Abstract
Exosomes have recently emerged as highly promising cancer biomarkers because they are abundant in biofluids, carry proteins and RNA reflecting their originating cells and are stable over weeks. Beyond abundance and stability, detailed exosome analyses could be clinically useful for diagnosing and profiling cancers. Despite their clinical potential, simple, reliable and sensitive approaches for rapidly quantifying exosomes and their molecular information has been challenging. Therefore, there is a clear need to develop next-generation sensing technologies for exosome detection and analysis. In this critical review, we will describe three nanotechnology sensing platforms developed for analysis of exosomal proteins and RNAs directly from clinical specimens and discuss future development to facilitate their translation into routine clinical use.
Graphical abstract

I. Introduction
Along the roadmap to highly precise and personalized cancer therapy, a critical unmet need is to develop reliable and specific modalities for early detection and real time treatment monitoring1–3. Tissue biopsies remain the gold standard, but often fail to capture the heterogeneity and temporal evolution of tumor4 from invasiveness and limited-sampling. Conventional imaging techniques could offer non-invasive alternatives, but they are costly when serially used and insensitive to detect subtle invasion, micrometastases and early stages of cancer formation5–7. Liquid biopsies, based on novel biomarkers in circulation (e.g. circulating tumor cells8,9, proteins10, DNA11,12), promise to open a new horizon in cancer management. Liquid biopsies are fast, minimally painful, and repeatable, yet can produce rich molecular information about primary tumors and metastases.
Exosomes or extracellular vesicles (EV) present new opportunities for cancer diagnosis and treatment monitoring13. These cell-derived membrane-bound vesicles (50–200 nm in diameter) are abundant in biological fluids (e.g. >109 vesicles per mL of blood) and carry cell-specific cargos (lipids, proteins and genetic materials), which can be harnessed as a minimally invasive means to probe the molecular status of tumors14–17. Furthermore, the number of tumor vesicles and their molecular profiles have correlated with tumor burden and treatment efficacy15,17.
Despite such clinical potential, routine and reliable analyses of exosomes remain challenging due to their small sizes18. Current obstacles include: 1) lengthy and extensive processing for EV isolation and 2) low sensitivity of conventional analytical methods (e.g., Western blotting, ELISA) that require large amount of samples (>500 μL per marker). Most prior exosome studies thus focused on RNA analysis to harness the power of PCR amplification. Conversely, exosome proteomic analyses through conventional approaches, where no amplification safety net exists, have been facing technical hurdles. Several recent studies have demonstrated exosomal protein screening using flow cytometry19–21, but have involved specialized high-end equipments to handle exosomes' small, but dispersed size22. More recent studies have applied novel nanomaterials (e.g. graphene oxide23 nanorod particles24) or microfluidic analytical systems25–27 to detect exosomes and identify their protein contents. Development and advancement of such diverse, ultrasensitive detection technologies could offer additional insight into understanding the heterogeneity and production dynamics of exosomes and other EV subtypes.
From the translational research perspective, exosomes represent novel diagnostic biomarkers poised for further exploitation. Furthermore, their integral roles in cell to cell communication28,29, creation of the pre-metastatic niche30, and high potential as drug delivery carriers31 offer interdisciplinary opportunities to generate novel platforms that align with patient preferences (e.g. liquid biopsies) and biorepository needs (e.g. precious specimen amounts).
In this critical review, we will describe three new platforms developed for analysis of exosomal proteins and RNAs directly from clinical specimens; factors that could facilitate their translation into routine clinic use are also discussed.
II. Nanoplasmonic sensing
Surface plasmon resonance (SPR) sensors have been widely used for detection of analytes as well as characterization of molecular interactions between antibody-antigens, proteins and small molecules32,33. These sensors detect local refractive index changes upon the binding of target substances to a sensing surface. Because a secondary label for detection is not required, rapid, label-free sensing with minimal sample processing can be achieved.
SPR represents a new exosome detection technology for rapid label-free exosome analyses15,34–37. Notably, SPR sensors' high sensitivity for exosome detection is attributed to close matching between their nanoscale sensing range and exosome size. This renders them suitable for simple but sensitive exosome analysis. The recently developed nano-plasmonic exosome (nPLEX) system demonstrated fully quantitative detection and proteomic profiling of exosomes with sensitivities >100 times better than ELISA and Western Blotting15. The nPLEX affords high throughput (12 different protein markers within 30 min) exosome profiling through multi-channel microfluidics to capture marker-specific exosomes with antibodies immobilized in each channel.
Sensing Principle
The nPLEX employs metallic nanohole structures and operates in a collinear light transmission mode while conventional SPR systems are based on total internal reflection known as the Kretschmann configuration. The nanohole structures simplify the optical setup and enable system miniaturization and scaling of sensing arrays in any given chip38–40.
The sensor comprises a periodic nanohole lattice patterned onto gold film and standard glass slides (Fig. 1a). In the first prototype, 12 × 3 sensing arrays were integrated with multichannel microfluidics; 12 different exosomal protein markers could be measured in triplicate. To capture marker-specific exosomes, antibodies were grafted onto a polyethylene glycol (PEG) polymer-coated sensing surface. Following target-specific exosome binding, the nPLEX sensor displays spectral shifts proportional to the level of target marker. For exosomes collected from ovarian cancer cell lines, the nPLEX correlated well with the gold standard ELISA across various transmembrane exosome markers. The nPLEX showed a detection threshold of 3,000 exosomes (670 attomolar) — about 100-fold more sensitive than ELISA and 10,000-fold more sensitive than Western Blotting. Importantly, nPLEX analyses showed good correlation between exosome protein profiles and their corresponding cell lines, which underscored the use of exosomes as likely cellular surrogates.
Figure 1. Nanoplasmonic exosome (nPLEX) platform.
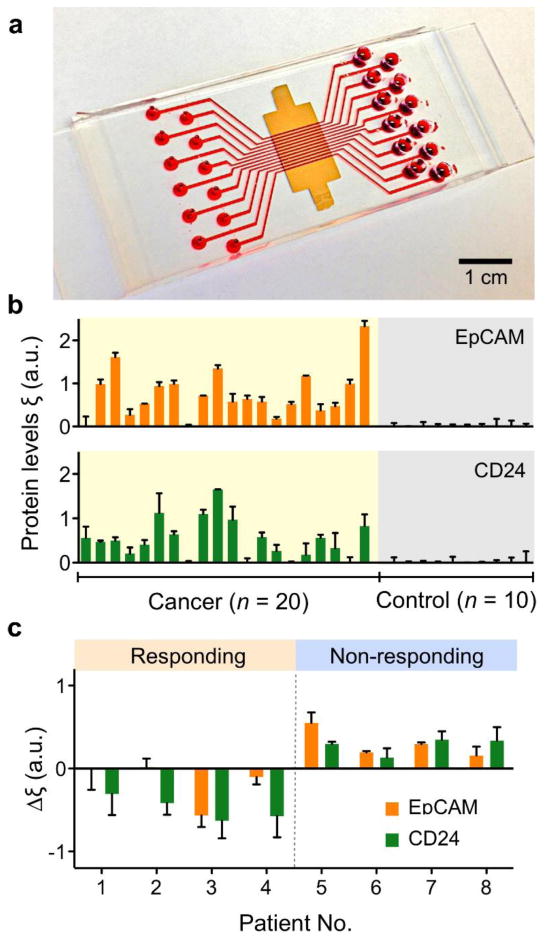
(a) A photograph of nPLEX chip integrated with a 12-channel microfluidic cell. (b) EpCAM and CD24 levels in exosomes from clinical ascot samples were measured by nPLEX. The elevated EpCAM and CD24 expression were measured in ovarian cancer patients (n = 20) while the levels were negligible in non-cancer control patient (n = 10). (c) The ascites exosome analysis shows that measuring temporal changes in exosomal expressions of EpCAM and CD24 could distinguish treatment response of ovarian cancer patients after chemotherapy. Reproduced with permission from Ref. 14.
Exosome Analysis in Human Clinical Samples
The nPLEX system supported the high potential of exosomes as cancer diagnostic and treatment monitoring markers. For example, using exosomes collected from 30 patients with abdominal fluid buildup (ascites), elevated levels of EpCAM and/or CD24 were noted on exosomes from ovarian cancer ascites compared to exosomes from ascites due to advanced liver disease (non-cancer). Using EpCAM and CD24 profiles, detection accuracy reached 97% for the 30 patient samples (Fig. 1b). Furthermore, levels of exosomal EpCAM, CD24 or both decreased among clinically responding patients, whereas levels of these markers increased in patients not responding to standard chemotherapy. In a small subset (n = 8), serial exosome analyses demonstrated the feasibility of frequent assessments of patient status during treatment (Fig. 1c).
Next Generation of nPLEX
There are areas where the first generation nPLEX system can be further improved to augment research and clinical applications. First, high throughput chip fabrication would be important to meet the patient volume demands of clinical use. The serial writing method used in the first prototype could be too slow and expensive for such clinical needs. Interference lithography reflects one way to fabricate periodic nanostructures in a wafer-scale batch process41,42. Combined with conventional semiconductor fabrication processes, a large number of chips can be fabricated at decreased cost as production rates scale up15,43. In addition, it will allow integrating a vast number of sensing arrays not unlike protein microarray sensors. Second, the sensitivity of the system could be further improved to enable single exosome analysis. While the current nPLEX system shows much better sensitivity than other currently available methods, it is still limited to bulk analyses (i.e. measuring averaged signals from certain exosome populations). Single exosome analysis will open up new avenues in EV research to better understand exosome biology, their role in cell-to-cell communications and tumor heterogeneity29,30. Third, current nPLEX detection is based on capturing whole exosomes on sensor surfaces using antibodies targeting markers residing on EV membranes. Therefore, it currently lacks the capability to analyze proteins and RNA markers present inside exosomes. Accessing proteins regardless of location as well as RNA would expand the use of exosomes to serially probe dynamic pathway alterations in response to targeted therapies.
III. Electrochemical sensing
The use of new nanosensing technologies has not yet extended into clinical settings for routine testing. For exosome analysis, it is due in part to the additional procedures and equipment needed for exosome isolation and the technical complexities and high costs associated with chip fabrication and analytical instruments. Electrochemical sensing approaches that provide rapid assays and affordable readout systems could be an effective detection modality. Electrochemical sensors operate by reacting with the target substance of interest and producing an electrical signal proportional to the target material. Miniaturized electrochemical sensors have been used for decades to routinely detect toxic gases and chemicals as well as biomolecules. They can prove highly sensitive when combined with certain enzyme reporters for signal amplification.
A recently developed integrated magnetic-electrochemical exosome (iMEX) platform is a miniaturized analytical system that can rapidly isolate and detect exosomes in clinical specimens44. It combines magnetic isolation of exosomes with magnetic microbeads and electrochemical sensing of exosomal proteins (Fig. 2a). The iMEX can isolate cell-specific exosomes directly from plasma samples, achieve high detection sensitivity, and be expanded for parallel measurements. A handheld iMEX system was able to detect exosomes at a sensitivity of <105 vesicles using 10 μL of samples within 1 hour.
Figure 2. Integrated magnetic-electrochemical exosome (iMEX) platform.
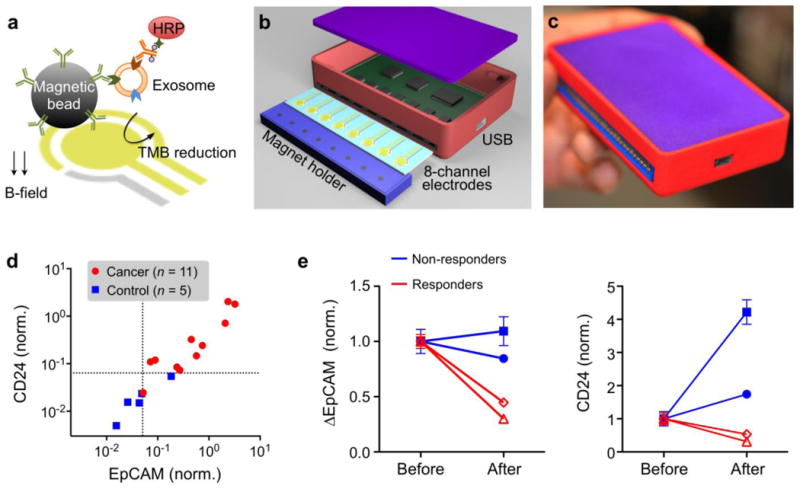
(a) Assay schematic. Exosomes are captured on magnetic beads and labeled with HRP enzyme for electrochemical detection. Magnetic beads are coated with capture antibodies (e.g., CD63 antibody). HRP, horseradish peroxidase; TMB, 3,3′,5,5′-tetramethylbenzidine. (b) An iMEX detector is designed to simultaneously measure signals from eight electrodes. A small cylindrical magnet is located below each electrode to concentrate immunomagnetically captured exosomes. (c) The packaged iMEX device has a small form factor (9 × 6 × 2 cm3). (d) Plasma samples from ovarian cancer patients (n = 11) and healthy controls (n = 5) were analyzed by the iMEX assay. EpCAM and CD24 levels were higher in cancer patients. (e) Plasma samples from four ovarian cancer patients were analyzed before and after drug treatment. EpCAM and CD24 levels in responders were decreased significantly. All measurements were in duplicate. All signals were normalized against CD63 signal. Reproduced with permission from Ref. 44.
Sensing principle
The handheld iMEX system contains 8 independent sensing elements (Fig. 2b). Each sensor is equipped with a potentiostat for instant signal readouts through metal electrodes (50 msec per sensor). A magnet holder, placed underneath the electrode, concentrates magnetic beads to the electrode surface for improved detection sensitivity. Compared to commercial equipment, the iMEX system showed comparable performance, but at much smaller size and cost (<$50). For detection, exosomes are first captured onto immuno-magnetic beads coated with antibodies against CD63, a marker widely reportedly as enriched in exosomes. Secondary antibodies with an oxidizing enzyme (horseradish peroxidase; HRP) are then applied followed by mixture of beads with a 3,3′,5,5′-tetramethylbenzidine (TMB) substrate that generates electrical current in the presence of HRP.
Exosome protein analyses with human clinical samples
The iMEX assay isolates exosomes directly from plasma, and profiles them in a rapid, high-throughput manner — key for successful integration into the clinical workflow. In the iMEX assay, clinical plasma samples were aliquoted without any purification, and each aliquot (10 μL per marker) was incubated with magnetic beads for exosome capture (15 min), followed by magnetic washing. The bead-bound exosomes were consecutively labeled for target markers (15 min) and HRP (15 min), and loaded onto the device. A single-time point measurement of plasma samples from ovarian cancer and healthy controls showed elevated express levels of EpCAM and CD24 in ovarian cancer patient exosomes (Fig. 2c), in concordance with previous studies using nPLEX45. In addition, serial exosome testing at two time points (2 months apart) in ovarian cancer patients undergoing drug treatment showed that exosomal expression levels of EpCAM and CD24 increased in non-responding patients while both markers decreased in responding patients (Fig. 2d).
Future perspective
The low cost, portability and integrated assay for exosome isolation and detection render the iMEX system highly poised for clinical exosome analyses. A unique feature of iMEX is the integration of vesicle isolation and detection into a single platform. Both magnetic selection and exosome enrichment simplify assay procedures and improve detection sensitivities. The electrochemical sensing facilitates high-throughput screening and sensor miniaturization. There are multiple directions to further advance the iMEX technology. First, assay throughput can be improved by increasing the number of detection sites. The sensing elements (electrodes) can be readily microfabricated into a large array format, and signals (electrical currents) can be read out by compact electronics with high-speed multiplexing. Second, detection sensitivity can be enhanced by increasing sensor surface areas and optimizing enzymes used for signal amplification. Third, the target of interest can be expanded to exosomal RNAs. Electrochemical sensing detects nucleic acids with high sensitivity while obviating the need for PCR amplification. Multiplex sensing of exosomal RNA and proteins should provide in-depth and complementary information to more accurately access tumor status at a given time point.
IV. IMER (Immuno-Magnetic Exosome RNA) analysis
In addition to proteins, nucleic acids in exosomes are regarded as markers that highly reflect the underlying disease. Recently, various studies indicate that exosomes contain non-coding RNAs (including miRNA) in addition to DNA.46,47 Skog et al. found that serum exosomes from glioblastoma (GBM) patients contain characteristic mRNA mutants that could be used to provide diagnostic information. 48. Moreover, many other studies have identified specific exosomal miRNA markers for different cancer subtypes49,50. The heterogeneous mixture and contents of exosomes challenge their reliable and specific evaluation against the backdrop of diverse tumor genotype, phenotype, and physiologic responses to stimuli from the local and external milieu.
To detect and analyze nucleic acids in exosomes, a microfluidic platform, immuno-magnetic exosome RNA (iMER), has been developed and tested for on-chip enrichment, purification, and analysis of exosomal RNA. The iMER system integrates three features on the single microfluidic chip: immunomagnetic tumor exosome enrichment, RNA purification, and real-time RT-qPCR. The iMER platform can capture exosomes directly from 100 μL of serum with >93% capture efficiency. A single chip achieves an integrated work flow, from EV isolation to RNA analysis by qPCR, within two hours.17
Working principle
The iMER chip uses magnetic microbeads (3 μm in diameter) with anit-EGFR/EGFRvIII coating (in the GBM example) to capture and enrich cancer-specific exosomes. Beads are incubated with media or blood samples; only magnetic beads with specific exosomes are isolated in the chip. The enriched and purified populations of exosomes are lysed in the chip and introduced to a glass bead filter. Next, mRNAs in the exosomes can be absorbed onto a glass bead via electrostatic interaction between the glass substrate and mRNAs. The isolated mRNAs are then reverse transcribed, amplified, and measured by qPCR (Fig. 3a). The platform uses torque-activated valves to control buffer flow and isolation at each step (Fig. 3b)17.
Figure 3. Immuno-magentic exosome RNA (iMER) platform.
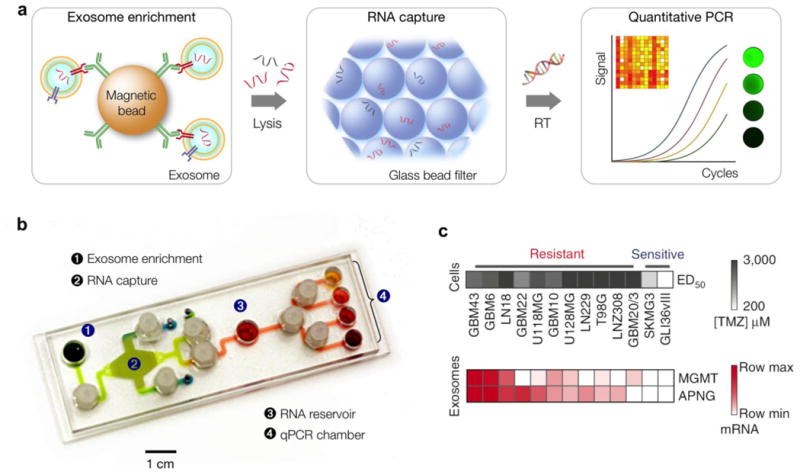
(a) Schematic of iMER assay. Cancer exosomes are first isolated by immuno-mangetic microbeads. After exosome lysis, exosomal RNA are captured on purified by a glass bead filter and analyzed through quantitative on-chip PCR. (b) Photograph of the microfluidic iMER prototype. Torque-activated valves were used to control the flow of solutions. (c) iMER analysis showed that the levels of MGMT, APNG or both were elevated in exosomes from resistant cell lines, whereas they were both low in sensitive ones. The exosomal MGMT and APNG mRNA levels correlate with in vitro TMZ sensitivity. Reproduced with permission from Ref. 17.
Exosomal mRNA analysis for drug efficacy
Analyses of exosomes have examined their relationship with parental cell origin and their potential application as cancer diagnostic markers. As described earlier, exosomal proteins have been examined with micro/nano sensing platforms44,45,51. Here, iMER platform leverages mRNA as predictive markers such as in drug resistance monitoring. Since detection of exosomal proteins related to drug resistance poses technical challenges due to their scant amount, the iMER platform uses mRNA as the counterpart to proteins. In a pilot clinical study17, the iMER platform showed that key exosomal mRNA markers potentially predicted drug resistance. Specifically, the study compared the mRNA profiles of GBM-derived exosomes against those of their cells of origin and followed dynamic sequential changes on treatment initiation. The study identified key exosomal mRNA markers (MGMT, APNG) potentially predictive of Temozolimide resistance and showed the capacity of exosomal RNA for probing the epigenetic status of primary tumors. The platform was also applied to profile exosomes in blood from GBM patients and healthy controls.
Future perspective
The simple and miniaturized on-chip processing capacity of the iMER platform affords various advantages including rapid sample analysis, minimal sample volume requirement and high sensitivity. Moreover, the platform can be adopted to both predictive and diagnostic applications. To expand its functionality and usage, some aspects of the system could be further modified. First, mass produced and disposable cartridges will be desired. From the cost perspective, microfluidic chips need to be manufactured using mass production processes such as injection molding or roll-to-roll fabrication. Second, current fluidic network designs can be modified to small and array-type features for highly sensitive and parallel measurements. Third, using cocktails of capturing antibodies could enable the platform to detect heterogeneous and variable cancer subtypes. Furthermore, combining iMER's mRNA detection modalities with exosomal protein detection can provide comprehensive snapshots of tumor. Such potential advances will facilitate both clinical applications and biological studies of exosomes.
Conclusions
Current analytical tools are often not amenable to comprehensive exosome profiling given their requirements of extensive purification, large sample volumes, or different assay schemes per detection targets. Besides the platforms reviewed here, many biosensors are being developed to facilitate exosome preparation and analyses24,27,37,52–55 (Table 1). Such technology development will help deepening our understanding of exosome biology as well as assessing exosomes' clinical value.
Table 1. Comparative analysis of nanosensing technology for exosome detection.
| Technology | Principle | Limit of detection | Throughput | Diseases | Diagnostic accuracy | Ref. |
|---|---|---|---|---|---|---|
| nPLEX | SPR | 3,000 EVs | 36 arrays in 30 min | Ovarian cancer | 93% (n = 30) | 14 |
| iMEX | Electrochemical sensing | < 105 EVs | 8 channels in 1 h | Ovarian cancer | - | 44 |
| iMER | microfluidic qPCR | 100 μl serum | multiplexed mRNA analysis in 2 h | Glioblastoma multiforme | 90% (n = 32) | 17 |
| Nano-IMEX | Fluorogenic ELISA | 50 EVs/μl | 5 channels | Ovarian cancer | p < 0.001 (n = 12) | 23 |
| nPES | Light scattering | 0.2 ng/μl | - | Pancreatic cancer | 96% (n = 145)94% sensitivity;85% specificity | 24 |
| FLOWER | Optical resonator | Single particle | - | - | - | 26 |
| μMED | Smartphone ELISA | 107 EVs | Single channel in 1 h | Mild traumatic brain injury | 73% sensitivity; 71% specificity | 52 |
| ExoSearch | Fluorescence | 7.5×105 EVs | 3 channels in 3 h | Ovarian cancer | 100% (n = 20) | 27 |
| Au@Ag NRs | SERS | 1,200 EVs | 2 h | - | - | 55 |
Further driving the widespread adoption of exosome diagnostics into practice, however, would require more than proof-of-concept systems; assay automation, minimal hands-on processing and data analytics are some key components that should be integrated. For clinical translation and commercialization, a host of other key issues should be addressed in prototype systems. These include cost and protocols that align with clinical workflow. With respects to cost, significant up keeping requirements or on-site servicing often clash with budgetary realities. As such, developing robust, modular platforms that require little maintenance and used by end users of various skillsets, could help justify purchasing. Drastic interruptions to clinical workflow will incur end user frustration and reduced productivity. Assays that require highly specific pre-analytical handling from unique specimen collection kits to sample processing, could also limit widespread clinical adoption. Developing platforms in silos without attention to clinical or other practical realities, could derail global efforts. For rigorous clinical studies, it is also important to establish the standardization of sample collection protocol, statistical consideration for designing training and validation sets, and randomized clinical trials in large patient cohorts. Practically, multi-institutional or international efforts could accelerate clinical translation of new nanotechnology.
It is thus encouraging to witness the convergence of clinical medicine, materials science, engineering, biology, computer science, and biophysics among other disciplines in exosome research; it surely will propel miniaturized exosome technologies into the clinical mainstream.
Acknowledgments
This work was supported in part by US NIH Grants R21-CA205322 (H.L.), R01-HL113156 (H.L.), R01CA204019 (R.W.), P01CA069246 (R.W.), K12CA087723-11A1 (C.M.C.), K99CA201248 (H.I.), the Andrew L. Warshaw, M.D. Institute Pilot Research grant (H.I.), and MGH Physician Scientist Development Award (C.M.C.).
References
- 1.Bidard FC, Weigelt B, Reis-Filho JS. Sci Transl Med. 2013;5:207ps14. doi: 10.1126/scitranslmed.3006305. [DOI] [PubMed] [Google Scholar]
- 2.Pantel K, Alix-Panabieres C. Cancer Res. 2013;73:6384. doi: 10.1158/0008-5472.CAN-13-2030. [DOI] [PubMed] [Google Scholar]
- 3.Basik M, Aguilar-Mahecha A, Rousseau C, Diaz Z, Tejpar S, Spatz A, Greenwood CM, Batist G. Nat Rev Clin Oncol. 2013;10:437. doi: 10.1038/nrclinonc.2013.101. [DOI] [PubMed] [Google Scholar]
- 4.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. N Engl J Med. 2012;366:883. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissleder R, Pittet MJ. Nature. 2008;452:580. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brindle K. Nat Rev Cancer. 2008;8:94. doi: 10.1038/nrc2289. [DOI] [PubMed] [Google Scholar]
- 7.Condeelis J, Weissleder R. Cold Spring Harbor Perspect Biol. 2010;2:a003848. doi: 10.1101/cshperspect.a003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petricoin EF, III, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC. The Lancet. 2002;359:572. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 11.Alix-Panabieres C, Schwarzenbach H, Pantel K. Annu Rev Med. 2012;63:199. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- 12.Dawson SJ, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B. N Engl J Med. 2013;368:1199. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 13.Théry C. Nature. 2015;523:161. doi: 10.1038/nature14626. [DOI] [PubMed] [Google Scholar]
- 14.Alderton GK. Nat Rev Cancer. 2015;15:453. doi: 10.1038/nrc3990. [DOI] [PubMed] [Google Scholar]
- 15.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Nat Biotechnol. 2014;32:490. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahbari M, Rahbari N, Reissfelder C, Weitz J, Kahlert C. Langenbeck's Archives of Surgery. 2016;401:1097. doi: 10.1007/s00423-016-1468-2. [DOI] [PubMed] [Google Scholar]
- 17.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R. Nat Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liga A, Vliegenthart ADB, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M. Lab Chip. 2015;15:2388. doi: 10.1039/c5lc00240k. [DOI] [PubMed] [Google Scholar]
- 19.Arraud N, Gounou C, Turpin D, Brisson AR. Cytometry Part A. 2016;89:184. doi: 10.1002/cyto.a.22669. [DOI] [PubMed] [Google Scholar]
- 20.Nolte ENM, van der Vlist EJ, Aalberts M, Mertens HCH, Bosch BJ, Bartelink W, Mastrobattista E, van Gaal EVB, Stoorvogel W, Arkesteijn GJA. Nanomed Nanotechnol Biol Med. 2012;8:712. doi: 10.1016/j.nano.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Der Vlist EJ, Nolte ENM, Stoorvogel W, Arkesteijn GJA, Wauben MHM. Nat Protoc. 2012;7:1311. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- 22.Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E. J Extracell Vesicles. 2015;4:25530. doi: 10.3402/jev.v4.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, He M, Zeng Y. Lab Chip. 2016;16:3033. doi: 10.1039/c6lc00279j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH. Nat Biomed Eng. 2017;1:0021. doi: 10.1038/s41551-016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Son KJ, Rahimian A, Shin DS, Siltanen C, Patel T, Revzin A. Analyst. 2016;141:679. doi: 10.1039/c5an01648g. [DOI] [PubMed] [Google Scholar]
- 26.Su J, Goldberg AFG, Stoltz BM. Light: Sci Appl. 2016;5:e16001. doi: 10.1038/lsa.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Z, Yang Y, Zeng Y, He M. Lab Chip. 2016;16:489. doi: 10.1039/c5lc01117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tkach M, Théry C. Cell. 2016;164:1226. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 29.Pucci F, Garris C, Lai CP, Newton A, Pfirschke C, Engblom C, Alvarez D, Sprachman M, Evavold C, Magnuson A. Science. 2016;352:242. doi: 10.1126/science.aaf1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H. Nat Cell Biol. 2015;17:816. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andaloussi SEL, Mäger I, Breakefield XO, Wood MJA. Nat Rev Drug Discovery. 2013;12:347. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 32.Cooper MA. Nat Rev Drug Discov. 2002;1:515. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- 33.Homola J. Chem Rev. 2008;108:462. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 34.Grasso L, Wyss R, Weidenauer L, Thampi A, Demurtas D, Prudent M, Lion N, Vogel H. Anal Bioanal Chem. 2015;407:5425. doi: 10.1007/s00216-015-8711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rupert DLM, Lässer C, Eldh M, Block S, Zhdanov VP, Lotvall JO, Bally M, Höök F. Anal Chem. 2014;86:5929. doi: 10.1021/ac500931f. [DOI] [PubMed] [Google Scholar]
- 36.Sina AAI, Vaidyanathan R, Dey S, Carrascosa LG, Shiddiky MJA, Trau M. Sci Rep. 2016;6:30460. doi: 10.1038/srep30460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu L, Wang K, Cui J, Liu H, Bu X, Ma H, Wang W, Gong H, Lausted C, Hood L. Anal Chem. 2014;86:8857. doi: 10.1021/ac5023056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brolo AG. Nat Photonics. 2012;6:709. [Google Scholar]
- 39.Coskun AF, Cetin AE, Galarreta BC, Alvarez DA, Altug H, Ozcan A. Sci Rep. 2014;4:6789. doi: 10.1038/srep06789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahlin AB. Analyst. 2015;140:4748. doi: 10.1039/c4an02258k. [DOI] [PubMed] [Google Scholar]
- 41.Menezes JW, Barea LAM, Chillcce EF, Frateschi N, Cescato L. IEEE Photonics J. 2012;4:544. [Google Scholar]
- 42.Yanik AA, Cetin AE, Huang M, Artar A, Mousavi SH, Khanikaev A, Connor JH, Shvets G, Altug H. Proc Natl Acad Sci U S A. 2011;108:11784. doi: 10.1073/pnas.1101910108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Im H, Lee SH, Wittenberg NJ, Johnson TW, Lindquist NC, Nagpal P, Norris DJ, Oh SH. ACS Nano. 2011;5:6244. doi: 10.1021/nn202013v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H. ACS Nano. 2016;10:1802. doi: 10.1021/acsnano.5b07584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Nat Biotechnol. 2014;32:490. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 47.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Nat Cell Biol. 2008;10:1470. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryant RJ, Pawlowski T, Catto JWF, Marsden G, Vessella RL, Rhees B, Kuslich C, Visakorpi T, Hamdy FC. Br J Cancer. 2012;106:768. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva J, García V, Zaballos A, Provencio M, Lombardía L, Almonacid L, García JM, Domínguez G, Peña C, Diaz R. Eur Respir J. 2011;37:617. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 51.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Nat Med. 2012;18:1835. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ko J, Hemphill MA, Gabrieli D, Wu L, Yelleswarapu V, Lawrence G, Pennycooke W, Singh A, Meaney DF, Issadore D. Sci Rep. 2016;6:31215. doi: 10.1038/srep31215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JH, Kim JA, Jeong S, Rhee WJ. Biosens Bioelectron. 2016;86:202. doi: 10.1016/j.bios.2016.06.058. [DOI] [PubMed] [Google Scholar]
- 54.Lee K, Shao H, Weissleder R, Lee H. ACS Nano. 2015;9:2321. doi: 10.1021/nn506538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zong S, Wang L, Chen C, Lu J, Zhu D, Zhang Y, Wang Z, Cui Y. Anal Methods. 2016;8:5001. [Google Scholar]


