Summary
CCR5 is the primary chemokine receptor utilized by HIV to infect leukocytes, whereas CCR5 ligands inhibit infection by blocking CCR5 engagement with HIV gp120. To guide the design of improved therapeutics, we solved the structure of CCR5 in complex with chemokine antagonist [5P7]CCL5. Several structural features appeared to contribute to the anti-HIV potency of [5P7]CCL5, including the distinct chemokine orientation relative to the receptor, the near-complete occupancy of the receptor binding pocket, the dense network of intermolecular hydrogen bonds, and the similarity of binding determinants with the FDA-approved HIV inhibitor Maraviroc. Molecular modeling indicated that HIV gp120 mimicked the chemokine interaction with CCR5, providing an explanation for the ability of CCR5 to recognize diverse ligands and gp120 variants. Our findings reveal that structural plasticity facilitates receptor-chemokine specificity and enables exploitation by HIV, and provide insight into the design of small molecule and protein inhibitors for HIV and other CCR5-mediated diseases.
Keywords: Chemokine antagonist, HIV entry inhibitor, CCL5/RANTES, CCR5-gp120 interaction, G protein-coupled receptor (GPCR), viral mimicry, membrane protein structure, maraviroc, two-site model, V3 loop
Introduction
Shortly after the discovery that the G protein-coupled receptor (GPCR) CCR5 serves as a coreceptor that enables HIV to infect cells, it was recognized that its endogenous ligands, including CCL5, could function as HIV entry inhibitors by blocking the engagement of the HIV glycoprotein, gp120. However, native chemokines are not efficient inhibitors, in part because they only bind with high affinity to the subpopulation of CCR5 that is G protein-coupled, whereas HIV gp120 is indiscriminate (Colin et al., 2013; Jin et al., 2014). Capitalizing on knowledge that N-terminal modifications can alter chemokine pharmacological properties and binding affinity (Proudfoot et al., 1996; Simmons et al., 1997), efforts to identify more potent chemokine-based HIV inhibitors ensued. Such inhibitors included CCL5 analogues identified by phage display and selected for high affinity receptor binding (Gaertner et al., 2008), with one of them currently under pre-clinical development as a topical inhibitor of HIV (Cerini et al., 2016; Dorgham et al., 2016). To guide the design of improved CCR5 inhibitors, and to understand how they compete with gp120, the molecular interactions between these CCL5 variants and CCR5 need to be understood.
However, structure determination of GPCRs, and particularly complexes with protein ligands is challenging (Qin et al., 2015). As a consequence, only two structures of receptor-chemokine complexes have been reported, that of human CXCR4 in complex with a viral chemokine antagonist, vMIP-II (Qin et al., 2015), and viral chemokine receptor US28 in complex with the human chemokine CX3CL1 (Burg et al., 2015). Additional structures are needed that are purely human in origin, and representative of the four different chemokine subfamilies (CXC, CC, CX3C and XC) to explain receptor-chemokine recognition specificity (Qin et al., 2015). Structures are also needed to understand how chemokines stabilize different receptor functional states, and the molecular basis for biased signaling by different ligands of the same receptor (Scholten et al., 2011; Steen et al., 2014).
Taking advantage of the high affinity, G protein-independent nature of one of the engineered CCL5 variants, the potent anti-HIV antagonist [5P7]CCL5 (Gaertner et al., 2008), herein we successfully solved the structure of this chemokine with CCR5. While maintaining an architecture consistent with the general concepts of "the two-site model" (Scholten et al., 2011), the structure revealed interaction epitopes not predicted by the model and not observed in the two previously solved receptor-chemokine structures (Burg et al., 2015; Qin et al., 2015). In contrast to the other two structures, each of which contains a viral component with broad specificity across human chemokine subfamilies, the CCR5-[5P7]CCL5 structure represents a human receptor-chemokine complex where both ligand and receptor are specific to a single (CC) subfamily. The structure provided insight into receptor-chemokine recognition and signaling, as well as HIV inhibition, and also suggested clever mechanisms by which gp120 mimics chemokine interactions with CCR5.
Results
Engineered [5P7]CCL5 forms stable complexes with CCR5 that are amenable to crystallization
To understand the molecular basis for their exceptional anti-HIV potency, several N-terminally modified CCL5 variants from the phage-display study (Gaertner et al., 2008) were selected as potential candidates for crystallization with CCR5. The variants were coexpressed with CCR5 in insect cells, the complexes were purified, and their thermostabilities determined by differential scanning fluorimetry (DSF) (Alexandrov et al., 2008). From this screen, a complex between CCR5 with an intracellular loop 3 (ICL3) rubredoxin fusion ((Tan et al., 2013), referred to as CCR5) and the antagonist variant [5P7]CCL5 (containing a 10-residue N-terminal sequence 0-QGPPLMALQS-9 in place of the 9-residue sequence 1-SPYSSDTTP-9 in WT CCL5) emerged as having one of the highest melting temperatures (Tm ≈ 68°C, Figure S1A). [5P7]CCL5 also co-eluted with CCR5 during purification, indicative of its slow dissociation rate from the receptor, and the complex was monodisperse by analytical size-exclusion chromatography (Figure S1B). It crystallized in lipidic cubic phase (Figure S1C-F), and the structure was determined to 2.2 Å resolution. Data collection and refinement statistics are shown in Table S1.
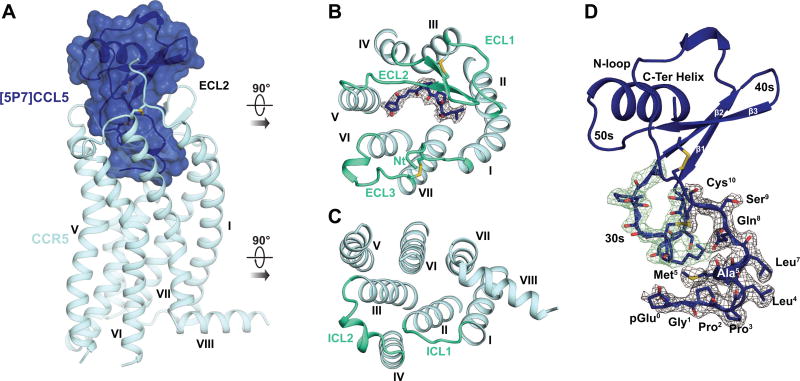
In the CCR5-[5P7]CCL5 complex structure, CCR5 adopted the canonical topology of a GPCR with seven transmembrane (TM) helices connected by three extracellular and three intracellular loops (ECL and ICL, Figure 1A-C), and an eighth helix parallel to the membrane on the intracellular side. Its extracellular N-terminus was connected to ECL3 via a conserved disulfide bond that is critically important for chemokine function (Blanpain et al., 1999; Howard et al., 1999). A second disulfide, highly conserved in Class A receptors, linked helix III to the base of the β-hairpin-shaped ECL2.
Figure 1. The structure of the complex between CCR5 and the anti-HIV chemokine variant [5P7]CCL5.
(A) Structure of [5P7]CCL5-bound CCR5, viewed parallel to the membrane plane.
(B-C) Structure viewed from the extracellular (B) and intracellular (C) side. Loops connecting the TM helices are lime green. Residues 0–4 of the chemokine are shown as sticks, with the electron density of the Fo-Fc omit map contoured at 3σ.
(D) View of [5P7]CCL5 with residues 0–9 and the 30s loop residues shown as sticks. The electron density of the Fo-Fc omit maps is shown and contoured at 3σ for the engineered N-terminus (gray mesh) and 30s loop (green mesh), respectively.
See also Figure S1 and Table S1.
[5P7]CCL5 had the characteristic tertiary structure of CC chemokines, with a flexible N-terminus stapled to the globular core via two intramolecular disulfide bonds (Figure 1D). The globular core sat on the extracellular side of CCR5, cradled by the receptor N-terminus and extracellular loops, while the chemokine N-terminus penetrated the binding pocket of the receptor (Figure 1A). With the exception of S68 and the side-chain of M67, all chemokine residues were well-resolved in the electron density (Figure 1B and 1D). As expected from prior pharmacological characterization of [5P7]CCL5, CCR5 in the complex showed clear signatures of an inactive conformation (Dalton et al., 2015).
The structure reveals receptor-chemokine interaction epitopes beyond the two-site model
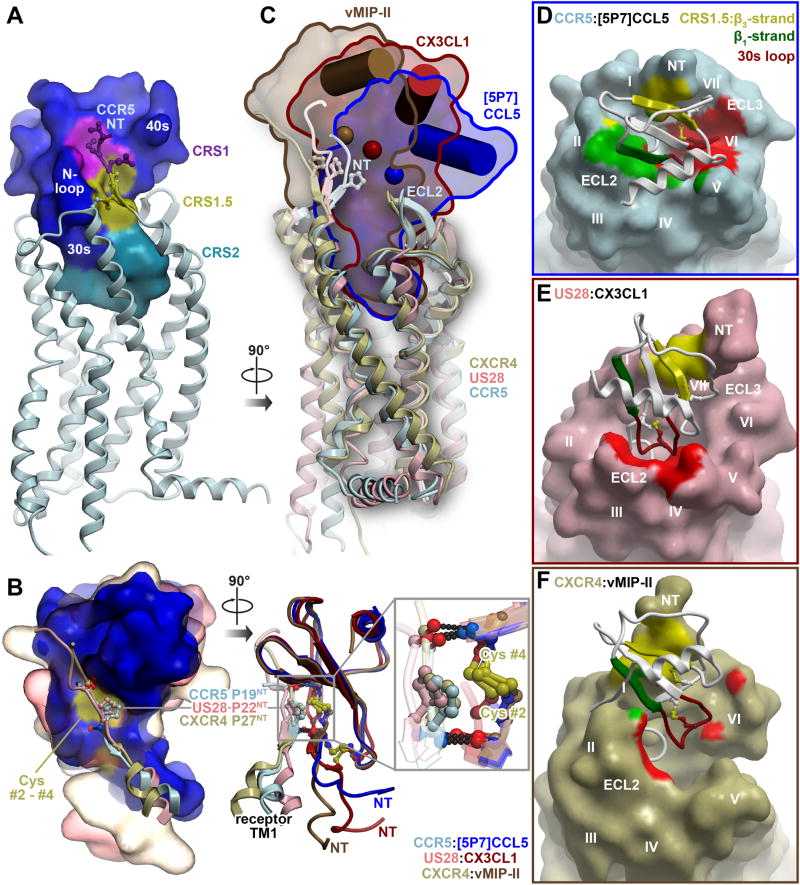
CCR5-[5P7]CCL5 and the two previously solved receptor-chemokine complex structures, US28-CX3CL1 and CXCR4-vMIPII (Burg et al., 2015; Qin et al., 2015), share three interaction epitopes that are emerging as conserved elements of receptor-chemokine architecture: (i) chemokine recognition site 1 (CRS1) where the N-terminus of the receptor binds to the groove between the N-loop and 40s loop of the chemokine (see Figure 1D for nomenclature); (ii) CRS2 where the flexible N-terminus of the chemokine reaches into the TM domain of the receptor, and (iii) the intermediate CRS1.5 where the conserved 19-PC-20 (Pro19-Cys20) motif of the receptor packs against the conserved disulfide of the chemokine (Figure 2A). The geometry and hydrogen-bonding pattern in CRS1.5 is similar between all three complexes (Figure 2B), where it serves as an anchor point that orients the receptor N-terminus into the CRS1 binding groove on the chemokine while simultaneously directing the chemokine N-terminus into the CRS2 receptor binding pocket.
Figure 2. The CCR5-[5P7]CCL5 complex shows additional, distinct interaction epitopes in the context of the conserved two-site architecture.
(A) Three epitopes observed in earlier receptor-chemokine structures are highlighted on the structure of CCR5-[5P7]CCL5: CRS1 (purple), CRS1.5 (gold), and CRS2 (cyan).
(B) Comparison of the CRS1.5 interaction geometry and hydrogen bonding pattern between CCR5-[5P7]CCL5 and earlier receptor-chemokine structures. Chemokines are superimposed by their globular cores and shown as molecular surfaces on the left and ribbons on the right. The fragments involving the N-terminus and helix I of each receptor are shown as ribbons with the conserved Pro of the 19-PC-20 motif in sticks. The Pro invariably packs against the conserved disulfide (yellow surface on left and yellow sticks on right) of the chemokine, and the flanking residues form invariable backbone hydrogen bonds to the chemokine proximal N-terminus and β3-strand.
(C) The diverse positions of the chemokine globular cores with respect to the TM domains of the receptors are shown for CCR5-[5P7]CCL5, US28-CX3CL1 and CXCR4-vMIP-II. The TM domains are superimposed and shown in ribbons, chemokines are shown as molecular surfaces and contoured for clarity. The C-terminal helix and Cα atom of the central residue of the β-sheet (V40 in CCL5) of each chemokine are shown as a cylinder and a sphere, respectively.
(D-F) The interactions formed by the [5P7]CCL5 β1-strand with ECL2 of CCR5 (green) and its 30s loop with CCR5 ECL3 and helices V-VII deep within the CCR5 binding pocket (red), are highlighted and compared with similar regions of US28-CX3CL1 and CXCR4-vMIP-II in (E and F). The β1-strand interaction is virtually absent from both US28-CX3CL1 (E) and CXCR4-vMIPII (F) complexes. The chemokine 30s loop contacts ECL2 in the US28-CX3CL1 complex (E) but shows almost no interaction in the CXCR4-vMIPII complex (F).
See also Figure S2.
Further analysis demonstrated that CRS1.5 acted as a pivot between CRS1 and CRS2, and despite its structural uniformity in the three complexes, enabled a unique arrangement and interactions between CCR5 and [5P7]CCL5. Although the N-termini of all three chemokines penetrated the receptor binding pocket to similar depths, the globular core of [5P7]CCL5 was tilted down towards receptor ECL2, thus sitting much deeper in the binding pocket (4 Å and 7 Å deeper than CX3CL1 and vMIPII, respectively, Figure 2C) and resulting in RMSDs of 10.2 Å and 15.6 Å relative to CX3CL1 and vMIPII, respectively. In this deep-seated pose, [5P7]CCL5 buried 1700 Å2 of surface area (compared to 1300 Å2 for both CX3CL1 and vMIPII) (Figure 2D-2F). As part of this buried interface, the chemokine β1-strand tightly packed against ECL2 of the receptor, while the 30s loop was submerged into the receptor binding pocket where it directly interacted with helix V and activation-associated helices VI and VII (Figure 2D). These interaction epitopes have not been previously observed in receptor-chemokine structures. For example, in US28-CX3CL1, the chemokine 30s loop was much higher in the pocket and packed against ECL2 of the receptor (Figure 2E), and in CXCR4-vMIPII, it made almost no contacts with the receptor (Figure 2F). The US28-CX3CL1 and CXCR4-vMIPII structures also showed no interactions involving the chemokine β1-strand, unlike CCR5-[5P7]CCL5 (Figure 2D-2F). These differences reflect the ability of the chemokines to pivot around CRS1.5 and differentially engage the receptor ECLs, which may be key for the specificity of receptor-chemokine recognition.
Extensive hydrophobic and polar interactions in CRS2 explain high affinity of [5P7]CCL5
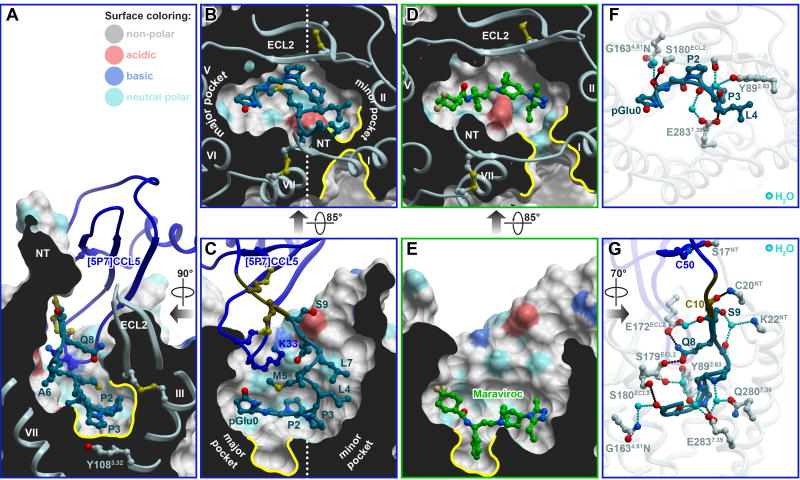
The engineered N-terminus of [5P7]CCL5 was observed to bind in the receptor CRS2 – the wide, polar binding pocket that is conventionally divided into the minor subpocket (helices I-III and VII) and the major subpocket (helices III-VII) (Figure 3A-C). The conformation and interactions of that N-terminus were dramatically different from both the CXCR4-vMIPII and US28-CX3CL1 complexes (Figure S2). The proximal N-terminal residues 4-LMAL-7 (Leu4, Met5, Ala6, and Leu7) of [5P7]CCL5 were found to form a loose helix and to pack against the top parts of receptor helices I and II in the minor subpocket (Figure 3B-C and Figure S2A-D). The distal N-terminus (residues 0-pGluGPP-3 containing pGlu0, the cyclized pyroglutamate form of Q0, followed by Gly1, Pro2, and Pro3) occupied the bottom of the CRS2 pocket, running roughly parallel to the membrane plane, and spanned both the minor and the major subpockets of CCR5, with pGlu0 reaching towards helices V and VI (Figure 3B-C, Figure S2A-D). This binding mode spatially overlapped and resembled that of Maraviroc, the FDA approved HIV entry inhibitor (Tan et al., 2013) (Figure 3D and 3E), although there were important differences as well. A comparison of the residue contact patterns of Maraviroc and the distal N-terminus of the chemokine is shown in Figure S3.
Figure 3. Structural determinants of [5P7]CCL5 affinity in CRS2, and overlap with the HIV inhibitor Maraviroc.
(A-C) Steric packing of the chemokine N-terminus (ribbon and sticks), in the TM domain pocket of the receptor (cut-away surfaces colored by residue sidechain properties). (A) and (C) are views along the plane of the membrane ((C) is designated as “front”, (A) is “right”). (B) shows a view across the plane of the membrane from the extracellular side. Prominent non-polar subpockets mentioned in the text are highlighted with yellow contours.
(D-E) Steric packing of the small molecule antagonist Maraviroc in complex with CCR5 is shown with views and orientations identical to those in (B) and (C).
(F-G) Polar interactions of residues 0–4 (F) and residues 0–9 (G) of [5P7]CCL5 with CCR5. Direct and water-mediated hydrogen bonds are shown in black and cyan, respectively. (F) presents a view across the plane of the membrane from the extracellular side, identical to (B) and (D). (G) is a view along the plane of the membrane from the “left”.
[5P7]CCL5 induced a more compact conformation of CCR5 than Maraviroc (Figure 3B and 3D). Both ECL2 and the N-terminus closed onto the binding pocket, and the extracellular parts of helices I and VII were brought together to block the bilayer-directed channel (Figure 3B and 3D) observed in antagonist-bound structures of CCR5 (Tan et al., 2013) and the closely homologous CCR2 (Zheng et al., 2016). In this differently shaped binding pocket, the distal N-terminus of [5P7]CCL5 made numerous steric contacts, including docking of P2 under the receptor ECL2 (not utilized by Maraviroc, Figure 3D), and stacking of P3 against the π-system of Y1083.32 (Figure 3A and 3B, the superscript indicates receptor residue number according to Ballesteros–Weinstein nomenclature (Ballesteros and Weinstein, 1992)). The unique non-polar subpocket formed by residues Y1083.32, F1093.33, F1123.36, I1985.42 and W2486.48 appeared to be a critical hydrophobic anchoring determinant for Maraviroc (Figure 3E), but was not utilized by [5P7]CCL5 (Figure 3C). Overall, this structural comparison suggested possibilities for rational structure-based optimization of both molecules.
Despite its highly apolar nature, the N-terminus of [5P7]CCL5 made 15 hydrogen bonds to CCR5, eleven of which were mediated by six visible waters (Figure 3F and 3G, and Table S2). In fact, with the exception of G1, every N-terminal residue of [5P7]CCL5 was involved in hydrogen bonding with the receptor: the non-polar residues 0-pGluGPPLMAL-7 through their backbones, and the polar 8-QS-9 through both backbones and sidechains (Figure 3F and 3G). Residues Q8 and S9 also appeared to contribute to the closure of the receptor binding pocket by simultaneously hydrogen bonding to E172 and S179 in CCR5 ECL2 and to the backbone of K22 in its N-terminus. E2837.39, which resides at the boundary between the minor and the major subpocket of CCR5 (also utilized by Maraviroc), hydrogen bonded to four chemokine residues: L4 (directly), and P2, M5, and A6 (through water molecules). Consistent with this data, [5P12]CCL5, a variant with one-residue difference from [5P7]CCL5, showed an 80-fold loss of HIV inhibitory potency against CCR5 containing an E2837.39A mutation (Choi et al., 2012).
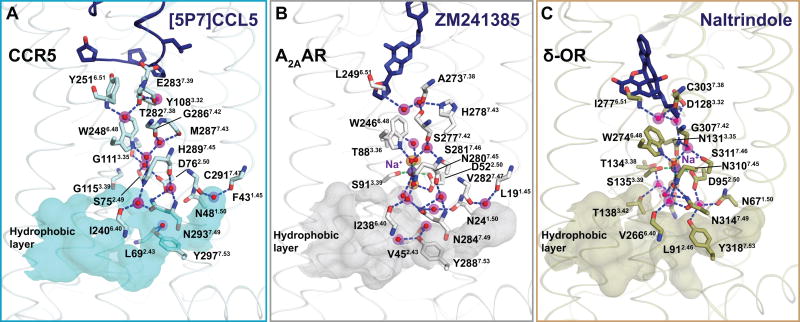
Altogether, the observed extensive hydrophobic, direct polar, and water-mediated interactions between the receptor and the chemokine appeared to contribute to the high affinity of [5P7]CCL5. The water-mediated contacts may also enable efficient recognition of other, diverse natural and engineered chemokine sequences by CCR5 (Steen et al., 2014), and contribute to their signaling capacity and bias. Visible water molecules also continued from the receptor-binding pocket into the TM domain where they appeared to play a role in stabilizing the inactive conformation (Figure 4), similar to the inactive state structures of the adenosine A2A and δ-opioid receptors (Fenalti et al., 2014; Liu et al., 2012).
Figure 4. Water molecules in the TM domain of the CCR5-[5P7]CCL5 structure compared to other GPCR complexes.
Within the TM domain, the water-mediated network is shown traversing from the antagonist binding site (dark blue ribbon/sticks), through the allosteric sodium site (purple spheres), to the hydrophobic layer (matte surface) at the intracellular end of the receptors. Water is shown as red sticks overlaid on magenta spheres.
(A) Water network in the CCR5-[5P7]CCL5 structure. The figure shows that despite the missing sodium ion in the CCR5 structure (possibly due to incompatibility with residues G3.35 and G3.39), the overall polar network is maintained.
(B) In the adenosine A2A receptor (PDB ID: 4EIY), residues D2.50 and S3.39 coordinate a transmembrane sodium ion along with three water molecules (orange).
(C) In the δ-opioid receptor (PDB ID: 4N6H), residues D2.50, N3.35, S3.39, and two water molecules (orange) participate in sodium coordination.
CCL5 signaling epitopes are not confined to its N-terminus and include the N-loop and 30s loop
The classic "two-site model" of receptor-chemokine interaction suggests that globular core of the chemokine is effectively a docking domain for the receptor N-terminus (CRS1) that provides binding affinity whereas the chemokine N-terminus is the signaling trigger that engages the receptor binding pocket (CRS2). In the CCR5-[5P7]CCL5 structure, however, integral parts of the globular core (the chemokine N-loop, β1-strand, and 30s loop) were found to interact with the TM domain of the receptor in ways not predicted by the two-site model.
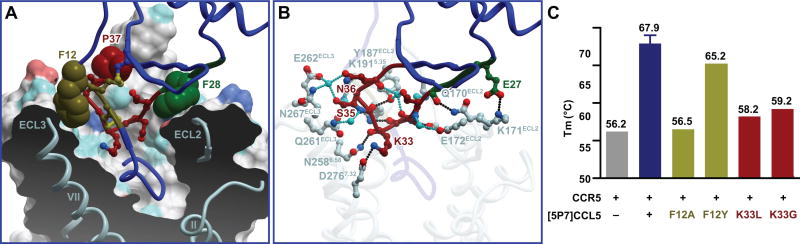
The 30s loop (30-TSGKCSNPA-38) was submerged in the binding pocket (Figure 5A and 2D) where it contributed to CRS2 interactions. Chemokine residues F12 (N-loop), F28 (β1-strand), and P37 (30s loop) mediated aromatic π-stacking interactions with the ECLs of the receptor (the backbone amide of N268-C269 in ECL3, the side-chain of Y187 at the helix V-ECL2 junction, and the backbone amide of E172-G173 at the tip of ECL2, respectively), and served to orient the 30s loop into the receptor-binding pocket (Figure 5A). There, the 30s loop made extensive polar contacts including 17 hydrogen bonds, 12 of which were mediated by five water molecules (Figure 5B and Table S2). This orientation enabled [5P7]CCL5 residue K33 to reach towards receptor helix VII where it formed a salt bridge with D2767.32, while simultaneously hydrogen-bonding (through the backbone) to N2586.58 in helix VI (Figure 5A and 5B).
Figure 5. N-loop and 30s loop interactions represent new motifs in receptor-chemokine interactions.
(A) Steric interactions of the N-loop (brown), 30s loop (red), and β1-strand (green) of [5P7]CCL5 (blue ribbon except for the highlighted parts) with the TM domain and ECLs of CCR5. Residues involved in aromatic stacking interactions are shown as spheres. The receptor is shown as a cut-away surface colored as in Figure 3.
(B) Polar interactions of [5P7]CCL5 30s loop (red), and β1-strand (green) with the CCR5 binding pocket and ECLs. Direct and water-mediated receptor-chemokine hydrogen bonds are shown in black and cyan, respectively.
(C) The effect of mutations of [5P7]CCL5 residues K33 and F12 on the thermostability of CCR5-[5P7]CCL5 complexes.
See also Table S2 and Table S3.
To confirm that the observed N-loop (F12) and 30s loop (K33) residues are indeed important for high-affinity binding of [5P7]CCL5 to CCR5, we mutated them and tested the mutants for their ability to stabilize CCR5 using DSF (Alexandrov et al., 2008). As shown in Figure 5C, K33L and K33G mutations destabilized the complex by more than 10°C relative to "WT" [5P7]CCL5. Substitution of F12 with an Ala (but not Tyr) decreased the melting temperature of the complex to the level of the apo receptor. These data suggested that the N-loop and 30s loop epitopes are important for the affinity between CCR5 and [5P7]CCL5.
Although [5P7]CCL5 is an antagonist, these interactions also appear to play critical roles in the context of the endogenous agonist, WT CCL5. Mutation of F12 in CCL5 caused a 5000-fold reduction in binding affinity and a >90% reduction in calcium flux (Table S3) (Pakianathan et al., 1997), demonstrating the importance of F12 stacking against the extracellular loop connecting the activation-associated helices VI and VII of CCR5. The equivalent of F12 is also found in other endogenous ligands of CCR5 (CCL3 and CCL4, Figure S4), and at least in the case of CCL4, mutation to Ala almost completely abrogates signaling (Laurence et al., 2000).
In contrast to F12, modifications of K33 strongly modulate chemokine signaling and anti-HIV potency with only mild impact on binding affinity. Acetylation of K33 completely abrogates signaling of WT CCL5 (Martin et al., 2001), while a K33L mutation in the CCL5 variant, PSC-RANTES, produces an ~30-fold loss in signaling potency, a significant loss of ability to internalize CCR5, and a remarkable 1000-fold loss in anti-HIV potency, all while showing only a ~6-fold loss in binding affinity. On the contrary, introduction of a lysine (L33K) in the homologous position of an N-terminally modified CCL4 variant produces no change in binding affinity but leads to ~10-fold improvement in signaling potency, significant improvement in internalization capacity, and ~200-fold improvement in anti-HIV potency (Gaertner et al., 2008). The structure suggested that this occurs through loss (or, in the case of CCL4 L33K, gain) of interactions with D2767.32 in the activation-associated CCR5 helix VII. Corroborating this hypothesis, substitution of an Ala for CCR5 residue D2767.32 also causes a significant loss of CCL5 binding and signaling (Navenot et al., 2001; Thiele et al., 2011) (Table S4).
These findings suggest that the N-loop and 30s loop interactions are not only important affinity determinants, but that they also play roles in signaling and the anti-HIV capacity of chemokines. Therefore, for some receptor-chemokine systems, it may be possible to optimize chemokine affinity and pharmacology by engineering mutations outside of the chemokine N-terminus. More broadly, our findings demonstrate that chemokine signaling is not confined to its N-terminus, which has been the existing paradigm for many years.
Modeling of CCR5 with WT CCL5 provides insight into receptor sulfotyrosine recognition and activation
Next, we sought to gain insight into the interactions of CCR5 with WT agonist chemokine CCL5 through modeling (see Methods). Such models would facilitate structure-based interpretation of a large body of mutagenesis data on CCR5-CCL5 (Table S3 and S4). As presented in the previous section, the crystallographically observed interactions and mutagenesis of the globular core of [5P7]CCL5 were in striking agreement with prior mutagenesis of WT CCL5 (Figure 5A-C, Table S3 and S4). Along with other architectural features conserved amongst receptor-chemokine complexes (Figure 2), this suggested that the orientation of the globular core of WT CCL5 with respect to CCR5 is similar to that of [5P7]CCL5. On the other hand, the details of both CRS2 and CRS1 interactions of CCR5 with WT CCL5 were uncertain and could benefit from modeling: the former (CRS2) because of N-terminal sequence differences and the agonist nature of CCL5, and the latter (CRS1) because the sulfotyrosinated N-terminus of the receptor was disordered in the crystal structure.
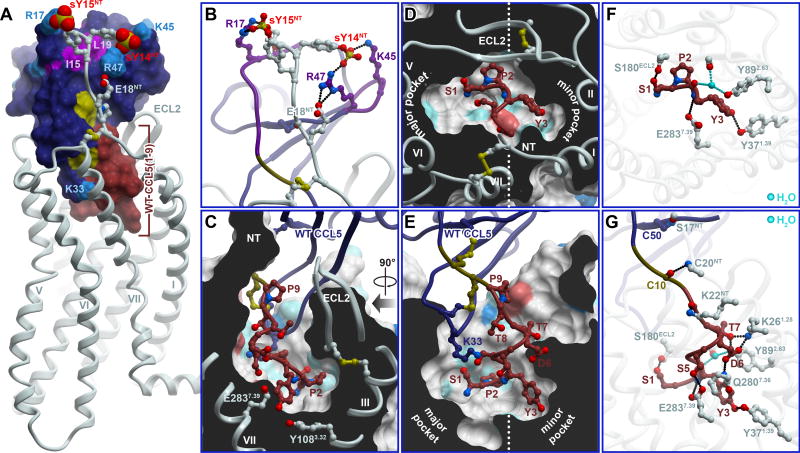
The predicted CRS1 interactions between CCR5 and WT CCL5 (also relevant for CCR5-[5P7]CCL5) are shown in Figure 6A and 6B. The models consistently suggested engagement of E18NT and sulfated Y14NT (sY14NT) of CCR5 in a salt bridge network with chemokine 40s loop residues K45 and R47, while CCR5 residue sY15NT docked near chemokine N-loop residue R17. CCR5 residue E18NT and sulfation of Y14NT and Y15NT were previously shown to be important for CCL5 binding (Bannert et al., 2001; Navenot et al., 2001). Similarly, CCL5 40s loop residues K45 and R47 were demonstrated to be important for CCR5 activation (Martin et al., 2001) and chemotaxis (Proudfoot et al., 2003). Finally, our CRS1 models were in agreement with studies of the N-loop residues I15 and L19 (Figure 6A) which showed an 8-fold and > 5000-fold reduction in affinity for CCR5 upon mutation (Pakianathan et al., 1997).
Figure 6. Modeling provides insights into CCR5 interaction with its endogenous chemokine agonist CCL5.
(A) Overall view of the CCR5-CCL5 model. Residues 1–9 that differ between CCL5 and [5P7]CCL5 are highlighted in saddle brown. Tyrosine sulfates on the receptor N-terminus are shown as spheres. Residues important for the CRS1 interaction (by mutagenesis) are labeled and highlighted in color.
(B) Predicted polar interactions in CRS1.
(C-E) Predicted steric packing of the CCL5 N-terminus (ribbon and sticks) in the TM domain pocket of the CCR5 (cut-away surfaces colored by residue sidechain properties, as in Figure 3). (C) and (E) are two views along the plane of the membrane, “right” and “front”, respectively, as in Figure 3. (D) shows a view across the plane of the membrane from the extracellular side.
(F-G) Predicted polar interactions of the distal N-terminus (residues 1-3, F) and the complete N-terminus (residues 1-9, G) of CCL5 with receptor residues. Direct and water-mediated receptor-chemokine hydrogen bonds are shown in black and cyan, respectively. (F) presents a view identical to (D). (G) is the “front” view, identical to (E).
See also Figure S4, Table S3 and Table S4.
By contrast with CRS1, CRS2 interactions were expected to show the greatest structural differences between the CCR5-[5P7]CCL5 and CCR5-CCL5 complexes, because (i) WT CCL5 is an agonist and (ii) the N-terminal sequence of WT CCL5 is primarily polar (1-SPYSSDTTP-9) while that of [5P7]CCL5 is primarily non-polar (0-pGluGPPLMALQS-9). Nevertheless, the model suggested some important similarities as well. Both peptides followed a similar helical path into the receptor pocket. CCL5 P2 packed under ECL2 of the receptor and just above receptor residues Y1083.32 and F1093.33, similar to P2 of [5P7]CCL5 (Figure 6C-E and 3A-C). As the dominant non-polar interaction between the receptor and WT CCL5, this explained the deleterious effects of a CCL5 P2A mutation (Pakianathan et al., 1997), as well as CCR5 Y1083.32A and F1093.33A mutations (Kondru et al., 2008) on the affinity of the CCR5-CCL5 complex (Table S3 and S4). As in the [5P7]CCL5 complex, the backbone of WT CCL5 was within hydrogen bonding distance from CCR5 residue E2837.39 (Figure 6F-G), which is consistent with a 20-fold reduction in CCR5-CCL5 affinity (Kondru et al., 2008) and the significant decrease in CCL5-induced receptor activation (Thiele et al., 2011) with an E2837.39A mutant (Table S4).
Notwithstanding the above similarities, N-terminal sequence differences between WT CCL5 and [5P7]CCL5 dictated different interactions with the receptor. The sidechain of WT CCL5 S1 formed a hydrogen bond with receptor S180ECL2, while the sidechains of Y3, S5, and D6 participated in an intricate network engaging receptor residues Y371.39, Q2807.36, E2837.39 (Figure 6F-G) and, indirectly, T2847.40. Receptor residue K22NT was held in place by a salt bridge with D2767.32 (both residues are important for high affinity CCL5 binding (Navenot et al., 2001)) and coordinated the proximal N-terminus of CCL5 through hydrogen bonding to the backbone of T8. Receptor residue K261.28 (also important for CCL5 affinity (Navenot et al., 2001)) formed a salt-bridge with chemokine D6 and hydrogen bonded to the side-chain of T7. Finally, receptor residue Y892.63 interacted with the backbones of chemokine P2 and receptor C178ECL2 via a three-way water mediated hydrogen bond (Figure 6F and 6G), as in CCR5-[5P7]CCL5.
With the exception of P2, mutations of individual residues in the CCL5 N-terminus have modest effects (0–60% decrease) on CCR5 binding and signaling (Table S3). Furthermore, high affinity and even agonist activity can be achieved with many N-terminal residue combinations (Gaertner et al., 2008), which contrasts with the more strict signaling requirements of e.g. CXCR4-CXCL12 (Hanes et al., 2015). The tolerance to chemokine N-terminal substitutions may be due to the extensive use of hydrogen bonding and water-mediated interactions, as reflected in the [5P7]CCL5-bound structure and the CCL5-bound model.
Although CCL5 modeling was performed in the context of the inactive receptor structure, it suggested structural determinants of receptor activation. By comparison with the [5P7]CCL5-bound structure, these determinants appeared to involve the interaction of Y3 and D6 of the chemokine with a cluster of CCR5 minor pocket residues including the polar network of K261.28, Y371.39, Y892.63, Q2807.36, E2837.36, and T2847.40, and the non-polar A291.31, L331.35, W862.60, A902.64, and M2877.43. Most prominently, contact of Y3 with Y371.39 and M2877.43 appeared capable of initiating a chain of signal transmission-mediating conformational changes in TM helices VI and VII, in a manner similar to CXCR4 (Wescott et al., 2016). In particular, a bulky residue in chemokine position 3 and polar residues in positions 4–7 are found in almost all agonist CCL5 variants from the 2008 phage display study (Gaertner et al., 2008), whereas in most potent antagonists (including [5P7]CCL5), residue 3 is a Pro and residues 4–7 are bulky hydrophobics unable to engage in the minor pocket polar network (Figure S4B and S4C). Also corroborating the important role of the helix I and VII cluster in CCL5 signaling, CCR5 A291.31S does not efficiently bind CCL5 or promote a chemotactic response (Howard et al., 1999). Finally, residue Y371.39 has not been studied in the context of CCL5-induced signaling but CCR5 mutant Y371.39A shows no chemotactic response to CCL3 (Hall et al., 2009).
Overall, despite the fact that the CCR5-[5P7]CCL5 complex is an inactive state structure, using it as a template enabled a more complete and accurate CCR5-CCL5 model than could be achieved using more distant templates (Tamamis and Floudas, 2014). The model showed consistency with a large body of mutagenesis data, and provided insight into the activation of CCR5 by CCL5 in CRS2 and interactions of sulfotysosine residues in CCR5 with CCL5 in CRS1. The CCR5-CCL5 model and the modeling-enhanced CCR5-[5P7]CCL5 structure also provided a basis for considering the interaction of CCR5 with gp120.
The interaction of gp120 with CCR5 mimics the two-site architecture of CCR5-chemokine complex
We next sought to use the structure and models in the context of prior mutagenesis studies to gain insight into the mechanisms by which CCR5 interacts with HIV gp120, and to better understand how chemokines inhibit this interaction.
The determinants of CCR5 recognition of gp120 are concentrated in its 3rd variable (V3) loop as well its 4th conserved (C4) region (Table S5 and Figure S5A-C). The interaction is dependent on the sulfo-tyrosinated N-terminus of the receptor (CRS1). In fact, isolated sulfopeptides containing CCR5 residues D2-E18 or Y10-E18 bind gp120 (JRFL strain) with appreciable affinity (Cormier et al., 2001), and alanine mutations of CCR5 N-terminal residues Y10, D11, Y14, Y15, S17, and E18 (Blanpain et al., 1999; Doranz et al., 1997; Dragic et al., 1998; Farzan et al., 1998; Rabut et al., 1998), as well as elimination of sulfation on Y10 or Y14 (Cormier et al., 2000), abrogate or strongly reduce binding and CCR5-mediated cell entry for numerous R5-tropic HIV strains (Table S6). As demonstrated by binding experiments with a sulfated N-terminal CCR5 peptide (residues D2-E18), the CRS1-mimicking interaction depends on basic and other polar residues in the gp120 C4 region and in the “stem” part of the V3 loop (Figure S5C-E)(Cormier and Dragic, 2002; Cormier et al., 2001). By contrast, V3 “crown” residues are important for binding to full-length CCR5 (Cormier and Dragic, 2002; Cormier et al., 2001; Rizzuto and Sodroski, 2000; Rizzuto et al., 1998) and are likely recognized by CRS2 (Figure S5C-E).
The two-site architecture suggested by these experiments (Cormier and Dragic, 2002) is similar to that of receptor-chemokine complexes. Specifically, it appears that the basic residues in the V3 stem and C4 of gp120 bind the sulfated N-terminus of CCR5 (thus mimicking the CRS1 interaction) and that the V3 crown of CCR5 reaches into the TM binding pocket of CCR5 (CRS2). This latter CRS2-mimicking interaction critically depends on the conserved P311 in the V3 crown (Cormier and Dragic, 2002; Cormier et al., 2001) (Figure S5D and S5E) as well as on CCR5 residue Y1083.32 (Tam et al., 2016) (Garcia-Perez et al., 2011; Maeda et al., 2006) at the base of the CRS2 pocket (Figure 3A). Based on these findings, we hypothesized that gp120 P311 interacts with CCR5 Y1083.32 in the same manner as P2 of both WT CCL5 and [5P7]CCL5 (Figure 3A, Figure 6C, and Figure S5E).
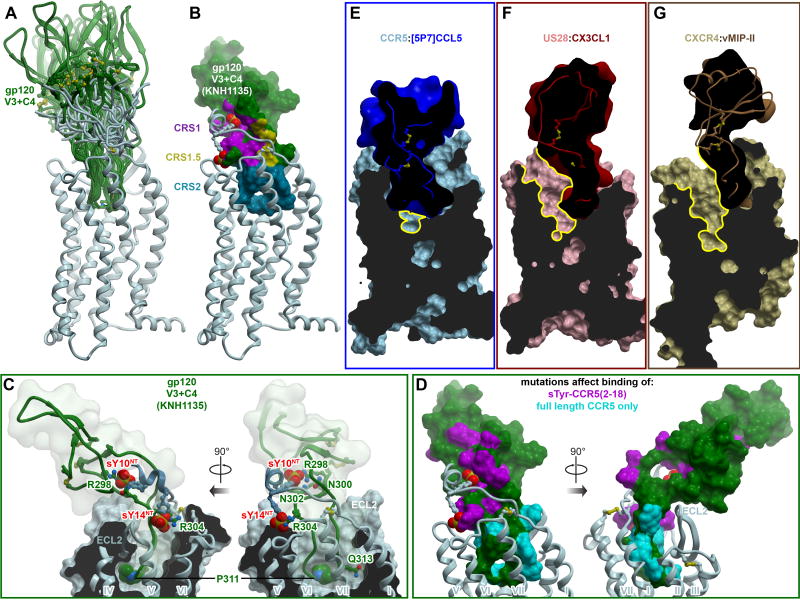
Guided by these insights, we constructed an ensemble of models of CCR5-gp120 complexes (Figure 7A, see Methods) with gp120 sequences from commonly used R5-tropic strains (YU2, JRFL, and KNH1135, Figure S5E). By contrast with earlier predictions (Tamamis and Floudas, 2014; Tan et al., 2013), our docking simulation included not only the V3 loop itself but also the C4 region (Figure S5C and S5D). As with receptor-chemokine recognition, the models suggested numerous possible orientations of the folded core of gp120 with respect to the receptor TM domain (Figure 7A). Conformational variation was achieved though remarkable flexibility of both the V3 stem and receptor N-terminus. However, all predicted geometries were consistent with the two-site architecture of the complex (Figure 7B) and with the membrane orientation of an open, CD4-bound gp120 trimer (Figure S5). Models alternatively suggested interaction of (i) sY14 with R298V3stem and N302V3stem with sY10 reaching towards R327C4, or (ii) sY10 with R298V3stem and N302V3stem with sY14 contacting R304V3stem (Figure 7C). The latter binding mode parallels V3 interactions with “V2” that has been described as a mimic of CCR5 (Cimbro et al., 2016). In some high-scoring models, the N-terminus of CCR5 (residues I9-sY14) folded into an α-helix (Figure 7C-D) as previously predicted based on peptide NMR in the presence of gp120 (Huang et al., 2007). In CRS2, gp120 P311V3crown packed under the receptor ECL2 and just above Y1083.32 (Figure 7C) whereas the critically important polar residue Q/R313 in the GPG[RQ] motif at the tip of the V3 crown formed a polar network with E2837.39 and Q2807.36, the latter also indispensable for gp120 entry (YU2 and 89.6 strains (Farzan et al., 1998)). Overall, steric feasibility of the model and its consistency with receptor and gp120 mutagenesis suggested that CCR5-gp120 largely mimics chemokine interactions and utilizes similar recognition epitopes of the receptor, including CRS1 and CRS2 (Figure 7B and 7D).
Figure 7. Insights into CCR5 interaction with HIV gp120 and inhibition of this interaction by chemokines.
(A) Ensemble of models of complexes between CCR5 and a fragment of HIV gp120 containing its V3 loop and C4 region. The ensemble illustrates multiple possible orientations of gp120 with respect to the receptor, enabled via the inherent flexibility of the interaction interface.
(B) Representative model of CCR5 (ribbon) in complex with the V3/C4 fragment of HIV gp120 (strain KNH1135, molecular surface). Interaction sites for the receptor N-terminus, the conserved 19-PC-20 motif, and the TM binding pocket are highlighted to illustrate architectural similarity to complexes with chemokines.
(C) Same model as in (B), showing interactions of important residues. CCR5 is shown in ribbon and a cut-away molecular surface, gp120 in ribbon surrounded by a transparent simplified molecular surface. Residues mentioned in the text are shown in sticks. Residue P311 in the conserved GPG[RQ] motif of the gp120 V3 crown, as well as the CCR5 N-terminal tyrosine sulfates are shown as spheres.
(D) Prior mutagenesis of gp120 mapped onto the surface of the gp120 V3/C4 fragment using the same model as in (B). Residues that affect binding of a sulfo-tyrosinated CCR5 N-terminal peptide (2–18) are colored in magenta. Residues whose mutations show effects only in the context of full-length CCR5 are show in cyan. The predicted architecture and topology of the complex are highly consistent with mutagenesis.
(E-F) Among chemokine crystallized with receptors, [5P7]CCL5 is unique in its ability to entirely fill the binding pocket of CCR5 (E). By contrast, binding of CX3CL1 to US28 (F) or vMIPII to CXCR4 (G) leads to only partial occupancy of the pocket, with much of the major subpocket remaining accessible (yellow contours). Receptors are shown as cut-away surfaces, chemokines as simplified cut-away surfaces and ribbons.
See also Figure S5, Table S5 and Table S6.
Discussion
The present structure of CCR5 with a potent chemokine inhibitor of HIV entry elucidated the architecture, motifs, and epitopes explored by this receptor when binding to diverse ligands. It revealed an "extended two-site” architecture as a versatile, structurally adaptable framework that enables receptor interactions not only with distantly homologous chemokines but with unrelated proteins such as gp120. The structure also suggested that the interaction versatility in CRS2 is dependent on the polar nature of the receptor TM pocket and is aided by networks of direct and water-mediated hydrogen bonds. The resulting binding pocket plasticity may play to the advantage of gp120 by enabling productive CCR5-mediated viral fusion to be triggered by a diverse repertoire of gp120 sequences.
HIV gp120 appears to creatively utilize the extended two-site architecture to mimic chemokines in its interactions with the receptor. Therefore, it is not surprising that chemokines can function as HIV entry inhibitors. But not all chemokines are created equal; in fact, WT CCL5 and other endogenous chemokine agonists are rather weak inhibitors while engineered variants like [5P7]CCL5 can be exceptionally potent. This is despite current evidence suggesting that WT CCL5 and [5P7]CCL5 share many similarities in the way they interact with CCR5.
The reason for the poor anti-HIV properties of endogenous agonist chemokines lies partly in their conformational selectivity, as they only bind with high affinity to the subpopulation of receptors that is G protein-coupled, whereas gp120 is indiscriminate and targets the entire receptor pool (Colin et al., 2013). This drawback can be alleviated with chemokine N-terminal engineering. Indeed, efforts to improve anti-HIV potency resulted in numerous chemokine variants that appear to act through two distinct mechanism: one mediated by receptor internalization and sequestration (such variants are frequently receptor agonists, e.g. [PSC]CCL5), and another by high affinity binding to the entire CCR5 population and thus by competitive inhibition of gp120 (such variants are typically receptor antagonists, e.g. [5P12]CCL5) (Colin et al., 2013).
The crystallized variant [5P7]CCL5 is highly similar, both in its sequence (one amino acid difference) and its pharmacology, to [5P12]CCL5, and therefore likely acts through this latter competitive displacement mechanism. The current study provided insight into the structural basis of its high affinity antagonism and potency against HIV. The antagonist affinity determinants included packing of the bulky hydrophobic residues 4–7 of [5P7]CCL5 in the minor subpocket of the receptor with simultaneous extension of the engineered pGlu0 into the major subpocket, both reminiscent of Maraviroc. At the same time, the polar residues in the proximal N-terminus of the chemokine (Q8 and S9, absent in WT CCL5) ensured closure of the extracellular parts of the receptor and engagement of ECL2 residues critical for gp120 interaction (like E172ECL2 (Maeda et al., 2006; Tam et al., 2016)). Finally, immersion of the [5P7]CCL5 30s loop along with its N-terminus into the receptor binding pocket effectively filled it (Figure 7E-G) and completed the gp120 blockade.
Our biochemical experiments revealed a slow rate of [5P7]CCL5 dissociation from CCR5, which likely contributes to its anti-HIV potency. Slow off-rate is also a defining feature of Maraviroc (Swinney et al., 2014). However, [5P7]CCL5 may have additional advantages over Maraviroc with respect to the development of HIV drug resistance. In response to prolonged exposure to Maraviroc, gp120 accumulates mutations that allow it to utilize Maraviroc-bound CCR5 for entry, by relying more heavily on interactions outside the CRS2 pocket of the receptor (Roche et al., 2016; Tilton et al., 2010). Such resistance mutants would be unlikely to evolve in the presence of [5P7]CCL5 challenge (Nedellec et al., 2011) because, as the structure and models demonstrate, [5P7]CCL5 occupies virtually all epitopes required for gp120 binding (Figure 7E). The structure showed that regions other than the chemokine N-terminus can be engineered for the development of highly effective HIV entry inhibitors and suggested ways in which small molecules such as Maraviroc can also be further optimized.
Our findings have broad implications with respect to principles of receptor-chemokine recognition, which become apparent by comparison of CCR5-[5P7]CCL5 with the structures of CXCR4-vMIP-II and US28-CX3CL1. The different orientations of the chemokines with respect to their receptors, and the exclusive involvement of the 30s loop in the receptor binding pocket and N-loop in ECL interactions in the CCR5-[5P7]CCL5 complex, illustrate potential mechanisms for recognition specificity and fine-tuning of signaling properties. The dense network of waters observed in the CCR5-[5P7]CCL5 interface may also be a common characteristic of chemokine complexes, and play a role in the ability of selected receptors to accommodate and differentially signal in response to multiple chemokine ligands (Steen et al., 2014). Because CCR5 is involved not only in HIV but also in many inflammatory diseases and cancer (Martin-Blondel et al., 2016; Weitzenfeld and Ben-Baruch, 2014), its structures with diverse small molecule and chemokine ligands provide important insight into strategies for developing drugs against these diseases.
STAR*Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Tracy M. Handel (thandel@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Spodoptera frugiperda (Sf9) cells (ATCC, CRL-1711™) were used to produce recombinant CCR5:[5P7]CCL5. The cells were maintained in ESF 921 Protein-Free Insect Cell Culture Medium (Expression systems, 96-001-01) at 27°C with shaking at 140 rpm, and passaged every 72h.
Sf9 cells were obtained from and authenticated by ATCC. Sex of cells is not relevant.
METHOD DETAILS
Overview of CCR5-chemokine complex screening pipeline
In broad overview, N-terminally modified variants of CCL5 were coexpressed with engineered CCR5 in Sf9 cells, and the complexes were purified by TALON affinity chromatography through a His tag on the receptor. To identify complexes suitable for crystallization trials, the purified samples were analyzed by SDS-PAGE gel for the presence and abundance of co-purified chemokine, by analytical size exclusion chromatography to evaluate complex homogeneity, and by differential scanning fluorimetry to determine relative thermostabilities. Details follow in the next sections.
Design of CCL5 expression construct
Eleven potent anti-HIV variants of CCL5 from the phage display study by (Gaertner et al., 2008) were cloned and characterized: 5P1, 5P2, 5P3, 5P4, 5P7, 5P9, 5P12, 6P4, 6P10, 7P17, and 7P19. To ensure proper processing and secretion of the chemokine variants, the expression constructs were designed to contain the signal sequence of human CCL25 (MNLWLLACLVAGFLGAWAPAVHT). The design capitalized on the fact that all studied chemokine variants contained QG as the first two amino acids, as does CCL25. Plasmid construction was carried out in two steps as follows: First, three fragments were ligated together -- fragment A (two annealed oligonucleotides coding for the signal peptide and QGPP with 5’ BamHI and 3’ NcoI cutting site overhangs), fragment B (amplified CCL5 coding sequence from residue C10 to S68 plus a stop codon, with 5’ NcoI and 3’ XhoI overhangs), and fragment C (pFastBac1 vector (Invitrogen), linearized by double digestion with BamHI and XhoI). Next, the resulting plasmid was used to insert the variable sequences after QGPP and before C10 using the SLIM method (Chiu et al., 2008). For crystallization, an additional modification to the best-behaving constructs involved insertion of a 10XHis tag at the C-terminus of the chemokine coding sequence to reduce its loss during purification. The final CCL5-5P7 expression vector contained in order: the CCL25 signal sequence, the [5P7]CCL5 open reading frame (ORF) of the mature chemokine sequence, a 3C protease cutting site (LEVLFQGP) and a 10XHis tag all under the control of polyhedron promotor in pFastbac1. All final expression vectors were verified by sequencing.
Design of the CCR5 expression construct
The expression construct for CCR5 was the same one used to solve the Maraviroc bound structure (Tan et al., 2013). In order from 5' to 3', it contains a cleavable signal sequence from influenza hemagglutinin (HA signal sequence, encoding MKTIIALSYIFCLVFA), the CCR5-rubredoxin ORF containing 4 stabilizing mutations (C58Y, G163N, A233D, K303E) as described in (Tan et al., 2013), a 3C protease cleavage site, a 10xHis tag and a FLAG tag followed by a stop codon, all under the control of the polH promoter. All final expression vectors were verified by sequencing.
Coexpression in Sf9 cells
The generation of high-titer recombinant baculovirus (>109 viral particles per mL) was achieved using the Bac-to-Bac Baculovirus Expression System (Invitrogen). Initial transfection was carried out by adding a pre-mixture containing 5 µL recombinant target gene bacmid, 3 µL of Xtreme Gene Transfection Reagent (Roche), and 100 µL of Transfection Medium (Expression Systems) into 2.5 mL of Sf9 cells at a density of 1.2×106 cells/mL. Cell suspensions were incubated at 27 °C for 96 h with shaking. P0 viral stocks were then isolated by centrifugation and used to seed P1 viral stocks. Viral titers were quantified by flow cytometry following cell staining with PE-conjugated anti-gp64 antibody (Expression Systems). Sf9 cells at a density of 2–2.6×106 cells/mL were co-infected with P1 virus of both CCR5-rubredoxin and chemokine, each at an MOI of 5. Biomass was harvested by centrifugation between 44 and 48 h post infection and stored at −80 °C until further use.
Purification of CCR5-chemokine complexes
Biomass was thawed and lysed in hypotonic buffer A (10 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl, and EDTA-free protease inhibitor cocktail (Roche)). Purified membranes were prepared by repeated (4×) dounce homogenization of the samples (40 strokes per round) followed by centrifugation at 50,000×g at 4 °C for 30 min. Membrane pellets were resuspended in buffer A during round 1–2 and in a high salt buffer B (buffer A plus 1 M NaCl) during rounds 3–4. Following the last centrifugation, membranes were resuspended and homogenized in hypotonic buffer A supplemented with 30% glycerol (v/v) and flash-frozen for storage at −80 °C until further use.
CCR5-chemokine complexes were purified with a one-step Talon affinity chromatography method using the His tag on the receptor (in the initial construct screening) or on both the receptor and the chemokine (for the final crystallization studies). Washed membranes were resuspended in buffer containing 2 mg/ml iodoacetamide, and EDTA-free complete protease inhibitor cocktail tablets, and incubated at 4 °C for 1 h before solubilization. The membranes were then solubilized in equal volumes of 2X Buffer C (50 mM HEPES pH 7.5, 400 mM NaCl, 1% (w/v) n-dodecyl-β-d-maltopyranoside (DDM, Anatrace), 0.2% (w/v) cholesteryl hemisuccinate (CHS, Sigma)) at 4 °C for 3 h. The supernatant was isolated by centrifugation at 50,000×g for 30 min, and incubated with TALON IMAC resin (Clontech) overnight at 4 °C. After binding, the resin was washed with ten column volumes of Wash I Buffer (50 mM HEPES pH 7.5, 400 mM NaCl, 10% (v/v) glycerol, 0.1% (w/v) DDM, 0.02% (w/v) CHS, 10 mM imidazole), followed by four column volumes of Wash II Buffer (50 mM HEPES pH 7.5, 400 mM NaCl, 10% (v/v) glycerol, 0.02% (w/v) DDM, 0.004% (w/v) CHS, 50 mM imidazole). The protein was then eluted with three column volumes of Elution Buffer (50 mM HEPES pH 7.5, 400 mM NaCl, 10% (v/v) glycerol, 0.02% (w/v) DDM, 0.004% (w/v) CHS, 250 mM imidazole). PD MiniTrap G-25 columns (GE Healthcare) were used to remove imidazole. Complexes were then treated overnight with His-tagged 3C protease (produced in house) to cleave the C-terminal His-tag and Flag-tag on the receptor and His-tag on the chemokine. 3C protease and the cleaved C-terminal tag fragments from both receptor and the chemokine were removed by binding to TALON IMAC resin for 2 h at 4 °C and collecting the flow through. The final protein complex was concentrated to approximately 40 mg/mL using a 100 kDa MWCO Amicon concentrator (Millipore).
Screening and characterization of CCR5-chemokine complexes
Complexes were analyzed for their thermostability, which was quantified by a differential scanning fluorimetry assay adapted from a previous publication (Alexandrov et al., 2008) using a RotorGene Q 6-plex RT–PCR instrument (Qiagen). Briefly, for a 20 µL assay volume, 1–5 µg of protein was mixed with 3 µM 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) dye (2.5 mM stock in DMSO) in 25 mM HEPES pH 7.5, 400 mM NaCl, 0.02% DDM, 0.004% CHS, 10% glycerol. After a 5 min incubation at room temperature, the samples were gradually heated from 28 °C to 90 °C at a rate of 0.8 °C/ min, with CPM fluorescence (excitation 365 nm, emission 460 nm) recorded every 1 °C. The melting temperature (Tm) was determined from the first derivative of the denaturation curve, using Rotor-Gene Q – Pure Detection software (version 2.0.3). [5P7]CCL5 was selected for crystallization based on its ability to stabilize CCR5-rubredoxin to a high Tm (68 °C). The complexes were further analyzed for homogeneity by analytical size exclusion chromatography using a Sepax SRT-C 300 column.
Lipidic cubic phase crystallization
Purified CCR5-[5P7]CCL5 was reconstituted into lipidic cubic phase (LCP) by mixing with molten lipid using a mechanical syringe mixer (Caffrey and Cherezov, 2009). The protein–LCP mixture contained 40% (w/w) receptor-chemokine complex solution, 54% (w/w) monoolein (Sigma) and 6% (w/w) cholesterol (Sigma). Crystallization trials were performed in 96-well glass sandwich plates (Hampton Research) using a Mosquito LCP robot (TTP Labtech) by dispensing 35 nL of protein-laden LCP and 800 nL of precipitant solution per well. Plates were incubated at 20 °C and imaged using a Rock Imager 2 (Formulatrix). The final crystallization condition contained 30 % (v/v) PEG 400, 140 mM lithium citrate, 1.2 % (w/v) 1,5-diaminopentane dihydrochloride, 100 mM 2-(N-morpholino)ethanesulfonic acid at pH 6.3. Crystals usually grew to a maximum size of 70 µm×20 µm×10 µm in 10 days, after which they were harvested directly from the LCP matrix using MiTeGen micromounts and flash cooled in liquid nitrogen.
Data collection and structure determination
X-ray diffraction data were collected using a 5 µm collimated minibeam at a wavelength of 1.0332 Å with a Pilatus3 6M direct detector on the 23ID-D beamline (GM/CA CAT) of the Advanced Photon Source at the Argonne National Laboratory. Crystals were located and aligned by the rastering strategy (Cherezov et al., 2009). Most crystals diffracted to 2.2–2.5 Å resolution when exposed to 0.3 s of unattenuated beam using 0.3° oscillations. To mitigate radiation damage, a continuous (helical vector) data collection mode was employed (Stepanov et al., 2011). A 99.1% complete data set at 2.2 Å resolution was obtained by merging data from 10 crystals, using XDS (Kabsch, 2010) and Aimless (Winn et al., 2011). As the data showed strong anisotropy, the UCLA Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/) was used to performs ellipsoidal truncation of the data to the recommended resolution limits (2.3 Å, 2.9 Å, and 2.2 Å along the a*, b*, and c* axes, respectively), to anisotropically scale the data, and to apply negative isotropic B-factor correction (Strong et al., 2006). Initial phase information was obtained by molecular replacement with the program Phaser (McCoy et al., 2007) using three separate components, the receptor structure (PDB accession number 4MBS), the rubredoxin structure (PDB accession number 1IRO), and the [1P2]CCL5 chemokine structure (PDB accession number 2VXW) as search models. The correct molecular replacement solution (translation function Z-score = 27.9) contained one CCR5-rubredoxin molecule and one [5P7]CCL5 molecule in the asymmetric unit. Refinement was performed with Refmac (Winn et al., 2011) and Phenix (Adams et al., 2010) followed by manual examination and rebuilding of the refined coordinates in the program COOT (Emsley et al., 2010) using both |2Fo| − |Fc| and |Fo| − |Fc| maps. The final model included 295 residues (16–223 and 227–316) of the 352 residues of CCR5, residues 1–54 of rubredoxin, and 5P7[CCL5] from residue pGlu0 to M67. The remaining N- and C-terminal residues of the receptor and the C-terminal residue of [5P7]CCL5 were disordered and therefore not built. Data collection and refinement statistics are shown in Table S1. Placement of key regions of [5P7]CCL5, such as the first 11 residues of the N-terminus and 30s loop (Figure 1D), was additionally verified by separately omitting these regions and generating composite omit maps in Phenix, using simulated annealing with bulk solvent exclusion.
Buried surface area calculation
Buried surface area in CCR5-[5P7]CCL5, US28-CX3CL1, and CXCR4-vMIPII complexes was calculated using the PISA software (Krissinel and Henrick, 2007) which is also a part of the CCP4 package (Winn et al., 2011). The reported number is the average buried surface area per molecule. Hydrogen atoms were not included in the calculation.
Molecular modeling of the CCR5-CCL5 complex
Models of the CCR5 complex with WT CCL5 were built in the ICM software package (Abagyan and Totrov, 1994). The stabilizing G1634.61N mutation was reverted back to WT prior to modeling.
The position and orientation of the globular domain of WT CCL5 with the respect to the receptor was replicated from the CCR5-[5P7]CCL5 complex. Full conformational flexibility (backbone and sidechain) was allowed for the chemokine N-terminus and 30s loop, as well as for the sulfo-tyrosinated receptor N-terminus. Models of CRS1 and CRS2 interactions were built separately, each in two stages: one employing 3D grid potentials and another full-atom representations of all components.
For CRS2 modeling, the receptor binding pocket was first represented with a set of grid interaction potentials, including the potentials for van der Waals, electrostatic, hydrogen bonding and apolar surface interactions (Totrov and Abagyan, 2001). The N-terminus and the 30s loop of the chemokine (residues 1-SPYSSDTTPCC-11 and 31-SGKCSNP-37, respectively) were built ab initio. An explicit disulfide bond was imposed between C10 and C34; residues C10, C11, S31, C34 and P37 were tethered to the corresponding residues in the template. Five water molecules coordinated by the 30s loop residues, one coordinated by Y89 and C178 of the receptor, and one coordinated by S180 were copied into the model from the template, with their oxygen atoms tethered to template positions. The system including the N-terminus, 30s loop, and water molecules was thoroughly sampled in the receptor potential grids to generate a stack of diverse energetically favorable conformations. For the second stage, the obtained conformational stack was merged with the full atom model of the receptor, and at least 108 steps of Monte Carlo optimization were performed allowing for the same level of flexibility in the chemokine fragments and water molecules with added full flexibility of receptor binding pocket sidechains. Three to five top scoring conformations were inspected visually. The selection of the final conformation took into account consistency between multiple simulations, compatibility with one-residue N-terminal extension of the chemokine, and agreement with prior mutagenesis.
For CRS1 modeling, the chemokine was first represented with a set of 3D grid interaction potentials. The receptor N-terminus (residues 13-NsYsYTSEPC-20) was built ab initio and its conserved cysteine (C20) was tethered to the corresponding residue in the template. The N-terminus was thoroughly sampled in the chemokine potential grids to generate a stack of diverse energetically favorable conformations. The obtained stack was merged with the full atom model of the chemokine, and another 108 steps of Monte Carlo optimization were performed, allowing for the same level of flexibility in the receptor N-terminus with added flexibility of chemokine interface sidechains.
For final model assembly, top scoring conformations of receptor and chemokine fragments were merged with the remaining parts of both molecules into an intact complex, and another short round of side-chain refinement was performed to remove steric conflicts resulting from the merge.
Inaccuracies are to be expected in the CCR5-CCL5 model because of the exclusion, for practical reasons, of water molecules at the bottom of the binding pocket, as well as the use of fixed positions of the receptor helices in an inactive state structure. Despite these caveats, the final model recapitulates a large body of mutagenesis data. The benefit of having the CCR5-[5P7]CCL5 structure as a template enables a more complete and accurate CCR5-CCL5 model than could be obtained by NMR studies focused on CRS1 interactions (Duma et al., 2007) or by homology modeling based on more distant templates (Tamamis and Floudas, 2014).
Molecular modeling of the CCR5-gp120 complex
Models of CCR5 complex with a fragment of HIV gp120 were also built in ICM (Abagyan and Totrov, 1994). As for CCR5-CCL5 model generation, the stabilizing G1634.61N mutation was reverted back to WT prior to modeling.
Construction of the ensemble of CCR5-gp120 models was hypothesis-driven and based on the observed features of the CCR5-[5P7]CCL5 structure. A full atom docking module was assembled from (i) the N-terminal part of the gp120 V3 loop (residues C296-G310, residue numbering as in Figure S5), (ii) the C-terminal part of the gp120 V3 loop (residues G312-C330), (iii) gp120 residues C378-C385, (iv) the gp120 C4 region (residues C418-C445), and (v) the sulfotyrosinated N-terminus of CCR5 (residues 8–20). Explicit disulfide bonds were imposed between gp120 residues C296 and C330, C378 and C445, C385 and C418. Five backbone hydrogen bonding distance restraints were set to maintain the β-sheet arrangement between the N-terminal part of V3 (residues C296-R298) and the surrounding strands of the gp120 module, as observed in CD4-bound gp120 (C296 O to C445 N, R298 backbone N/O to I443 backbone O/N, respectively, and T297 backbone N/O to H329 O/N, respectively). The backbone atoms of V3 residues G310 and G312 (except the G312 carbonyl) were tethered to the corresponding atoms of residues G1 and P3 of [5P7]CCL5. The side-chain of residue C20 of the CCR5 N-terminal peptide was tethered to its position in the CCR5-[5P7]CCL5 structure. For receptor sulfotyrosines, a set of soft harmonic distance restraints was imposed in two alternative modes: one to promote proximity of the sulfate groups on CCR5 sY10 and sY14 to the sidechains of gp120 R298 and R304, respectively, and another to promote proximity of the sulfate group on CCR5 sY14 to the sidechain of gp120 R298. The remaining parts of the receptor (residues 21–316) were represented with grid potentials as above.
In the course of the sampling, full conformational flexibility was allowed for residues C296-R326 of gp120 and the entire receptor N-terminus; for all other residues in the gp120 module, only side chain flexibility was allowed. The system was built using gp120 sequences of three representative strains: YU2, BaL, and KNH1135. The initial conformation of the gp120 module was replicated from the CD4-bound structure of gp120 (PDB 2QAD (Huang et al., 2007)). The system was thoroughly sampled (at least 1010 Monte Carlo optimization steps) in internal coordinates using two random starts for each strain. Top scoring conformations were inspected visually.
QUANTIFICATION AND STATISTICAL ANALYSIS
Prism v.5.0 (GraphPad Software Inc.) and Rotor-Gene Q – Pure Detection v. 2.0.3 (Qiagen) were used to analyze DSF data.
DATA AND SOFTWARE AVAILABILITY
The coordinates and structure factors for the CCR5-[5P7]CCL5 complex structure have been deposited in the Protein Data Bank under accession code 5UIW. Coordinates of the CCR5-CCL5 complex model are provided as supplementary data S1. Coordinates of CCR5-gp120 complexes are available from the authors upon request.
Supplementary Material
Atomic coordinates for the predicted complex between CCR5 and WT CCL5, in PDB format.
Figures S1–S5 and Tables S1–S6.
Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| gp64-PE antibody | Expression Systems | Cat# 97-201 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 4693159001 |
| Iodoacetamide | Sigma | Cat# I1149 |
| n-dodecyl-beta-D-maltopyranoside (DDM) | Anatrace | Cat# D310 |
| Cholesterol hemisucinate (CHS) | Sigma | Cat# C6512 |
| TALON IMAC resin | Clontech | Cat# 635507 |
| 1-Oleoyl-rac-glycerol (monoolein) | Sigma | Cat# M7765 |
| Cholesterol | Sigma | Cat# C8667 |
| N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) | Invitrogen | Cat# D10251 |
| Tetracycline | Sigma | Cat# T7660 |
| Geneticin | Life Technologies | Cat# 10131-035 |
| LB Agar plates with Gentamicin 7, Kanamycin 50, Tetracycline 10 Bluo gal 100, IPTG 100mm plates | Teknova | Cat# L1919 |
| ESF921 | Expression Systems | Cat# 96-001-01 |
| X-tremeGENE™ HP DNA Transfection Reagent | Roche | Cat# 6366244001 |
| His-tagged 3C protease | GenScript | Cat# Z03092-500 |
| Critical Commercial Assays | ||
| Bac-to-Bac Baculovirus Expression system | Invitrogen | Cat# A11100 |
| QuikChange site-directed mutagenesis | Agilent Technologies | Cat# 200519 |
| Deposited Data | ||
| CCR5-[5P7]CCL5 | This paper | PDB: 5UIW |
| Experimental Models: Cell Lines | ||
| Sf9 cells | ATCC | Cat# CRL-1711 |
| ESF 921 Protein-Free Insect Cell Culture Medium | Expression Systems | Cat# 96-001-01 |
| Software and Algorithms | ||
| AIMLESS | (Winn et al., 2011) | www.ccp4.ac.uk/html/scala.html |
| COOT | (Emsley et al., 2010) | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| ICM | (Abagyan and Totrov, 1994) | http://www.molsoft.com |
| Phaser | (McCoy et al., 2007) | http://www.ccp4.ac.uk |
| Phenix | (Adams et al., 2010) | https://www.phenix-online.org |
| PISA | (Krissinel and Henrick, 2007) | http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html |
| Prism v.5.0 | GraphPad Software Inc. | https://www.graphpad.com/scientific-software/prism/ |
| REFMAC | (Murshudov et al., 2011) | http://www.ccp4.ac.uk |
| Rotor-Gene Q – Pure Detection (v. 2.0.3) | Qiagen | https://www.qiagen.com/us/ |
| XDS | (Kabsch, 2010) | http://xds.mpimf-heidelberg.mpg.de/ |
| Other | ||
| 100 kDa molecular weight cut-off Vivaspin 6 concentrator | Sartorius Stedim | Cat# VS0642 |
| PD MiniTrap G-25 columns | GE Healthcare | Cat# 28-9180-07 |
| Amicon™ Ultra-0.5 Centrifugal Filter Units Ultra | EMD Millipore | Cat# UFC510096 |
| 96-well LCP glass sandwich set | Hampton Research | Cat# HR3-151 |
Acknowledgments
We thank L. Qin for help during early stages of the project, M. Gustavsson and T. Kawamura for helpful suggestions, A. Ishchenko, H. Zhang, and M. Audet for help with FRAP experiments and X-ray data collection, R. Sanishvili, N. Venugopalan, and staff at beamline 23ID at GM/CA CAT Advanced Photon Source.
Funding was provided by National Institutes of Health grants R01 AI118985, R01 GM117424, R21 AI121918, R21 AI122211, and R01 GM071872. GM/CA@APS has been funded from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated by Argonne National Laboratory under contract number DE-AC02-06CH11357.
Footnotes
Author Contributions
I.K. and T.M.H. designed the study and coordinated experiments. Y.Z. conducted all experiments and was in charge of all aspects of the structure determination. I.K. designed and performed all molecular modelling and computational analyses. V.C. and R.C.S. assisted with crystallization. G.W.H. assisted with structure determination and refinement. R.A. assisted with structure analysis and modelling. B.W. contributed the CCR5-rubredoxin fusion construct. Y.Z., I.K., and T.M.H. wrote the paper.
References
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Microscale Fluorescent Thermal Stability Assay for Membrane Proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Ballesteros JA, Weinstein H. Analysis and refinement of criteria for predicting the structure and relative orientations of transmembranal helical domains. Biophysical Journal. 1992;62:107–109. doi: 10.1016/S0006-3495(92)81794-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Craig S, Farzan M, Sogah D, Santo NV, Choe H, Sodroski J. Sialylated OGlycans and Sulfated Tyrosines in the NH<sub>2</sub>-Terminal Domain of CC Chemokine Receptor 5 Contribute to High Affinity Binding of Chemokines. J Exp Med. 2001;194:1661–1674. doi: 10.1084/jem.194.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Doranz BJ, Vakili J, Rucker J, Govaerts C, Baik SSW, Lorthioir O, Migeotte I, Libert F, Baleux F, et al. Multiple Charged and Aromatic Residues in CCR5 Amino-terminal Domain Are Involved in High Affinity Binding of Both Chemokines and HIV-1 Env Protein. J Biol Chem. 1999;274:34719–34727. doi: 10.1074/jbc.274.49.34719. [DOI] [PubMed] [Google Scholar]
- Burg JS, Ingram JR, Venkatakrishnan AJ, Jude KM, Dukkipati A, Feinberg EN, Angelini A, Waghray D, Dror RO, Ploegh HL, et al. Structural basis for chemokine recognition and activation of a viral G protein–coupled receptor. Science. 2015;347:1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerini F, Gaertner H, Madden K, Tolstorukov I, Brown S, Laukens B, Callewaert N, Harner JC, Oommen AM, Harms JT, et al. A scalable low-cost cGMP process for clinical grade production of the HIV inhibitor 5P12-RANTES in Pichia pastoris. Protein Expr Purif. 2016;119:1–10. doi: 10.1016/j.pep.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Hanson MA, Griffith MT, Hilgart MC, Sanishvili R, Nagarajan V, Stepanov S, Fischetti RF, Kuhn P, Stevens RC. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 µm size X-ray synchrotron beam. J R Soc Interface. 2009;6:S587–S597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J, Tillett D, Dawes IW, March PE. Site-directed, Ligase-Independent Mutagenesis (SLIM) for highly efficient mutagenesis of plasmids greater than 8kb. J Microbiol Methods. 2008;73:195–198. doi: 10.1016/j.mimet.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Choi W-T, Nedellec R, Coetzer M, Colin P, Lagane B, Offord RE, Hartley O, Mosier DE. CCR5 Mutations Distinguish N-Terminal Modifications of RANTES (CCL5) with Agonist versus Antagonist Activity. J Virol. 2012;86:10218–10220. doi: 10.1128/JVI.00353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimbro R, Peterson FC, Liu Q, Guzzo C, Zhang P, Miao H, Van Ryk D, Ambroggio X, Hurt DE, De Gioia L, et al. Tyrosine-sulfated V2 peptides inhibit HIV-1 infection via coreceptor mimicry. EBioMedicine. 2016 doi: 10.1016/j.ebiom.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin P, Benureau Y, Staropoli I, Wang Y, Gonzalez N, Alcami J, Hartley O, Brelot A, Arenzana-Seisdedos F, Lagane B. HIV-1 exploits CCR5 conformational heterogeneity to escape inhibition by chemokines. Proc Natl Acad Sci USA. 2013;110:9475–9480. doi: 10.1073/pnas.1222205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier EG, Dragic T. The Crown and Stem of the V3 Loop Play Distinct Roles in Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Interactions with the CCR5 Coreceptor. J Virol. 2002;76:8953–8957. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier EG, Persuh M, Thompson DAD, Lin SW, Sakmar TP, Olson WC, Dragic T. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc Natl Acad Sci USA. 2000;97:5762–5767. doi: 10.1073/pnas.97.11.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier EG, Tran DN, Yukhayeva L, Olson WC, Dragic T. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J Virol. 2001;75:5541–5549. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton JA, Lans I, Giraldo J. Quantifying conformational changes in GPCRs: glimpse of a common functional mechanism. BMC Bioinformatics. 2015;16:124. doi: 10.1186/s12859-015-0567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doranz BJ, Lu ZH, Rucker J, Zhang TY, Sharron M, Cen YH, Wang ZX, Guo HH, Du JG, Accavitti MA, et al. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorgham K, Cerini F, Gaertner H, Melotti A, Rossitto-Borlat I, Gorochov G, Hartley O. Generating Chemokine Analogs with Enhanced Pharmacological Properties Using Phage Display. In: Handel TM, editor. Methods in Enzymology. Academic Press; 2016. pp. 47–72. [DOI] [PubMed] [Google Scholar]
- Dragic T, Trkola A, Lin SW, Nagashima KA, Kajumo F, Zhao L, Olson WC, Wu L, Mackay CR, Allaway GP, et al. Amino-Terminal Substitutions in the CCR5 Coreceptor Impair gp120 Binding and Human Immunodeficiency Virus Type 1 Entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duma L, Häussinger D, Rogowski M, Lusso P, Grzesiek S. Recognition of RANTES by Extracellular Parts of the CCR5 Receptor. J Mol Biol. 2007;365:1063–1075. doi: 10.1016/j.jmb.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, et al. A Tyrosine-Rich Region in the N Terminus of CCR5 Is Important for Human Immunodeficiency Virus Type 1 Entry and Mediates an Association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenalti G, Giguere PM, Katritch V, Huang X-P, Thompson AA, Cherezov V, Roth BL, Stevens RC. Molecular control of delta-opioid receptor signalling. Nature. 2014;506:191–196. doi: 10.1038/nature12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner H, Cerini F, Escola J-M, Kuenzi G, Melotti A, Offord R, Rossitto-Borlat In, Nedellec R, Salkowitz J, Gorochov G, et al. Highly potent, fully recombinant anti-HIV chemokines: Reengineering a low-cost microbicide. Proc Natl Acad Sci U S A. 2008;105:17706–17711. doi: 10.1073/pnas.0805098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez J, Rueda P, Alcami J, Rognan D, Arenzana-Seisdedos F, Lagane B, Kellenberger E. Allosteric Model of Maraviroc Binding to CC Chemokine Receptor 5 (CCR5) J Biol Chem. 2011;286:33409–33421. doi: 10.1074/jbc.M111.279596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SE, Mao A, Nicolaidou V, Finelli M, Wise EL, Nedjai B, Kanjanapangka J, Harirchian P, Chen D, Selchau V, et al. Elucidation of Binding Sites of Dual Antagonists in the Human Chemokine Receptors CCR2 and CCR5. Mol Pharmacol. 2009;75:1325–1336. doi: 10.1124/mol.108.053470. [DOI] [PubMed] [Google Scholar]
- Hanes MS, Salanga CL, Chowdry AB, Comerford I, McColl SR, Kufareva I, Handel TM. Dual Targeting of the Chemokine Receptors CXCR4 and ACKR3 with Novel Engineered Chemokines. J Biol Chem. 2015;290:22385–22397. doi: 10.1074/jbc.M115.675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard OMZ, Shirakawa A-K, Turpin JA, Maynard A, Tobin GJ, Carrington M, Oppenheim JJ, Dean M. Naturally Occurring CCR5 Extracellular and Transmembrane Domain Variants Affect HIV-1 Co-receptor and Ligand Binding Function. J Biol Chem. 1999;274:16228–16234. doi: 10.1074/jbc.274.23.16228. [DOI] [PubMed] [Google Scholar]
- Huang C-c, Lam SN, Acharya P, Tang M, Xiang S-H, Hussan SS-u, Stanfield RL, Robinson J, Sodroski J, Wilson IA, et al. Structures of the CCR5 N Terminus and of a Tyrosine-Sulfated Antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Colin P, Staropoli I, Lima-Fernandes E, Ferret C, Demir A, Rogee S, Hartley O, Randriamampita C, Scott MG, et al. Targeting spare CC chemokine receptor 5 (CCR5) as a principle to inhibit HIV-1 entry. J Biol Chem. 2014;289:19042–19052. doi: 10.1074/jbc.M114.559831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondru R, Zhang J, Ji C, Mirzadegan T, Rotstein D, Sankuratri S, Dioszegi M. Molecular Interactions of CCR5 with Major Classes of Small-Molecule Anti-HIV CCR5 Antagonists. Mol Pharmacol. 2008;73:789–800. doi: 10.1124/mol.107.042101. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Laurence JS, Blanpain C, Burgner JW, Parmentier M, LiWang PJ. CC Chemokine MIP-1β Can Function As a Monomer and Depends on Phe13 for Receptor Binding. Biochem. 2000;39:3401–3409. doi: 10.1021/bi9923196. [DOI] [PubMed] [Google Scholar]
- Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, Ijzerman AP, et al. Structural Basis for Allosteric Regulation of GPCRs by Sodium Ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Das D, Ogata-Aoki H, Nakata H, Miyakawa T, Tojo Y, Norman R, Takaoka Y, Ding J, Arnold GF, et al. Structural and Molecular Interactions of CCR5 Inhibitors with CCR5. J Biol Chem. 2006;281:12688–12698. doi: 10.1074/jbc.M512688200. [DOI] [PubMed] [Google Scholar]
- Martin-Blondel G, Brassat D, Bauer J, Lassmann H, Liblau RS. CCR5 blockade for neuroinflammatory diseases [mdash] beyond control of HIV. Nat Rev Neurol. 2016;12:95–105. doi: 10.1038/nrneurol.2015.248. [DOI] [PubMed] [Google Scholar]
- Martin L, Blanpain C, Garnier P, Wittamer V, Parmentier M, Vita C. Structural and Functional Analysis of the RANTES-Glycosaminoglycans Interactions. Biochem. 2001;40:6303–6318. doi: 10.1021/bi002670n. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallographica Section D: Biological Crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navenot J-M, Wang Z-x, Trent JO, Murray JL, Hu Q-x, DeLeeuw L, Moore PS, Chang Y, Peiper SC. Molecular anatomy of CCR5 engagement by physiologic and viral chemokines and HIV-1 envelope glycoproteins: differences in primary structural requirements for RANTES, MIP-1α, and vMIP-II binding. J Biol Chem. 2001;313:1181–1193. doi: 10.1006/jmbi.2001.5086. [DOI] [PubMed] [Google Scholar]
- Nedellec R, Coetzer M, Lederman MM, Offord RE, Hartley O, Mosier DE. Resistance to the CCR5 Inhibitor 5P12-RANTES Requires a Difficult Evolution from CCR5 to CXCR4 Coreceptor Use. PLoS ONE. 2011;6:e22020. doi: 10.1371/journal.pone.0022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakianathan DR, Kuta EG, Artis DR, Skelton NJ, Hebert CA. Distinct but overlapping epitopes for the interaction of a CC-chemokine with CCR1, CCR3 and CCR5. Biochem. 1997;36:9642–9648. doi: 10.1021/bi970593z. [DOI] [PubMed] [Google Scholar]
- Proudfoot AEI, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TNC, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci U S A. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot AEI, Power CA, Hoogewerf AJ, Montjovent M-O, Borlat F, Offord RE, Wells TNC. Extension of Recombinant Human RANTES by the Retention of the Initiating Methionine Produces a Potent Antagonist. J Biol Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, Fenalti G, Wu H, Han GW, Cherezov V, et al. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut GE, Konner JA, Kajumo F, Moore JP, Dragic T. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage-and dualtropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:3464–3468. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16:741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- Rizzuto CD, Wyatt R, Hernández-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A Conserved HIV gp120 Glycoprotein Structure Involved in Chemokine Receptor Binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- Roche M, Borm K, Flynn JK, Lewin SR, Churchill MJ, Gorry PR. Molecular Gymnastics: Mechanisms of HIV-1 Resistance to CCR5 Antagonists and Impact on Virus Phenotypes. Curr Top Med Chem. 2016;16:1091–1106. doi: 10.2174/1568026615666150901114724. [DOI] [PubMed] [Google Scholar]
- Scholten DJ, Canals M, Maussang D, Roumen L, Smit MJ, Wijtmans M, de Graaf C, Vischer HF, Leurs R. Pharmacological modulation of chemokine receptor function. Br J Pharmacol. 2011;165:1617–1643. doi: 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Clapham PR, Picard L, Offord RE, Rosenkilde MM, Schwartz TW, Buser R, Wells TN, Proudfoot AE. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- Steen A, Larsen O, Thiele S, Rosenkilde MM. Biased and G protein-independent signaling of chemokine receptors. Frontiers in Immunology. 2014;5 doi: 10.3389/fimmu.2014.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov S, Makarov O, Hilgart M, Pothineni SB, Urakhchin A, Devarapalli S, Yoder D, Becker M, Ogata C, Sanishvili R, et al. JBluIce-EPICS control system for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2011;67:176–188. doi: 10.1107/S0907444910053916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney DC, Beavis P, Chuang KT, Zheng Y, Lee I, Gee P, Deval J, Rotstein DM, Dioszegi M, Ravendran P, et al. A study of the molecular mechanism of binding kinetics and long residence times of human CCR5 receptor small molecule allosteric ligands. Br J Pharmacol. 2014;171:3364–3375. doi: 10.1111/bph.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam K, Schultz M, Reyes-Robles T, Vanwalscappel B, Horton J, Alonzo F, Wu B, Landau NR, Torres VJ. Staphylococcus aureus Leukocidin LukED and HIV-1 gp120 Target Different Sequence Determinants on CCR5. mBio. 2016;7 doi: 10.1128/mBio.02024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamis P, Floudas CA. Elucidating a Key Anti-HIV-1 and Cancer-Associated Axis: The Structure of CCL5 (Rantes) in Complex with CCR5. Scientific Reports. 2014;4:5447. doi: 10.1038/srep05447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Li J, et al. Structure of the CCR5 Chemokine Receptor-HIV Entry Inhibitor Maraviroc Complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele S, Steen A, Jensen PC, Mokrosinski J, Frimurer TM, Rosenkilde MM. Allosteric and Orthosteric Sites in CC Chemokine Receptor (CCR5), a Chimeric Receptor Approach. J Biol Chem. 2011;286:37543–37554. doi: 10.1074/jbc.M111.243808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilton JC, Wilen CB, Didigu CA, Sinha R, Harrison JE, Agrawal-Gamse C, Henning EA, Bushman FD, Martin JN, Deeks SG, et al. A Maraviroc-Resistant HIV-1 with Narrow Cross-Resistance to Other CCR5 Antagonists Depends on both N-Terminal and Extracellular Loop Domains of Drug-Bound CCR5. J Virol. 2010;84:10863–10876. doi: 10.1128/JVI.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totrov M, Abagyan R. Protein-ligand docking as an energy optimization problem. In: Raffa RB, editor. Drug-Receptor Thermodynamics: Introduction and Applications. Wiley; 2001. pp. 603–624. [Google Scholar]
- Weitzenfeld P, Ben-Baruch A. The chemokine system, and its CCR5 and CXCR4 receptors, as potential targets for personalized therapy in cancer. Cancer Letters. 2014;352:36–53. doi: 10.1016/j.canlet.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Wescott MP, Kufareva I, Paes C, Goodman JR, Thaker Y, Puffer BA, Berdougo E, Rucker JB, Handel TM, Doranz BJ. Signal transmission through the CXC chemokine receptor 4 (CXCR4) transmembrane helices. Proc Natl Acad Sci U S A. 2016;113:9928–9933. doi: 10.1073/pnas.1601278113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Qin L, Ortiz Zacarías NV, de Vries H, Han GW, Gustavsson M, Dabros M, Zhao C, Cherney RJ, Carter P, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Atomic coordinates for the predicted complex between CCR5 and WT CCL5, in PDB format.
Figures S1–S5 and Tables S1–S6.