Abstract
Bisphenol A (BPA), bisphenol AF (BPAF), and bisphenol S (BPS) are well known endocrine disruptors. Previous in vitro studies showed that these compounds antagonize androgen receptor (AR) transcriptional activity; however, the mechanisms of action are unclear. In the present study, we investigated interactions of coregulator peptides with BPA, BPAF, or BPS at the AR complexes using Micro Array for Real-time Coregulator Nuclear Receptor Interaction (MARCoNI) assays and assessed the binding of these compounds on AR by molecular dynamics (MD) simulations. The set of coregulator peptides that are recruited by BPA-bound AR, either positively/or negatively, are different from those recruited by the agonist R1881-bound AR. Therefore, the data indicates that BPA shows no similarities to R1881 and suggests that it may recruit other coregulators to the AR complex. BPAF-bound AR recruits about 70- 80% of the same coregulator peptides as BPA-bound AR. Meanwhile, BPS-bound AR interacts with only few peptides compared to BPA or BPAF-bound AR. MD results show that multiple binding sites with varying binding affinities are available on AR for BPA, BPAF, and BPS, indicating the availability of modified binding surfaces on AR for coregulator interactions. These findings help explain some of the distinct AR-related toxicities observed with bisphenol chemicals and raise concern for the use of substitutes for BPA in commercial products.
Keywords: BPA analogues, androgen receptor (AR), coregulators peptide, binding sites
1. Introduction
Bisphenols (BPA, BPAF and BPS) are reported to be endocrine disruptor chemicals (EDCs), causing a variety of human health issues (Rochester 2013; Rochester and Bolden 2015). The vast majority of studies have focused on the agonistic effects of BPA analogues on the estrogen-signaling pathway because they can bind to the estrogen receptors alpha and beta (ERα and β) and activate estrogen responsive genes. Contrary to the positive modulation on ER, BPA analogues antagonize the transcriptional activity of the androgen receptor (AR), which could affect the AR signaling pathway (Safe 2002; Teng et al. 2013). (Rehan et al 2015) Androgens, upon binding to the AR, play a major role in male sex organ development and are critical for maintaining male reproductive function (Luccio-Camelo and Prins 2011) (Sidorkiwicz et al. 2017). In addition, they affect normal physiology of a wide variety of tissues and deregulate AR function, which may potentially lead to cancer (Rochester 2013; Tan et al. 2015) (Dai et al., 2017).
The AR is a member of the nuclear receptor superfamily and is consistent with the characteristic nuclear receptor modular structure (Helsen and Claessens 2014). However, the 538 amino acid stretch at the N-terminus domain bears (NTD) little homology with other nuclear receptors, showing an intrinsically disordered structure, which is in contrast to the DNA-binding domain (DBD) and ligand-binding domain (LBD) (McEwan 2012). Ligand binding to the C-terminal of the AR leads to conformation changes and creates two protein-protein interaction surfaces for coregulators to bind to the AF2 and BF3 sites. Importantly, the BF3 site can bind small molecules and allosterically regulate protein-protein interactions at the AF2 site (Estebanez-Perpina et al. 2007). Furthermore, mutations at this location occur frequently in prostate cancer patients (Munuganti et al. 2014). The functional activity of the ligand-bound AR requires interactions between the N- and C- terminus of the receptor protein as well as protein-protein interactions with coregulators (Kumar and McEwan 2012). Based on cell types, target genes, and the types of ligands bound to the receptor, transcription of the target genes could become enhanced or repressed, depending on the coregulator or corepressor. (Millard et al. 2013; O'Malley and McKenna 2008). Hundreds of nuclear receptor (NR) coregulators have been identified (http://www.NURSA.org) and many have been revealed with the proteomic analysis approach (Foulds et al. 2013). The transcription levels of NR target genes are dictated by the coregulator proteins that are recruited to the ligand bound NR. This, in turn, modifies the chromatin structure to either favor transcriptional activation or repression (Millard et al. 2013). The conserved interaction motifs of the coregulators and NR, with or without ligands, are LxxLL (the NR box; (Heery et al. 1997)) and LxxH/IIxxxI/L (CoRNR box; (Hu and Lazar 1999; Nagy et al. 1999). In general, coregulators possess enzymatic activity, which modifies chromatin structure via various mechanisms and influences the rate of transcription (Millard et al. 2013).
In the present study, the objective was to search for corregulator protein recruitment by the AR when BPA analogues were bound. We used the MARCoNI peptide screening method to identify coregulators in BPA analogue-bound AR complexes. Furthermore, we used molecular dynamic (MD) simulation techniques that have emerged as a common tool to characterize the strength of small molecule protein interactions to investigate how the interaction surfaces of the coregulators can be modified due to various ligand binding. This is the first report to describe the recruitment of coregulators to the BPA analogue-AR complexes and to show that the BPA analogues may have the potential to bind at multiple locations on the AR complex.
2. Materials and methods
2.1. Reagents
The synthetic androgen, methyltrienolone (R1881, CASRN: 965-93-5), was purchased from RTI International (Durham, NC). The three EDCs, BPA (bisphenol A, Chemical Abstracts Services Registry Number, CASRN: 80-05-7), BPAF (bisphenol AF, CASRN: 1478- 61-1), BPS (bisphenol S, CASRN: 80-08-1), and the synthetic anti-androgen CPA (cyproterone acetate, CASRN: 2098-66-0) were obtained from the Midwest Research Institute (Kansas City, MO) through a contract with the National Toxicology Program. DHT (5α-dihydrotestosterone, CASRN: 521-18-6) was purchased from Sigma-Aldrich (St. Louis, MO).
2.2. Cell line and tissue culture
Recombinant U2OS (human bone osteosarcoma epithelial) cells stably expressing EGFP-tagged human AR (GenBank Acc. NM_000044) fused to the C-terminus of enhanced green fluorescent protein (EGFP) were purchased from Thermo Fisher Scientific (Pittsburgh, PA). Cells were maintained in phenol red–free Dulbecco’s modified essential medium (DMEM; Gibco, Thermo Fisher) supplemented with 10% fetal bovine serum (FBS; Gemini Bio Products, West Sacramento, CA), 5 mM L-glutamine (Invitrogen, Carlsbad, CA), and streptomycin (Gibco, Thermo Fisher).
2.3.Cell lysate preparation
Snap-frozen cell pellets of U2OS cells stably expressing EGFP-tagged human AR were provided on dry ice by NIH/NCATS and stored at −80°C. Cell pellets were slowly thawed on ice and suspended in ice cold lysis buffer supplemented with Halt protease inhibitor cocktail (Pierce Biotechnology, Rockford, Il). A volume of 150 μl was used for 1x107 cells. The suspended cells were transferred to a PTFE membrane and directly placed in a shaking container containing a Tungsten sphere. Thereafter, the cells were snap-frozen by submerging the container in liquid nitrogen. Mechanical lysis was performed for 60 seconds using a Mikro-Dismembrator S (Sartorius) at a frequency of 1000 min−1. The shaking step was repeated to ensure complete visual pulverization of the frozen cell suspension. The container was thawed at room temperature and centrifuged for 3 minutes at 1250 x g. The supernatant was transferred to a fresh Eppendorf tube on ice and centrifuged for 45 minutes at 16000 x g at 4°C to remove any remaining insoluble material. The supernatant was divided in aliquots and stored at -80°C. A Bradford assay was used to quantify the protein concentration of the cell lysates and a GFP ELISA was performed to identify the presence of the AR protein (Abcam, Cambridge, MA).
2.4. Microarray Assay for Real-time Coregulator-Nuclear Receptor Interaction (MARCoNI)
Ligand-modulated coregulator interactions of the EGFP-tagged AR in the lysates was assessed using a PamChip® plate (PamGene International B.V. Wolvenhoek, Netherland), which allowed for the comparison of 96 identical peptide microarrays in parallel (Aarts et al. 2013; Houtman et al. 2012; Koppen et al. 2009). Each array, which included 154 coregulator-derived motifs (PamChip #88101, PamGene), was incubated in the assay mixture containing the lysate and EGFP-AR. The detection of the antibody was measured in the absence or presence of the compound(s). Assay mixtures (in PBS, 1% BSA, 0.25% v/v TWEEN-20) were prepared on ice and contained an empirically optimized amount of lysate, 25 nM of Alexa488-conjugated GFP-antibody (Invitrogen) and 0.2 mM test compound (pre-diluted in DMSO with a final assay concentration of 2% (v/v). An aliquot of 25 μl was used from all conditions and were analyzed using multiple technical replicates (arrays). Incubation was performed in a fully automated microarray processing platform, PamStation®96 (PamGene) at 20°C for 80 cycles (two cycles per minute). After removal of the unbound receptor using 25 μl TBS to wash each array, the image was taken by the CCD camera from PamStation®96. Tiff images were analyzed for quantification of NR binding using Bionavigator software (PamGene). Data analysis and visualization were performed by R software ([Anonymous] 1980).
2.5. MD simulations
Starting structures of human AR with various ligands (DHT, R1881, BPA, BPAF, and BPS) bound at the ligand binding site were modeled using the available X-ray crystal structure of the LBD of human AR with DHT bound at the ligand binding site (pdb ID: 3L3X). The peptide mimicking the SRC3 coactivator bound to the AR in the X-ray crystal structure was removed. R1881, BPA, and its analogues were placed in the binding site using the OpenEye software suite. Protons were added and the resulting complex structures were solvated in a box of water for each system. The amount of water molecules varied from 10000 to 15000 for these systems. Various ligands such as tolfenamic acid (that were similar in structure with two aromatic rings to BPA) were identified that bind to the AF2 and BF3 sites on the AR in the X-ray structures (Estebanez-Perpina et al. 2007). Using the OpenEye software, ligands were placed in the AF2 and BF3 sites (one ligand occupying one site at a time) while DHT was occupying the actual ligand binding site on the AR. These systems were prepared for molecular dynamic calculations by solvating them in water boxes as described above. Prior to equilibration, each system was subjected to the following steps: (1) a nanosecond of constrained molecular dynamics with the peptide and the ligand constrained to the original position with a force constant of 10 kcal/mol/nm, (2) minimization, (3) low temperature constant pressure MD simulation to assure a reasonable starting density, (4) minimization, (5) stepwise heating MD at constant volume, and (6) a constant volume MD run for another 9 nanoseconds. All final unconstrained trajectories were calculated at 300K under constant volume MD (50 ns total dynamics at 1fs time step) using PMEMD from the Amber.14 program to accommodate long-range interactions. The amino-acid parameters were taken from the Amber SB14 force field. The charges of atomic positions of the ligands were derived using the ChelpG scheme with the 6–31g* basis, set at the B3LYP level. The other ligand force field parameters were generic parameters listed in the Amber force field. Additional MD simulations were performed with the same protocol for each ligand in water to establish the solvation behavior and energies of each ligand. A 50 ns MD simulation was performed for ligand-free human AR to obtain a reference state under the same conditions for comparison.
3. Results
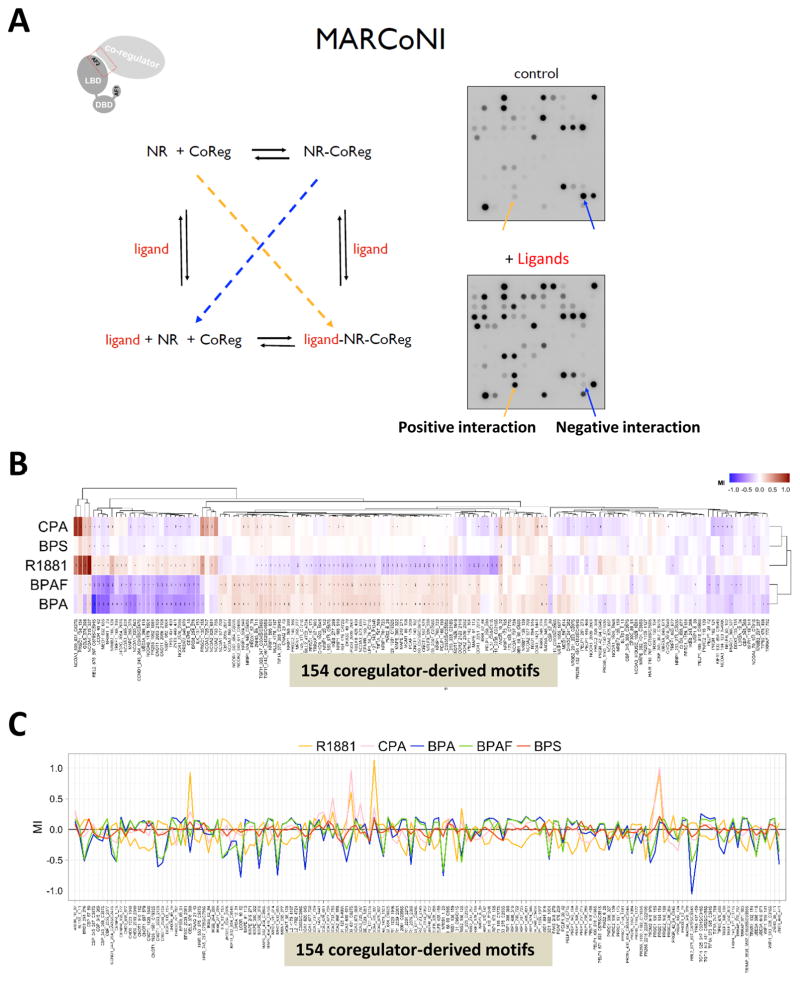
Previous studies showed that BPA represses androgen-induced reporter activity in transiently transfected CV-1 cells through multiple molecular mechanisms (Teng et al. 2013). To understand the coregulator interactions with BPA analogues bound to the AR, a MARCoNI assay was applied. This assay was used to mimic NR-coregulator interactions in vitro (Wang et al. 2013). The MARCoNI assay consists of 154 immobilized coregulator-derived binding peptides, including 129 highly conserved helical LxxLL (NR-box, coactivators), 3 LXXXIXXXL (CoRNR-box, corepressors), and 22 random motifs (McKenna et al. 1999). A working model of the MARCoNI assay is shown in Fig. 1A. This system detects the positive interactions (increased binding, yellow arrow) and negative interactions (decreased binding, blue arrow) while the peptides are in the ligand-AR complex (comparing the spot intensities of control and + ligand in Fig. 1A, right panels).
Fig. 1.
The profiles of coregulator peptides interacting with AR (bound to R1881, CPA, BPA, BPAF, and BPS) using a MARCoNI assay. (A) Working model of the MARCoNI assay. (B) The heatmap of the peptide screening (Red color bar shows positive interactions and blue color bar shows negative interactions) (C) Modulation Index (MI) in response to R1881, CPA, BPA, BPAF, and BPS binding to the AR.
To investigate and compare the mechanisms of action by which compounds modulate AR activity, the AR binding profile was quantified in the presence of R1881 (an AR agonist, a synthetic androgen), CPA (an AR antagonist), BPA, BPAF, BPS, and the solvent control (2% DMSO). Compound response profiles (i.e. compound-induced log-fold change of AR binding modulation index (MI)) were subjected to hierarchical clustering and demonstrated clear differences between compounds and their abilities to modify AR conformation, as shown by the heat-map (Fig. 1B). The red color depicts the positive interactions (increased binding: darker red is stronger and lighter red is weaker) and the blue color depicts the negative interactions (decreased binding: darker blue is stronger and lighter blue is weaker). Clustering of the motifs of groups 1, 2, and 3 clearly showed that BPA and BPAF have similar profiles to CPA (antagonist) but not to R1881 (agonist). Fig. 1C presents the quantification of binding with a plus (+) or minus (−). R1881, CPA, and BPA analogues bound AR differently and modulate it either positively or negatively, as indicated in both heat-map and MI analyses. However, the antagonist profile of CPA-bound AR has more in common with that of R1881- bound AR.
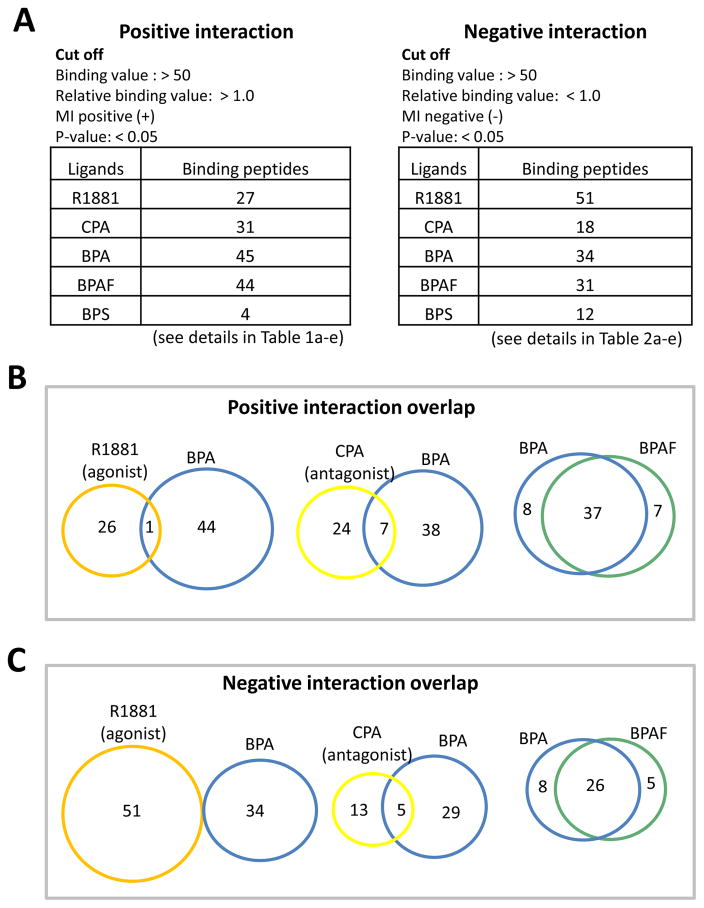
With a >50 binding value cut off for positive interactions, we found 27 peptides (from 23 different coregulators) for R1881-bound AR, 31 peptides (from 22 different coregulators) for CPA-bound AR, 45 peptides (from 28 different coregulators) for BPA-bound AR, and 44 peptides (from 30 different coregulators) for BPAF-bound AR, which all had increased binding values. However, only 4 peptides for BPS-bound AR showed increased binding values (Fig. 2A). It was also discovered that reduced binding (negative interactions) occurred in R1881- bound AR (51 peptides from 28 different coregulators), CPA-bound AR (18 peptides from 12 different coregulators), BPA-bound AR (34 peptides from 30 different coregulators), BPAFbound AR (31 peptides from 28 different coregulators), and BPS-bound AR (12 peptides from 9 different coregulators) (Fig. 2B). A complete list of the peptides that bind to the R1881, CPA, BPA, BPAF, and BPS in the AR complexes, either positively or negatively, is shown in Tables 1 and 2. This data indicates that the coregulators recruited by the BPA-, BPAF-, and BPS-AR complexes are distinct from R1881-bound AR and differ from the known androgen antagonist CPA-bound AR.
Fig. 2.
Comparison of the coregulatory-derived binding peptides when ligands R1881, CPA, BPA, BPAF, and BPS are bound to the AR. (A) Positive interactions. (B) Negative interactions. (C) Overlapping positive interactions. (D) Overlapping negative interactions.
Table 1a.
27 peptides that positively interact with R1881
| Peptide names (27) | Relative binding value | Description of the proteins (23) |
|---|---|---|
| ANDR_10_32 | 1.94 | AR: Androgen receptor |
|
| ||
| BRD8_254_276 | 1.43 | p120: Thyroid hormone-receptor conceiving protein |
|
| ||
| CBP_57_80 | 1.46 | CBP: CREB-binding protein |
|
| ||
| CNOT1_2086_2108 | 1.25 | NOT1H: CCR4-Not transcription complex |
|
| ||
| EP300_69_91 | 1.33 | p300: Histone acetyltransferase p300 |
|
| ||
| GELS_376_398 | 8.46 | AGEL: Gelsolin precursor |
|
| ||
| JHD2C_2054_2076 | 1.39 | TRIP-8: Thyroidreceptor-interacting protein 8 |
|
| ||
| LCOR_40_62 | 1.31 | LCoR: Ligand-dependent corecpressor |
|
| ||
| MED1_632_655 | 1.36 | PBP: PPAR-binding protein |
|
| ||
| NCOA1_620_643 | 1.29 | |
| NCOA1_737_759 | 1.53 | SRC-1/RIP160: Nuclear receptor co-activator 1 |
|
| ||
| NCOA2_628_651 | 1.71 | SRC-2/NCoA-2: Nuclear receptor co-activator 2 |
|
| ||
| NCOA3_609_631 | 1.41 | |
| NCOA3_609_631_C627S | 4.10 | SRC-3/NCoA-3 |
|
| ||
| NCOA6_1479_1501 | 1.22 | NCoA-6: Nuclear receptor co-activator 6 |
|
| ||
| NCOR1_2376_2398 | 1.30 | N-CoR1: Nuclear receptor corepressor 1 |
|
| ||
| NR0B1_1_23 | 1.28 | |
| NR0B1_68_90_C69S | 1.46 | DAX-1: Nuclear receptor subfamily member 1 |
|
| ||
| NR0B2_106_128 | 2.18 | DAX-2: Nuclear receptor subfamily member 2 |
|
| ||
| PR285_1062_1084 | 1.23 | PDIP1: PPAR-gamma DBD-interacting protein 1 |
|
| ||
| PRDM2_948_970 | 1.33 | PRDM2: PR domain zine finger protein 2 |
|
| ||
| PRGC1_130_155 | 2.92 | |
| PRGC1_134_154 | 8.44 | PGC1α: PPAR-g co-activator 1-alpha |
|
| ||
| RBL2_875_897_C879S/C894S | 1.29 | p130/RBR-2: Retinoblastome-like protein 2 |
|
| ||
| TF65_437_459 | 1.26 | NF-kB p65: Transcription factor p65 |
|
| ||
| TREF1_168_190 | 1.43 | Trep-132: Transcriptional-regulating factor 1 |
|
| ||
| ZNT9_449_471 | 1.27 | ZnT-9: Zinc transporter 9 |
Table 2b.
18 peptides that negatively interact with CPA
| Peptide names (18) | Relative binding value | Description of the proteins (12) |
|---|---|---|
| IKBB_277_299 | 0.58 | IkB-B/TRIP-9: I-kappa-B-eta/TR-interacting protein 9 |
|
| ||
| MEN1_255_277 | 0.89 | Menin |
|
| ||
| NCOR1_2376_2398 | 0.90 | N-CoR1: Nuclear receptor corepressor 1 |
|
| ||
| NR0B1_1_23 | 0.25 | SHP: Nuclear receptor subfamily member 2 |
|
| ||
| NRIP1_121_143_P124R | 0.32 | |
| NRIP1_488_510 | 0.37 | |
| NRIP1_8_30 | 0.39 | |
| NRIP1_805_831 | 0.14 | NR-interacting protein 1/nuclear factor RIP140 |
|
| ||
| PELP1_142_164 | 0.34 | |
| PELP1_20_42 | 0.97 | |
| PELP1_571_593_C575S/C581S | 0.11 | Proline-, glutamic acid- and leucine-rich protein 1 |
|
| ||
| PPRC1_151_173 | 0.38 | PRC: PGC-1-related coactivator |
|
| ||
| PR285_1062_1084 | 0.26 | PPAR-gamma DBD-interacting protein 1 |
|
| ||
| PRGR_102_124 | 0.57 | |
| PRGR_42_64_C64S | 0.84 | Progesterone receptor |
|
| ||
| TGFI1_325_347_C334S/C346S | 0.11 | Transforming growth factor beta-1-induced transcript 1 |
|
| ||
| TREF1_168_190 | 0.30 | Transcriptional-regulating factor 1 |
|
| ||
| UBE3A_649_671 | 0.93 | Ubiquitin-protein ligase E3A |
To further understand the different effects of EDCs on AR function, the binding peptides were compared between the BPA, BPAF, and BPS systems. Among the positive bindings, R1881-bound AR robustly recruited peptides from many of the known coactivators, such as Gelsolin (Nishimura et al. 2003), NCoA, and PGC-1α (review and references therein (van de Wijngaart et al. 2012). These binding values are several-fold higher than peptides derived from other coactivators (Table 1a). There were only two positively recruited peptides derived from the corepressor proteins, LCOR_40_62 and NCOR1_2376_2398. Interestingly, many of the coactivator peptides were also recruited to the CPA-AR complex in regarding to positive interactions (Table 1b). Analysis of the peptides with positive interactions showed 3 peptides overlapping between 27 R1881 and 31 CPA (Fig. 2C, 1st Venn diagram) whereas 9 peptides were shown to be overlapping between 51 R1881 and 18 CPA in the negative interaction (Fig. 2D, 1st Venn diagram). These results suggest that the assay system can identify the differences between agonists (R1881) and the antagonists (CPA). When comparing the overlapping peptides between R1881- and BPA-bound AR, only one peptide was identified in the positive interactions and none in the negative interactions (Fig. 2C and 2D, 2nd Venn diagram). There were more interactions between CPA and BPA in both positive (7 overlapping) and negative (5 overlapping) interactions (Fig. 2C and 2D, 3rd Venn diagram). This data suggests that BPA could act as an antagonist to the AR. Over 80% of the peptides interfiacting with BPA- or BPAF-bound AR overlapped in either positively or negatively (Fig. 2C and 2D, 4th Venn diagram), suggesting that similar coregulators were recruited by BPA and BPAF to the AR complex. In this binding profile, about 27% of the peptides from known corepressors, such as NCoR and RIP140, were positively bound to the BPA-AR or BPAF-AR complex (Tables 1c and 1d). But, the BPS-AR complex recruited only 4 peptides (Table 1e).
Table 1b.
31 peptides that positively interact with CPA
| Peptide names (31) | Relative binding value | Description of the proteins (22) |
|---|---|---|
| ANDR_10_32 | 2.05 | AR: Androgen receptor |
|
| ||
| BL1S1_1_11 | 19.84 | BLOC-1: BLOC1subunit 1/GCN5-like protein |
|
| ||
| BRD8_254_276 | 5.5 | p120: Thyroid hormone-receptor conceiving protein |
|
| ||
| CBP_2055_2077 | 6.6 | |
| CBP_345_368_C367S | 13.7 | CBP: CREB-binding protein |
|
| ||
| CENPR_159_177 | 1.25 | CENP-R: Centromere protein R |
|
| ||
| CHD9_1023_1045 | 1.17 | |
| CHD9_2018_2040 | 1.17 | CHD-9: Chromodomain-helicase-DNA-binding protein 9 |
|
| ||
| CNOT1_1929_1951_C1932S | 1.23 | |
| CNOT1_2083_2105 | 2.57 | |
| CNOT1_2086_2108 | 2.89 | |
| CNOT1_557_579 | 1.6 | NOT1: CCR4-NOT transcription complex subunit 1 |
|
| ||
| DHX30_241_262 | 7.14 | DHX30: Putative ATP-dependent RNA helicase |
|
| ||
| EP300_2039_2061 | 1.64 | |
| EP300_69_91 | 2.75 | p300: Histone acetyltransferase p300 |
|
| ||
| GNAQ_21_43 | 1.9 | Guanine nucleotide-binding protein G subunit alpha |
|
| ||
| IKBB_244_266 | 4.01 | |
| IKBB_62_84 | 3.42 | IkB-B/TRIP-9: I-kappa-B-eta/TR-interacting protein 9 |
|
| ||
| KIF11_832_854_C854S | 2.4 | KIF11: Kinesin-like protein KIF11 |
|
| ||
| MAPE_91_113 | 1.43 | OIP4: Melanoma antigen preferentially expressed in tumor |
|
| ||
| MED1_591_614 | 4.04 | PBP: PPAR-binding protein |
|
| ||
| MLL2_4702_4724 | 1.57 | MLL2: Mixed-lineage leukemia protein 2 |
|
| ||
| MTA1S_388_410_C393S/C396S | 5.05 | MTA1: Metastasis-associated protein |
|
| ||
| NCOA2_628_651 | 1.6 | |
| NCOA2_677_700 | 4.24 | SRC-2/NCoA-2: Nuclear receptor co-activator 2 |
|
| ||
| NCOA3_609_631 | 1.18 | |
| NCOA3_MOUSE_1029_1051 | 3.51 | SRC-3/NCoA-3: Nuclear receptor co-activator 3 |
|
| ||
| NR0B1_136_159 | 2.63 | DAX-1: Nuclear receptor subfamily member 1 |
|
| ||
| NR0B2_237_257 | 1.16 | SHP: Nuclear receptor subfamily member 2 |
|
| ||
| NRBF2_128_150 | 2.2 | NRBF2: Nuclear receptor-binding factor 2 |
|
| ||
| PNRC2_118_139 | 1.26 | PNRC2: Proline-rich nuclear receptor coactivator 2 |
Table 1c.
45 peptides that positively interact with BPA
| Peptide names (45) | Relative binding value | Description of the proteins (28) |
|---|---|---|
| ANDR_10_32 | 1.36 | AR: Androgen receptor |
|
| ||
| CENPR_159_177 | 1.58 | CENP-R: Centromere protein R |
|
| ||
| CHD9_1023_1045 | 1.41 | CHD-9: Chromodomain-helicase-DNA-binding protein 9 |
|
| ||
| CNOT1_140_162 | 1.30 | |
| CNOT1_1929_1951_C1932S | 1.29 | |
| CNOT1_557_579 | 1.39 | NOT1: CCR4-NOT transcription complex subunit 1 |
|
| ||
| DHX30_49_70 | 1.55 | DHX30: Putative ATP-dependent RNA helicase |
|
| ||
| EP300_2039_2061 | 1.58 | p300: Histone acetyltransferase p300 |
|
| ||
| GNAQ_21_43 | 1.40 | Guanine nucleotide-binding protein G subunit alpha |
|
| ||
| HAIR_553_575_C567S | 1.38 | Protein hairless |
|
| ||
| IKBB_277_299 | 1.45 | IkB-B/TRIP-9: I-kappa-B-eta/TR-interacting protein 9 |
|
| ||
| MAPE_249_271 | 1.32 | |
| MAPE_300_322 | 1.25 | Melanoma antigen preferentially expressed in tumor |
|
| ||
| MGMT_86_108 | 1.29 | Methylated-DNA-protein-cysteine methytransferase |
|
| ||
| MLL2_4175_4197 | 1.54 | Mixed-lineage leukemia protein 2 |
|
| ||
| NCOA1_677_700 | 1.32 | SRC-1/RIP160: Nuclear receptor co-activator 1 |
|
| ||
| NCOA3_673_695 | 1.50 | SRC-3/NCoA-3: Nuclear receptor co-activator 1 |
|
| ||
| NCOA4_315_337 | 1.56 | NCoA-4: Nuclear receptor co-activator 4 |
|
| ||
| NCOR1_2039_2061_C2056S | 1.43 | |
| NCOR1_2251_2273 | 1.43 | |
| NCOR1_662_684_C662S | 1.60 | N-CoR1: Nuclear receptor corepressor 1 |
|
| ||
| NCOR2_649_671_C649S | 1.60 | N-CoR2: Nuclear receptor corepressor 2 |
|
| ||
| NELFB_328_350 | 1.46 | |
| NELFB_428_450 | 1.55 | NELF-B: Negative elongation factor B |
|
| ||
| NRIP1_121_143_P124R | 1.53 | |
| NRIP1_368_390 | 1.50 | |
| NRIP1_488_510 | 1.44 | |
| NRIP1_700_722 | 1.38 | |
| NRIP1_701_723 | 1.29 | |
| NRIP1_805_831 | 1.63 | |
| NRIP1_924_946 | 1.40 | |
| NRIP1_924_946_C945S | 1.38 | Nuclear receptor-interacting protein 1/nuclear factor RIP140 |
|
| ||
| PAK6_248_270 | 1.38 | PAK6: p21-activatedkinase 6 |
|
| ||
| PELP1_168_190 | 1.39 | |
| PELP1_56_78_C71S | 1.52 | |
| PELP1_571_593_C575S/C581S | 1.22 | Proline-, glutamic acid- and leucine-rich protein 1 |
|
| ||
| PPRC1_151_173 | 1.20 | PRC: PGC-1-related coactivator |
|
| ||
| RAD9A_348_370 | 1.44 | Cell cycle checkpoint control protein RAD9A |
|
| ||
| TGFI1_325_347_C334S/C346S | 1.80 | |
| TGFI1_443_461_C452S/C455S | 1.59 | Transforming growth factor beta-1-induced transcript 1 |
|
| ||
| TIF1A_373_395_C394S | 1.48 | |
| TIF1A_747_769 | 1.55 | TIF1-alpha: transcription intermediary factor 1-alpha |
|
| ||
| TRIP4_149_171_C171S | 1.33 | TRIP4: thyroid receptor-interacting protein 4 |
|
| ||
| TRRAP_3535_3557_C3535S/C3555S | 1.52 | Transcription domain-associated protein |
|
| ||
| ZNHI3_89_111 | 1.53 | Zinc finger HIT domain-containing protein 3 |
Table 1d.
44 peptides that positively interact with BPAF
| Peptide names (44) | Relative binding value | Description of the proteins (30) |
|---|---|---|
| ANDR_10_32 | 1.47 | AR: Androgen receptor |
|
| ||
| CBP_2055_2077 | 1.50 | CBP: CREB-binding protein |
|
| ||
| CENPR_159_177 | 1.46 | CENP-R: Centromere protein R |
|
| ||
| CHD9_1023_1045 | 1.41 | CHD-9: Chromodomain-helicase-DNA-binding protein 9 |
|
| ||
| CNOT1_140_162 | 1.33 | |
| CNOT1_1929_1951_C1932S | 1.29 | CCR4-NOT transcription complex subunit 1 |
|
| ||
| DHX30_49_70 | 1.54 | Putative ATP-dependent RNA helicase DHX30 |
|
| ||
| EP300_2039_2061 | 1.53 | p300: Histone acetyltransferase p300 |
|
| ||
| GNAQ_21_43 | 1.42 | Guanine nucleotide-binding protein G subunit alpha |
|
| ||
| HAIR_553_575_C567S | 1.30 | Protein hairless |
|
| ||
| IKBB_277_299 | 1.46 | IkB-B/TRIP-9: I-kappa-B-eta/TR-interacting protein 9 |
|
| ||
| MAPE_454_476_C472S | 1.50 | Melanoma antigen preferentially expressed in tumor |
|
| ||
| MEN1_255_277 | 1.19 | Menin |
|
| ||
| MGMT_86_108 | 1.30 | Methylated-DNA-protein-cysteine methytransferase |
|
| ||
| MLL2_4175_4197 | 1.50 | |
| MLL2_4702_4724 | 1.28 | Mixed-lineage leukemia protein 2 |
|
| ||
| NCOA1_677_700 | 1.41 | SRC-1/RIP160: Nuclear receptor co-activator 1 |
|
| ||
| NCOA3_673_695 | 1.40 | SRC-3/NCoA-3: Nuclear receptor co-activator 3 |
|
| ||
| NCOA4_315_337 | 1.36 | NCoA-4 |
|
| ||
| NCOR1_2039_2061_C2056S | 1.51 | |
| NCOR1_2251_2273 | 1.47 | |
| NCOR1_662_684_C662S | 1.59 | N-CoR1: Nuclear receptor corepressor 1 |
|
| ||
| NCOR2_649_671_C649S | 1.54 | N-CoR2: Nuclear receptor corepressor 2 |
|
| ||
| NELFB_328_350 | 1.30 | NELF-B: Negative elongation factor B |
| NRIP1_121_143_P124R | 1.47 | |
|
| ||
| NRIP1_368_390 | 1.44 | |
| NRIP1_488_510 | 1.44 | |
| NRIP1_700_722 | 1.37 | |
| NRIP1_701_723 | 1.23 | |
| NRIP1_805_831 | 1.60 | |
| NRIP1_924_946 | 1.39 | |
| NRIP1_924_946_C945S | 1.40 | Nuclear receptor-interacting protein 1/nuclear factor RIP140 |
|
| ||
| PAK6_248_270 | 1.40 | PAK6: p21-activatedkinase 6 |
|
| ||
| PELP1_142_164 | 1.26 | |
| PELP1_168_190 | 1.36 | |
| PELP1_56_78_C71S | 1.50 | Proline-, glutamic acid- and leucine-rich protein 1 |
|
| ||
| PIAS2_6_28 | 1.24 | Protein inhibitor of activated STAT2 |
|
| ||
| RAD9A_348_370 | 1.44 | Cell cycle checkpoint control protein RAD9A |
| TGFI1_325_347_C334S/C346S | 1.48 | |
| TGFI1_443_461_C452S/C455S | 1.46 | Transforming growth factor beta-1-induced transcript 1 |
|
| ||
| TIF1A_747_769 | 1.45 | TIF1-alpha: transcription intermediary factor 1-alpha |
|
| ||
| TRIP4_149_171_C171S | 1.52 | TRIP4: thyroid receptor-interacting protein 4 |
|
| ||
| TRRAP_3535_3557_C3535S/C3555S | 1.46 | Transcription domain-associated protein |
|
| ||
| ZNHI3_89_111 | 1.53 | Zinc finger HIT domain-containing protein 3 |
Table 1e.
4 peptides that positively interact with BPS
| Peptide names (4) | Relative binding value | Description of the proteins (4) |
|---|---|---|
| ANDR_10_32 | 1.22 | AR: Androgen receptor |
| CBP_57_80 | 1.50 | CBP: CREB-binding protein |
| NCOA4_315_337 | 1.53 | NCoA-4: Nuclear receptor coactivator 4 |
| TGFI1_325_347_C334S/C346S | 1.12 | Hic-5: Hydrogen peroxide-inducible clone 5 protein |
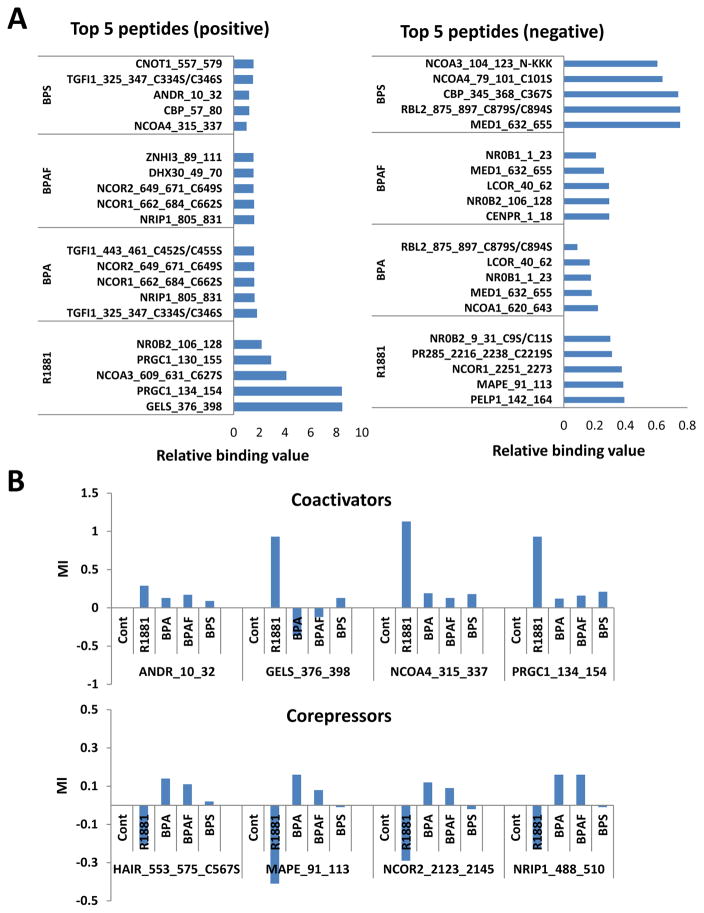
Since negative interactions indicate reduced binding, many of the peptides from known corepressors appear in the R1881 negative binding peptide list. However, the opposite is true for the BPA and BPAF lists, which include the coactivator-derived peptides (compare Table 2a–2e). Comparing the top 5 peptides that positively or negatively interacted with each compound, it not only showed the difference in binding strength, but also displayed the type of peptide from which the specific-coregulator protein was derived (Fig. 3A). To further illustrate the differences in peptides that were recruited to the AR complexes (R1881, CPA, BPA, BPAF, and BPS), we plotted the relative binding strengths of the known AR coactivators and corepressors (Fig. 3B). R1881 strongly bound coactivators but this was not observed in the BPA analogues in the AR complexes (Fig. 3B, upper panel). The opposite binding trends for R1881 and the BPA analogues were observed for corepressors (Fig. 3B, lower panel). These results suggest that the BPA analogue-AR complexes assimilate into a molecular structure that is in favor of the corepressor binding and provide a possible explanation for the antagonist action of these compounds. Since it was reported that certain compounds can interact with alternate sites on the AR surface, in the current study we attempted to answer the question: “Can BPA analogues alter coactivator or corepressor interactions with the AR by binding to alternate sites such as AF2 and BF3 on the AR molecular surface?” The AF2 surface that has been targeted in drug development (Feng et al. 1998) comprises the residues from helices 3, 5, and 12. The BF3 surface is situated next to the AF2 surface and residues Arg-840, Asn-727, Phe-826, and Glu- 829 are mainly involved in creating this binding site. Chemical binding to the BF3 site introduces disorder to coregulator peptides bound at the AF2 site (Estebanez-Perpina et al. 2007). As described in the methods section, multiple simulations were completed with DHT, R1881, BPA, BPAF, and BPS present in the natural ligand-binding site (LBS). Additional simulations were performed with all ligands to identify their binding affinity to the BF3 and AF2 sites, located on the AR, while DHT was bound at the natural LBS. Root mean square deviations (RMSDs) of the backbone protein atoms were used in MD simulations to measure the stability and protein refolding due to ligand binding. The stabilities of the MD simulations were established by analyzing RMSDs of the backbone AR atoms and the RMSDs were calculated from the evenly spaced 500 configurations collected from the last 40ns of each trajectory. The X-ray crystal structure of the AR was used as the reference configuration and the results are displayed in Supplemental Figs. 1–3. By observing the figs, all RMSDs are well within the range of fluctuations for stable protein conformations of this size once they were subjected to solution simulations. The results display no drastic shifts, indicating rather stable ligand bound conformations and no large conformation changes (or refolding events) in the protein associated with ligand binding at surface sites. Therefore, it is safe to conclude that the MD simulations created ensembles of reasonably stable ligand-bound conformations of the AR while interacting with various ligands at different locations on the surface of the receptor. Using these configurations, the binding affinities can be estimated for each ligand at the natural LBS, including the selected alternate sites, AF2 and BF3, on the AR. The interaction energies of each ligand with water molecules (once each ligand was individually solvated in water) were calculated to facilitate the calculation for binding affinities. The summary of binding enthalpies along with their fluctuations were given in Table 3. Comparison of the ligand binding energies of the ligands with the AR in solution with the ligand-solvent interaction energies provided a measure to obtain additional stabilization/or destabilization energy of the ligand when bound to the AR in solution, giving rise to a way in estimating binding affinities.
Table 2a.
51 peptides that negatively interact with R1881
| Peptide names (51) | Relative binding value | Description of the proteins (28) |
|---|---|---|
| CENPR_159_177 | 0.60 | CENP-R: Centromere protein R |
|
| ||
| CHD9_1023_1045 | 0.54 | CHD-9: Chromodomain-helicase-DNA-binding protein 9 |
|
| ||
| CNOT1_140_162 | 0.49 | |
| CNOT1_1626_1648 | 0.44 | |
| CNOT1_1929_1951_C1932S | 0.51 | |
| CNOT1_557_579 | 0.52 | CCR4-NOT transcription complex subunit 1 |
|
| ||
| DHX30_49_70 | 0.55 | Putative ATP-dependent RNA helicase DHX30 |
|
| ||
| EP300_2039_2061 | 0.54 | p300: Histone acetyltransferase p300 |
|
| ||
| GNAQ_21_43 | 0.70 | Guanine nucleotide-binding protein G subunit alpha |
|
| ||
| HAIR_553_575_C567S | 0.61 | Protein hairless |
|
| ||
| IKBB_277_299 | 0.60 | |
| IKBB_62_84 | 0.42 | IkB-B/TRIP-9: I-kappa-B-eta/TR-interacting protein 9 |
|
| ||
| MAPE_249_271 | 0.57 | |
| MAPE_300_322 | 0.54 | |
| MAPE_454_476_C472S | 0.55 | |
| MAPE_91_113 | 0.38 | Melanoma antigen preferentially expressed in tumor |
|
| ||
| MEN1_255_277 | 0.61 | Menin |
|
| ||
| MGMT_86_108 | 0.56 | Methylated-DNA-protein-cysteine methytransferase |
|
| ||
| MLL2_4175_4197 | 0.72 | |
| MLL2_4702_4724 | 0.71 | Mixed-lineage leukemia protein 2 |
|
| ||
| NCOA3_673_695 | 0.62 | SRC-3/NCoA-3: Nuclear receptor co-activator 3 |
|
| ||
| NCOR1_2039_2061_C2056S | 0.51 | |
| NCOR1_2251_2273 | 0.38 | N-CoR1: Nuclear receptor corepressor 1 |
| NCOR2_2123_2145 | 0.51 | N-CoR2: Nuclear receptor corepressor 2 |
|
| ||
| NELFB_328_350 | 0.47 | |
| NELFB_428_450 | 0.59 | NELF-B: Negative elongation factor B |
|
| ||
| NR0B2_9_31_C9S/C11S | 0.30 | SHP: Nuclear receptor subfamily member 2 |
|
| ||
| NRIP1_120_142 | 0.57 | |
| NRIP1_121_143_P124R | 0.60 | |
| NRIP1_368_390 | 0.70 | |
| NRIP1_488_510 | 0.59 | |
| NRIP1_700_722 | 0.55 | |
| NRIP1_701_723 | 0.50 | |
| NRIP1_805_831 | 0.66 | Nuclear receptor-interacting protein 1/nuclear factor RIP140 |
|
| ||
| PCAF_178_200 | 0.49 | Histone acetyltransferase PCAF |
|
| ||
| PELP1_142_164 | 0.39 | |
| PELP1_168_190 | 0.51 | |
| PELP1_258_280 | 0.43 | |
| PELP1_446_468 | 0.64 | |
| PELP1_56_78_C71S | 0.61 | |
| PELP1_571_593_C575S/C581S | 0.50 | Proline-, glutamic acid- and leucine-rich protein 1 |
|
| ||
| PIAS2_6_28 | 0.45 | Protein inhibitor of activated STAT2 |
|
| ||
| PPRC1_151_173 | 0.66 | PRC: PGC-1-related coactivator |
|
| ||
| PR285_2216_2238_C2219S | 0.31 | ATP-dependent helicase PRIC285 protein |
|
| ||
| TGFI1_325_347_C334S/C346S | 0.78 | |
| TGFI1_443_461_C452S/C455S | 0.78 | Transforming growth factor beta-1-induced transcript 1 |
|
| ||
| TIF1A_373_395_C394S | 0.68 | TIF1-alpha: Transcription intermediary factor 1-alpha |
|
| ||
| TREF1_850_872 | 0.61 | Transcriptional-regulating factor 1 |
|
| ||
| TRRAP_3535_3557_C3535S/C3555S | 0.44 | |
|
| ||
| TRRAP_971_993 | 0.57 | Transcription domain-associated protein |
|
| ||
| ZNHI3_89_111 | 0.81 | Zinc finger HIT domain-containing protein 3 |
Table 2e.
12 peptides that negatively interact with BPS
| Peptide names (12) | Relative binding value | Description of the proteins (9) |
|---|---|---|
| CBP_345_368 | 0.77 | |
| CBP_345_368_C367S | 0.74 | CBP: CREB-binding protein |
|
| ||
| CNOT1_2083_2105 | 0.81 | |
| CNOT1_2086_2108 | 0.82 | |
| CNOT1_557_579 | 0.88 | NOT1H: CCR4-Not transcription complex |
|
| ||
| MED1_632_655 | 0.75 | PBP: PPAR-binding protein |
|
| ||
| NCOA3_104_123_N-KKK | 0.61 | SRC-3/NCoA-3: Nuclear receptor co-activator 3 |
|
| ||
| NCOA4_79_101_C101S | 0.64 | NCoA-4: Nuclear receptor co-activator 4 |
|
| ||
| NR0B1_1_23 | 0.77 | DAX-1: Nuclear receptor subfamily member 1 |
|
| ||
| RBL2_875_897_C879S/C894S | 0.75 | p130/RBR-2: Retinoblastome-like protein 2 |
|
| ||
| UBE3A_396_418 | 0.80 | Ubiquitin-protein ligase E3A |
|
| ||
| ZNT9_449_471 | 0.80 | Znt-9: Zinc transporter 9 |
Fig. 3.
Primary binding peptides and coregulators. (A) Top 5 positive and negative peptides. (B) Top 5 positive and negative coactivators and corepressors.
Table 3.
Binding enthalpies (kcal/mol) of the ligands on AR calculated using MD simulations
| Ligand | Ligand binding site (LBS) | AF2 site | BF3 site | In solution (water) |
|---|---|---|---|---|
| R1881 | -68.3±2.8 | −59.8±3.8 | −55.2±3.6 | −50.6±4.4 |
| DHT | −67.8±2.9 | −59.6±3.8 | −61.1±4.4 | −53.7±5.1 |
| BPA | −52.3±3.0 | −52.7±3.6 | −53.3±4.7 | −46.4±3.9 |
| BPAF | −54.2±3.4 | −50.1±3.1 | −53.4±4.4 | −49.2±3.7 |
| BPS | −57.7±4.3 | −64.7±5.4 | −65.3±4.4 | −62.2±5.3 |
DHT, a natural ligand of the AR, and R1881, the synthetic counterpart, showed a greater affinity at its natural binding site on the AR when compared to values in solution (Table 3). An extra stability of 14.1 kcal/mol was observed for DHT due to the AR binding at the natural ligand binding site in comparison to the value of R1881 which is about 17.7 kcal/mol. Both BPA and BPAF showed smaller gains of 5.9 and 5.0 kcal/mol due to AR interactions. In contrast, under the current force field used in this study, it was observed that there was a slight energy penalty when BPS occupied the natural ligand-binding site of the AR when compared to being solubilized in water. BPS showed an energy loss of 4.5 kcal/mol if bound at the natural binding site (but this is within the error bars of the calculations). For both DHT and R1881 ligands, the AF2 and BF3 sites were also energetically preferred over the solution, but to a much lesser extent (Table 3) when compared with the natural LBS. However, both BPA and BPAF showed rather similar preferences for all three sites when the stabilization energies were compared. Meanwhile, BPS was seen to preferentially bind to either AF2 or BF3 sites over the natural LBS of the AR.
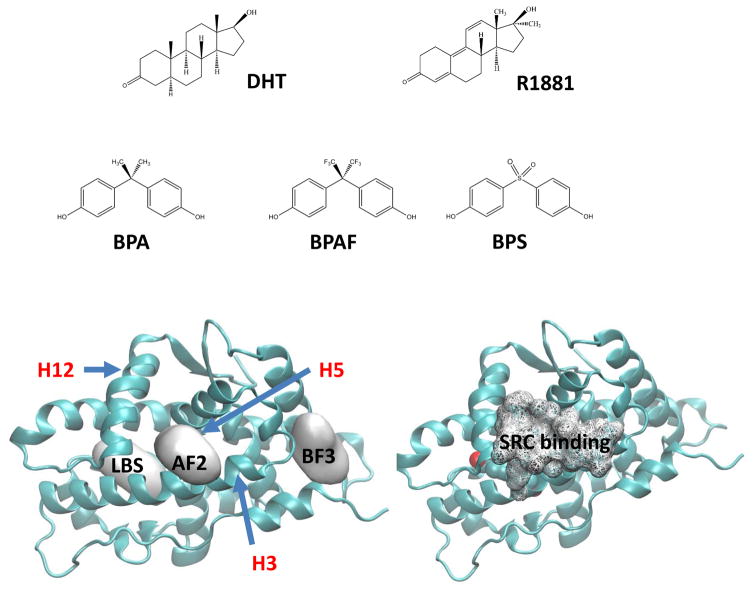
We now turn to the characterization of structures of the AR in the presence of ligands at the natural LBS and two other potential ligand binding sites, AF2 and BF3, described above. The locations of various LBSs on the AR-LBD along with the chemical structures of DHT, R1881, BPA, BPAF, and BPS are presented in Fig. 4. The AF2 site is naturally reserved for the cofactor binding and is a hydrophobic surface located in the area bracketed by Helix 3, 5, and 12. For comparison, the SRC binding (wire frame in the bottom right fig.) on the agonist-bound AR from the X-ray crystal of the pdb ID 3L3X is shown. However, the potential small molecule binding at this site was described in the work of Estebanez-Perpina et. al. (Ref). The BF3 site is found near Helix 1 and can preferentially bind compounds such as Triac with two phenyl rings. It has been reported that mutations of residues such as Gln-670, Ile-672, and Leu- 723 in the Helix 1 were associated with prostate cancer (Buchanan et al. 2001; Shi et al. 2002). According to the observation made by Estebanez-Perpina (Estebanez-Perpina et al. 2007), such mutations could weaken the coactivator binding to the AF2 site due to the large moment involving Arg-726.
Fig. 4.
Chemical structures of the ligands (agonists and BPA analogues) used in the MD simulations. Various binding sites of BPA analogues on AR tested in the present MD study are shown in the bottom-left. For comparison, the SRC binding site of DHT bound AR from the X-ray crystal structure (pdb ID 3L3X) is given in the bottom-right. The three helices, H3, H5 (behind AF2), and H12 that create the binding surface of coregulators are also shown.
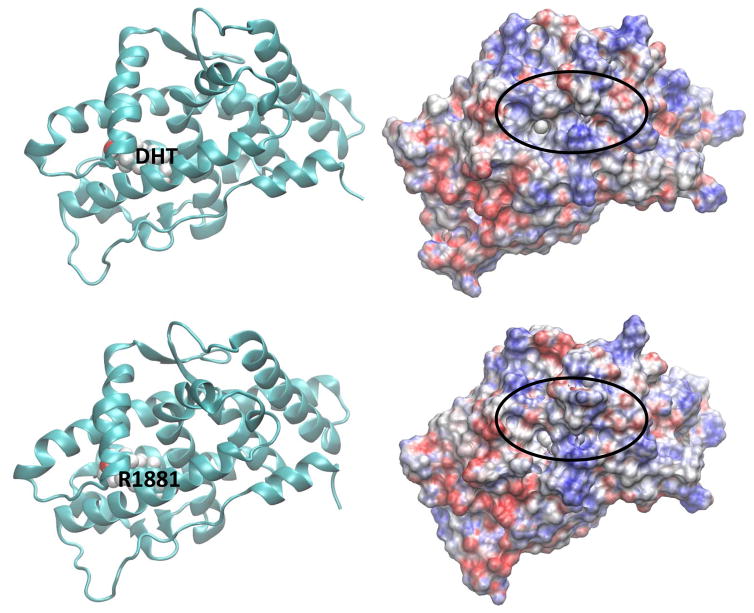
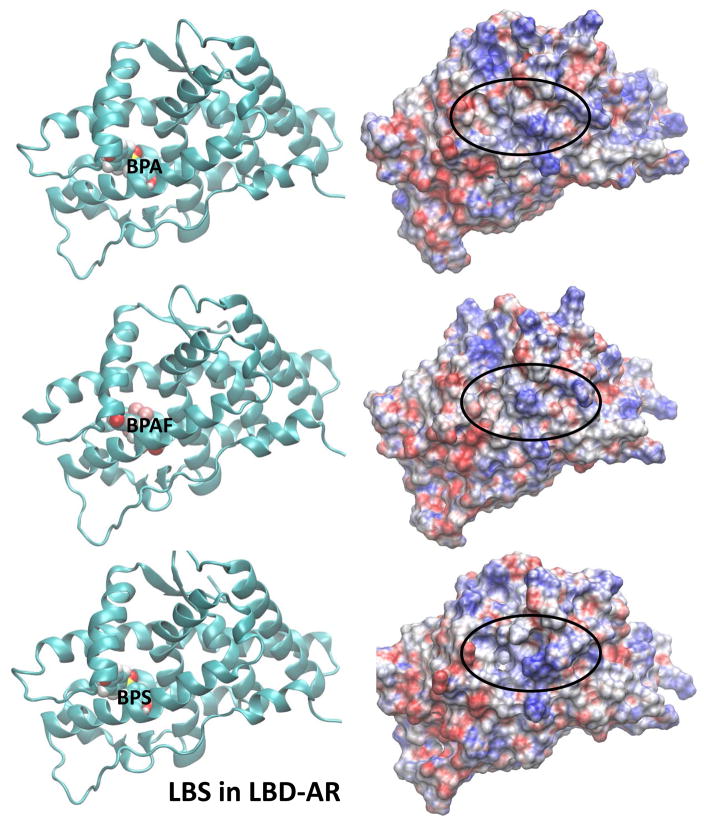
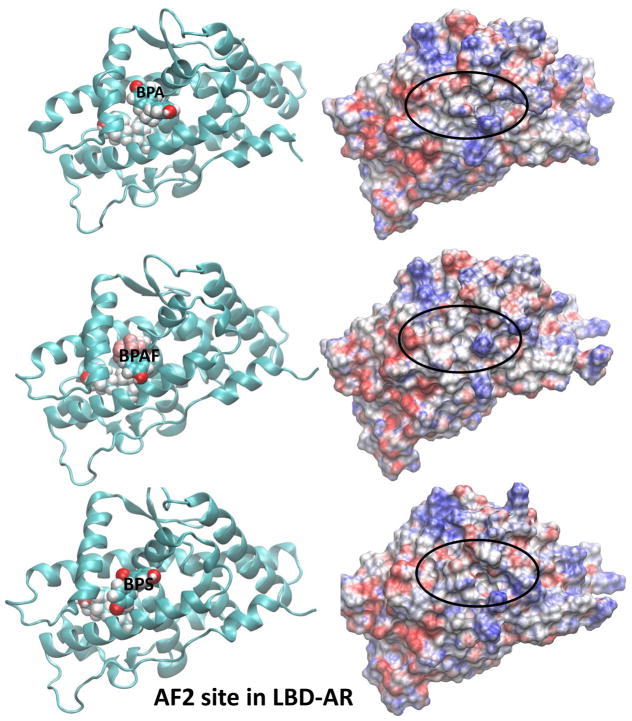
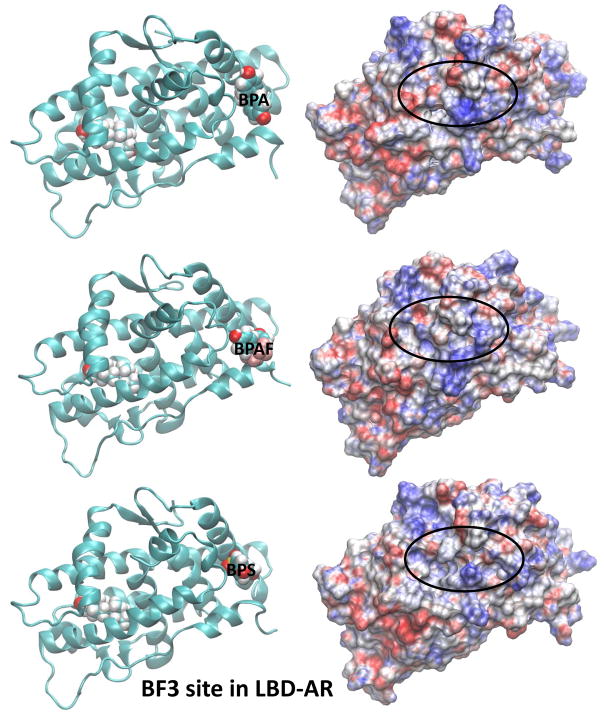
The electrostatic potential calculated at the surface of a molecule is a powerful tool in characterizing how the molecular surface can facilitate (or obstruct) intermolecular association. Areas displayed in red on the molecular surface represent negative electrostatic potentials whereas the blue represent the positive electrostatic potentials. Electrostatic potential surfaces (EPS) of ligand-bound AR along with the representative binding poses for the two agonists, DHT and R1881, located in the LBS of AR are shown in Fig. 5. All AR orientations are similar to facilitate the comparison. The corresponding EPSs and the binding poses for the BPA analogues bound at LBS of the AR are also displayed in Fig. 5. Similar features for structures and electrostatic potentials are observed for all three ligands, if the ligands are located at LBS. However, slight variations in the EPSs can yield significant changes in the populations of cofactors due to variations in their interaction strengths. In this case, we have looked at the competitive ligand binding at the LBS. However, once DHT, a natural ligand, is present at LBS, there is potential for BPA analogues to bind at the AF2 or BF3 site, altering coregulator bindings. The ligand binding poses on electrostatic potential surfaces of BPA analogues at the AF2 site are given in Fig. 6. Note that the SRC binding site partially covered by ligand and therefore the cofactor binding may be inhibited in this situation. Variations in the EPSs are apparent on the SRC binding surface. In Fig. 7, EPSs and ligand binding poses of BPA analogues at the BF3 site are displayed. The ligand binding takes place just right of the SRC binding surface and more changes are brought onto the SRC binding surface in terms of variations in electrostatic potential.
Fig. 5.
Representative solution structures (left) from the latter part of the MD simulations of ligandbound AR where the ligand is at the natural LBS and the corresponding electrostatic potential surface (EPS) (right) of ligand-bound AR. Locations of ligands are labelled. Orientation of LBD-AR in all cases is preserved to facilitate the comparison.
Fig. 6.
Representative solution structures (left) of BPA analogues-bound AR from MD simulations and the corresponding EPS (right) of ligand-AR complexes where the BPA analogues are at theAF2 site of LBD-AR. In all cases, a DHT molecule is placed at LBS.
Fig. 7.
Representative solution structures (left) of BPA analogues-bound AR from MD simulations and the corresponding EPS (right) of ligand-AR complexes where the BPA analogues are at the BF3 site of LBD-AR. In all cases, a DHT molecule is placed at LBS.
The calculated mean position fluctuations (or B-factors) of the residues within the protein were a good measure for the flexibility and variability around the average protein structure. The calculated B-factors (averaged over all backbone atoms and given in Supplemental Fig. 4A, 4C, and 4E) were averaged over sidechain heavy atoms (shown in Supplemental Fig. 4B, 4D, and 4F) for each residue plotted against the residue numbers. At a global level, all profiles were quite similar, but some local variability gave insight into measuring the variations for cofactor selectivity. For example, the residues in the 720–740 range participated in the SRC binding and caused variable fluctuations in the average positions (Supplemental Fig. 4). These variations were observed even when each ligand was bound in the natural ligand binding site, pointing to possible variations of binding affinity of cofactors for each ligand in this area. In fact, the peak is somewhat subdued (Supplementary Fig. 4C) when the ligands were bound to the residues in this area. When the ligands occupied the AF2 site, AR residues: Leu712, V713, V715, V716, M734, I737, Q738, W741, M894, I898, and Q902 were involved in the ligand binding. Meanwhile, residues: F673, P723, G724, N727, L830, N833, Y834, E837, and R840 were involved in ligand binding to the BF3 site. Note that some of the residues listed above participate in cofactor binding. When ligands were bound to either of the surface sites, only one of the ligand oxygens seemed to interact with some of these residues while the other was rather solvent exposed. Specifically, when BPS was at the AF2 site, large variations were observed in the C-terminus of AR.
Discussion
Dynamic communication between the structure of the ligand-bound AR and the regulator recruitment has been found to be crucial for its function (He and Wilson 2003; Xu et al. 2011). In the present study, the coregulator recruitment of BPA analogues was compared with that of R1881 (agonist) and CPA (antagonist) when bound to the AR. We found that there were significant differences in recruited coregulators between BPA analogues and R1881 or CPA. In addition, results from MD simulations of AR with BPA, BPAF and BPS showed that these compounds could interact with previously identified alternative surface binding sites on the AR and potentially modulate the normal AR function.
As of today, molecular mechanisms of antagonism of the AR function are still unclear. Studies from known androgen antagonists, such as cyproterone acetate, flutamide, nilutamide, and bicalutamide have demonstrated that antagonists inhibit AR function by competitive binding with the natural androgens. These antagonists modulate the nuclear translocation and fail to form stable complexes on DNA (McEwan 2013). Moreover, certain antagonists have been known to recruit corepressors (Hodgson et al. 2005). From high throughput screening (HTS) of the Tox21 project and the previous reports that BPA and BPAF block the AR reporter transactivation in cell-based assays (Teng et al. 2013), it was determined that the binding of BPA interferes with AR function in multiple ways. BPA is a known antagonist and competes for AR binding. It is also known to slow down nuclear transportation and form non-functional foci in the nucleus. The coregulator peptide recruitment assay in the present study reinforced the observation that these BPA analogues were unable to activate the AR due to conformational changes that favor corepressor binding. In the positive binding profile, peptides from gelsolin and PGC1α (two strong coactivators of the AR) that interact with the R1881- bound AR with high affinity and were found to have no interaction with BPA analogues. Instead, peptides from corepressors, such as RIP140 and NCoR, bound favorably. In the negative binding profiling, the opposite is true. The R1881-AR complex had reduced binding of the corepressor peptides while the BPA analogue-AR complexes had reduced binding of the coactivator peptides. When comparing the coregulator recruitment profiles of the AR complex with CPA and BPA analogues, discrepancies were seen in the peptide profiling. Therefore, this suggests that the mechanism of antagonism by these compounds may be different. The basis for corepressor recruitment by the BPA-AR complex is unknown and the precise mechanism of antagonism remains to be clarified. In general, distorting the AF-2 site in the AR-LBD complex, with the use of antagonists, interferes with coactivator binding and allows the corepressor that carries larger and bulky corepressor nuclear receptor boxes (CoRNR boxes) to enter the binding grove (Hodgson et al. 2008; Osguthorpe and Hagler 2011). In contrast to other NRs, the AR also requires an amino/carboxy-terminal (N/C) interaction for full activation function (He and Wilson 2003). After androgen binding, the AR-LBD preferentially interacts with its own FxxLF motif at the AF1 site in the NTD region and subsequently recruits coactivators. Whether the FxxLF motif in the gelsolin contributes to the strong binding to the R1881-AR complex remains to be answered. Interestingly, antagonists of the AR did not recruit cofactors that contained the FxxLF motif (Kazmin et al. 2006). Thus, different ligands influenced the recruitment of coregulators that contained various types of peptide motifs. In addition, coregulator recruitment is also affected by the mutations in the AR-LBD region. It was reported that a single point mutation at the AR-LBD (AR-T877A) in the human prostate cancer LNCaP cells and a human prostate cancer patient caused significant increases in the activity of the AR by anti-androgens (Brooke et al. 2008). Such mutations also cause dramatic changes of BPA effects on prostate cancer progression and interfere with the therapeutic intervention treatment (Wetherill et al. 2002). It was also reported that the AR mutant T877A, T877S and H874Y preferentially bind to LxxLL motifs in the presence of the anti-androgen CPA. But binding of FxxLF-like motifs becomes preferred when the anti-androgen hydroxyflumtamide becomes present (Brooke et al. 2008; Xu et al. 2011). Overall, this suggests that distinct receptor conformations, due to different ligands and mutations in the ligand-binding pocket have prominent effects on preferential binding of the LxxLL and FxxLF-like motifs. BPA analogues could bind to alternative AR surface sites and interfere with this critical step. Indeed, the results from the MD simulations showed the potential interactions of BPA analogues with AF2 and BF3 surface sites of the AR. No clear advantage was presented energetically to be bound at the natural ligand binding site as opposed to the two surface sites, for both BPA and BPAF. Meanwhile, in the ligand binding competition study, it was determined that BPS did not significantly compete with R1881 binding to the LBD (Teng et al. 2013). In fact, the present MD calculations predicted that BPS interacted preferentially with the AR at the alternative surface sites than of its nature LBS. These findings may help explain some of the distinct AR-related toxicological phenotypes observed with the bisphenol chemicals when compared to pharmacological agents specifically designed to target the AR and raises concern for the use of BPS as a substitute for BPA in commercial products.
In conclusion, this is the first report to describe the recruitment of coregulators when the BPA analogues-bound AR is assembled. BPA and BPAF were found to recruit similar coregulators while BPS recruited less in the AR complex. The BPA analogues were predicted to bind to multiple sites on the AR surface. BPA analogues were found to interfere, not only with the estrogen-signaling pathway, but also with the androgen-signaling pathway. From the results of the coregulator peptide recruitment study and MD simulations, the effects of BPA analogues on AR function are evident. Effects of BPA analogues on human health are complex and could depend on the gender, age, dose, physiological condition, and cell-type. The present study provides additional information on EDC’s effects to the human health.
Supplementary Material
Table 2c.
34 peptides that negatively interact with BPA
| Peptide names (34) | Relative binding value | Description of the proteins (30) |
|---|---|---|
| BRD8_254_276 | 0.31 | p120: Thyroid hormone-receptor conceiving protein |
|
| ||
| CBP_345_368_C367S | 0.65 | |
| CBP_57_80 | 0.54 | CBP: CREB-binding protein |
|
| ||
| CCND1_243_264_C243S/C247S | 0.39 | G1/S-specific cyclin-D1 (PRAD1 oncogene) |
|
| ||
| CENPR_1_18 | 0.29 | CENP-R: Centromere protein R |
| CNOT1_2083_2105 | 0.25 | |
| CNOT1_2086_2108 | 0.28 | CCR4-NOT transcription complex subunit 1 |
|
| ||
| GELS_376_398 | 0.42 | AGEL: Gelsolin precursor |
|
| ||
| ILK_131_153 | 0.34 | ILK-1: Integrin-liked protein kinase |
|
| ||
| JHD2C_2054_2076 | 0.39 | TRIP-8: Thyroidreceptor-interacting protein 8 |
|
| ||
| LCOR_40_62 | 0.17 | LCoR: Ligand-dependent corecpressor |
|
| ||
| MAPE_356_378 | 0.23 | Melanoma antigen preferentially expressed in tumor |
|
| ||
| MED1_632_655 | 0.18 | PBP: PPAR-binding protein |
|
| ||
| NCOA1_620_643 | 0.22 | SRC-1/RIP160: Nuclear receptor co-activator 1 |
|
| ||
| NCOA3_609_631 | 0.30 | SRC-3/NcoA-3: Nuclear receptor co-activator 3 |
|
| ||
| NCOA4_79_101_C101S | 0.58 | NcoA-4: Nuclear receptor co-activator 4 |
|
| ||
| NCOA6_1479_1501 | 0.36 | NcoA-6: Nuclear receptor co-activator 6 |
|
| ||
| NCOR1_2376_2398 | 0.27 | N-CoR1: Nuclear receptor corepressor 1 |
|
| ||
| NR0B1_1_23 | 0.18 | |
| NR0B2_106_128 | 0.37 | DAX-1: Nuclear receptor subfamily member 1 |
|
| ||
| NRIP1_8_30 | 0.77 | Nuclear receptor-interacting protein 1/nuclear factor RIP140 |
|
| ||
| NSD1_982_1004 | 0.33 | Histone-lysine N-methyltransferase |
|
| ||
| PNRC1_306_327 | 0.46 | Proline-rich nuclear receptor coactivator 1 |
| PR285_1062_1084 | 0.40 | |
| PR285_1160_1182_C1163S | 0.59 | ATP-dependent helicase PRIC285 protein |
|
| ||
| PRDM2_948_970 | 0.28 | PR domain zinc finger protein 2 |
|
| ||
| PRGC1_130_155 | 0.43 | PGC1α: PPAR-g co-activator 1-alpha |
|
| ||
| RBL2_875_897_C879S/C894S | 0.09 | PRB2/p130: Retinoblastoma-like protein 2 |
|
| ||
| TF65_437_459 | 0.32 | NF-kB: Transcription factor p65 |
|
| ||
| TIP60_476_498 | 0.52 | Tip60: Histone acetyltransferase HTATIP |
|
| ||
| TREF1_168_190 | 0.40 | TReP-132: Transcriptional-regulating factor 1 |
|
| ||
| TRRAP_770_792 | 0.50 | STAF40: Transformation domain-associated protein |
|
| ||
| UBE3A_396_418 | 0.38 | Ubiquitin-protein ligase E3A |
|
| ||
| ZNT9_449_471 | 0.27 | ZnT-9: Zinc transporter 9 |
Table 2d.
31 peptides that negatively interact with BPAF
| Peptide names (31) | Relative binding value | Description of the proteins (28) |
|---|---|---|
| BRD8_254_276 | 0.32 | p120: Thyroid hormone-receptor conceiving protein |
|
| ||
| CCND1_243_264_C243S/C247S | 0.52 | G1/S-specific cyclin-D1 (PRAD1 oncogene) |
|
| ||
| CENPR_1_18 | 0.29 | CENP-R: Centromere protein R |
|
| ||
| CNOT1_2083_2105 | 0.35 | |
| CNOT1_2086_2108 | 0.33 | CCR4-NOT transcription complex subunit 1 |
|
| ||
| DDX5_133_155 | 0.50 | Probable ATP-dependent RNA helicase DDX5 |
|
| ||
| EP300_69_91 | 0.55 | p300: Histone acetyltransferase p300 |
|
| ||
| GELS_376_398 | 0.77 | AGEL: Gelsolin precursor |
|
| ||
| ILK_131_153 | 0.34 | ILK-1: Integrin-liked protein kinase |
|
| ||
| JHD2C_2054_2076 | 0.44 | TRIP-8: Thyroidreceptor-interacting protein 8 |
|
| ||
| KIF11_832_854_C854S | 0.48 | Kinesin-like protein KIF11 |
|
| ||
| L3R2A_12_34 | 0.56 | Loss of heterozygosity 3 chromosomal region 2 gene A protein |
|
| ||
| LCOR_40_62 | 0.29 | LCoR: Ligand-dependent corecpressor |
|
| ||
| MAPE_356_378 | 0.36 | Melanoma antigen preferentially expressed in tumor |
|
| ||
| MED1_632_655 | 0.26 | PBP: PPAR-binding protein |
|
| ||
| NCOA1_620_643 | 0.47 | SRC-1/RIP160: Nuclear receptor co-activator 1 |
|
| ||
| NCOA3_609_631 | 0.48 | SRC-3/NcoA-3: Nuclear receptor co-activator 3 |
|
| ||
| NCOA6_1479_1501 | 0.38 | NcoA-6: Nuclear receptor co-activator 6 |
|
| ||
| NCOR1_2376_2398 | 0.39 | N-CoR1: Nuclear receptor corepressor 1 |
|
| ||
| NR0B1_1_23 | 0.21 | |
| NR0B2_106_128 | 0.29 | DAX-1: Nuclear receptor subfamily member 1 |
|
| ||
| NRIP1_8_30 | 0.84 | Nuclear receptor-interacting protein 1/nuclear factor RIP140 |
|
| ||
| NSD1_982_1004 | 0.35 | Histone-lysine N-methyltransferase |
|
| ||
| PELP1_20_42 | 0.43 | Proline-, glutamic acid- and leucine-rich protein 1 |
|
| ||
| PNRC1_306_327 | 0.58 | Proline-rich nuclear receptor coactivator 1 |
|
| ||
| PR285_1062_1084 | 0.43 | ATP-dependent helicase PRIC285 protein |
|
| ||
| PRDM2_948_970 | 0.36 | PR domain zinc finger protein 2 |
|
| ||
| RBL2_875_897_C879S/C894S | 0.29 | PRB2/p130: Retinoblastoma-like protein 2 |
|
| ||
| TF65_437_459 | 0.47 | NF-kB: Transcription factor p65 |
|
| ||
| UBE3A_396_418 | 0.36 | Ubiquitin-protein ligase E3A |
|
| ||
| ZNT9_449_471 | 0.43 | ZnT-9: Zinc transporter 9 |
Acknowledgments
We thank Vicki Sutherland, Lars Pedersen, and Jeffrey Tyler Ramsey for critical review. Research support was provided by Intramural Research Programs of the National Toxicology Program, National Institute of Environmental Health sciences (NIEHS), National Institutes of Health (NIH), and contract with PamGene International. This research was also supported by Research Project Number Z01-ES043010 (L.P) in the Intramural Research Program of NIEHS, NIH.
Abbreviations
- BPA
bisphenol A
- BPAF
bisphenol AF
- BPS
bisphenol S
- AR
androgen receptor
- MARCoNI
Microarray Assay for Real-Time Coregulator-Nuclear Receptor Interaction
- MD
molecular dynamics
Footnotes
Conflict of interests
The authors declare that they have no conflict of interests.
References
- Anonymous. Revision of the standards for the assessment of hormone receptors in human breast cancer; report of the second e.O.R.T.C. Workshop, held on 16–17 march, 1979, in the netherlands cancer institute. European journal of cancer. 1980;16:1513–1515. doi: 10.1016/0014-2964(80)90064-x. [DOI] [PubMed] [Google Scholar]
- Aarts JM, Wang S, Houtman R, van Beuningen RM, Westerink WM, Van De Waart BJ, et al. Robust array-based coregulator binding assay predicting eralpha-agonist potency and generating binding profiles reflecting ligand structure. Chemical research in toxicology. 2013;26:336– 346. doi: 10.1021/tx300463b. [DOI] [PubMed] [Google Scholar]
- Brooke GN, Parker MG, Bevan CL. Mechanisms of androgen receptor activation in advanced prostate cancer: Differential co-activator recruitment and gene expression. Oncogene. 2008;27:2941–2950. doi: 10.1038/sj.onc.1210955. [DOI] [PubMed] [Google Scholar]
- Buchanan G, Yang M, Harris JM, Nahm HS, Han G, Moore N, et al. Mutations at the boundary of the hinge and ligand binding domain of the androgen receptor confer increased transactivation function. Molecular endocrinology. 2001;15:46–56. doi: 10.1210/mend.15.1.0581. [DOI] [PubMed] [Google Scholar]
- Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, et al. A surface on the androgen receptor that allosterically regulates coactivator binding. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16074–16079. doi: 10.1073/pnas.0708036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Ribeiro RC, Wagner RL, Nguyen H, Apriletti JW, Fletterick RJ, et al. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747– 1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- Foulds CE, Feng Q, Ding C, Bailey S, Hunsaker TL, Malovannaya A, et al. Proteomic analysis of coregulators bound to eralpha on DNA and nucleosomes reveals coregulator dynamics. Molecular cell. 2013;51:185–199. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Wilson EM. Electrostatic modulation in steroid receptor recruitment of lxxll and fxxlf motifs. Molecular and cellular biology. 2003;23:2135–2150. doi: 10.1128/MCB.23.6.2135-2150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Molecular and cellular endocrinology. 2014;382:97–106. doi: 10.1016/j.mce.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Hodgson MC, Astapova I, Cheng S, Lee LJ, Verhoeven MC, Choi E, et al. The androgen receptor recruits nuclear receptor corepressor (n-cor) in the presence of mifepristone via its n and c termini revealing a novel molecular mechanism for androgen receptor antagonists. The Journal of biological chemistry. 2005;280:6511–6519. doi: 10.1074/jbc.M408972200. [DOI] [PubMed] [Google Scholar]
- Hodgson MC, Shen HC, Hollenberg AN, Balk SP. Structural basis for nuclear receptor corepressor recruitment by antagonist-liganded androgen receptor. Molecular cancer therapeutics. 2008;7:3187–3194. doi: 10.1158/1535-7163.MCT-08-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtman R, de Leeuw R, Rondaij M, Melchers D, Verwoerd D, Ruijtenbeek R, et al. Serine-305 phosphorylation modulates estrogen receptor alpha binding to a coregulator peptide array, with potential application in predicting responses to tamoxifen. Molecular cancer therapeutics. 2012;11:805–816. doi: 10.1158/1535-7163.MCT-11-0855. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. The cornr motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Kazmin D, Prytkova T, Cook CE, Wolfinger R, Chu TM, Beratan D, et al. Linking ligand-induced alterations in androgen receptor structure to differential gene expression: A first step in the rational design of selective androgen receptor modulators. Molecular endocrinology. 2006;20:1201– 1217. doi: 10.1210/me.2005-0309. [DOI] [PubMed] [Google Scholar]
- Koppen A, Houtman R, Pijnenburg D, Jeninga EH, Ruijtenbeek R, Kalkhoven E. Nuclear receptor-coregulator interaction profiling identifies trip3 as a novel peroxisome proliferator-activated receptor gamma cofactor. Molecular & cellular proteomics : MCP. 2009;8:2212–2226. doi: 10.1074/mcp.M900209-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, McEwan IJ. Allosteric modulators of steroid hormone receptors: Structural dynamics and gene regulation. Endocrine reviews. 2012;33:271–299. doi: 10.1210/er.2011-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luccio-Camelo DC, Prins GS. Disruption of androgen receptor signaling in males by environmental chemicals. The Journal of steroid biochemistry and molecular biology. 2011;127:74–82. doi: 10.1016/j.jsbmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan IJ. Intrinsic disorder in the androgen receptor: Identification, characterisation and drugability. Molecular bioSystems. 2012;8:82–90. doi: 10.1039/c1mb05249g. [DOI] [PubMed] [Google Scholar]
- McEwan IJ. Androgen receptor modulators: A marriage of chemistry and biology. Future medicinal chemistry. 2013;5:1109–1120. doi: 10.4155/fmc.13.69. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: Cellular and molecular biology. Endocrine reviews. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Millard CJ, Watson PJ, Fairall L, Schwabe JW. An evolving understanding of nuclear receptor coregulator proteins. Journal of molecular endocrinology. 2013;51:T23–36. doi: 10.1530/JME-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munuganti RS, Hassona MD, Leblanc E, Frewin K, Singh K, Ma D, et al. Identification of a potent antiandrogen that targets the bf3 site of the androgen receptor and inhibits enzalutamide-resistant prostate cancer. Chem Biol. 2014;21:1476–1485. doi: 10.1016/j.chembiol.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Love JD, Li C, Banayo E, Gooch JT, et al. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes & development. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Ting HJ, Harada Y, Tokizane T, Nonomura N, Kang HY, et al. Modulation of androgen receptor transactivation by gelsolin: A newly identified androgen receptor coregulator. Cancer research. 2003;63:4888–4894. [PubMed] [Google Scholar]
- O'Malley BW, McKenna NJ. Coactivators and corepressors: What's in a name? Molecular endocrinology. 2008;22:2213–2214. doi: 10.1210/me.2008-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osguthorpe DJ, Hagler AT. Mechanism of androgen receptor antagonism by bicalutamide in the treatment of prostate cancer. Biochemistry. 2011;50:4105–4113. doi: 10.1021/bi102059z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester JR. Bisphenol a and human health: A review of the literature. Reproductive toxicology. 2013;42:132–155. doi: 10.1016/j.reprotox.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Rochester JR, Bolden AL. Bisphenol s and f: A systematic review and comparison of the hormonal activity of bisphenol a substitutes. Environmental health perspectives. 2015;123:643–650. doi: 10.1289/ehp.1408989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S. Environmental estrogens: Roles in male reproductive tract problems and in breast cancer. Reviews on environmental health. 2002;17:253–262. doi: 10.1515/reveh.2002.17.4.253. [DOI] [PubMed] [Google Scholar]
- Shi XB, Ma AH, Xia L, Kung HJ, de Vere White RW. Functional analysis of 44 mutant androgen receptors from human prostate cancer. Cancer research. 2002;62:1496–1502. [PubMed] [Google Scholar]
- Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36:3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C, Goodwin B, Shockley K, Xia M, Huang R, Norris J, et al. Bisphenol a affects androgen receptor function via multiple mechanisms. Chemico-biological interactions. 2013;203:556– 564. doi: 10.1016/j.cbi.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wijngaart DJ, Dubbink HJ, van Royen ME, Trapman J, Jenster G. Androgen receptor coregulators: Recruitment via the coactivator binding groove. Molecular and cellular endocrinology. 2012;352:57–69. doi: 10.1016/j.mce.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Wang S, Houtman R, Melchers D, Aarts J, Peijnenburg A, van Beuningen R, et al. A 155- plex high-throughput in vitro coregulator binding assay for (anti-)estrogenicity testing evaluated with 23 reference compounds. ALTEX. 2013;30:145–157. doi: 10.14573/altex.2013.2.145. [DOI] [PubMed] [Google Scholar]
- Wetherill YB, Petre CE, Monk KR, Puga A, Knudsen KE. The xenoestrogen bisphenol a induces inappropriate androgen receptor activation and mitogenesis in prostatic adenocarcinoma cells. Molecular cancer therapeutics. 2002;1:515–524. [PubMed] [Google Scholar]
- Xu X, Yang W, Wang X, Li Y, Wang Y, Ai C. Dynamic communication between androgen and coactivator: Mutually induced conformational perturbations in androgen receptor ligand-binding domain. Proteins. 2011;79:1154–1171. doi: 10.1002/prot.22951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.