or included (red) probesets compared with control-fed conditions, respectively. Hierarchical ordered partitioning and collapsing hybrid clustering analysis of the log2 probeset expression values from the differentially expressed probesets from animals fed for 2 (C) or 8 wk (D). Red and blue colors in the heatmaps indicate more highly included or excluded probesets, respectively. (E) Three-dimensional scatter-plot of the first 3 PCs of the log2 expression values of differentially expressed probesets. (F) Differentially expressed probeset summary statistics. n = 4/group. FIRMA, finding isoforms by using the robust multichip analysis; MiDAS, microarray detection of alternative splicing; PC, principal component.
GO and KEGG pathway mapping of genes containing differentially expressed probesets.
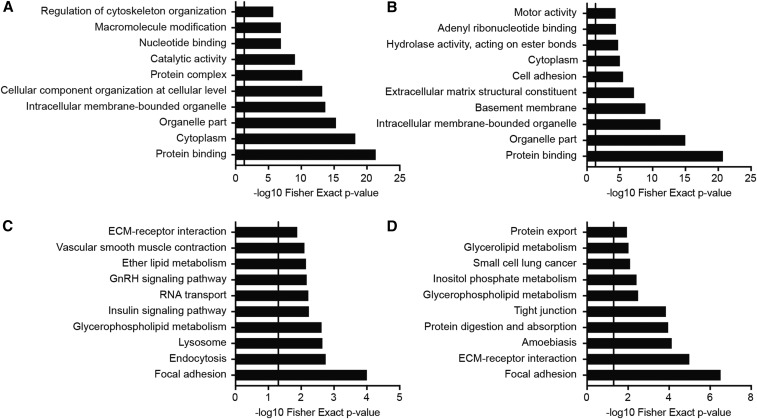
The top 10 most significant GO-Elite terms from rats fed a 10% or 60% fat diet for 2 or 8 wk are shown in Figure 2A, B, respectively. Four of the top 10 GO categories (protein binding, cytoplasm, organelle part, and intracellular membrane–bounded organelle) are common to both time points and had highly significant Fisher’s exact P values. KEGG pathway analysis of genes containing differentially expressed probesets revealed enrichment of several lipid metabolism pathways at 2 and 8 wk (Figure 2C, D, respectively).
FIGURE 2.
Top 10 most significant GO-Elite and KEGG terms from rats fed a 10% or 60% fat diet for 2 or 8 wk. GO-Elite (A and B) or KEGG Pathway (C and D) analysis of genes containing differentially expressed probesets from rats fed experimental diets for 2 (A and C) or 8 wk (B and D). The vertical line within each panel indicates P = 0.05. ECM, extracellular matrix; GnRH, gonadotropin-releasing hormone; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes;
Up- and downregulated probesets.
The top 10 up- and downregulated probesets from rats fed 60% fat diets for 2 wk are listed in Table 1. Probeset 6548887 within exon 2 of the stearoyl-CoA desaturase 1 gene (Scd1) was the most highly downregulated from rats fed a 60% fat diet for 2 wk. Interestingly, probeset 5932570 within exon 6 of Scd1 was among the top 10 upregulated probesets, suggesting that the Scd1 pre-mRNA is subject to multiple alternative splicing events in response to consumption of a high-fat diet. The top 10 up- and downregulated probesets from rats fed 60% fat diets for 8 wk are listed in Table 2. Probeset 5667005 in exon 2 of the netrin 4 gene contained the most highly downregulated probeset from these rats. The top upregulated probeset from these rats was listed as being from an uncharacterized gene. Probeset 6023981 in exon 2 of the RNA binding protein 18 gene was the second most highly upregulated in rats fed 60% fat diets for 8 wk.
TABLE 1.
Top 10 up- and downregulated probesets of characterized genes from rats fed a 10% or 60% fat diet for 2 wk based on FIRMA fold changes
| Symbol | Description | Probeset | FIRMA fold change | Regulation | MiDAS P |
| Scd1 | Stearoyl-CoA desaturase 1 | 6548887 | 2.94 | Downregulated | 0.0421 |
| Spag7 | Sperm associated antigen 7 | 5639798 | 2.78 | Downregulated | 0.0288 |
| Postn | Periostin | 6468013 | 2.68 | Downregulated | 0.0396 |
| Psd3 | Pleckstrin and Sec7 domain containing 3 | 5780988 | 2.54 | Downregulated | 0.0454 |
| Crbn | Protein cereblon | 6241641 | 2.38 | Downregulated | 0.0445 |
| Aifm1 | Apoptosis-inducing factor 1, mitochondrial | 6502947 | 2.31 | Downregulated | 0.0404 |
| Ubr4 | E3 ubiquitin-protein ligase UBR4 | 5965788 | 2.29 | Downregulated | 0.0254 |
| Rasa1 | Ras GTPase-activating protein 1 | 5930788 | 2.28 | Downregulated | 0.0425 |
| Sdhc | Succinate dehydrogenase complex, subunit C | 6712937 | 2.27 | Downregulated | 0.0479 |
| Zcchc2 | Zinc finger CCHC domain-containing protein 2 | 6194754 | 2.27 | Downregulated | 0.0123 |
| Col27a1 | Collagen α-1(XXVII) chain | 6520506 | 3.02 | Upregulated | 0.0409 |
| Hbb | Hemoglobin subunit β-1 | 6509338 | 2.54 | Upregulated | 0.0345 |
| Mrpl50 | 39S ribosomal protein L50, mitochondrial | 6316017 | 2.40 | Upregulated | 0.0447 |
| Wtap | Wilms’ tumor 1-associating protein isoform a | 6681494 | 2.32 | Upregulated | 0.0379 |
| Scd1 | Stearoyl-CoA desaturase 1 | 5932570 | 2.27 | Upregulated | 0.0357 |
| Chodl | Chondrolectin | 5622067 | 2.25 | Upregulated | 0.0156 |
| Srpk1 | Serine/threonine-protein kinase | 5789741 | 2.24 | Upregulated | 0.0468 |
| Map4 | Microtubule-associated protein 4 | 6665260 | 2.23 | Upregulated | 0.0336 |
| Adamts9 | A disintegrin and metalloproteinase with thrombospondin motifs 9 | 6498275 | 2.18 | Upregulated | 0.0428 |
| Scd1 | Stearoyl-CoA desaturase 1 | 5991178 | −2.17 | Upregulated | 0.0462 |
FIRMA, finding isoforms by using the robust multichip analysis; MiDAS, microarray detection of alternative splicing; UBR4, ubiquitin Protein Ligase E3 Component N-Recognin 4.
TABLE 2.
Top 10 up- and down-regulated probesets of characterized genes from rats fed a 10% or 60% fat diet for 8 wk based on FIRMA fold changes
| Symbol | Description | Probeset | FIRMA fold change | Regulation | MiDAS P |
| Ddx18 | ATP-dependent RNA helicase DDX18 | 6097108 | 2.40 | Downregulated | 0.0264 |
| Csf2rb | Cytokine receptor common subunit β | 5802847 | 2.35 | Downregulated | 0.0410 |
| Mme | Neprilysin | 6701046 | 2.32 | Downregulated | 0.0419 |
| Pqlc3 | PQ-loop repeat-containing protein 3 | 6101856 | 2.29 | Downregulated | 0.0384 |
| Tnnt3 | Troponin T, fast skeletal muscle | 6500616 | 2.28 | Downregulated | 0.0468 |
| Stac3 | SH3 and cysteine-rich domain containing protein 3 | 6203961 | 2.25 | Downregulated | 0.0457 |
| Thumpd2 | THUMP domain containing 2 isoform 1 | 5653530 | 2.22 | Downregulated | 0.0354 |
| Slc6a8 | Sodium- and chloride-dependent transporter 1 | 6358207 | 2.22 | Downregulated | 0.0235 |
| Kdm2b | Lysine-specific demethylase 2B | 6588441 | 2.20 | Downregulated | 0.0483 |
| Slc9a2 | Sodium/hydrogen exchanger 2 | 5796646 | 2.15 | Downregulated | 0.0227 |
| Rbm18 | Probable RNA-binding protein 18 | 6023981 | 3.16 | Upregulated | 0.0456 |
| Col9a2 | Collagen α-2(IX) chain | 6435486 | 3.09 | Upregulated | 0.0352 |
| Rab36 | ras-Related protein Rab-36 | 6172077 | 2.92 | Upregulated | 0.0379 |
| Dpf1 | Zinc finger protein nero-d4 | 5926640 | 2.66 | Upregulated | 0.3154 |
| Psme3 | Glucose-6-phosphatase | 6000094 | 2.63 | Upregulated | 0.0342 |
| Elmo3 | Engulfment and cell motility protein 3 | 6733359 | 2.45 | Upregulated | 0.0245 |
| Phf15 | Protein Jade-2 | 6376305 | 2.41 | Upregulated | 0.0154 |
| Sema4d | Sema domain, Ig domain transmembrane domain and short cytoplasmic domain, (semaphorin) 4D | 5780532 | 2.38 | Upregulated | 0.0469 |
| Clcn1 | Chloride channel protein 1 | 5768692 | 2.37 | Upregulated | 0.0436 |
| Inppl1 | Phosphatidylinositol-3,4,5-triphosphate 5-phosphate 2 | 5814771 | 2.37 | Upregulated | 0.0357 |
FIRMA, finding isoforms by using the robust multichip analysis; MiDAS, microarray detection of alternative splicing; SH3, Src homology 3; THUMP, thiouridine synthase, RNA methylase and pseudouridine synthase.
High-fat-diet–induced changes in Tnnt3 alternative splicing.
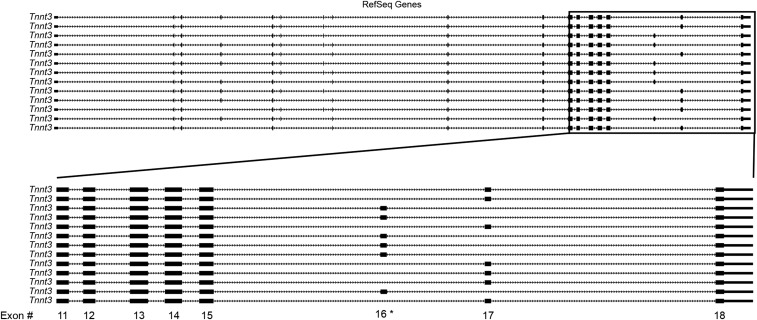
Probeset 6500616 within Tnnt3 was among the most highly downregulated from rats fed a 60% fat diet for 8 wk (Table 2). Known RefSeq Tnnt3 splice forms are shown in Figure 3 and illustrate that probeset 6500616 falls within exon 16 of Tnnt3. Exons 16 and 17 of Tnnt3 are mutually exclusive (30), and splice forms containing these exons are referred to as Tnnt3α and Tnnt3β splice forms, respectively. The FIRMA fold score for probeset 6500616 was similar at 2 and 8 wk (2.23 and 2.28, respectively), indicating that the high-fat diet induced rapid and sustained changes in Tnnt3 alternative splicing of exon 16.
FIGURE 3.
RefSeq transcript variants of Tnnt3 highlighting exons 16 and 17. Thirteen known full-length Tnnt3 RefSeq transcripts from the rat genome assembly on University of California Santa Cruz Genome Browser [July 2014 (RGSC 6.0/rn6)]. The lower portion shows the expanded section of Tnnt3 displaying exons 13–18. *Location of probeset 6500616. RefSeq, reference sequence; Tnnt3, troponin T3.
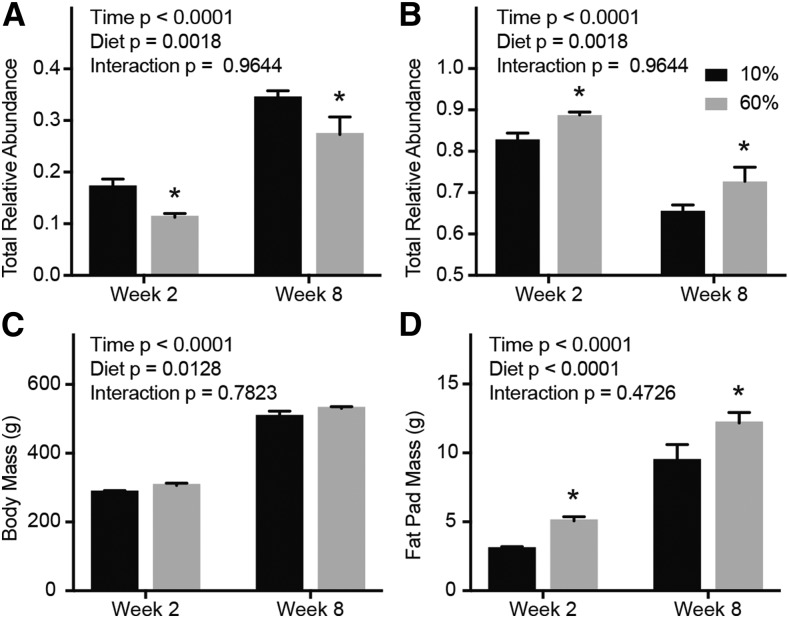
To confirm results from the exon array, we quantified Tnnt3 alternative splicing using fluorescence-based PCR as described in Methods (10). Consumption of a 60% fat diet caused a significant change in 9 of the 12 Tnnt3 splice forms (Table 3 and Supplemental Table 4). Indeed, rats that consumed a 60% fat diet for 2 wk expressed significantly fewer total Tnnt3α splice forms and more total Tnnt3β splice forms than did rats fed the 10% fat diet (Figure 4A, B, respectively). A similar effect was observed after 8 wk of consuming a 60% fat diet (Figure 4A, B). Diet treatments did not affect body mass (Figure 4C), but rats consuming a 60% fat diet had significantly heavier epididymal fat pads at both 2 and 8 wk of treatment (Figure 4D), indicating that these rats were developing obesity relative to the 10% fat control rats. Together, these data support the exon array data and demonstrate that consumption of a 60% fat diet reduced the expression of Tnnt3 exon 16–containing transcripts (Tnnt3α splice forms).
TABLE 3.
Effects of consumption of a 60% fat diet and time on Tnnt3 splice forms in the muscle of rats fed for 2 or 8 wk1
| Effect of diet |
Effect of time |
Diet × time |
||||
| Tnnt3 splice form | F | P | F | P | F | P |
| α1 | 12.63 | 0.0014 | 84.48 | <0.0001 | 0.04363 | 0.8361 |
| α2 | 6.345 | 0.0178 | 60.49 | <0.0001 | 0.01778 | 0.8949 |
| α3 | 8.596 | 0.0066 | 6.678 | 0.0153 | 1.156 | 0.2916 |
| β1 | 26.96 | <0.0001 | 33.19 | <0.0001 | 6.853 | 0.0141 |
| β2 | 3.266 | 0.0815 | 0.002181 | 0.9631 | 0.03275 | 0.8577 |
| β3 | 0.2153 | 0.6462 | 3.912 | 0.0579 | 0.9482 | 0.3385 |
| β4 | 7.186 | 0.0122 | 2.394 | 0.1331 | 1.541 | 0.2247 |
| β5 | 6.078 | 0.0201 | 9.637 | 0.0043 | 1.848 | 0.1849 |
| β6 | 6.275 | 0.0183 | 20.25 | 0.0001 | 0.03208 | 0.8591 |
| β7 | 6.892 | 0.0139 | 116.2 | <0.0001 | 3.112 | 0.0886 |
| β8 | 1.693 | 0.2039 | 3.001 | 0.0942 | 0.2668 | 0.6096 |
| β9 | 5.78 | 0.0231 | 36.32 | <0.0001 | 0.3347 | 0.5675 |
| Total Tnnt3α | 11.95 | 0.0018 | 76.79 | <0.0001 | 0.002029 | 0.9644 |
| Total Tnnt3β | 11.95 | 0.0018 | 76.79 | <0.0001 | 0.002029 | 0.9644 |
Results of a 2-factor ANOVA. n = 11/group at 2 wk and n = 5/group at 8 wk. Statistics were performed on arcsine-transformed relative abundance values. Tnnt3, troponin T3.
FIGURE 4.
Tnnt3 splice forms and morphology of rats fed a 10% or 60% fat diet for 2 or 8 wk. The sum of Tnnt3α (A) and Tnnt3β (B) splice forms, body mass (C), and epididymal fat mass (D). Values are means ± SEMs. *Different means by 2-factor ANOVA with the use of the Holm-Sidak correction for multiple comparisons, compared with respective controls at the given time point, P < 0.05. n = 11/group at 2 wk and n = 5/group at 8 wk. Tnnt3, troponin T3.
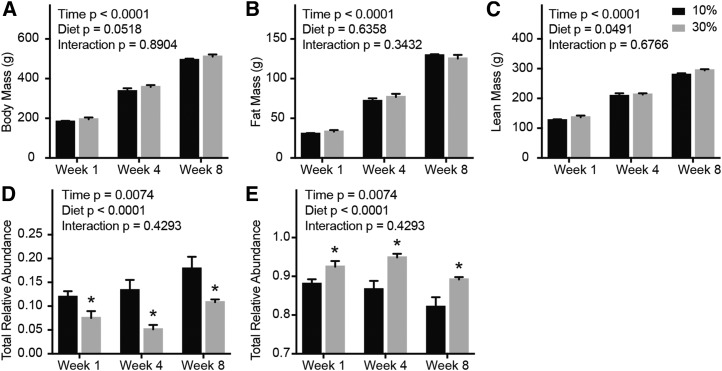
Feeding a moderate-fat diet delays diet-induced increases in body mass (31). Thus, to minimize differential body fat accumulation in high-fat-fed rats, we instead fed rats a 30% fat diet for 1, 4, or 8 wk and examined the pattern of Tnnt3 alternative splicing. A whole-body NMR-based body composition analyzer was used to obtain a more accurate measure of body composition. The body morphology of rats fed a 10% or 30% fat diet for 1, 4, or 8 wk indicated that body mass increased with time, but there was no statistically significant difference in body mass between dietary treatments (Figure 5A). In addition, no statistically significant differences were observed for either fat or lean mass (Figure 5B, C). Analogous to the Tnnt3 alternative splicing changes observed in rats fed a 10% compared with a 60% fat diet, consumption of a 30% fat diet caused a significant change in 10 of the 12 Tnnt3 splice forms (Table 4 and Supplemental Table 5). Rats that consumed a 30% fat diet for 1, 4, or 8 wk exhibited a significant decrease in the relative abundance of the Tnnt3α splice forms with a concomitant increase in the Tnnt3β splice forms (Figure 5D, E). Interestingly, at the 8-wk time point the proportion of the Tnnt3α and Tnnt3β splice forms from the 30% fat–fed rats resembled the much lighter 10% fat–fed rats at 1 wk (Figure 5D, E). The observation that consumption of a high-fat diet alters the pattern of Tnnt3 alternative splicing independent of differences in body mass and composition strongly suggests that the effect may be due to excessive consumption of FAs.
FIGURE 5.
Morphology and Tnnt3 splice forms of rats fed a 10% or 30% fat diet for 1, 4, or 8 wk. Body mass (A), fat mass (B), lean mass (C), and the sum of Tnnt3α (D) and Tnnt3β (E) splice forms. Values are means ± SEMs. *Different means by 2-factor ANOVA with the use of the Holm-Sidak correction for multiple comparisons, compared with respective controls at the given time point, P < 0.05. n = 4/group. Tnnt3, troponin T3.
TABLE 4.
Effects of consumption of a 30% fat diet and time on Tnnt3 splice forms in the muscle of rats fed for 1, 4, or 8 wk1
| Effect of diet |
Effect of time |
Diet × time |
||||
| Tnnt3 splice form | F | P | F | P | F | P |
| α1 | 25.88 | <0.0001 | 10.97 | 0.0008 | 0.9652 | 0.3998 |
| α2 | 19.11 | 0.0004 | 3.724 | 0.0443 | 0.3337 | 0.7206 |
| α3 | 39.2 | <0.0001 | 1.2 | 0.3240 | 3.377 | 0.0568 |
| β1 | 16.62 | 0.0007 | 2.671 | 0.0965 | 0.4266 | 0.6592 |
| β2 | 16.83 | 0.0007 | 5.882 | 0.0108 | 2.266 | 0.1325 |
| β3 | 6.224 | 0.0225 | 49.42 | <0.0001 | 2.712 | 0.0934 |
| β4 | 2.42 | 0.1372 | 1.353 | 0.2834 | 0.5022 | 0.6134 |
| β5 | 13.44 | 0.0018 | 10.21 | 0.0011 | 1.731 | 0.2053 |
| β6 | 6.454 | 0.0205 | 6.929 | 0.0059 | 0.3195 | 0.7305 |
| β7 | 6.309 | 0.0218 | 3.8 | 0.0420 | 1.928 | 0.1744 |
| β8 | 0.4001 | 0.5350 | 3.939 | 0.0409 | 1.833 | 0.1885 |
| β9 | 7.842 | 0.0118 | 27.91 | <0.0001 | 0.3434 | 0.7139 |
| Total Tnnt3α | 26.65 | <0.0001 | 6.533 | 0.0074 | 0.8865 | 0.4293 |
| Total Tnnt3β | 26.65 | <0.0001 | 6.533 | 0.0074 | 0.8865 | 0.4293 |
Results of a 2-factor ANOVA. n = 4/group. Statistics were performed on arcsine-transformed relative abundance values. Tnnt3, troponin T3.
Fat type–induced changes in Tnnt3 alternative splicing.
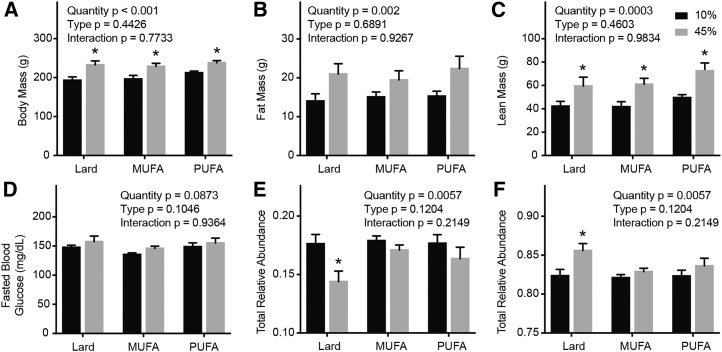
Lard was used as the primary fat source in the diet in the aforementioned experiments. The high-fat diets enriched with lard contain similar proportions of SFAs, MUFAs, and PUFAs (Supplemental Table 3). Therefore, to determine the contribution of unsaturated FAs to the high-fat-diet–induced changes in Tnnt3 alternative splicing, we compared the effects of purified versions of a high-fat diet containing fat sources primarily from MUFAs or PUFAs with those enriched with lard. After 2 wk of consuming diets enriched with varying fat quantity and type, rats fed the 45% fat diets weighed significantly more than their respective controls (Figure 6A). There were no statistically significant differences in fat mass between groups (Figure 6B), but rats fed 45% fat diets had significantly more lean mass compared with their respective control fat–fed rats (Figure 6C). There were no statistically significant differences in fasting blood glucose between groups (Figure 6D).
FIGURE 6.
Body composition, morphology, and Tnnt3 splicing of rats fed 10% or 45% fat diets varying in FA composition for 2 wk. Body mass (A), fat mass (B), lean mass (C), fasting blood glucose (D), and the sum of Tnnt3α (E) and Tnnt3β (F) splice forms. Values are means ± SEMs. *Different means by 2-factor ANOVA with the use of the Holm-Sidak correction for multiple comparisons, compared with respective controls for a given diet type, P < 0.05. n = 6/group. Tnnt3, troponin T3.
Consumption of a 45% fat diet led to significant changes in 6 of the 12 Tnnt3 splice forms, whereas the fat type significantly affected 4 of the 12 Tnnt3 splice forms (Table 5 and Supplemental Table 6). Consistent with the aforementioned data, high-fat diets enriched with lard led to a significant reduction in the total relative abundance Tnnt3α splice forms and a concomitant increase in Tnnt3β splice forms (Figure 6E, F). Interestingly, no significant differences in the total relative abundance of Tnnt3α or Tnnt3β splice forms were observed in rats fed the 45% diets enriched with either MUFAs or PUFAs (Figure 6E, F). These data suggest that Tnnt3 alternative splicing is sensitive to changes in dietary fat content and further that the changes in alternative splicing may be driven by saturated fats in the diet.
TABLE 5.
Effects of consumption of 45% fat diets varying in fat type on Tnnt3 splice forms in the muscle of rats fed for 2 wk1
| Effect of fat quantity |
Effect of fat type |
Quantity × type |
||||
| Tnnt3 splice form | F | P | F | P | F | P |
| α1 | 5.787 | 0.0225 | 1.817 | 0.1800 | 2.896 | 0.0708 |
| α2 | 6.209 | 0.0185 | 1.66 | 0.2071 | 0.1297 | 0.8788 |
| α3 | 22.63 | <0.0001 | 3.613 | 0.0393 | 1.506 | 0.2381 |
| β1 | 0.1033 | 0.7501 | 0.1822 | 0.8343 | 3.942 | 0.0302 |
| β2 | 7.874 | 0.0087 | 0.6268 | 0.5411 | 0.6557 | 0.5263 |
| β3 | 0.8253 | 0.3709 | 0.7709 | 0.4715 | 6.35 | 0.0050 |
| β4 | 21.57 | <0.0001 | 5.002 | 0.0133 | 1.68 | 0.2034 |
| β5 | 0.001491 | 0.9694 | 5.159 | 0.0119 | 1.727 | 0.1950 |
| β6 | 5.236 | 0.0293 | 2.222 | 0.1259 | 1.515 | 0.2362 |
| β7 | 0.151 | 0.7003 | 0.6775 | 0.5155 | 3.589 | 0.0401 |
| β8 | 2.076 | 0.1599 | 4.89 | 0.0145 | 1.443 | 0.2521 |
| β9 | 2.677 | 0.1123 | 2.137 | 0.1356 | 3.24 | 0.0532 |
| Total Tnnt3α | 8.865 | 0.0057 | 2.273 | 0.1204 | 1.619 | 0.2149 |
| Total Tnnt3β | 8.865 | 0.0057 | 2.273 | 0.1204 | 1.619 | 0.2149 |
Results of a 2-factor ANOVA. n = 6/group. Statistics were performed on arcsine-transformed relative abundance values. Tnnt3, troponin T3.
Discussion
Previous studies have shown that the expression of genes involved in energy metabolism, lipid metabolism and signaling, and β oxidation is altered in obese compared with lean animals as well as in animals consuming a high-fat compared with a control diet (6, 17, 18, 32). The results of the present study extend those observations to show that, compared with a 10% fat diet, consumption of a 60% fat diet leads to rapid changes in alternative splicing of pre-mRNAs, encoding proteins involved in lipid metabolism. For example, differential expression of 3 different probesets within the Scd1 pre-mRNA occurred within 2 wk of feeding rats a 60% diet compared with one with a 10% fat diet. Scd1 is a rate-limiting enzyme in lipid and energy metabolism that catalyzes the desaturation of palmitic acid and stearic acid to the corresponding MUFAs, palmitoleic acid, and oleic acid, respectively (33). Transgenic mice that overexpress Scd1 in skeletal muscle exhibit increased TG PUFA content, decreased fasting plasma glucose concentrations, and increased FA oxidation (34). Moreover, Scd1 expression is elevated in muscle after a single bout of aerobic exercise (35), and its expression is higher in athletes compared with sedentary individuals (36). Overall, these studies suggest a correlation between Scd1 expression and insulin sensitivity in skeletal muscle. Probesets 6548887, 5991178, and 5932570 fall within exon 2, 4, and 6, respectively, of the Scd1 transcript. Exon 2 encodes the N-terminal cytoplasmic domain, a full and partial section of 2 helical endoplasmic reticulum transmembrane domains, and an endoplasmic reticulum luminal portion of the Scd1 protein. Exons 4 and 6 encode cytoplasmic portions of the Scd1 protein, and exon 6 contains several iron-binding sites. Interestingly, mutation of the iron-binding amino acid sites within exon 6 abolished enzyme activity of the protein (37). Whether the alternatively spliced forms of Scd1 detected in the exon array analysis in the present study exhibit differential enzymatic activity is currently unknown and is a topic for investigation in future studies.
Another exon that was alternatively spliced in response to feeding a high-fat diet is exon 16 of the Tnnt3 pre-mRNA. Previous studies from our laboratory (10, 38) and others (11) show that the abundance of this exon in Tnnt3 transcripts increases in direct proportion to natural or experimentally induced increases in body mass, and that the effect is conserved across species. The results of the present study are in agreement with the previous ones and show that the abundance of exon 16 in Tnnt3 transcripts is higher in heavier than in lighter animals, independent of diet. However, Tnnt3 exon 16 abundance was lower in the muscle of rats consuming a high-fat compared with a low-fat diet, regardless of body weight or fasting blood glucose concentrations. Indeed, although rats fed a 30% fat diet for 8 wk had 2.5 times more body mass than rats fed a 10% fat diet for 1 wk, the proportion of Tnnt3 transcripts containing exon 16 was similar. In a previous study, a similar impairment to the normal response of Tnnt3 pre-mRNA alternative splicing to increased body mass was observed in obese compared with lean Zucker rats, i.e., the pattern of splicing in obese rats was similar to that observed in much lighter animals (10). Although the Zucker rats in that study were not fed a high-fat diet, the fat composition of their diet was primarily lard. Thus, because obese Zucker rats are hyperphagic and consume dramatically more calories per day compared with lean littermates (39), the amount of lard they consumed was correspondingly increased compared with that of control animals. Combined, the results of this study and our previous one (10) suggest that an increased consumption of lard, rather than obesity or blood glucose concentrations per se, leads to an impaired sensing of load by skeletal muscle.
Lard is composed of a mixture of SFAs and unsaturated FAs, with palmitate and oleate being the primary SFA and MUFA, respectively (Supplemental Table 3). Notably, in previous studies, palmitate and oleate were found to have differential effects on the alternative splicing of the X-box binding protein 1 (XBP-1) mRNA (21, 40), suggesting that FA saturation concentration differentially affects pre-mRNA alternative splicing. The results of the present study provide additional support for this conclusion because the diet containing lard, but not ones enriched in either oleate or the PUFA linoleate, affected alternative splicing of Tnnt3 exon 16. The mechanism through which SFAs might act to modulate alternative splicing of pre-mRNAs is unknown, but it is tempting to speculate that the effect might be mediated by lipotoxic metabolites, such as ceramides. Ceramides modulate alternative splicing in muscle (41) and aberrantly accumulate in the skeletal muscle of obese individuals and those with metabolic diseases (42, 43). An alternative mechanism through which diets enriched in SFAs compared with MUFAs or PUFAs might act to modulate alternative splicing is through their differential effects on inflammation (40). Indeed, genes involved in RNA processing (e.g., splice factors) have altered expression in inflamed skeletal muscle (44) and in the skeletal muscle of high-fat-fed mice and obese humans (45). Additionally, dietary FAs alter DNA methylation (46, 47) and remodel chromatin (48), which are known to play a role in alternative splicing (49, 50). Assessment of these and other possible mechanisms are a topic for future investigations.
Overall, the results of the present study show that consumption of a high-fat diet causes changes in alternative splicing of pre-mRNAs that encode proteins involved in metabolism and muscle function and contraction. A caveat of studying the transcript diversity engendered by pre-mRNA alternative splicing is that some transcripts may be expressed at such low amounts as to be physiologically insignificant. Secondly, the Affymetrix Rat Exon 1.0 ST array provides data only on probeset expression and does not discern how exons in a transcript are spliced together. Therefore, this array does not give any indication of the proportion of expressed transcripts that were alternatively spliced or how many splice forms are present for a given transcript. However, using the Tnnt3 as a marker gene, we show that diet-induced changes in alternative splicing can occur rapidly and manifest before the development of obesity. We also show that high-fat-diet–induced changes in Tnnt3 pre-mRNA alternative splicing are not observed when rats are fed diets enriched in MUFAs or PUFAs but only when the animals are fed a lard-based diet, suggesting that the effect is due to the consumption of SFAs. Given the known relation between Tnnt3α splice form expression and the sensitivity of muscle to calcium-induced contraction (51–54), it is tempting to speculate that one of the consequences of high-fat-diet–induced changes in alternative splicing is the development of a divergence of body weight and muscle performance, leading to impaired mobility and functional disability in obese individuals.
Acknowledgments
We thank Lydia Kutzler, Holly Lacko, Sharon Rannels, and Paul Hsu for their technical assistance. The authors’ responsibilities were as follows—AJB, SR, LSJ, SRK, and RJS: designed the research; AJB, SR, and RJS: conducted the research; AJB and RJS: analyzed the data; AJB: wrote the manuscript; AJB, LSJ, SRK, and RJS: had primary responsibility for final content; and all authors: read and approved the final manuscript.
Footnotes
Abbreviations used: FIRMA, finding isoforms by using the robust multichip analysis; GO, gene ontology; HOPACH, hierarchical ordered partitioning and collapsing hybrid; KEGG, Kyoto Encyclopedia of Genes and Genomes; MiDAS, microarray detection of alternative splicing; PC, principal component; Scd1, stearoyl-CoA desaturase 1; Tnnt3, troponin T3.
References
- 1.Thiebaud D, Jacot E, DeFronzo RA, Maeder E, Jequier E, Felber JP. The effect of graded doses of insulin on total glucose uptake, glucose oxidation, and glucose storage in man. Diabetes 1982;31:957–63. [DOI] [PubMed] [Google Scholar]
- 2.DeFronzo RA, Tripathy D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 2009;32 Suppl 2:S157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuzefovych L, Wilson G, Rachek L. Different effects of oleate vs. palmitate on mitochondrial function, apoptosis, and insulin signaling in L6 skeletal muscle cells: role of oxidative stress. Am J Physiol Endocrinol Metab 2010;299:E1096–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner N, Kowalski GM, Leslie SJ, Risis S, Yang C, Lee-Young RS, Babb JR, Meikle PJ, Lancaster GI, Hesntridge DC, et al. Distinct patterns of tissue specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia 2013;56:1638–48. [DOI] [PubMed] [Google Scholar]
- 5.Møller AB, Kampmann U, Hedegaard J, Thorsen K, Nordentoft I, Vendelbo MH, Møller N, Jessen N. Altered gene expression and repressed markers of autophagy in skeletal muscle of insulin resistant patients with type 2 diabetes. Sci Rep 2017;7:43775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh TS, Yun JW. DNA microarray analysis reveals differential gene expression in the soleus muscle between male and female rats exposed to a high fat diet. Mol Biol Rep 2012;39:6569–80. [DOI] [PubMed] [Google Scholar]
- 7.Erskine RM, Tomlinson DJ, Morse CI, Winwood K, Hampson P, Lord JM, Onambélé GL. The individual and combined effects of obesity- and ageing-induced systemic inflammation on human skeletal muscle properties. Int J Obes (Lond) 2017;41:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizunoya W, Iwamoto Y, Shirouchi B, Sato M, Komiya Y, Razin FR, Tatsumi R, Sato Y, Nakamura M, Ikeuchi Y. Dietary fat influences the expression of contractile and metabolic genes in rat skeletal muscle. PLoS One 2013;8:e80152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eshima H, Tamura Y, Kakehi S, Kurebayashi N, Murayama T, Nakamura K, Kakigi R, Okada T, Sakurai T, Kawamori R, et al. Long-term, but not short-term high-fat diet induces fiber composition changes and impaired contractile force in mouse fast-twitch skeletal muscle. Physiol Rep 2017;5:e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilder RJ, Kimball SR, Marden JH, Jefferson LS. Body weight-dependent troponin T alternative splicing is evolutionarily conserved from insects to mammals and is partially impaired in skeletal muscle of obese rats. J Exp Biol 2011;214:1523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marden JH, Fescemyer HW, Saastamoinen M, MacFarland SP, Vera JC, Frilander MJ, Hanski I. Weight and nutrition affect pre-mRNA splicing of a muscle gene associated with performance, energetics and life history. J Exp Biol 2008;211:3653–60. [DOI] [PubMed] [Google Scholar]
- 12.Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter S, Watt S, Çolak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012;338:1587–93. [DOI] [PubMed] [Google Scholar]
- 13.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 2012;338:1593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giudice J, Loehr JA, Rodney GG, Cooper TA. Alternative splicing of four trafficking genes regulates myofiber structure and skeletal muscle physiology. Cell Reports 2016;17:1923–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedrotti S, Giudice J, Dagnino-Acosta A, Knoblauch M, Singh RK, Hanna A, Mo Q, Hicks J, Hamilton S, Cooper TA. The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function. Hum Mol Genet 2015;24:2360–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Díaz-Rúa R, García-Ruiz E, Caimari A, Palou A, Oliver P. Sustained exposure to diets with an unbalanced macronutrient proportion alters key genes involved in energy homeostasis and obesity-related metabolic parameters in rats. Food Funct 2014;5:3117–31. [DOI] [PubMed] [Google Scholar]
- 17.de Wilde J, Smit E, Mohren R, Boekschoten MV, de Groot P, van den Berg SA, Bijland S, Voshol PJ, van Dijk KW, de Wit NW, et al. An 8-week high-fat diet induces obesity and insulin resistance with small changes in the muscle transcriptome of C57BL/6J mice. J Nutrigenet Nutrigenomics 2009;2:280–91. [DOI] [PubMed] [Google Scholar]
- 18.de Wilde J, Mohren R, van den Berg S, Boekschoten M, Dijk KW, de Groot P, Muller M, Mariman E, Smit E. Short-term high fat-feeding results in morphological and metabolic adaptations in the skeletal muscle of C57BL/6J mice. Physiol Genomics 2008;32:360–9. [DOI] [PubMed] [Google Scholar]
- 19.Anderson AS, Haynie KR, McMillan RP, Osterberg KL, Boutagy NE, Frisard MI, Davy BM, Davy KP, Hulver MW. Early skeletal muscle adaptations to short-term high-fat diet in humans before changes in insulin sensitivity. Obesity (Silver Spring) 2015;23:720–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciapaite J, van den Berg SA, Houten SM, Nicolay K, van Dijk KW, Jeneson JA. Fiber-type-specific sensitivities and phenotypic adaptations to dietary fat overload differentially impact fast- versus slow-twitch muscle contractile function in C57BL/6J mice. J Nutr Biochem 2015;26:155–64. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Larade K, Jiang ZG, Ito S, Wang W, Zhu H, Bunn HF. The flavoheme reductase Ncb5or protects cells against endoplasmic reticulum stress-induced lipotoxicity. J Lipid Res 2010;51:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and domainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res 2010;38:W755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–64. [DOI] [PubMed] [Google Scholar]
- 24.AltAnalyze.org. AltAnalyze – comprehensive transcriptome analysis [Internet]. [cited 2017 Apr 20]. Available from: www.altanalyze.org.
- 25.Affymetrix. Identifying and validating alternative splicing events - an introduction to managing data provided by GeneChip® exon arrays. c2006. [cited 2016 Nov 14]. Available from: https://tools.thermofisher.com/content/sfs/brochures/id_altsplicingevents_technote.pdf.
- 26.Purdom E, Simpson KM, Robinson MD, Conboy JG, Lapuk AV, Speed TP. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics 2008;24:1707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Affymetrix. Alternative transcript analysis methods for exon arrays. Affymetrix gene chip exon array white paper collection. c2005. [cited 2017 Apr 12]. Available from: https://tools.thermofisher.com/content/sfs/brochures/exon_alt_transcript_analysis_whitepaper.pdf.
- 28.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA 2000;97:10101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Laan MJ, Pollard KS. A new algorithm for hybrid hierarchical clustering with visualization and the bootstrap. J Stat Plan Inference 2003;117:275–303. [Google Scholar]
- 30.Breitbart RE, Nguyen HT, Medford RM, Destree AT, Mahdavi V, Nadal-Ginard B. Intricate combinatorial patterns of exon splicing generate multiple regulated from a single gene. Cell 1985;41:67–82. [DOI] [PubMed] [Google Scholar]
- 31.Ghibaudi L, Cook J, Farley C, van Heek M, Hwa JJ. Fat intake affects adiposity, comorbidity factors, and energy metabolism of Sprague-Dawley rats. Obes Res 2002;10:956–63. [DOI] [PubMed] [Google Scholar]
- 32.Turner N, Bruce CR, Beale SM, Hoehn KL, So T, Rolph MS, Cooney GJ. Excess lipid availability increases mitochondrial fatty acid oxidative capacity in muscle: evidence against a role for reduced fatty acid oxidation in lipid-induced insulin resistance in rodents. Diabetes 2007;56:2085–92. [DOI] [PubMed] [Google Scholar]
- 33.Stamatikos AD, Paton CM. Role of stearoyl-CoA desaturase-1 in skeletal muscle function and metabolism. Am J Physiol Endocrinol Metab 2013;305:E767–75. [DOI] [PubMed] [Google Scholar]
- 34.Rogowski MP, Flowers MT, Stamatikos AD, Ntambi JM, Paton CM. SCD1 activity in muscle increases triglyceride PUFA content, exercise capacity, and PPARδ expression in mice. J Lipid Res 2013;54:2636–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schenk S, Horowitz JF. Acute exercise increases triglyceride synthesis in skeletal muscle and prevents fatty acid-induced insulin resistance. J Clin Invest 2007;117:1690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amati F, Dube JJ, Alvarez-Carnero E, Edreira MM, Chomentowski P, Coen PM, Switzer GE, Bickel PE, Stefanovic-Racic M, Toledo FG, et al. Skeletal muscle triglycerides, diacylglycerols, and ceramides in insulin resistance: another paradox in endurance-trained athletes? Diabetes 2011;60:2588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanklin J, Whittle E, Fox BG. Eight histidine residues are catalytically essential in a membrane associated iron enzyme, stearoyl-coA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 1994;33:12787–94. [DOI] [PubMed] [Google Scholar]
- 38.Ravi S, Schilder RJ, Berg AS, Kimball SR. Effects of age and hindlimb immobilization and remobilization on fast troponin T precursor mRNA alternative splicing in rat gastrocnemius muscle. Appl Physiol Nutr Metab 2016;41:142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kowalski TJ, Ster AM, Smith GP. Ontogeny of hyperphagia in the Zucker (fa/fa) rat. Am J Physiol 1998;275:R1106–9. [DOI] [PubMed] [Google Scholar]
- 40.Salvadó L, Coll T, Gómez-Foix AM, Salmerón E, Barroso E, Palomer X, Vázquez-Carrera M. Oleate prevents saturated-fatty-acid-induced ER stress, inflammation and insulin resistance in skeletal muscle cells through an AMPK-dependent mechanism. Diabetologia 2013;56:1372–82. [DOI] [PubMed] [Google Scholar]
- 41.Ghosh N, Patel N, Jiang K, Watson JE, Cheng J, Chalfant CE, Cooper DR. Ceramide-activated protein phosphatase involvement in insulin resistance via Akt, serine/arginine-rich protein 40, and ribonucleic acid splicing in L6 skeletal muscle cells. Endocrinology 2007;148:1359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bikman BT, Summers SA. Ceramides as modulators of cellular and whole-body metabolism. J Clin Invest 2011;121:4222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlinson DJ, Erskine RM, Morse CI, Winwood K, Onambele-Pearson G. The impact of obesity on skeletal muscle strength and structure through adolescence to old age. Biogerontology 2016;17:467–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong Z, Shaibani A, Li YP, Yan Y, Zhang S, Yang Y, Yang F, Wang H, Yang XF. Alternative splicing factor ASF/SF2 is down regulated in inflamed muscle. J Clin Pathol 2006;59:855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pihlajamäki J, Lerin C, Itkonen P, Boes T, Floss T, Schroeder J, Dearie F, Crunkhorn S, Burak F, Jimenez-Chillaron JC, et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab 2011;14:208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brøns C, Jacobsen S, Nilsson E, Rönn T, Jensen CB, Storgaard H, Poulsen P, Groop L, Ling C, Astrup A, et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J Clin Endocrinol Metab 2010;95:3048–56. [DOI] [PubMed] [Google Scholar]
- 47.Ma Y, Smith CE, Lai CQ, Irvin MR, Parnell LD, Lee YC, Pham LD, Aslibekyan S, Claas SA, Tsai MY, et al. The effects of omega-3 polyunsaturated fatty acids and genetic variants on methylation levels of the interleukin-6 gene promoter. Mol Nutr Food Res 2016;60:410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung A, Trac C, Du J, Natarajan R, Schones DE. Persistent chromatin modifications induced by high fat diet. J Biol Chem 2016;291:10446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science 2010;327:996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 2012;19:998–1004. [DOI] [PubMed] [Google Scholar]
- 51.Marden JH, Fitzhugh GH, Girgenrath M, Wolf MR, Girgenrath S. Alternative splicing, muscle contraction and intraspecific variation: associations between troponin T transcripts, Ca2+ sensitivity and the force and power output of dragonfly flight muscles during oscillatory contraction. J Exp Biol 2001;204:3457–70. [DOI] [PubMed] [Google Scholar]
- 52.Sancisi V, Germinario E, Esposito A, Morini E, Peron S, Moggio M, Tomelleri G, Danieli-Betto D, Tupler R. Altered Tnnt3 characterizes selective weakness of fast fibers in mice overexpressing FSHD region gene 1 (FRG1). Am J Physiol Regul Integr Comp Physiol 2014;306:R124–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reiser PJ, Greaser ML, Moss RL. Developmental changes in troponin T isoform expression and tension production in chicken single skeletal muscle fibres. J Physiol 1992;449:573–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaudhuri T, Mukherjea M, Sachdev S, Randall JD, Sarkar S. Role of the fetal and alpha/beta exons in the function of fast skeletal troponin T isoforms: correlation with altered Ca2+ regulation associated with development. J Mol Biol 2005;352:58–71. [DOI] [PubMed] [Google Scholar]