Abstract
Aging increases the risk of cardiovascular disease and metabolic syndrome. Alterations in epicardial fat play an important pathophysiological role in coronary artery disease and hypertension. We investigated the impact of normal aging on obesity-related genes in epicardial fat. Sex-specific changes in obesity-related genes with aging in epicardial fat (EF) were determined in young (6 months) and old (30/36 months) female and male, Fischer 344 × Brown Norway hybrid (FBN) rats, using a rat obesity RT2 PCR Array. Circulating sex hormone levels, body and heart weights were determined. Statistical significance was determined using two-tailed Student’s t test and Pearson’s correlation. Our results revealed sex-specific differences in obesity-related genes with aging. Dramatic changes in the expression profile of obesity-related genes in EF with aging in female, but not in male, FBN rats were observed. The older (30 months) female rats had more significant variations in the abundance of obesity-related genes in the EF compared to that seen in younger female rats or both age groups in male rats. A correlation of changes in obesity-related genes in EF to heart weights was observed in female rats, but not in male rats with aging. No correlation was observed to circulating sex hormone levels. Our findings indicate a dysfunctional EF in female rats with aging compared to male rats. These findings, with further functional validation, might help explain the sex differences in cardiovascular risk and mortality associated with aging observed in humans.
Keywords: Adipose dysfunction, Ectopic fat, Expression profile, Gender differences
Introduction
Although the risk factors for cardiovascular disease (CVD) such as aging, smoking, diet/alcohol, physical inactivity, and metabolic syndrome are similar between men and women, there still is higher mortality rates in women compared to men [33]. Obesity rates are increasing alarmingly in the USA. The prevalence of extreme obesity is much higher in women compared to men (35 vs 7%) [26].
The mass, distribution, and function of adipose tissue undergo dramatic changes throughout life [4]. Adipose-derived factors are key mediators of the alterations in body mass that is observed during aging [39]. Changes in visceral fat mass and function are known to play an important role in promotion of insulin resistance, obesity, and cardiovascular diseases through its secretion of adipocytokines (secretome) [23]. Epicardial or perivascular fat, the fat surrounding the heart and its major arteries, has recently gained importance in playing a role in CVD. Epicardial fat (EF) covers about 80% of the heart’s surface and represents 20% of the heart’s weight. Due to its close proximity to the heart and surrounding arteries, and a lack of fascia support, this fat participates directly in the physiology of the underlying cardiac and arterial tissues in a paracrine manner [17, 32, 34]. EF differs from other fat depots (omental, visceral/abdominal, or subcutaneous) by having significantly larger number of smaller adipocytes, a different fatty acid and protein composition, as well as different lipid metabolism (increased fatty acid synthesis and fatty acid breakdown) [17]. Several recent human studies have shown an association between obesity and increases in EF mass [37, 38]. Clinical studies have also shown that EF expresses increased inflammatory cytokines (TNFα and IL6) compared to other adipose depots in patients with coronary artery disease and other diseases [2, 18].
Changes in EF mass and function result in enhanced coronary calcification in postmenopausal women [7], alterations in lipid metabolism [29], enhanced risk of developing atherosclerosis with obesity and coronary artery disease [21], as well as correlation with other CVD risk factors [1]. Very little is known about changes in EF function during aging. We have earlier shown increased changes in EF adipokines in female rats during aging compared to male rats [9]. In the present study, we investigated if aging results in sex-specific changes in the expression profile of obesity-related genes in the EF, using an established rodent aging model, Fischer 344 × Brown Norway hybrid rats (FBN). Our studies showed significant changes in EF gene expression profile that correlated to changes in heart weight in female aging rats.
Materials and methods
Animals
Twenty-five Fischer 344 × Brown Norway hybrid (FBN) rats was obtained from the animal colony maintained by the National Institutes on Aging, USA. There were 16 female (young, 6 months old, n = 8, and aged, 30 months old, n = 8; body weights = 230 ± 14 and 320 ± 20 g, respectively) and 9 male (young, 6 months old, n = 5 and aged, 36 months old, n = 4; body weights = 422 ± 42 and 450 ± 35 g, respectively) FBN rats. Marshall University’s Institutional Animal Care and Use Committee (IACUC) approved all protocols, and the animals were treated in compliance with Marshall University IACUC Committee regulations. Probability of survival curves provided for the FBN hybrid rats by the National Institute on Aging were employed to select age groups corresponding roughly to humans in their third (6-month rats) and eighth (30/36-month rats; female/male, respectively) decade of life [44]. The differences in the advanced age groups chosen between the two sexes are due to differences in the survivability of male versus female FBN rats. The female FBN rats generally do not survive over the age of 30–32 months; however, the male FBN rats can reach the maximum age of 36 months. This phenomenon is very different from humans. We chose this latter time points in view of the fact that cardiovascular dysfunction in humans accelerates in this interval and because this age group represents one of the fastest growing segments of the aging population in the USA [24].
The animals were acclimated for 2 weeks in our animal facility. During this time, the animals were fed a standard laboratory diet (Laboratory Rodent Diet 5001; www.labdiet.com which consisted of protein min % = 23; crude fat min % = 4.5 and crude fiber max % = 6.0) and water ad libitum. Rats were kept under standard conditions: temperature 21.0 ± 2.0 °C, humidity 55.0 ± 5.0% with a 12:12-h light/dark cycle (07:00–19:00). Rats were carefully monitored, and weekly weights were taken to monitor signs of stress and weight loss. After 2 weeks of acclimation, the rats (not fasting) were sacrificed. Care was taken that all surgeries were performed at similar times of the day in order to minimize circadian changes. Rats from each group were sacrificed on the same day. The body and heart weights of all rats were recorded. EF was excised after anesthetizing using ketamine-xylazine (45:5 mg/kg i.p) and euthanizing by exsanguinations via a cardiac puncture. Epicardial fat isolation from the rats was performed under a stereotactic microscope. During the EF collection, care was taken to prevent any cross-contamination with the cardiac or fibrotic tissue. The samples were immediately collected in Tri-reagent (Sigma) and stored at −80 °C for RNA isolation.
RNA extraction
The RNA was extracted by homogenization of the adipose tissue under cold conditions in Tri-reagent (100 mg adipose tissue in 1mlTri-reagent), following the manufacturer’s protocol (T9424, Sigma). The isolated RNA concentration and purity were analyzed by Nanodrop model 1000 (Thermo Scientific, Nanodrop Technologies Inc), and its integrity was determined on a 1.2% agarose gel electrophoresis. Only RNA with a RIN number >7 (Agilent Bioanalyzer) was used for the PCR array analyses.
Rat obesity RT2 Profiler PCR array
The rat obesity RT2 Profiler PCR array was performed on RNA extracted from EF obtained from young (6 months) and aged (30/36 months) female/male FBN rats. The genes assayed in 6-month rats for both sexes were defined as control (CTRL). One microgram of purified RNA was used for amplification to cDNA using RT2 First Strand Kit (C-03, Superarray BioScience Corporation). The rat obesity PCR Array (PARN-017A, Superarray BioScience Corporation) that profiles the expression of 84 genes related to obesity was performed on all samples (EF from 25 rats). The obesity array included 17 orexigenic genes, 54 anorectic genes, and 13 genes involved in energy expenditure (http://www.sabiosciences.com/rt_pcr_product/HTML/PARN-017A.html) (Superarray BioScience Corporation). The 96-well plate array was performed using the MyiQ Bio-Rad Real Time PCR system (Bio-Rad) following the manufacturer’s instructions. The data obtained was interpreted using the Superarray PCR Array data analysis Web portal (http://www.superarray.com/pcrarraydataanalysis.php). Quality control of all the PCR arrays was measured by assessing the quality of internal controls such as reverse transcription control (RTC) and genomic DNA contamination control (GPC). An array with the RTC value ≤5 and GPC value ≥35 passes the quality control. All the arrays that were performed passed the quality control test. The array design and final data processing were consistent with the requirements of Minimum Information about a Microarray Experiment (MIAME 2.0) (http://www.mged.org/Workgroups/MIAME/miame.html). The variations between groups were defined as fold changes in gene expression in aged animals compared to gene expression in the younger (6 months) animals in the two sexes.
Validation of PCR array data with quantitative RT-qPCR
Real-time reverse transcriptase polymerase chain reaction (RT-qPCR) of selected genes was performed to validate their changes in gene expression observed in PCR array analysis. The levels of four candidate genes representing the three major groups (orexigenic, anorectic, and energy expenditure pathways) were randomly selected for validation using RT-qPCR (Ghsr: orexigenic gene; Lep and Sstr1: for anorectic genes and Thrb for energy expenditure genes). Purified RNA (1 μg) was utilized for the synthesis of complementary DNA (cDNA) using iScript cDNA synthesis kit (170-8890, Bio-Rad). Real-time PCR was carried out in 25 μl of a SYBR green reaction mixture containing 1 μl of cDNA iQSYBR Green Supermix (170-8882, Bio-Rad), and the respective primers in triplicates. The following primers were used: growth hormone secretagogue receptor (Ghsr) (NM_032075): 5′-ccatcgctcattgctctaca-3′, 3′-ctgcccatctggctctactc-5′; leptin (Lep) (NM_013076): 5′-tgacaccaaaaccctcatca-3′, 3′-atgaagtccaaaccggtgac-5′; somatostatin receptor 1 (Sstr1) (NM_012719): 5′-cttatgcaccctggtgtgtg-3′, 3′-tgtcactggaacaggagctg-5′; thyroid hormone receptor beta (Thrb) (NM_012672): 5′-gaggaatgggagctcatcaa-3′, 3′-gggtgcttgtccaatgtctt-5′; 18s was used as the housekeeping gene; 18 s (M11188): 5′-gcaattattccccatgaacg-3′, 3′-ggcctcactaaaccatccaa-5′.
Statistics
For the rat obesity RT2 Profiler PCR array, the statistical significance in fold changes between young (6 months) and aged animals (30/36 months) for each sex was automatically generated by the PCR array online data analysis data portal (http://www.superarray.com/pcrarraydataanalysis.php). Fold changes >3.0 or <0.3 were defined as fold increase or decrease and used for statistical analysis. The significant differences between the ΔCt values between groups were analyzed using two-tailed Student’s t test. For the validation studies using RT-qPCR analysis, one-way ANOVA was performed at the level of ΔCt, in order to exclude potential bias due to averaging of data transformed through the Pfaffl equation 2−(ΔΔCt) [30]. Significance was confirmed using post hoc analysis with Fisher’s least significant difference (Fisher’s LSD) test.
The correlation between obesity-related genes and the body weight or heart weights of aged group of rats was calculated by performing the Pearson’s correlation. For this, the average ΔCt value of individual genes from the array was correlated to the body or heart weights of the group. Pearson’s correlation coefficient (r) was first obtained. This value was then used to calculate the t statistic, by dividing the coefficient by standard error. Finally, the significance p value was calculated from the t-table. A two-tailed value of p < 0.05 indicated statistical significance.
Results
Expression profiling of epicardial fat reveals sex differences in obesity-related genes with aging
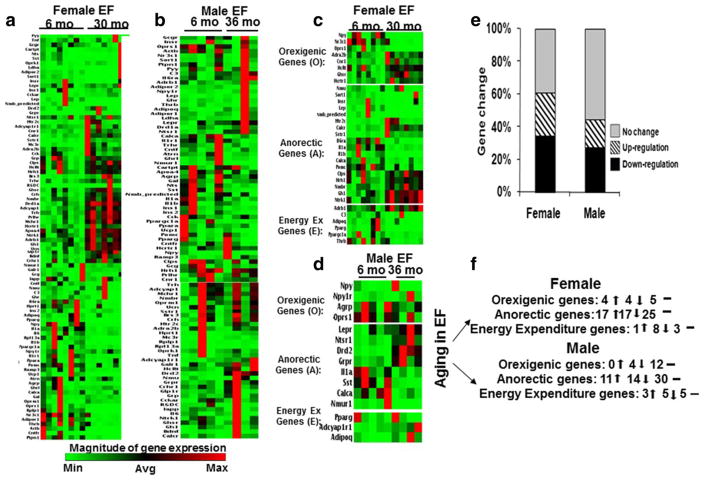
Expression profiling of obesity-related genes in EF was performed using RT2 Obesity PCR array in tissues obtained from both young (6 months old) and aged (30/36 months old) female and male FBN rats. Overall changes in the expression levels of all genes (84 genes) in EF of each age group and the two sexes are shown in the heat map (Fig. 1a–d). Detailed analysis of the heat map revealed that the differences in the gene expression were sex specific. In the female EF, from the total 84 genes detected in the array, the expression levels of 30 genes (35%) were upregulated, 21 genes (25%) were downregulated, and there was no change in 33 genes (40%). In contrast, in the EF obtained from the male rats, the expression levels of 14 genes (17%) were upregulated, 10 genes (12%) were downregulated, and 60 genes (71%) had no change (Fig. 1e, f).
Fig. 1.
Heat map of the RT2 Obesity PCR array in EF of young and old rats: RT2 Obesity PCR array was performed in EF obtained from young (6 months) (female, n = 8, male n = 5) and old (30/36 months) (female n = 8, male n = 4) FBN rats. The array consisted of 84 genes belonging to orexigenic (17 genes), anorectic (54 genes), and energy expenditure (13 genes) pathways. Heat map of all genes whose expression levels were altered in old rats compared to young rats were obtained after analyses: a female EF, b male EF. Heat map of significantly altered genes in EF of old rats compared to young rats were obtained after analyses: c female EF, d male EF. The percent changes in genes that were either upregulated (>3.0-fold), downregulated (<0.3-fold) or had no change in EF (e, f) in both sexes. ↑ Upregulation, ↓ downregulation, – no change. Significance was analyzed using two-tailed Student’s t test exhibited increased expression of pancreatic and gut-derived anorectic genes such as calcitonin receptor (Calcr), colipase, pancreatic (Clps), somatostatin receptor 1 (Sstr1), and apolipoprotein A-IV (Apoa4) (p < 0.01) in aging rats compared to younger controls. Moreover, the expression of several neuropeptides (brain-derived neurotrophic factor (Bdnf),
Sex differences in the expression of orexigenic genes with aging in epicardial fat
Figure 1c, d represents the heat map of the expression levels of all genes that reached statistical significance in female and male EF. Further evaluation of the heat map revealed unique age-dependent changes in specific obesity-related genes that were generally classified as orexigenic, anoroxigenic, and energy-expenditure genes. Table 1 shows that the expression levels of CNS-derived genes, such as adrenergic, alpha-2B-, receptor (Adrα2b), cannabinoid receptor 1 (brain) (Cnr1), melanin-concentrating hormone receptor 1 (Mchr1), hypocretin (HcRt), hypocretin (orexin) receptor 1 (Hcrtr1) (p < 0.05), and gut-derived genes, Ghsr (p < 0.005), were significantly increased in aged female EF. However, the expression levels of nuclear receptor subfamily 3, group C, member 1 (Nr3c1) and opioid receptor, sigma 1 (Oprs1) significantly decreased in aged female EF (p<0.05). No such changes were observed in male EF with aging.
Table 1.
The expression levels of orexigenic genes altered in EF from female and male FBN rats
| Orexigenic genes | Epicardial fat | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Female | Male | |||||
|
|
|
|||||
| Refseq | Symbol | Description | Fold | p value | Fold | p value |
| NM_138505 | Adrα2b | Adrenergic, alpha-2B-, receptor | 2.34 | 0.015 | 0.99 | 0.98 |
| NM_012784 | Cnr1 | Cannabinoid receptor 1 (brain) | 3.03 | 0.001 | 2.24 | 0.38 |
| NM_013179 | HcRt | Hypocretin | 3.63 | 0.034 | 2.45 | 0.217 |
| NM_013064 | Hcrtr1 | Hypocretin (orexin) receptor 1 | 3.71 | 0.002 | 2.21 | 0.151 |
| NM_032075 | Ghsr | Growth hormone secretagogue receptor | 4.65 | 0.003 | 1.36 | 0.62 |
| NM_031758 | Mchr1 | Melanin-concentrating hormone receptor 1 | 10.9 | 0.000 | 1.32 | 0.65 |
| NM_012576 | Nr3c1 | Nuclear receptor subfamily 3, group C, member 1 | 0.07 | 0.006 | 1.55 | 0.795 |
| NM_030996 | Oprs1 | Opioid receptor, sigma 1 | 0.13 | 0.024 | 0.45 | 0.377 |
The variations in expression profile of orexigenic genes between younger and aged rats are expressed as fold changes. Two tailed student’s t-test was used to analyze the statistical significance. The gene changes that reached statistical significance are in italics
Sex differences in the expression of anorectic genes with aging in epicardial fat
Figure 1c, d and Table 2 depict changes in the expression levels of anorectic genes in EF of both sexes. The female EF bombesin-like receptor 3 (Brs3), corticotropin-releasing hormone (Crh), corticotropin-releasing hormone receptor 1 (Crhr1), dopamine receptor D1A (Drd1a), dopamine receptor D2 (Drd2), growth hormone 1 (Gh1), glucagon-like peptide 1 receptor (Grpr), histamine receptor H 1 (Hrh1), 5-hydroxytryptamine (serotonin) receptor 2C (Htr2c), melanocortin 3 receptor (Mc3r), neuromedin B receptor (Nmbr), neurotrophic tyrosine kinase, receptor kinase 1 (Ntrk1), prolactin-releasing hormone receptor (Prlhr), thyrotropin-releasing hormone receptor (Trhr), urocortin (Ucn)) was also significantly (p < 0.05) upregulated during aging in female EF. However, several pancreatic and gut-derived anorectic genes, such as insulin receptor (Insr) (p < 0.05) and Lep (p < 0.05), and the neuropeptides, such as calcitonin/calcitonin-related polypeptide, alpha (Calcα), ciliary neurotrophic factor receptor (Cntfr), interleukin 1 alpha (IL1a), interleukin 1 beta (IL1b), interleukin 6 receptor, alpha (Il6rα), neuromedin B (Nmb), proopiomelanocortin (Pomc), and sortilin 1 (Sort1) (p < 0.05), were significantly downregulated. In contrast, in male EF, the expression levels of only few genes like Drd2, Grpr, leptin receptor (Lepr), and neurotensin receptor 1 (Ntsr1) (p < 0.05) were upregulated, whereas Calcα, IL1a, neuromedin U receptor 1 (Nmur1), and somatostatin (Sst) (p < 0.05) were downregulated.
Table 2.
The expression levels of Anorectic genes altered in EF in female and male FBN rats
| Anorectic genes | Epicardial fat | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Female | Male | |||||
|
|
|
|||||
| Refseq | Symbol | Description | Fold | p value | Fold | p value |
| NM_012737 | Apoa4 | Apolipoprotein A-IV | 3.74 | 0.005 | 0.47 | 0.130 |
| NM_012513 | Bdnf | Brain derived neurotrophic factor | 10.3 | 0.001 | 2.82 | 0.340 |
| NM_152845 | Brs3 | Bombesin-like receptor 3 | 16.1 | 0.0001 | 1.81 | 0.393 |
| NM_053816 | Calcr | Calcitonin receptor | 10.4 | 0.0001 | 1.74 | 0.372 |
| NM_013139 | Clps | Colipase, pancreatic | 3.78 | 0.031 | 0.78 | 0.599 |
| NM_031019 | Crh | Corticotropin-releasing hormone | 9.86 | 0.0004 | 1.49 | 0.517 |
| NM_030999 | Crhr1 | Corticotropin-releasing hormone receptor 1 | 8.10 | 0.001 | 1.99 | 0.297 |
| NM_012546 | Drd1a | Dopamine receptor D1A | 10.2 | 0.000 | 3.57 | 0.113 |
| NM_012547 | Drd2 | Dopamine receptor D2 | 6.71 | 0.059 | 22.4 | 0.000 |
| NM_0010348 | Gh1 | Growth hormone 1 | 6.13 | 0.0009 | 1.44 | 0.566 |
| NM_012728 | Glp1r | Glucagon-like peptide 1 receptor | 7.74 | 0.002 | 1.79 | 0.454 |
| NM_139193 | Prlhr | Prolactin-releasing hormone receptor | 10.1 | 0.000 | 1.39 | 0.665 |
| NM_012706 | Grpr | Gastrin-releasing peptide receptor | 10.5 | 0.004 | 4.66 | 0.017 |
| NM_017018 | Hrh1 | Histamine receptor H 1 | 3.51 | 0.030 | 0.79 | 0.493 |
| NM_012765 | Htr2c | 5-Hydroxytryptamine (serotonin) receptor 2C | 10.3 | 0.007 | 4.47 | 0.161 |
| NM_012596 | Lepr | Leptin receptor | 1.27 | 0.570 | 2.80 | 0.040 |
| NM_001025270 | Mc3r | Melanocortin 3 receptor | 29.6 | 0.0002 | 4.65 | 0.330 |
| XM_218815 | Nmb_ | Neuromedin B | 0.04 | 0.0001 | 0.39 | 0.399 |
| NM_012799 | Nmbr | Neuromedin B receptor | 3.77 | 0.003 | 1.02 | 0.968 |
| NM_023100 | Nmur1 | Neuromedin U receptor 1 | 2.23 | 0.394 | 0.05 | 0.021 |
| NM_021589 | Ntrk1 | Neurotrophic tyrosine kinase, receptor, type 1 | 4.11 | 0.004 | 1.07 | 0.896 |
| XM_345484 | Ntsr1 | Neurotensin receptor 1 | 1.67 | 0.171 | 7.62 | 0.002 |
| NM_012719 | Sstr1 | Somatostatin receptor 1 | 9.56 | 0.000 | 1.97 | 0.369 |
| NM_013046 | Trh | Thyrotropin-releasing hormone | 4.69 | 0.000 | 1.11 | 0.864 |
| NM_013047 | Trhr | Trh receptor | 6.70 | 0.003 | 0.31 | 0.115 |
| NM_019150 | Ucn | Urocortin | 9.87 | 0.000 | 1.24 | 0.717 |
| NM_017338 | Calcα | Calcitonin/calcitonin-related polypeptide, alpha | 0.32 | 0.015 | 0.19 | 0.040 |
| NM_001003929 | Cntfr | Ciliary neurotrophic factor receptor | 0.04 | 0.006 | 0.58 | 0.563 |
| NM_172092 | Gcgr | Glucagon receptor | 0.39 | 0.201 | 0.86 | 0.878 |
| NM_017019 | Il1a | Interleukin 1 alpha | 0.20 | 0.000 | 0.29 | 0.036 |
| NM_031512 | Il1b | Interleukin 1 beta | 0.10 | 0.022 | 0.18 | 0.298 |
| NM_017020 | Il6r α | Interleukin 6 receptor, alpha | 0.02 | 0.00005 | 5.51 | 0.170 |
| NM_017071 | Insr | Insulin receptor | 0.09 | 0.014 | 0.84 | 0.911 |
| NM_013076 | Lep | Leptin | 0.12 | 0.027 | 5.35 | 0.254 |
| NM_139326 | Pomc | Proopiomelanocortin | 0.20 | 0.005 | 0.75 | 0.812 |
| NM_020100 | Ramp3 | Receptor activity modifying protein 3 | 0.36 | 0.233 | 3.41 | 0.136 |
| XM_342317 | Sort1 | Sortilin 1 | 0.09 | 0.038 | 1.13 | 0.930 |
| NM_012659 | Sst | Somatostatin | 0.88 | 0.894 | 0.14 | 0.039 |
| NM_012675 | Tnf | Tumor necrosis factor | 0.32 | 0.217 | 1.70 | 0.500 |
The variations in expression profile of anorectic genes between younger and aged rats are expressed as fold changes. Two-tailed Student’s t test was used to analyze the statistical significance. The gene changes that reached statistical significance are in italics
Sex differences in the expression of energy expenditure genes with aging in epicardial fat
Sex-dependent changes in the expression of genes related to energy expenditure pathway were also observed in EF of aging rats (Fig. 1c, d and Table 3). The expression of CNS-derived genes such as adenylate cyclase activating polypeptide 1 (pituitary) (Adcyap1) and adrenergic, beta-1-, receptor (Adrβ1) significantly increased in aged female EF. However, the expression of adiponectin, C1Q, and collagen domain (Adipoq), (complement component 3 (C3) (p < 0.05), protein tyrosine phosphatase, nonreceptor type 1 (Ptpn1), and thyroid hormone receptor beta (Thrb) (p < 0.05) were significantly decreased in aged female EF. In the male rats, the expression of Adcyap1r1 and Adipoq were significantly increased (p < 0.05) in aging EF compared to younger controls.
Table 3.
The expression levels of Energy expenditure genes altered in young and aged rats
| Energy expenditure genes | Epicardial fat | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Female | Male | |||||
|
|
|
|||||
| Refseq | Symbol | Description | Fold | p value | Fold | p value |
| NM_016989 | Adcyap1 | Adenylate cyclase activating polypeptide 1 (pituitary) | 5.18 | 0.0003 | 0.97 | 0.946 |
| NM_133511 | Adcyap1r1 | Adenylate cyclase activating polypeptide 1 receptor type I | 2.34 | 0.194 | 3.96 | 0.011 |
| NM_012701 | Adrβ1 | Adrenergic, beta-1-, receptor | 1.91 | 0.004 | 2.25 | 0.285 |
| NM_144744 | Adipoq | Adiponectin, C1Q and collagen domain | 0.02 | 0.042 | 87.8 | 0.046 |
| NM_016994 | C3 | Complement component 3 | 0.05 | 0.018 | 8.73 | 0.129 |
| NM_012637 | Ptpn1 | Protein tyrosine phosphatase, nonreceptor type 1 | 0.06 | 0.001 | 0.67 | 0.759 |
| NM_012672 | Thrb | Thyroid hormone receptor beta | 0.25 | 0.047 | 1.82 | 0.477 |
The variations in expression profile of energy expenditure genes between younger and aged rats are expressed as fold changes. Two tailed student’s t-test was used to analyze the statistical significance. The gene changes that reached statistical significance are in italics
Validation of genes altered in PCR array using RT-qPCR
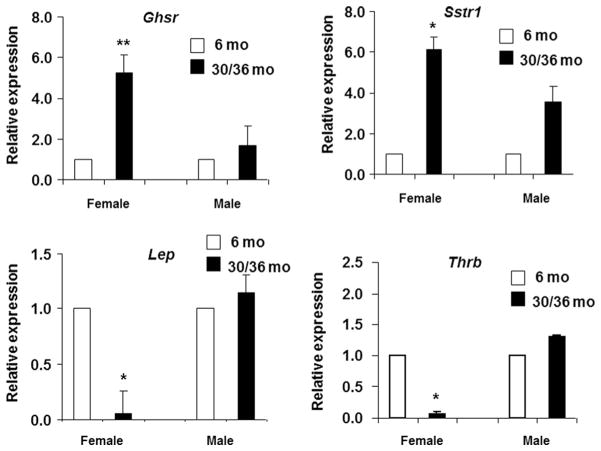
The quantitative real-time PCR confirmed the variation in levels of genes that were shown to be altered in the PCR array analyses. As seen in Fig. 2, similar to PCR array analysis, RT-qPCR also showed that the expression levels of Ghsr (p < 0.01) and Sstr1 (p < 0.05) were upregulated in female EF and not in males with aging. In contrast, the expression levels of Thrb (p < 0.05) and Lep were decreased in female but not in male EF with aging.
Fig. 2.
Validation of PCR array data using RT-qPCR in EF: real-time quantitative PCR was used to validate the changes observed in genes altered in the PCR array analysis. Expression levels of selected genes belonging to orexigenic (Ghsr), anorectic (Lep, Sstr1) and energy expenditure (Thrb) pathways were measured by RT-qPCR in triplicates in the EF of young and old FBN rats of both sexes: female EF (6 months, n = 8; 30 months, n = 8), male EF (6 months, n = 5; 36 months, n = 4). The data were expressed as relative expression ± standard error of the mean (SEM). The white bars represent 6 months (female and male), and the black bars represent 30-month (female) or 36-month (male) FBN rats, respectively. One-way ANOVA followed by the Fisher’s LSD test was used for calculating significance. *p < 0.05; **p < 0.01
Correlation of expression levels of obesity genes from EF to changes in heart and body weights with aging and not to circulating sex hormone levels
We had previously shown an increase in both body and heart weights with age in female rats but an increase in heart weight with minimal increase in body weight in male rats with age [9]. We investigated if the expression levels of obesity-related genes that were significantly altered in the EF correlated to the changes in heart and body weights in young and aged rats of both sexes. As shown in Table 4, IL1a and IL1b expression levels in EF had a significant negative association with the changes in body weight in young (6 months) female rats but not upon aging (30 months). Adrβ1, C3, Cnr1, Pomc, Sort1, and Thrb expression levels in EF were all negatively correlated to the heart weight in the young (6 months) female rats. The expression levels of Adcyap1, Apoa4, Crh, Mchr1, Hcrtr1, Il1a, Nmb, Nmbr, Trh, Trhr, and Ucn was positively correlated to heart weight in the aged (30 months) female rats.
Table 4.
The correlation of obesity-related genes in EF to the body and heart weights of female and male FBN rats
| Body weight | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Female | Young | Aging | Male | Young | Aging | ||||
| Genes | r | p value | r | p value | Genes | r | p value | r | p value |
| Ilia | −0.73 | 0.036 | NS | NS | Adcyaplrl | NS | NS | −0.96 | 0.018 |
| Mb | −0.76 | 0.028 | NS | NS | Grpr | NS | NS | −0.98 | 0.011 |
| Ilia | NS | NS | −0.96 | 0.043 | |||||
| Nmurl | NS | NS | −0.96 | 0.043 | |||||
| Heart weight | |||||||||
| Adcyap1 | NS | NS | 0.81 | 0.014 | Drd2 | 0.90 | 0.033 | NS | NS |
| Adrβ1 | −0.74 | 0.036 | NS | NS | |||||
| Apoa4 | NS | NS | 0.76 | 0.027 | |||||
| C3 | −0.87 | 0.004 | NS | NS | |||||
| Cnr1 | −0.74 | 0.035 | NS | NS | |||||
| Crh | NS | NS | 0.73 | 0.038 | |||||
| Mchr1 | NS | NS | 0.81 | 0.014 | |||||
| Hcrtr1 | NS | NS | 0.78 | 0.022 | |||||
| Il1a | NS | NS | 0.74 | 0.035 | |||||
| Nmb | NS | NS | 0.81 | 0.014 | |||||
| Nmbr | NS | NS | 0.81 | 0.014 | |||||
| Pomc | −0.88 | 0.003 | NS | NS | |||||
| Sort1 | −0.85 | 0.006 | NS | NS | |||||
| Thrb | −0.77 | 0.026 | NS | NS | |||||
| Trh | NS | NS | 0.81 | 0.014 | |||||
| Trhr | NS | NS | 0.79 | 0.020 | |||||
| Ucn | NS | NS | 0.81 | 0.014 | |||||
The obesity-related genes that were significantly altered in the EF were correlated to the body weights (average BW = female: 6 months = 230 ± 14 g; 30 months = 320 ± 20 g; male: 6 months = 422 ± 42 g; 36 months = 450 ± 35 g) and heart weights (average HW = female: 6 months = 0.7 ± 0.03 g; 30 months = 1.16 ± 0.03 g; male: 6 months = 1.08 ± 0.03 g; 36 months = 1.7 ± 0.13 g) [9] of the young and aging rats using Pearson’s correlation. The two-tailed p value <0.05 indicated statistical significance. The gene changes that reached statistical significance are emphasized in italics r correlation coefficient, NS no significance
In male rats, no significant correlation was found between the expression levels of the obesity-related genes in EF and body weight in young rats, but Adcyap1r1, Grpr, Il1a, and Nmur1 expression levels in EF were negatively correlated to the body weights in aged rats. Drd2 was the only EF gene whose expression was positively correlated to the heart weight in young rats, but not in aged, male rats.
Circulating testosterone and estradiol levels were measured in the young (6 months) and aged (30/36 months) female and male FBN rats using the Immulite 2000 Immunoassay system (Siemens, Deerfield, IL). Though the testosterone levels decreased in male rats with aging (2.2 vs. 0.5 ng/ml; 6 vs 36 months), there was no change in estradiol levels with aging in female rats (33.3 vs 29.3 pg/ml; 6 vs 30 months). No correlation was observed between circulating sex hormone levels and obesity-related genes.
Discussion
Aging is associated with both fat mass redistribution and changes in fat function. There is an increased accumulation of fat in ectopic sites such as the liver, muscle, and heart with aging[28, 46]. This increased fat mass results in changes in gene expression in the various fat depots, with some depots more vulnerable to aging than others [41]. Abdominal adiposity is the hallmark of obesity [11]. Though genome-wide studies have shown a unique loci for visceral fat distribution in women compared to men [10], this fat remains unaltered during aging [16]. Recent studies have described the importance of EF in the pathophysiology of heart and vascular function [5, 27]. Not much is known about changes in EF during aging. We have earlier shown sex-specific changes in EF adipokine expression with aging in FBN rats [9]. In the present study, we compared the expression profile of obesity-related genes in EF from old and young FBN rats. We observed sex-specific changes in these genes with aging. These changes were more predominant in the EF from older female rats compared to males of similar ages.
An in-depth assessment of the significantly altered genes revealed changes in the expression of several CNS-derived genes (Adcyap1, Bdnf, Brs3, Crh, Crhr1, Drd1a, Drd2, Grpr, Hrh1, Htr2c, Mc3r, Ntrk1, Prlhr, Trhr, Ucn, Nmbr), whose immediate relevance to EF function can only presently be speculated. However, given the role of adipose tissue in endocrinology and metabolism and it being directly regulated by the CNS, it is plausible that these genes might also exist in peripheral adipose tissues. Among the genes that were upregulated, the adrenergic receptor alpha (Adrα2) initiates prolipolytic effects and has been shown to be increased during aging [6, 22], in contrast to Adrβ1, has been shown to decrease with aging [13]. Our results showed an increased expression of both Adrα2b and Adrβ1 in aged female rats. These differences in adrenergic receptor changes with aging suggest that female EF expresses a more enhanced prolipolytic function than in male rats with aging. The reduction in growth hormone (Gh) production correlates with aging and results in a decline of the somatotrophic axis (somatopause) associated with a decrease in muscle mass and increase in adiposity [20]. Ghsr is implicated in the Gh secretion, orexigenic and fat lipolytic effects in a ligand-dependent manner [42]. Besides the CNS, Ghsr has been found to exist in a large variety of peripheral tissues including adipose tissue [42]. Somatostatin is a known Gh inhibitor [15], and its receptors (Sstr) are found to be expressed in adipose tissue [36]. Our data showed that both Gh1 and Ghsr were upregulated in both male and female EF. An increase in the Gh axis was accompanied with an increase in the levels of its inhibitor (Sstr1). This might indicate a compensatory reaction by the adipose tissue in response to the decline in the levels of Gh during aging [25]. This effect was also most likely a result of the imbalance between the levels of somatostatin and its receptor.
Among the downregulated genes, the expression level of Nr3c1 was decreased in aged female EF but exhibited a moderate increase in male rats with aging. Defects in Nr3c1 are associated with obesity, hyperinsulinemia, hypertension, and coronary artery disease [12]. Obesity is associated with chronic low-grade inflammation with an increase in factors such as IL1 and IL6 [31, 40]. We found a significant decrease in IL1a, IL1b, and IL6rα in female EF. On the contrary, we observed a dramatic increase in melanocortin 3 receptor (Mc3r) in female EF (30-fold). The knockout of Mc3r showed increased fat mass [3] but also exhibited maintenance of adiponectin levels and delayed inflammation in response to a high-fat diet [43]. The significant higher levels of Mc3r in EF of aged female rats might also be responsible for the lower expression of inflammatory factors in this fat depot. Insr is another gene that is linked to the aging process. For example, a mutation in insulin signaling pathway (insulin/IGF-1) is often used to increase the lifespan in animal models [14]. Moreover, a mutation of Insr could also ameliorate the age-related decrease in cardiac function [47]. We observed a decline in Insr in EF of both sexes, especially in female rats. There was also a decrease in the expression of Sort1, another key factor that mediates insulin signaling pathway in adipose tissue [19], and Ptpn1, which inhibits the insulin pathway [45] with aging. This reflects a protective response to aging-related decline in insulin signaling which seems more prominent in EF. With a reduction in body weight, Lep gene was highly expressed in females compared to male rats [35]. We also observed a more dramatic decline in Lep gene in EF of female rats than in males.
The physiological significance of the observed differences in obesity-related genes in EF of females than in males does need further investigation. However, the stronger correlation of genes in EF to heart weight supports the postulated paracrine function of EF on the heart. The inflammatory genes in EF were more likely regulated by body weight at least in younger rats. As shown in Table 4, in female rats, some of the obesity-related genes were negatively associated with heart weight in younger rats, but others were positively associated to heart weight with increasing age. This probably might suggest an age-dependent alteration in EF function. Prior studies have indicated that lipogenesis and lipid incorporation in EF is higher than in other fat depots [17]; however, it is not known if this function is altered with aging. Our data did show that the lipid regulatory genes such as Adrβ1 and Thrb exhibit an association with heart weight in younger but not in older rats. Similarly, there were sex differences in correlation of EF genes to body weight with aging. Though no conclusive correlation was seen between circulating sex hormone levels and the significantly altered obesity-related genes, the sex hormonal influence on these genes cannot be completely ruled out. This observation might be species related. In the FBN rats, there was no much change in estradiol levels with aging, and hence no correlation with EF-related genes. In humans, however, the investigators of the SWAN Cardiovascular fat ancillary study recently showed an increase in cardiac fat (which included epicardial, paracardial, perivascular, and total heart fat) after menopause, and this increase was correlated to a decrease in estradiol levels, but not with androgen levels [8]. However, it should be noted that these were only association studies and not causality, which still needs to be determined. Our results support the assumption that alterations in EF function result in increased risk of heart disease with aging. The sexual dimorphism observed in EF from male and female rats with aging might be beyond just differences in the levels of sex hormones.
Conclusions
In conclusion, our results indicate that the aging process resulted in a dramatic perturbation of obesity-related genes in female EF compared to males. These findings might be helpful in understanding the differences in pathophysiological role of this fat depot in cardiac and vascular dysfunction in a sex-dependent manner.
Acknowledgments
The authors acknowledge Dr. Eric Blough for his generosity in sharing tissues from the FBN rats and Jia Fei for his technical help. The authors acknowledge the partial support of funds from National Institutes of Health, HL074239 (NS), 1R15AG051062-01 (NS), and 3P20RR016477-09S2. The funding sources had no direct involvement in any aspect of this manuscript.
Footnotes
Compliance with ethical standards Marshall University’s Institutional Animal Care and Use Committee (IACUC) approved all protocols, and the animals were treated in compliance with Marshall University IACUC Committee regulations.
References
- 1.Aydin H, Toprak A, Deyneli O, Yazici D, Tarcin O, Sancak S, Yavuz D, Akalin S. Epicardial fat tissue thickness correlates with endothelial dysfunction and other cardiovascular risk factors in patients with metabolic syndrome. Metab Syndr Relat Disord. 2010;8:229–234. doi: 10.1089/met.2009.0080. [DOI] [PubMed] [Google Scholar]
- 2.Baker AR, Silva NF, Quinn DW, Harte AL, Pagano D, Bonser RS, Kumar S, McTernan PG. Human epicardial adipose tissue expresses a pathogenic profile of adipocytokines in patients with cardiovascular disease. Cardiovasc Diabetol. 2006;5:1. doi: 10.1186/1475-2840-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begriche K, Girardet C, McDonald P, Butler AA. Melanocortin-3 receptors and metabolic homeostasis. Prog Mol Biol Transl Sci. 2013;114:109–146. doi: 10.1016/B978-0-12-386933-3.00004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartwright MJ, Schlauch K, Lenburg ME, Tchkonia T, Pirtskhalava T, Cartwright A, Thomou T, Kirkland JL. Aging, depot origin, and preadipocyte gene expression. J Gerontol A Biol Sci Med Sci. 2010;65:242–251. doi: 10.1093/gerona/glp213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaowalit N, Lopez-Jimenez F. Epicardial adipose tissue: friendly companion or hazardous neighbour for adjacent coronary arteries? Eur Heart J. 2008;29:695–697. doi: 10.1093/eurheartj/ehm643. [DOI] [PubMed] [Google Scholar]
- 6.Coutinho LL, Bergen WG, Romsos DR, Merkel RA. Alpha-2 adrenergic receptor activity in porcine adipocytes is androgen and age/cell size dependent. Comp Biochem Physiol Comp Physiol. 1993;105:333–339. doi: 10.1016/0300-9629(93)90217-r. [DOI] [PubMed] [Google Scholar]
- 7.de Vos AM, Prokop M, Roos CJ, Meijs MF, van der Schouw YT, Rutten A, Gorter PM, Cramer MJ, Doevendans PA, Rensing BJ, Bartelink ML, Velthuis BK, Mosterd A, Bots ML. Pericoronary epicardial adipose tissue is related to cardiovascular risk factors and coronary artery calcification in post-menopausal women. Eur Heart J. 2008;29:777–783. doi: 10.1093/eurheartj/ehm564. [DOI] [PubMed] [Google Scholar]
- 8.El Khoudary SR, Shields KJ, Janssen I, Hanley C, Budoff MJ, Barinas-Mitchell E, Everson-Rose SA, Powell LH, Matthews KA. Cardiovascular fat, menopause, and sex hormones in women: the SWAN cardiovascular fat ancillary study. J Clin Endocrinol Metab. 2015;100:3304–3312. doi: 10.1210/JC.2015-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fei J, Cook C, Blough E, Santanam N. Age and sex mediated changes in epicardial fat adipokines. Atherosclerosis. 2010;212:488–494. doi: 10.1016/j.atherosclerosis.2010.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, Lohman K, Johnson AD, Foster MC, Greenawalt DM, Griffin P, Ding J, Newman AB, Tylavsky F, Miljkovic I, Kritchevsky SB, Launer L, Garcia M, Eiriksdottir G, Carr JJ, Gudnason V, Harris TB, Cupples LA, Borecki IB. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox CS, Massaro JM, Hoffmann U, Pou KM, Maurovich-Horvat P, Liu CY, Vasan RS, Murabito JM, Meigs JB, Cupples LA, D’Agostino RB, Sr, O’Donnell CJ. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation. 2007;116:39–48. doi: 10.1161/CIRCULATIONAHA.106.675355. [DOI] [PubMed] [Google Scholar]
- 12.Geer EB, Islam J, Buettner C. Mechanisms of glucocorticoid-induced insulin resistance: focus on adipose tissue function and lipid metabolism. Endocrinol Metab Clin N Am. 2014;43:75–102. doi: 10.1016/j.ecl.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gettys TW, Rohlfs EM, Prpic V, Daniel KW, Taylor IL, Collins S. Age-dependent changes in beta-adrenergic receptor subtypes and adenylyl cyclase activation in adipocytes from Fischer 344 rats. Endocrinology. 1995;136:2022–2032. doi: 10.1210/endo.136.5.7720650. [DOI] [PubMed] [Google Scholar]
- 14.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 15.Guillermet-Guibert J, Lahlou H, Cordelier P, Bousquet C, Pyronnet S, Susini C. Physiology of somatostatin receptors. J Endocrinol Investig. 2005;28:5–9. [PubMed] [Google Scholar]
- 16.Hughes VA, Roubenoff R, Wood M, Frontera WR, Evans WJ, Fiatarone Singh MA. Anthropometric assessment of 10-y changes in body composition in the elderly. Am J Clin Nutr. 2004;80:475–482. doi: 10.1093/ajcn/80.2.475. [DOI] [PubMed] [Google Scholar]
- 17.Iacobellis G. Local and systemic effects of the multifaceted epicardial adipose tissue depot. Nat Rev Endocrinol. 2015;11:363–371. doi: 10.1038/nrendo.2015.58. [DOI] [PubMed] [Google Scholar]
- 18.Iacobellis G, Barbaro G. The double role of epicardial adipose tissue as pro- and anti-inflammatory organ. Horm Metab Res. 2008;40:442–445. doi: 10.1055/s-2008-1062724. [DOI] [PubMed] [Google Scholar]
- 19.Kaddai V, Jager J, Gonzalez T, Najem-Lendom R, Bonnafous S, Tran A, Le Marchand-Brustel Y, Gual P, Tanti JF, Cormont M. Involvement of TNF-alpha in abnormal adipocyte and muscle sortilin expression in obese mice and humans. Diabetologia. 2009;52:932–940. doi: 10.1007/s00125-009-1273-3. [DOI] [PubMed] [Google Scholar]
- 20.Kappeler L, De Magalhaes Filho C, Dupont J, Leneuve P, Cervera P, Perin L, Loudes C, Blaise A, Klein R, Epelbaum J, Le Bouc Y, Holzenberger M. Brain IGF-1 receptors control mammalian growth and lifespan through a neuroendocrine mechanism. PLoS Biol. 2008;6:e254. doi: 10.1371/journal.pbio.0060254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karastergiou K, Fried SK. Multiple adipose depots increase cardiovascular risk via local and systemic effects. Curr Atheroscler Rep. 2013;15:361. doi: 10.1007/s11883-013-0361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobatake T, Watanabe Y, Matsuzawa Y, Tokunaga K, Fujioka S, Kawamoto T, Keno Y, Tarui S, Yoshida H. Age-related changes in adrenergic alpha 1, alpha 2, and beta receptors of rat white fat cell membranes: an analysis using [3H]bunazosin as a novel ligand for the alpha 1 adrenoceptor. J Lipid Res. 1991;32:191–196. [PubMed] [Google Scholar]
- 23.Lago F, Dieguez C, Gomez-Reino J, Gualillo O. Adipokines as emerging mediators of immune response and inflammation. Nat Clin Pract Rheumatol. 2007;3:716–724. doi: 10.1038/ncprheum0674. [DOI] [PubMed] [Google Scholar]
- 24.Lunenfeld B. An aging world—demographics and challenges. Gynecol Endocrinol. 2008;24:1–3. doi: 10.1080/09513590701718364. [DOI] [PubMed] [Google Scholar]
- 25.Michalakis K, Goulis DG, Vazaiou A, Mintziori G, Polymeris A, Abrahamian-Michalakis A. Obesity in the ageing man. Metabolism. 2013;62:1341–1349. doi: 10.1016/j.metabol.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogorodnikova AD, Khan UI, McGinn AP, Zeb I, Budoff MJ, Harman SM, Miller VM, Brinton EA, Manson JE, Hodis HN, Merriam GR, Cedars MI, Taylor HS, Naftolin F, Lobo RA, Santoro N, Wildman RP. Ectopic fat and adipokines in metabolically benign overweight/obese women: the Kronos Early Estrogen Prevention Study. Obesity (Silver Spring) 2013;21:1726–1733. doi: 10.1002/oby.20200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perissinotto E, Pisent C, Sergi G, Grigoletto F. Anthropometric measurements in the elderly: age and gender differences. Br J Nutr. 2002;87:177–186. doi: 10.1079/bjn2001487. [DOI] [PubMed] [Google Scholar]
- 29.Pezeshkian M, Mahtabipour MR. Epicardial and subcutaneous adipose tissue fatty acids profiles in diabetic and non-diabetic patients candidate for coronary artery bypass graft. Bioimpacts. 2013;3:83–89. doi: 10.5681/bi.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi L, Zhang C, van Dam RM, FBH Interleukin-6 genetic variability and adiposity: associations in two prospective cohorts and systematic review in 26,944 individuals. J Clin Endocrinol Metab. 2007;92:3618–3625. doi: 10.1210/jc.2007-0877. [DOI] [PubMed] [Google Scholar]
- 32.Rabkin SW. Epicardial fat: properties, function and relationship to obesity. Obes Rev. 2007;8:253–261. doi: 10.1111/j.1467-789X.2006.00293.x. [DOI] [PubMed] [Google Scholar]
- 33.Raggi P, Shaw LJ, Berman DS, Callister TQ. Gender-based differences in the prognostic value of coronary calcification. J Women’s Health (Larchmt) 2004;13:273–283. doi: 10.1089/154099904323016437. [DOI] [PubMed] [Google Scholar]
- 34.Sacks HS, Fain JN. Human epicardial adipose tissue: a review. Am Heart J. 2007;153:907–917. doi: 10.1016/j.ahj.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Samara A, Herbeth B, Aubert R, Berrahmoune H, Fumeron F, Siest G, Visvikis-Siest S. Sex-dependent associations of leptin with metabolic syndrome-related variables: the Stanislas study. Obesity (Silver Spring) 2010;18:196–201. doi: 10.1038/oby.2009.156. [DOI] [PubMed] [Google Scholar]
- 36.Seboek D, Linscheid P, Zulewski H, Langer I, Christ-Crain M, Keller U, Muller B. Somatostatin is expressed and secreted by human adipose tissue upon infection and inflammation. J Clin Endocrinol Metab. 2004;89:4833–4839. doi: 10.1210/jc.2004-0271. [DOI] [PubMed] [Google Scholar]
- 37.Shim IK, Cho KI, Kim HS, Heo JH, Cha TJ. Impact of gender on the association of epicardial fat thickness, obesity, and circadian blood pressure pattern in hypertensive patients. J Diabetes Res. 2015;2015:924539. doi: 10.1155/2015/924539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song do K, Hong YS, Lee H, Oh JY, Sung YA, Kim Y. Increased epicardial adipose tissue thickness in type 2 diabetes mellitus and obesity. Diabetes Metab J. 2015;39:405–413. doi: 10.4093/dmj.2015.39.5.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.St-Onge MP, Gallagher D. Body composition changes with aging: the cause or the result of alterations in metabolic rate and macronutrient oxidation? Nutrition. 2010;26:152–155. doi: 10.1016/j.nut.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strandberg L, Mellstrom D, Ljunggren O, Grundberg E, Karlsson MK, Holmberg AH, Orwoll ES, Eriksson AL, Svedberg J, Bengtsson M, Ohlsson C, Jansson JO. IL6 and IL1B polymorphisms are associated with fat mass in older men: the MrOS study Sweden. Obesity (Silver Spring) 2008;16:710–713. doi: 10.1038/oby.2007.95. [DOI] [PubMed] [Google Scholar]
- 41.Tchkonia T, Morbeck DE, Von Zglinicki T, Van Deursen J, Lustgarten J, Scrable H, Khosla S, Jensen MD, Kirkland JL. Fat tissue, aging, and cellular senescence. Aging Cell. 2010;9:667–684. doi: 10.1111/j.1474-9726.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson NM, Gill DA, Davies R, Loveridge N, Houston PA, Robinson IC, Wells T. Ghrelin and des-octanoyl ghrelin promote adipogenesis directly in vivo by a mechanism independent of the type 1a growth hormone secretagogue receptor. Endocrinology. 2004;145:234–242. doi: 10.1210/en.2003-0899. [DOI] [PubMed] [Google Scholar]
- 43.Trevaskis JL, Gawronska-Kozak B, Sutton GM, McNeil M, Stephens JM, Smith SR, Butler AA. Role of adiponectin and inflammation in insulin resistance of Mc3r and Mc4r knockout mice. Obesity (Silver Spring) 2007;15:2664–2672. doi: 10.1038/oby.2007.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–B501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- 45.Ukkola O, Rankinen T, Lakka T, Leon AS, Skinner JS, Wilmore JH, Rao DC, Kesaniemi YA, Bouchard C. Protein tyrosine phosphatase 1B variant associated with fat distribution and insulin metabolism. Obes Res. 2005;13:829–834. doi: 10.1038/oby.2005.95. [DOI] [PubMed] [Google Scholar]
- 46.van Herpen NA, Schrauwen-Hinderling VB. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol Behav. 2008;94:231–241. doi: 10.1016/j.physbeh.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 47.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]