Abstract
Objective
To assess if the reduction in HIV-1 RNA in CD4+ T cells is correlated with the persistence of immune activation following early antiretroviral therapy (ART)
Design
Clinical trial (NCT01285050).
Methods
Next-generation sequencing was used to study total RNA from activated CD4+ T cells (CD38 and HLA-DR expressing) collected from 19 treatment-naïve HIV-1/HCV infected patients before and early after ART initiation (≥12 weeks after plasma HIV RNA < 50 c/ml). To validate comparisons, pre- and post-ART measures were adjusted for input RNA and overall read number.
Results
As expected, ART use was associated with a median (IQR) 4.3 (2.2–8.3) reduction in the proportion of activated CD4+ T cells (P=0.0008). Whereas in those activated CD4+ T cells no consistent differences in overall gene expression were detected, interferon stimulated gene expression declined (P<2×10−16). Pre-ART, sorted activated CD4+ T cells contained a median (IQR) of 959 (252–1614) HIV-1 reads/107 reads compared to 72 (55–152) HIV-1 reads/107 reads after ≥12 weeks of suppressive ART (P=8×10−5). The decrease in HIV-1 reads in activated CD4+ T cells was associated with the change in plasma HIV-1 RNA levels (r=0.77, P=2×10−4) and the change in the proportion of activated CD4+ T cells (r=0.70, P=0.0016).
Conclusions
Months of ART led to a marked decrease in cell-associated HIV-1 RNA and ISG expression in activated CD4+ T cells that were strongly associated with the reduction in the proportion of activated CD4+ T cells.
Keywords: HIV-1, antiretroviral therapy, T cell activation, Immune activation, cell-associated HIV-1 RNA
INTRODUCTION
Antiretroviral therapy (ART) durably suppresses HIV-1, prevents progression to acquired immunodeficiency syndrome, and reduces mortality. However, even with ART HIV-1 infected adults remain at higher risk of cardiovascular disease, dyslipidemia, insulin resistance, liver, kidney and bone dysfunction, neurocognitive disorders, accelerated biological aging, and cancer [1, 2]. Although the precise pathophysiology of many of these disorders is poorly understood, a syndrome of chronic immune activation and inflammation is a common feature [3, 4].
With initial HIV-1 infection, there is activation of innate and adaptive immunity that alters T cell homeostasis as characterized by elevated HLA-DR+/CD38+, CD57, PD-1 and CTLA-4 expression, and lower expression of CD28 [5–13], as well as enhanced expression of many cytokines and chemokines including IFNα, IFNγ, TNF, IL-6, IL-8, and CXCL10 [14, 15]. Cellular activation of CD4+ and CD8+ T cells, defined by co-expression of the surface markers HLA-DR and CD38, has been most closely associated with disease pathogenesis [7, 8, 11, 16]. Even when HIV-1 viremia has been suppressed for years, the proportion of HLA-DR+/CD38+ T cells remains higher than in uninfected persons and is correlated with failure to restore circulating CD4+ T cells and development of disease [2, 7, 17].
A dominant mechanism for HIV-1 persistence is the generation of a ‘silent’ provirus in resting memory CD4+ T cells that is unaffected by ART or host-immune responses [18–22]. Some non-induced pro-viruses are capable of replicating if ART is stopped [23, 24]. However, the functional consequences of persistent intracellular viral RNAs remain unclear. It is plausible that cell-associated viral RNA, even in the absence of virion production and infection of new cells, could sustain host innate immune responses and contribute to immune activation [25, 26]. Here we show that, even after more than 12 weeks of undetectable viremia following ART, HIV-1 RNA persists in activated CD4+ T cells, and that HIV-1 transcription is associated with the reduction in CD4+ T cell activation and expression of host interferon-stimulated genes (ISG).
METHODS
Human subjects
Persons with HIV-1/HCV co-infection were enrolled in a prospective study designed to test the effect HIV-1 control on HCV that is extensively described elsewhere [27]. Persons with untreated chronic HIV-1 and HCV infections were recruited from the Johns Hopkins HIV-1 Clinic, the Baltimore City Sexually Transmitted Diseases Clinic, and other area clinics. HIV-1 infection was established by detection of HIV-1 antibodies and an HIV-1 RNA level >400 c/mL; chronic HCV infection was determined by detection of HCV antibodies and HCV RNA >100,000 IU/mL for >6 months. Subjects had received <24 months of ART over their entire lives and none within 6 months of enrollment. Subjects also were excluded if HBsAg was detected in plasma; they were pregnant; there was a history of severe depression or any uncontrolled disease; platelet count was <50,000/mm3; or if there was a contraindication to use of raltegravir, tenofovir disoproxil fumarate (TDF), or emtricitabine. Because therapy was judged too urgent to wait for study procedures as per current treatment guidelines for HIV-1 and HCV, persons at screening whose CD4+ T cell counts were <200/mm3 or who had cirrhosis were excluded. A total of 32 patients were screened to identify 20 study subjects, and 19 completed the study. All subjects gave written informed consent to the protocol as approved by the Johns Hopkins University School of Medicine Institutional Review Board.
All subjects had received a single injection of peginterferon alpha 2b (IFN) two weeks prior to ART initiation to study the response of HIV-1 and HCV to IFN prior to ART. Two weeks following IFN administration, after plasma HIV-1 RNA levels normalized, ART consisting of raltegravir, TDF, and emtricitabine was initiated. Participants were followed until their plasma HIV-1 RNA levels had been undetectable for ≥12 weeks after ART initiation. Peripheral blood mononuclear cells (PBMCs) were collected prior to ART initiation and at regular intervals for ≥12 weeks after ART initiation. The first specimen collected for sequencing (“Pre-Treatment”) was acquired before the first peginterferon injection and the second specimen (“ART”) was collected after ≥12 weeks of continuous suppressed viremia, HIV-1 RNA <50 cp/mL), more than 4 months later. All subjects gave informed consent for participation in the study, which was approved by The Johns Hopkins University School of Medicine Institutional Review Board.
HIV-1 RNA quantitative PCR (qPCR) testing was performed using the Abbott RealTime HIV-1 Assay (No. 02631–090). CD4+ T cell count was measured by flow cytometry of whole blood that was delivered to the Johns Hopkins Hospital clinical laboratory.
Isolation of activated T cells
PBMCs were separated from whole blood and frozen. Freshly thawed cells were washed and incubated with CD3-FITC (Biolegend), CD4-PECy7 (Biolegend), CD8-APC (BD Biosciences), HLA-DR-PE (Biolegend), and CD38-BV421 (Biolegend) for 40 minutes at 4°C per the manufacturer’s recommendation. Immediately before sorting, plasma membrane compromised cells were labeled with propidium iodide (Sigma). Fluorescence-activated cell sorting (FACS) was performed on a MoFlo Legacy Sorter (Beckman-Coulter) at the Johns Hopkins School of Public Health Flow Cytometry Core Facility. Gates were set using fluorescence minus one plus isotype control. Because flow and sorting had to be performed over several sessions, the gates were set each session using a reference participant’s PBMCs (selected based on availability of cells). The population of interest was sorted directly into ≥4 volumes of Quick-RNA® MicroPrep lysis buffer (Zymo Research) per the manufacturer’s recommendation. Sorting was stopped when the number of sorted cells reached 125,000 cells, although many samples did not reach this number. Flow cytometry analysis on two randomly selected post-sort samples revealed >95% purity. Sorted samples were vortexed, incubated for 10 minutes at room temperature, vortexed again, and frozen at −80°C until isolation.
RNA preparation and sequencing
Isolation was performed using the Quick-RNA® MicroPrep kit (Zymo Research) according the manufacturer’s protocol without the on-column DNAase treatment. The eluate was treated with DNAse-I (Qiagen) according to the manufacturer’s protocol, then purified and concentrated using the RNA Clean-up and Concentrator kit (Zymo Research) according the manufacturer. The high-sensitivity assay for RNA or DNA was performed on RNA isolations and cDNA libraries respectively using a 2100 Biolanalyzer (Agilent).
Complementary DNA (cDNA) libraries were produced using the Ovation® Single-cell RNAseq kit (NuGEN) according to the manufacturer’s specifications. Briefly, reverse transcription was carried out using a random hexamer to oligo dT ratio of 50:1 and unique barcodes for each individual’s samples were ligated to ~250bp enzymatically fragmented molecules. All samples were linearly amplified with 19 cycles of PCR using primer annealing sites contained within the adapters. All sequencing was performed on a HiSeq2500 (Illumina) at the Johns Hopkins Genetics Research Core Facility. All sequencing data are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession number SRP068424).
Human Sequence Mapping and Differential Expression Calculation
Reads were aligned to the hg19 reference genome and annotated transcripts with RSEM (version 1.2.9) [28]. Differential expression of genes and isoforms was calculated using EBseq (version 1.11.0) [29], an empirical Bayes hierarchical model for expression analysis of RNAseq data.30 The human sequence mapping and differential expression calculation was performed at the Johns Hopkins University Sidney Kimmel Comprehensive Cancer Center Next Generation Sequencing core.
HIV-1 Genome Sequence Mapping
The reads were first quality-trimmed using the Phred algorithm with seqtk trimfq (version 1.0, error rate threshold 0.05, https://github.com/lh3/seqtk), and sequences shorter than 31 base pairs, after quality trimming, were discarded. The remaining reads were mapped to the human genome (build hg19) with bowtie2 (version 2.2.6) [30] using local alignment as a filter. All reads that did not align to the human genome were then mapped against a pool of 2,153 validated HIV-1 genomes, plus the HIV-1 reference genome, that were listed in the NCBI Viral Genome Resource [31] as of July 18, 2016 (Table S1). The reads were extracted using samtools (version 1.3) [32] and aligned to an index compiled from the HIV genomes with bowtie2. The resulting alignments were cleaned of PCR duplicates using Picard MarkDuplicates (version 1.119, http://broadinstitute.github.io/picard). Percent identity and alignment length for the HIV reads were calculated from SAM tags, and only reads with at least 80% identity over the aligned sequence, and alignment length of 31 base pairs, were kept. The genome coverage was calculated with samtools mpileup. The data was loaded in R version 3.3.1 [33] and visualized using ggplot2 version 2.20 [34].
Statistical Analyses
Comparison values, including post-correction fold-change, post-correction probability of differential expression (PPDE), and post-correction probability of equal expression (PPEE) values, were further analyzed using ‘stats’ in R version 3.1.2. Measurements for which neither the PPDE nor PPEE were ≥0.95 were discarded as being of low confidence. In the determination of significance of the gene’s change across the cohort after ART, all fold-change calculations with PPEE ≥ 0.95 were set to 0, reflecting the certainty of the PPEE. If there were remaining measurements for a gene in >10 of the individuals, a two-sided one-sample T-test was performed on log2 transformed fold-change values. The resulting P-values were adjusted for multiple comparisons using the Benjamani-Hochberg method. Spearman rank-correlations were performed between fold-changes of the observed ISGs and intracellular HIV-1 RNA decline; with fold-changes with PPEE ≥ 0.95 retained as their original values.
RESULTS
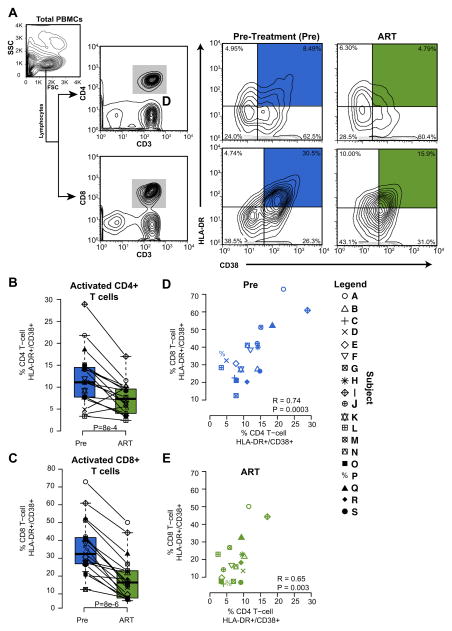
We collected samples from nineteen patients with HIV-1 and chronic HCV co-infection who completed the study (Table 1). The median (range) log plasma HIV-1 RNA and CD4+ T cell counts at baseline were 4.27 log c/ml (2.91–5.44 log c/ml) and 376 cells/mm3 (203–759 cells/mm3), respectively. ART resulted in suppressed viremia in all participants and a median (IQR) increase in CD4+ T cell count of 90 cells/mm3 (28 – 151 cells/mm3)[27]. Populations of activated CD4+ and CD8+ cells from total CD3+ T cells in PBMCs of patients before and after ART are shown in Fig. 1A. Prior to ART, CD4+/CD38+/HLA-DR+ cells constituted a median (IQR) of 11.3% (7.7%-14.5%) of total CD4+ T cells, while CD8+/CD38+/HLA-DR+ cells constituted 32.4% (26.90%-41.6%) of total CD8+ T cells. After ART initiation, CD4+ and CD8+ T cell activation decreased by a median (IQR) 4.3% (2.2–8.3) and 19.1% (14.1–22.5), respectively (Figs. 1B and C; P<0.001 for both), consistent with what has been seen previously [35–37]. In addition, CD4+ and CD8+ T cell activation were closely associated with each other (Fig. 1D and E).
Table 1.
Participant characteristics*
| Characteristic | N = 19 (100%) |
|---|---|
| Age,* | |
| median (range) | 49.2 (20.8–60.6) |
| Sex | |
| Male, n (%) | 15 (78.9) |
| Race | |
| Black, n (%) | 12 (63.2) |
| BMI, kg/m2* | |
| median (range) | 23.3 (17.7–39.0) |
| Plasma HIV-1 RNA, log10 copies/mL* | |
| median (range) | 4.27 (2.91–5.44) |
| CD4+ T-cell count, cells/mm3* | |
| median (range) | 376 (203–759) |
| Median ART duration (range), days | 175 (112–217) |
at baseline unless otherwise indicated
FIGURE 1. The proportion of activated T cells declines following ART.
(A) Flow cytometry data and sorting algorithm for isolation of activated CD4+ and CD8+ T cells from total PBMCs at baseline (Pre-ART, blue) and after ≥12 weeks of undetectable plasma HIV-1 RNA during ART (green). Boxplots showing the proportion of (B) CD4+ and (C) CD8+ T cells expressing both CD38 and HLA-DR. (D) Scatterplot comparing the degree of CD4+ and CD8+ activation by participant before and (E) after ART.
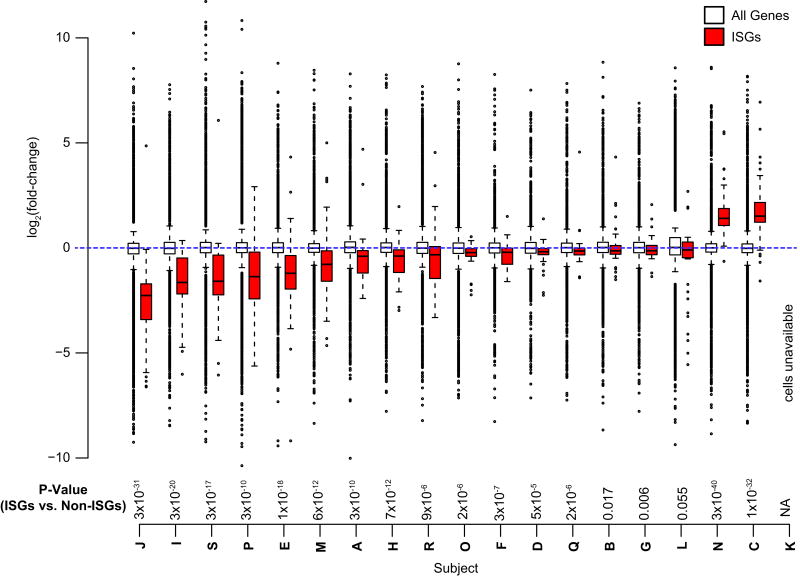
PBMCs were sorted before treatment was initiated and ≥12 weeks after plasma HIV-1 RNA was suppressed to isolate purified populations of activated CD4+ T cells, defined as CD3+/CD4+/CD38+/HLA-DR+. cDNA libraries were prepared from purified cells and a median (IQR) of 90.5 (85.4–107.6) million and 86.2 (83.5–99.6) million 100bp paired-end sequences were obtained before and following ART initiation, respectively (Table S2). We examined the change in the expression of a median (IQR) of 11,840 (10,080 – 13,180) genes following ART initiation in each individual’s activated CD4+ T cells (Fig. S1). After adjustment for multiple comparisons, no single gene was consistently differentially expressed pre- vs post-ART across the 19 individuals in the cohort. We considered interferon-stimulated genes (ISGs) as a separate group of genes whose expression is triggered in response to viral infections (Table S3). Of the 18 individuals in the cohort for whom cells were available, 17 had a significant change in ISGs (P-value range = 3×10−40 to 0.006). Fifteen of the individuals had a significant decrease in ISG expression while only 2 had an increase in ISG expression (Fig. 2). When the median fold-changes across the cohort were considered, ISGs were decreased compared to all other genes. Of the 96 ISGs examined, 84.4% were downregulated post-ART (P<2×10−16, for comparison with fold change of non-ISGs) and the median (IQR) magnitude of the changes among ISGs was −0.244 log2 fold (−0.438 to −0.0396) (Fig. S2).
FIGURE 2. Activated CD4+ T cells of patients following ART initiation have decreased expression of interferon stimulated genes (ISGs) compared to pre-ART.
Boxplots comparing the log2 fold-changes (y-axis) of all genes (unfilled) with ISGs (red) for each of the 19 subjects in the study. P-value shown is a Wilcoxon rank-sum test of the fold-changes of ISGs compared to the fold-changes of all other genes per individual.
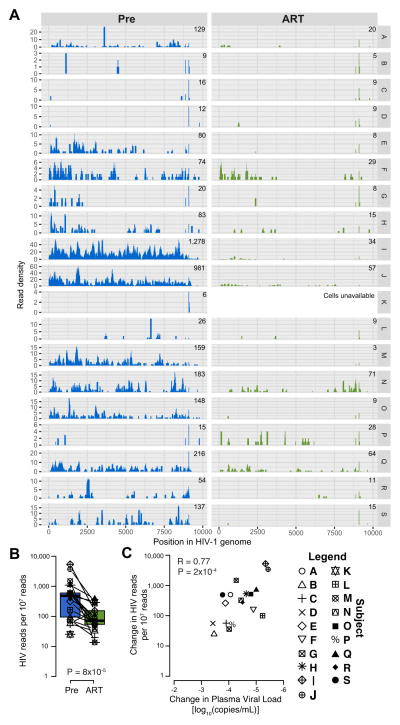
We next quantified HIV-1 RNA in activated CD4+ T cells by aligning RNA sequences to a reference panel of 2,153 HIV-1 genome sequences (Fig. 3A). Cell-associated HIV-1 RNA was detected both before and after ART initiation, but was less abundant during ART. A median (IQR) of 959 (252–1614) reads/107 reads and 72 (55–152) reads/107 reads mapped to HIV-1 before and after ART initiation, respectively (Fig. 3B) (P=8×10−5). In addition, pre-ART cell-associated HIV-1 RNAs were strongly associated with contemporaneous plasma HIV-1 RNA levels (R=0.79, P=5×10−5), and the decrease in cell-associated HIV-1 RNAs was associated with the suppression of plasma viremia (R=0.77, P=2×10−4)(Fig. 3C).
FIGURE 3. Intracellular HIV-1 reads in activated CD4+ T cells pre- and following ART initiation.
(A) Histograms displaying the HIV-1 read density (y axis) of each participant (designated at the far right, Y2, labels as A to S) for each position on the HIV-1 reference genome (x axis). (B) Boxplot with overlaid values comparing HIV-1 read quantitation pre- and following ART initiation. Lines connect values from the same individual. (C) Scatterplot comparing the change in the normalized number of reads mapping to HIV-1 from activated CD4+ T cells with ART and the change in plasma HIV RNA (with undetectable values while on ART set to zero).
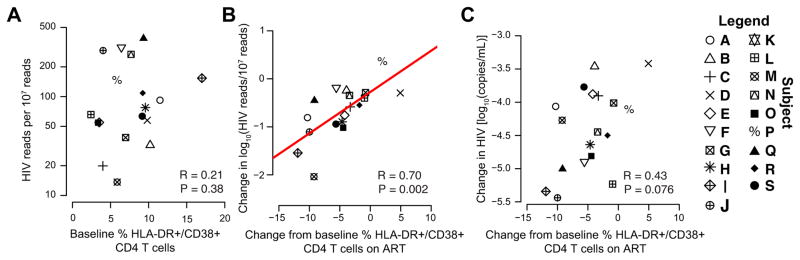
At baseline, cell-associated HIV-1 RNA and global CD4+ T cell activation were not correlated (Fig. 4A). However, the decrease in the proportion of CD3+/CD4+/CD38+/HLA-DR+ T cells after ART initiation was strongly associated with the decrease in intracellular HIV-1 RNA in the same activated cells (R=0.70, P=0.002; Fig. 4B). In contrast, the change in proportion of CD3+/CD4+/CD38+/HLA-DR+ T cells after ART initiation was weakly associated with the change in plasma HIV-1 RNA levels (R=.43 P=.076; Fig. 4C).
FIGURE 4. The change in intracellular HIV-1 RNA is associated with the change in proportion of activated CD4+ T cells.
(A) pre-ART HIV-1 RNA is not associated with the proportion of DR+/CD38+/CD4+ T cells as shown by scatter plot comparing the two values. (B) Scatterplot comparing the change in intracellular HIV-1 RNA and the change in the proportion of HLA-DR+/CD38+/CD4+ T cells. (C) Scatterplot comparing the change in plasma HIV-1 RNA (with undetectable values on ART set to zero) and the change in the proportion of HLA-DR+/CD38+/CD4+ T cells.
DISCUSSION
Despite the success of ART in improving clinical outcomes in patients with HIV-1, increasing non-AIDS morbidity and mortality have prompted efforts to understand the association between ART-suppressed virus persistence and host immunity. We found that the change in plasma and cell-associated HIV-1 RNA abundance following ART were strongly correlated, and that the decrease in cell-associated HIV-1 RNA was strongly associated with the reduction in CD38+ and DR+ CD4+ T cells. In addition, ART use was strongly associated with diminished expression of a panel of ISGs in activated CD4+ T cells. These findings are consistent with previous reports of decreases in cell-associated HIV-1 RNA in unsorted PBMCs [11]; however, in the present manuscript we measured cell-associated HIV-1 RNA strictly in activated CD4+ T cells, and linked this finding with the proportion of activated CD4+ T cells and ISG expression in those cells.
As in the case with all viral infections, ISGs are rapidly induced as a first line of host defenses in HIV-1 infected cells. ISGs are induced by type 1 interferons, which in turn are generated by the detection of viral nucleic acids and structural components by host cytosolic pattern recognition receptors. Proteins encoded by ISGs act in an autocrine and paracrine manner to promote an anti-viral state and the onset of adaptive immunity. Expression of ISGs in CD4+ T cells is one of the hallmarks of immune activation [38]. We found that the levels of 96 established ISGs in activated CD4+ T cells (Table S3) were significantly reduced en masse in patients on ART when compared to all evaluated genes (Fig. 2).
The finding of a correlation between the reduction in intracellular HIV RNA and activated CD4+ T cell change on ART is interesting in light of emerging data that suggest the virus itself may contribute to immune activation even in persons taking ART. While production of new, replication-competent virus appears to be aborted by ART, HIV-1 RNA has been previously identified in CD4+ T cells in patients taking ART [39]. Low copy numbers of HIV-1 RNA, as well as an excess of short abortive transcripts have been identified in latently infected resting memory CD4+ T cells in patients on therapy [24, 39]. Although the functional consequence of these nonproductive or dead RNAs remains unclear, our data could support that persistent inactive viral RNAs in activated cells may lead to innate immune signaling. However, because plasma HIV-1 changes were strongly correlated with cell-associated HIV-1 RNA changes, an alternative explanation is that those with the largest plasma HIV-1 reductions had the greatest restoration in CD4+ T cells not expressing DR or CD38. In light of findings by Murray et al. that ART leads to a biphasic decrease in CD4+/CD38+/HLA-DR+ T cells with a corresponding decline in total proviral DNA, we speculate that our findings may be related to the slow kinetics of turnover of the HIV-1 reservoir [40].
There were several challenges in our study. Because we employed a sorting strategy of activated CD4+ T cells, we were limited in the total amount of RNA that was available for individually targeted assays. Consequently, we did not fully characterize the HIV-1 RNAs or establish if they could propagate infection. Fortunately, on ART few/no of the RNAs would have been expected to propagate infection, and those limitations should not have affected our within-person comparisons. Another potential challenge was in our choice to study activated CD4+ T cells exclusively. Since activation itself promotes HIV-1 transcription, an expected but overgeneralized result would be to find less intracellular HIV-1 RNA during ART. However, while this would be true in a study of global CD4+ T cells, we specifically studied activated CD4+ T cells and adjusted for total transcription in those same cells. Another limitation is that we provide no mechanism explaining how CD8+ T cells would become activated, even though that cell type is strongly associated with the clinical immune activation outcomes. We note, however, that CD4+ and CD8+ T cell activation were closely related (Fig. 1D and E). Moreover, several studies have reported that CD4+ T cell activation also is associated with clinical outcomes [11, 16]. We also note that all participants in the present study were chronically co-infected with HCV, as is common among HIV-1 infected people who inject drugs in Baltimore [41]. It is possible other HIV-1 infected populations, including those without chronic HCV, infection might respond differently. However, our findings of a decrease in CD4+ and CD8+ T cell activation are consistent with previous studies in which HCV status was not presented [42].
All participants were treated with a single injection of pegylated interferon alpha 2b after acquisition of the ‘Pre-treatment’ samples and ≥12 weeks prior to the ‘ART’ specimen collection. While this could have led to an interferon refractory period, it would be unexpected for such a phenomenon to be present more than 4 months later. Indeed, ISG expression was observed to decrease to baseline values in rhesus macaques after 4 weeks despite weekly administration of exogenous pegylated interferon alpha [43].
We investigated the impact of 3 months of ART on HIV-1 cell-associated RNA activated CD4+ T cells. The reduction in activated CD4+ T cells was associated with the reduction in HIV-1 cell-associated RNAs and ISG expression. These findings provide new insights into the early effects of ART on a critical CD4+ T cell compartment.
Supplementary Material
Acknowledgments
FUNDING SOURCES: National Institutes of Health Grant R37 DA 013806, R01 DA 016078, UM1-AI-068613 and the Johns Hopkins Institute for Clinical and Translational Research (ICTR), which is funded in part by Grant Number UL1 TR 000424-06 through a Clinical and Translational Science Award (CTSA) from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research.
The authors would like to thank the participants of the study for contributing these invaluable samples; David Mohr of The Johns Hopkins Genetics Research Core Facility for expertise in library preparation and optimal sequencing; Hao Zhang, Zachary Freeman, and Rebecca Veenhuis for helpful discussion and assistance with designing FACS protocols; Paul Scherer and Neil Neumann for helpful discussion; the Next Generation Sequencing Center at the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center (NIH P30 CA006973) for their support with high-throughput sequencing experiments; and the organizers and instructors of the Quantitative Systems Immunology Summer School at Boston University and the Computational Immunology seminar series at Johns Hopkins University for introduction to the computational methods implemented in this investigation. Tenofovir disoproxil fumarate, emtricitabine, and Raltegravir were donated by Gilead. Peginterferon alfa 2b was donated by Merck.
ABBREVIATIONS
- HIV-1
human immunodeficiency virus
- HCV
epatitis C virus
- IQR
interquartile range
- qPCR
quantitative polymerase chain reaction
- RNAseq
RNA sequencing
- LLOD
lower limit of detection
- RT
reverse transcription
- Ct
Cycle Threshold
- IDU
injection drug user
- ALT
alanine aminotransferase
- U
units
- IU
international units
Footnotes
CONFLICT OF INTEREST: The authors indicate no conflict of interest
AUTHORS’ CONTRIBUTIONS
Study concept and design: RE, JB, SR, DT, AB. Acquisition of data: RE. Analysis and interpretation of data: RE, FB, MS, AS, JB, SR, SW, DT, AB. Statistical analysis: RE, FB, AS, SW, DT, AB. Drafting of the manuscript: RE, GS, DT, AB. All authors participated in the review and final approval of the manuscript.
References
- 1.Warriner AH, Burkholder GA, Overton ET. HIV-related metabolic comorbidities in the current ART era. Infect Dis Clin North Am. 2014;28:457–476. doi: 10.1016/j.idc.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Lederman MM, Funderburg NT, Sekaly RP, Klatt NR, Hunt PW. Residual immune dysregulation syndrome in treated HIV infection. Adv Immunol. 2013;119:51–83. doi: 10.1016/B978-0-12-407707-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aounallah M, Dagenais-Lussier X, El-Far M, Mehraj V, Jenabian MA, Routy JP, et al. Current topics in HIV pathogenesis, part 2: Inflammation drives a Warburg-like effect on the metabolism of HIV-infected subjects. Cytokine Growth Factor Rev. 2016;28:1–10. doi: 10.1016/j.cytogfr.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Nou E, Lo J, Grinspoon SK. Inflammation, immune activation, and cardiovascular disease in HIV. AIDS. 2016;30:1495–1509. doi: 10.1097/QAD.0000000000001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klatt NR, Chomont N, Douek DC, Deeks SG. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev. 2013;254:326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Younas M, Psomas C, Reynes J, Corbeau P. Immune activation in the course of HIV-1 infection: Causes, phenotypes and persistence under therapy. HIV Med. 2016;17:89–105. doi: 10.1111/hiv.12310. [DOI] [PubMed] [Google Scholar]
- 7.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 8.Robbins GK, Spritzler JG, Chan ES, Asmuth DM, Gandhi RT, Rodriguez BA, et al. Incomplete reconstitution of T cell subsets on combination antiretroviral therapy in the AIDS Clinical Trials Group protocol 384. Clin Infect Dis. 2009;48:350–361. doi: 10.1086/595888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauce D, Larsen M, Fastenackels S, Pauchard M, Ait-Mohand H, Schneider L, et al. HIV disease progression despite suppression of viral replication is associated with exhaustion of lymphopoiesis. Blood. 2011;117:5142–5151. doi: 10.1182/blood-2011-01-331306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tassiopoulos K, Landay A, Collier AC, Connick E, Deeks SG, Hunt P, et al. CD28-negative CD4+ and CD8+ T cells in antiretroviral therapy-naive HIV-infected adults enrolled in adult clinical trials group studies. J Infect Dis. 2012;205:1730–1738. doi: 10.1093/infdis/jis260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatano H, Jain V, Hunt PW, Lee TH, Sinclair E, Do TD, et al. Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. J Infect Dis. 2013;208:50–56. doi: 10.1093/infdis/jis630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone SF, Price P, French MA. Dysregulation of CD28 and CTLA-4 expression by CD4 T cells from previously immunodeficient HIV-infected patients with sustained virological responses to highly active antiretroviral therapy. HIV Med. 2005;6:278–283. doi: 10.1111/j.1468-1293.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 13.Jensen SS, Tingstedt JL, Larsen TK, Brandt L, Gerstoft J, Kronborg G, et al. HIV-Specific CD8+ T Cell-Mediated Viral Suppression Correlates With the Expression of CD57. J Acquir Immune Defic Syndr. 2016;71:8–16. doi: 10.1097/QAI.0000000000000837. [DOI] [PubMed] [Google Scholar]
- 14.Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson J, Kinloch-de-Loes S, Granath A, Sonnerborg A, Goh LE, Andersson J. Early immune activation in gut-associated and peripheral lymphoid tissue during acute HIV infection. AIDS. 2007;21:565–574. doi: 10.1097/QAD.0b013e3280117204. [DOI] [PubMed] [Google Scholar]
- 16.Balagopal A, Asmuth DM, Yang WT, Campbell TB, Gupte N, Smeaton L, et al. Pre-CRT Elevation of CRP and CD4+ T-cell Immune Activation Associated with HIV Clinical Progression in a Multinational Case-Cohort Study. J Acquir Immune Defic Syndr. 2015;70:163–171. doi: 10.1097/QAI.0000000000000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M, Pantazis N, Martin GE, Hickling S, Hurst J, Meyerowitz J, et al. Exhaustion of Activated CD8 T Cells Predicts Disease Progression in Primary HIV-1 Infection. PLoS Pathog. 2016;12:e1005661. doi: 10.1371/journal.ppat.1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 19.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 21.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 22.Strain MC, Gunthard HF, Havlir DV, Ignacio CC, Smith DM, Leigh-Brown AJ, et al. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci U S A. 2003;100:4819–4824. doi: 10.1073/pnas.0736332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’Doherty U, Paxinos EE, et al. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasternak AO, Lukashov VV, Berkhout B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology. 2013;10:41. doi: 10.1186/1742-4690-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balagopal A, Kandathil AJ, Higgins YH, Wood J, Richer J, Quinn J, et al. Antiretroviral therapy, interferon sensitivity, and virologic setpoint in human immunodeficiency virus/hepatitis C virus coinfected patients. Hepatology. 2014;60:477–486. doi: 10.1002/hep.27158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brister JR, Ako-Adjei D, Bao Y, Blinkova O. NCBI viral genomes resource. Nucleic Acids Res. 2015;43:D571–577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. http://www.R-project.org/ [Google Scholar]
- 34.Wickham H. Springer Science and Business Media. 2009. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 35.Koblavi-Deme S, Maran M, Kabran N, Borget MY, Kalou M, Kestens L, et al. Changes in levels of immune activation and reconstitution markers among HIV-1-infected Africans receiving antiretroviral therapy. AIDS. 2003;17(Suppl 3):S17–22. doi: 10.1097/00002030-200317003-00003. [DOI] [PubMed] [Google Scholar]
- 36.Hunt PW, Cao HL, Muzoora C, Ssewanyana I, Bennett J, Emenyonu N, et al. Impact of CD8+ T-cell activation on CD4+ T-cell recovery and mortality in HIV-infected Ugandans initiating antiretroviral therapy. AIDS. 2011;25:2123–2131. doi: 10.1097/QAD.0b013e32834c4ac1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng L, Taiwo B, Gandhi RT, Hunt PW, Collier AC, Flexner C, et al. Factors associated with CD8+ T-cell activation in HIV-1-infected patients on long-term antiretroviral therapy. J Acquir Immune Defic Syndr. 2014;67:153–160. doi: 10.1097/QAI.0000000000000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sedaghat AR, German J, Teslovich TM, Cofrancesco J, Jr, Jie CC, Talbot CC, Jr, et al. Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J Virol. 2008;82:1870–1883. doi: 10.1128/JVI.02228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lassen KG, Bailey JR, Siliciano RF. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J Virol. 2004;78:9105–9114. doi: 10.1128/JVI.78.17.9105-9114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray JM, Zaunders JJ, McBride KL, Xu Y, Bailey M, Suzuki K, et al. HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy. J Virol. 2014;88:3516–3526. doi: 10.1128/JVI.03331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas DL. The challenge of hepatitis C in the HIV-infected person. Annu Rev Med. 2008;59:473–485. doi: 10.1146/annurev.med.59.081906.081110. [DOI] [PubMed] [Google Scholar]
- 42.Jain V, Hartogensis W, Bacchetti P, Hunt PW, Hatano H, Sinclair E, et al. Antiretroviral therapy initiated within 6 months of HIV infection is associated with lower T-cell activation and smaller HIV reservoir size. J Infect Dis. 2013;208:1202–1211. doi: 10.1093/infdis/jit311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, et al. Type 1 interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.