Abstract
Precise control of cell death is essential for the survival of all organisms. Arabidopsis thaliana BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 (BAK1) and somatic embryogenesis receptor kinase 4 (SERK4) redundantly and negatively regulate cell death through elusive mechanisms. By deploying a genetic screen for suppressors of cell death triggered by virus-induced gene silencing of BAK1/SERK4 on Arabidopsis knockout collections, we identified STT3a, a protein involved in N-glycosylation modification, as an important regulator of bak1/serk4 cell death. Systematic investigation of glycosylation pathway and endoplasmic reticulum (ER) quality control (ERQC) components revealed distinct and overlapping mechanisms of cell death regulated by BAK1/SERK4 and their interacting protein BIR1. Genomewide transcriptional analysis revealed the activation of members of cysteine-rich receptor-like kinase (CRK) genes in the bak1/serk4 mutant. Ectopic expression of CRK4 induced STT3a/N-glycosylation-dependent cell death in Arabidopsis and Nicotiana benthamiana. Therefore, N-glycosylation and specific ERQC components are essential to activate bak1/serk4 cell death, and CRK4 is likely to be among client proteins of protein glycosylation involved in BAK1/SERK4-regulated cell death.
Plant receptor-like kinases (RLKs) regulate diverse biological processes ranging from plant growth, development, symbiosis to immunity1,2. BRI1 is a receptor for plant brassinosteroid hormones involved in plant growth and development3 and FLS2 is a receptor for bacterial flagellin or flg22 (a 22-amino-acid peptide derived from flagellin) involved in plant immunity4. Both BRI1 and FLS2 interact with a subgroup of RLKs, the SERKs, which consist of five members in Arabidopsis. Except for SERK5, which is likely to be a non-functional kinase5, SERK1, SERK2, SERK3 and SERK4 are involved in a wide range of physiological responses6. SERK1 and SERK2 play a crucial and redundant role in male gametophyte development7,8. SERK3, also known as BAK1, and SERK4 function in plant immunity by association with FLS2 and other immune receptors9–12. SERK1, BAK1 and SERK4 function in brassinosteroid signalling by association with BRI15,13,14. SERK1, SERK2, BAK1 and SERK4 regulate stomatal patterning by means of EPF peptide ligand-induced association with ERECTA family RLKs15. In addition, SERK1, SERK2 and BAK1 regulate PSK peptide hormone-mediated root growth by association with its receptor PSKR16,17.
In addition, BAK1 and SERK4 negatively regulate the plant cell death process18,19. The bak1-4/serk4-1 null mutant is post-embryonic seedling lethal associated with spontaneous cell death and constitutive H2O2 production18. In contrast to the well-defined signalling framework of BAK1/SERK-mediated plant growth and immunity, the mechanisms underlying BAK1/SERK4-regulated cell death control are poorly understood. Notably, BIR1, a BAK1-interacting RLK, also negatively regulates cell death and the bir1 mutant exhibits post-embryonic seedling lethality, which depends on another RLK SUPPRESSOR OF BIR1,1 (SOBIR1)20. It remains unknown whether the same or distinct mechanisms operate bir1 and bak1/serk4 cell death. As no viable seeds are produced by the bak1-4/serk4-1 null mutant plants, the conventional forward genetic screen of bak1/ serk4 cell death suppressors is not feasible. Here, we have developed an Agrobacterium-mediated tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) system21 with the TRV-RNA2 vector harbouring fragments of both BAK1 and SERK4. Mutants that suppress bak1/serk4 cell death were identified using VIGS screening of Arabidopsis transfer (T)-DNA insertion lines. We report that the protein glycosylation pathway and specific ERQC components are essential for bak1/serk4 cell death. Transcriptomic analysis revealed that the plasma membrane-associated genes, including members of cysteine-rich receptor-like kinase (CRK) genes, were highly enriched among upregulated genes in bak1-4/serk4-1. Further biochemical and genetic investigations have suggested that CRK4 is one of the client proteins of protein glycosylation involved in the BAK1/ SERK4-regulated cell death process.
Results
Arabidopsis wild-type (WT) Col-0 plants silenced with BAK1/SERK4 showed severe growth defects with chlorotic leaves and dwarfism two weeks after Agrobacterium-inoculation, which resembled bak1-4 plants silenced with SERK4 (Fig. 1a). Analysis using polymerase chain reaction with reverse transcription (RT-PCR) revealed the reduced transcripts of BAK1 and SERK4, but not SERK1 or SERK2 in BAK1/SERK4-silenced plants (Fig. 1b). Trypan blue and 3,3′-diaminobenzidine (DAB) staining indicated that WT plants following silencing of BAK1/SERK4 or the bak1-4 mutant silenced with SERK4 displayed spontaneous cell death and the elevated H2O2 accumulation (Fig. 1c). In addition, these plants showed increased levels of PR1 and PR2 gene expression (Fig. 1d). The bak1-4/serk4-1 cell death is partially dependent on the plant defence hormone salicylic acid18. Consistently, the transgenic plants carrying the bacterial salicylate hydroxylase gene NahG and the sid2 mutant that is deficient in salicylic acid biosynthesis partially suppressed the cell death and H2O2 production triggered by VIGS of BAK1/SERK4 (Fig. 1e and Supplementary 1a). Taken together, these results demonstrate that silencing of BAK1/SERK4 by means of VIGS phenocopies cell death observed in the bak1-4/serk4-1 null mutant.
Figure 1. The sobir1 mutant did not suppress bak1/serk4 cell death.
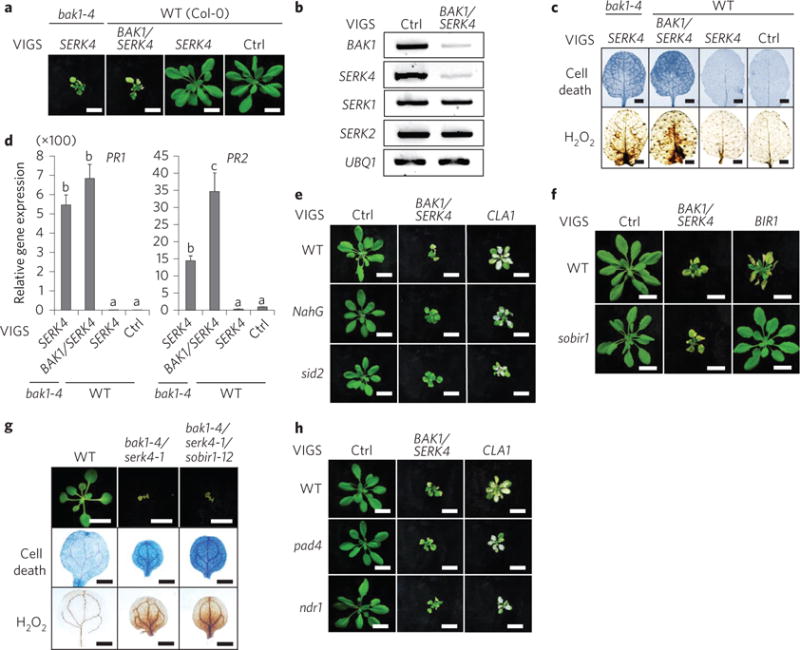
a, Silencing of BAK1/SERK4 by VIGS causes plant dwarfism and leaf chlorosis. Plants were photographed 2 weeks after inoculation of Agrobacterium carrying the indicated VIGS vectors in bak1-4 or WT plants. Scale bar, 1 cm. b, RT-PCR analysis of BAK1/SERK4-silenced plants after VIGS. BAK1, SERK4, SERK1 and SERK2 expression was analysed by RT-PCR from WT plants 2 weeks after VIGS. UBQ1 was used as an internal control. c, Silencing of BAK1/SERK4 by VIGS triggers cell death and H2O2 production. Trypan blue staining for cell death (upper panel) and DAB staining for H2O2 production (lower panel) are shown for true leaves of VIGS plants. Scale bar, 2 mm. d, Silencing of BAK1/SERK4 by VIGS induces PR1 and PR2 expression. The data are shown as mean ± s.e.m. from three independent repeats. The different letters denote a statistically significant difference according to one-way analysis of variance (ANOVA) followed by the Tukey test (P < 0.05). e, BAK1/SERK4-regulated cell death is partially dependent on salicylic acid. Plants silenced with CLA1 (Cloroplastos alterados 1) showed the albino phenotype as a visual marker of VIGS efficiency. NahG and sid2 plants after VIGS of BAK1/SERK4 remained greener with reduced leaf chlorosis than WT. Scale bar, 1 cm. f, BAK1/SERK4-regulated cell death is independent of SOBIR1. Scale bar, 1 cm. g, The sobir1-12 mutant did not suppress bak1-4/serk4-1 cell death in the bak1-4/serk4-1/sobir1-12 mutant. Seedlings were grown on a ½MS plate and photographed at 16 days after germination (top; scale bar, 1 cm). Cotyledons were stained with trypan blue for cell death (middle) and DAB for H2O2 accumulation (bottom). Scale bar, 1 mm in middle and bottom panels. h, BAK1/SERK4-regulated cell death is independent of PAD4 and NDR1. Scale bar, 1 cm. The above experiments were repeated at least three times with similar results.
We tested whether bak1/serk4 cell death depends on SOBIR1, which is required for bir1 cell death. Similar to the bir1 null mutant, cell death could be observed in WT plants silenced with BIR1 by means of VIGS (Fig. 1f). The dwarfism and leaf chlorosis associated with silencing of BIR1 were almost completely suppressed in sobir1-12 (Fig. 1f). Interestingly, the sobir1-12 mutant did not affect cell death triggered by VIGS of BAK1/SERK4 (Fig. 1f). We further generated the bak1-4/serk4-1/sobir1-12 triple mutant, which showed the same level of seedling lethality, elevated cell death and H2O2 accumulation with bak1-4/serk4-1 (Fig. 1g). Activation of resistance (R) protein-mediated defence is a common mechanism of plant cell death. However, the pad4 and ndr1 mutants, which had impaired R protein pathways, did not significantly suppress the cell death caused by VIGS of BAK1/SERK4 (Fig. 1h), suggesting that activation of R protein-mediated defences may not play a major role in bak1/serk4 cell death. In contrast, the pad4 mutant largely alleviated cell death caused by silencing of BIR1 (Supplementary Fig. 1b). Consistently, BAK1/SERK4-regulated cell death does not require R proteins RPS2 and RPM1 (Supplementary Fig. 1c). Together, the data indicate distinct mechanisms underlying BAK1/SERK4- and BIR1-regulated cell death.
To identify components involved in BAK1/SERK4-regulated cell death, we carried out a VIGS-based genetic screen of a sequence-indexed library of Arabidopsis T-DNA insertion lines. After screening ~6,000 homozygous lines, a series of mutants were isolated based on the suppression of cell death by silencing of BAK1/ SERK4. One mutant (line CS800052 with an annotated T-DNA insertion at STAUROSPORIN AND TEMPERATURE SENSITIVE3 (STT3a), stt3a-2 (Supplementary Fig. 2a)) largely suppressed dwarfism and leaf chlorosis triggered by VIGS of BAKl/SERK4 (Fig. 2a). MEKK1, a MAP kinase (MAPK) kinase kinase downstream of BAK1/SERK4 in flagellin signalling, is also involved in cell death regulation22. Silencing of MEKK1 by VIGS in WT plants resulted in severe dwarfism and cell death (Fig. 2a). However, the stt3a-2 mutant did not suppress MEKKl-regulated cell death (Fig. 2a), suggesting different mechanisms underlying BAK1/SERK4- and MEKKl-regulated cell death. The data also suggest that stt3a-2 did not affect the gene silencing machinery. RT-PCR analysis demonstrated a similar silencing efficiency of BAK1/SERK4 by VIGS in WT and stt3a-2 mutant (Supplementary Fig. 2b). Furthermore, the cell death and elevated H2O2 accumulation caused by VIGS of BAK1/SERK4 were almost completely abolished in stt3a-2 (Fig. 2b). Compared with WT, stt3a-2 showed much reduced accumulation of PR1 and PR2 genes by VIGS of BAK1/ SERK4 (Fig. 2c). Another T-DNA insertion mutant stt3a-1, which is in the ecotype C24 background, also suppressed cell death by silencing of BAK1/SERK4 (Fig. 2d). Furthermore, transformation of a genomic fragment containing the STT3a gene into stt3a-2 restored BAK1/SERK4-regulated cell death (Fig. 2e and Supplementary Fig. 2c). To investigate if stt3a-2 could genetically suppress bak1-4/serk4-1 seedling lethality and defence activation, we generated the bak1-4/serk4-1/stt3a-2 triple mutant. The bak1-4/serk4-1/stt3a-2 triple mutant overcame seedling lethality of bak1-4/serk4-1 and resembled WT plants at the two-week-old stage when grown on ½ Murashige and Skoog medium (½MS) medium plates (Fig. 2f and Supplementary Fig. 2d). In addition, cell death, H2O2 accumulation, and PR1 and PR2 expression were significantly ameliorated in bak1-4/serk4-1/stt3a-2 compared with those in bak1-4/serk4-1 (Fig. 2g,h). The stt3a-2 mutant also partially suppressed VIGS of BIR1-induced cell death (Supplementary Fig. 2e,f). The elevated expression of PR1 and PR2 was also significantly reduced in stt3a-2 silenced with BIR1 (Supplementary Fig. 2g). This is consistent with STT3a being required for the activation of defence responses in the bir1 genetic mutant23.
Figure 2. The stt3a mutants suppress BAK1/SERK4-regulated cell death.
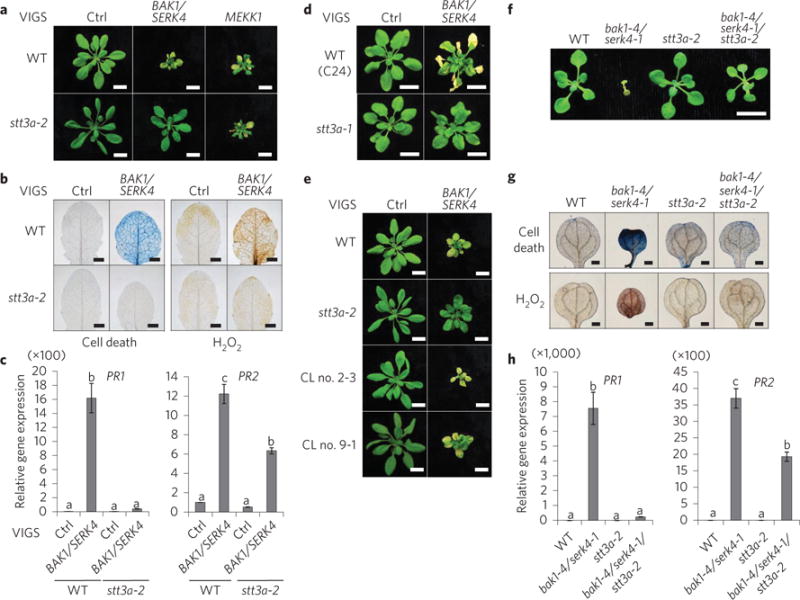
a, The stt3a-2 mutant suppresses growth defects triggered by VIGS of BAK1/SERK4 but not MEKK1. Scale bar, 1 cm. b, The stt3a-2 mutant suppresses cell death (left panel) and H2O2 production (right panel) by VIGS of BAK1/SERK4. Scale bar, 2 mm. c, The stt3a-2 mutant suppresses PR1 and PR2 expression by VIGS of BAK1/SERK4. d, The stt3a-1 mutant suppresses growth defects by VIGS of BAK1/SERK4. Plant phenotypes are shown from WT (C24) and stt3a-1 2 weeks after VIGS. Scale bar, 1 cm. e, Complementation of the stt3a-2 mutant with STT3a restores growth defects by VIGS of BAK1/SERK4. CL no. 2-3 and CL no. 9-1 are two homozygous complementation lines. Scale bar, 1 cm. f, The stt3a-2 mutant rescues the seedling lethality of the bak1-4/serk4-1 mutant. Seedlings were grown on ½MS plate and photographed at 16 days after germination. Scale bar, 1 cm. g, The alleviated cell death and H2O2 accumulation in bak1-4/serk4-1/stt3a-2. Scale bar, 2 mm. h, The reduced PR1 and PR2 expression in bak1-4/serk4-1/stt3a-2. The data in c and h are shown as mean ± s.e.m. from three independent repeats. The different letters denote a statistically significant difference according to one-way ANOVA followed by the Tukey test (P < 0.05). The above experiments were repeated three times with similar results.
Plant cell death and defence activation are often modulated by temperature and other environmental factors24. When grown on ½MS medium plates, seedling lethality of bak1-4/serk4-1 was largely ameliorated at 30 °C when compared with that at 23 °C (Supplementary Fig. 3a,b). The elevated expression of PR1 and PR2 in bak1-4/serk4-1 was also reduced when grown at 30 °C (Supplementary Fig. 3c). The alleviation of seedling lethality and growth defects of bak1-4/serk4-1 by the elevated temperature was a relatively subtle effect for plants grown on soil (Supplementary Fig. 3d). However, the bak1-4/serk4-1/stt3a-2 triple mutant grew significantly better on soil at 30 °C than that at 23 °C (Supplementary Fig. 3d). The bak1-4/serk4-1/stt3a-2 mutant could ultimately develop to maturity and occasionally produce some viable seeds when grown on soil at 30 °C.
STT3 is the catalytic subunit of the oligosaccharyltransferase (OST) complex that is involved in protein N-glycosylation modifications (Fig. 3a)25. There are two family members of STT3, STT3a and STT3b, in Arabidopsis. The mutations in STT3b (stt3b-2 and stt3b-3) did not affect cell death by silencing of BAK1/SERK4, suggesting a specific function of STT3a in this process (Fig. 3b). This is consistent with the observation that STT3a, but not STT3b, is involved in Arabidopsis salt and osmotic stresses25. In addition to the catalytic STT3 subunit, the OST complex also consists of several non-catalytic subunits that regulate substrate specificity, stability or complex formation26. The highly conserved subunit OST3/6 regulates overall protein glycosylation and is involved in plant immunity by controlling biogenesis of the EF-Tu receptor EFR (ref. 26). The ost3/6 mutant also suppressed BAK1/SERK4-regulated cell death (Fig. 3b).
Figure 3. Control of BAK1/SERK4-regulated cell death by protein N-glycosylation and specific components of ERQC.
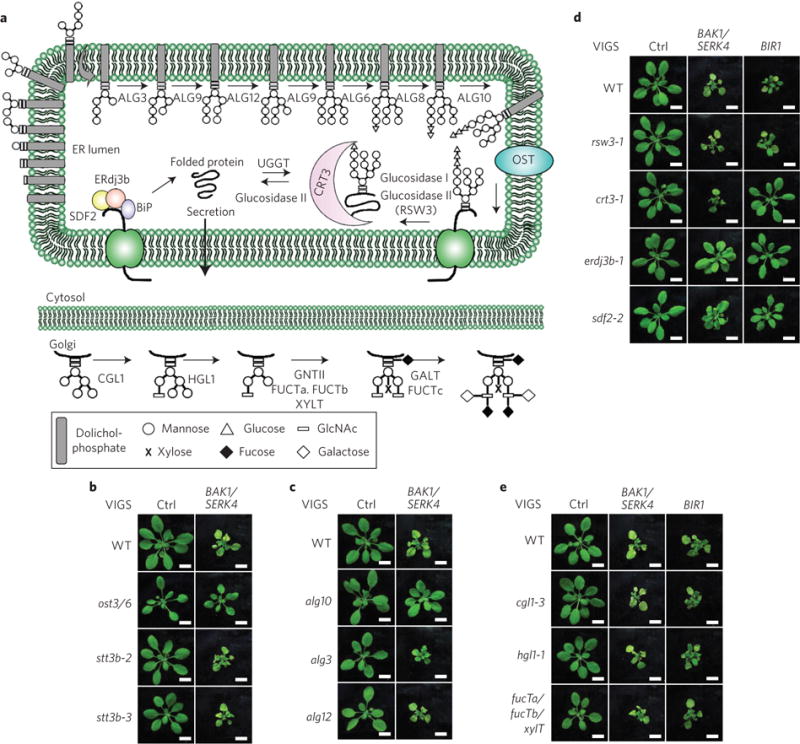
a, A schematic overview of protein N-glycan assembly in the ER and modification in the Golgi apparatus and ERQC system. The OST complex transfers the dolichol-linked N-glycan precursor preassembled by different ALG glucosyltransferases to the acceptor proteins in the ER. The N-glycan precursor is then modified by ER glucosidase, and recognized by CNX and CRT3 for proper protein folding. The misfolded proteins undergo additional CNX/CRT cycles by UGGT and glucosidase for another round of folding. The BiP-ERdj3-SDF2 complex also controls proper protein folding. The correctly folded proteins will be exported to the Golgi apparatus to form complex N-glycans catalysed by different enzymes. b, The ost3/6, but not stt3b, mutant suppresses cell death by VIGS of BAK1/SERK4. Phenotypes of different plants are shown 2 weeks after VIGS. Scale bar, 1 cm. c, The alg10, but not the alg3 or alg12, mutant suppresses cell death by VIGS of BAK1/SERK4. Scale bar, 1 cm. d, Differential ERQC components are required for BAK1/SERK4- and BIR1-regulated cell death. The erdj3b-1 and sdf2-2 mutants partially suppressed BAK1/SERK4- and BIR1-regulated cell death by VIGS. However, the crt3-1 mutant only suppressed BIR1- but not BAK1/SERK4-regulated cell death by VIGS. Scale bar, 1 cm. e, The N-glycan modification in the Golgi apparatus may not be required for BAK1/SERK4-regulated cell death. Various mutants impaired in N-glycan modification in the Golgi apparatus did not suppress cell death by VIGS of BAK1/SERK4 or BIR1. Scale bar, 1 cm. The above experiments were repeated three times with similar results.
The OST complex transfers the lipid (dolichol)-linked N-glycan precursor Glc3Man9GlcNAc2 (Glc, glucose; Man, mannose; GlcNAc, N-acetylglucosamine) to the acceptor proteins in the ER (Fig. 3a)27,28. The N-glycan precursor is preassembled by a series of glucosyltransferases encoded by asparagine-linked glycosylation (ALG) proteins (Fig. 3a). Loss-of-function of some ALG proteins prevented assembly of the N-glycan precursor and reduced protein N-glycosylation efficiency in Arabidopsis29,30. The alg10, but not alg3 or alg12 mutant, suppressed BAK1/SERK4 silencing-mediated cell death (Fig. 3c), suggesting that proper assembly of the N-glycan precursor is essential in bak1/serk4 cell death. ALG10 catalyses the last step of N-glycan precursor assembly by transferring the terminal glucose residue to the precursor, an essential step for OST complex recognition. The Arabidopsis alg10 mutant had a severe defect in protein N-glycosylation and increased sensitivity to salt stress29.
After the N-glycan precursor was transferred to the acceptor proteins by the OST complex, the two terminal glucose residues of the oligosaccharides were subsequently removed by glucosidase I and glucosidase II (RSW3 in Arabidopsis), and then recognized by the ER chaperone-like lectins calnexin (CNX) and calreticulin (CRT3) for proper protein folding and secretion (Fig. 3a)27,28. The incompletely folded or misfolded proteins will be recognized by UDP-glucose:glycoprotein glucosyltransferase (UGGT) for additional CNX/CRT cycles and another round of folding (Fig. 3a)27,28,31. ER-localized HSP70 proteins, BiPs and their associated factors ERdj3 and SDF2 also play important roles to prevent export of misfolded proteins (Fig. 3a)31. Some specific ERQC components, such as CRT3, UGGT, ERdj3b and SDF2, were genetically implicated in the protein folding and degradation of mutated bri1 receptor and immune receptor EFR and in BIR1-mediated cell death23,32–40. We observed that erdj3b-1 and sdf2-2, but not crt3-1 or rsw3-1, partially suppressed BAK1/SERK4 silencing-mediated cell death (Fig. 3d). RT-PCR analysis indicated a similar silencing efficiency of BAK1/SERK4 by VIGS in WT and different mutants (Supplementary Fig. 4a). As reported for genetic mutants32, crt3-1, erdj3b-1 and sdf2-2 suppressed BIR1 silencing-mediated cell death (Fig. 3d). We further generated genetic mutants of bak1-4/serk4-1/ crt3-1, bak1-4/serk4-1/erdj3b-1 and bak1-4/serk4-1/sdf2-2. Similar to VIGS assays, bak1-4/serk4-1/erdj3b-1 and bak1-4/serk4-1/sdf2-2, but not bak1-4/serk4-1/crt3-1, alleviated bak1-4/serk4-1 seedling lethality and H2O2 production (Supplementary Fig. 4b). The data further support that distinct mechanisms, but with certain overlapping features, control BAK1/SERK4- and BIR1-regulated cell death.
Proteins with native conformation after ERQC will be exported to the Golgi apparatus for further modifications, such as formation of complex and hybrid N-glycans catalysed by stepwise enzymatic reactions, including β1,2-N-acetylglucosaminyltransferase I (CGL1/GNTI), α-mannosidase II (HGL1/MANII), GNTII, β1,2-xylosyltransferase (XYLT), α1,3-fucosyltransferase (FUCTa and FUCTb), β1,3-galactosyltransferase (GALT) and α1,4-fucosyltransferase (FUCTc) (Fig. 3a)41. To investigate if N-glycan maturation in the Golgi apparatus also plays a role in bak1/serk4 cell death, we silenced BAK1/ SERK4 in the cgl1-3 and hgl1-1 single mutants and fucTa/fucTb/xylT triple mutant (Fig. 3e). All these mutants showed similar levels of cell death and growth retardation as WT plants after silencing of BAK1/SERK4 by VIGS (Fig. 3e). Apparently, these mutants also did not affect the BIR1 silencing-mediated cell death (Fig. 3e). These data suggest that protein glycosylation modification for proper folding in ER, but not N-glycan modification in the Golgi apparatus, is essential for the initiation of bak1/serk4 and bir1 cell death. N-glycan modification in the Golgi apparatus is also not required for EFR maturation in plant immunity37.
Preventing protein N-glycosylation often leads to protein misfolding, a major contributor to ER stress, thereby resulting in unfolded protein response (UPR)31. Defects in protein glycosylation and ERQC in the stt3a, erdj3b and sdf2 mutants is likely to induce ER stress and UPR. The Arabidopsis UPR signalling pathway is composed of two arms: one involving the bifunctional protein kinase/ RNA ribonuclease IRE1 and its target RNA bZIP60, and another involving ER membrane-associated transcription factors, such as bZIP28 (ref. 31). To test if UPR may contribute to bak1/serk4 cell death, we silenced BAK1/SERK4 in the ire1a/ire1b and bzip28/ bzip60 double mutants, both of which are deficient in UPR induction42. Neither ire1a/ire1b nor bzip28/bzip60 affected cell death by VIGS of BAK1/SERK4 (Supplementary Fig. 4c), suggesting that UPR may not directly link to BAK1/SERK4-regulated cell death.
In contrast to the negative regulation in cell death, BAK1 and SERK4 positively regulate plant immunity and brassinosteroid signalling5,9–14. We tested whether the mutation in stt3a also suppressed bak1 or bak1/serk4 deficiency in flagellin and brassinosteroid signalling. Flg22 triggers rapid phosphorylation of MAPKs and receptor-like cytoplasmic kinase BIK1 (ref. 43). The bak1-4 and bak1-4/serk4-1 mutants displayed compromised flg22-induced MAPK activation (Fig. 4a,b) and BIK1 phosphorylation (Fig. 4c). The stt3a mutant did not affect flg22 signalling as previously reported (Fig. 4a)37. The compromised flg22-induced MAPK activation and BIK1 phosphorylation remained the same in bak1-4/stt3a-2 and bak1-4/serk4-1/stt3a-2 as those in bak1-4 or bak1-4/serk4-1 (Fig. 4a–c). Similar to bak1-4/serk4-1, bakl-4/ serk4-1/stt3a-2 showed compromised expression of flg22-induced genes, WRKY30, MYB15 and LOX4, compared with WT or stt3a-2 (Fig. 4d). Taken together, these results suggest that stt3a-2 did not suppress bakl-4 or bak1-4/serk4-1 deficiency in flagellin-mediated immune signalling. When grown in the dark, the hypocotyls of the bak1-4/serk4-1 mutant elongated slightly, but were significantly shorter than those of WT plants (Fig. 4e,f). In the presence of brassinazole (BRZ), an inhibitor of brassinosteroid biosynthesis, bak1-4/serk4-1 displayed shorter hypocotyl elongation than WT. The stt3a-2 mutant exhibited hypocotyl elongation similar to WT plants. The hypocotyl elongation of bak1-4/serk4-1/stt3a-2 was similar to that of bak1-4/serk4-1 in the absence or presence of BRZ (Fig. 4e,f), suggesting that stt3a-2 did not interfere with the responsiveness of bak1-4/serk4-1 to BRZ.
Figure 4. Uncoupled BAK1 functions in cell death control, immunity and brassinosteroid signalling.
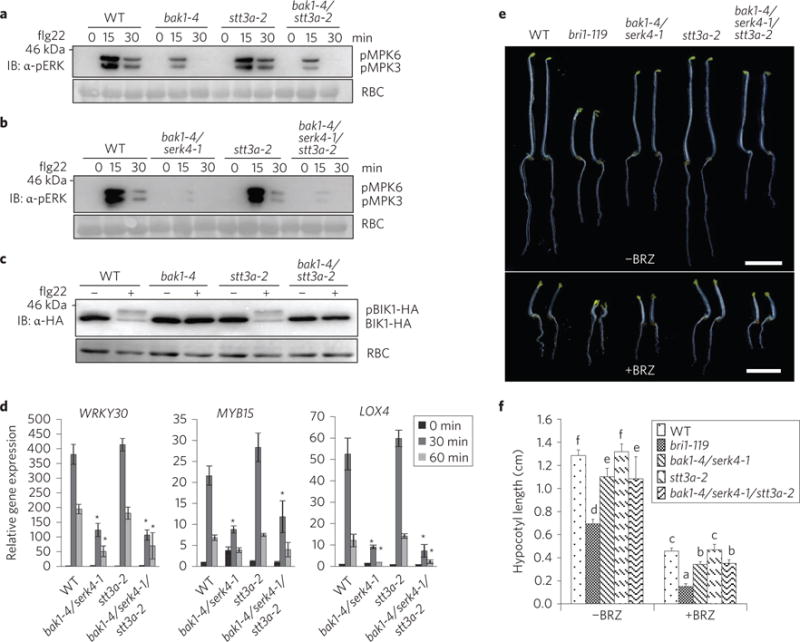
a, The stt3a-2 mutation did not interfere with the compromised flg22-induced MAPK activation in the bak1-4 mutant. Ten-day-old seedlings were treated without or with 100 nM flg22 for 15 and 30 min. MAPK activation was analysed by immunoblotting with α-pERK antibody (top panel); protein loading is shown by Ponceau S staining for Rubisco (RBC) (bottom panel). b, The stt3a-2 mutation did not interfere with the compromised flg22-induced MAPK activation in the bak1-4/serk4-1 mutant. c, The stt3a-2 mutation did not affect the compromised flg22-induced BIK1 phosphorylation in the bak1-4 mutant. Protoplasts from different plants were transfected with haemagglutinin (HA)-tagged BIK1 and treated with 100 nM flg22 for 10 min. BIK1-HA proteins were detected by immunoblotting using the α-HA antibody (top panel); protein loading is shown by Ponceau S staining for RBC (bottom panel). d, The stt3a-2 mutation did not affect the compromised flg22-induced marker gene expression in the bak1-4/serk4-1 mutant. Ten-day-old seedlings were treated without or with 100 nM flg22 for 30 or 60 min for qRT-PCR analysis. The data are shown as mean ±s.e.m. from three independent repeats. Asterisks indicate statistically significant differences from the WT within the same time point according to two-way ANOVA followed by the Tukey test (P < 0.05). e,f, The bak1-4/serk4-1 sensitivity to BRZ still occurs in the bak1-4/serk4-1/stt3a-2 mutant. The seedlings of indicated genotypes were grown in the dark for 5 days on ½MS plates with or without 2 μM BRZ (e), and hypocotyl lengths were quantified (f). The data are shown as mean ± s.e.m. from 20 seedlings. Scale bar, 0.5 cm. The different letters denote a statistically significant difference according to two-way ANOVA followed by the Tukey test (P < 0.05). The experiments in a–d were repeated three times and in e,f the experiments were repeated twice with similar results.
As PR genes are highly upregulated in the bak1/serk4 cell death process, we determined the genome-wide transcriptional changes in bak1-4/serk4-1 by RNA-sequencing (RNA-seq) analysis. Among 24,572 detectable transcripts (Fig. 5a and Supplementary Table 1), we identified 3,637 differentially expressed genes (fold change ≥ 2 and false discovery rate (FDR) < 0.1), including 1,920 induced genes (Supplementary Table 2) and 1,717 reduced genes (Supplementary Table 3) in bak1-4/serk4-1 compared with WT plants (Fig. 5b). Interestingly, gene ontology enrichment analysis indicated that genes encoding proteins associated with membrane, especially plasma membrane, were highly enriched among the upregulated genes in bak1-4/serk4-1 (Fig. 5c). Remarkably, among 44 CRK genes, the expression of 22 CRKs was upregulated in bak1-4/ serk4-1 (Fig. 5d and Supplementary Table 4). The induction of CRK4, CRK5, CRK7, CRK8, CRK19 and CRK20 in bak1-4/serk4-1 was confirmed by quantitative RT-PCR (qRT-PCR) (Fig. 5e).
Figure 5. Members of CRK genes are upregulated in the bak1/serk4 mutant.
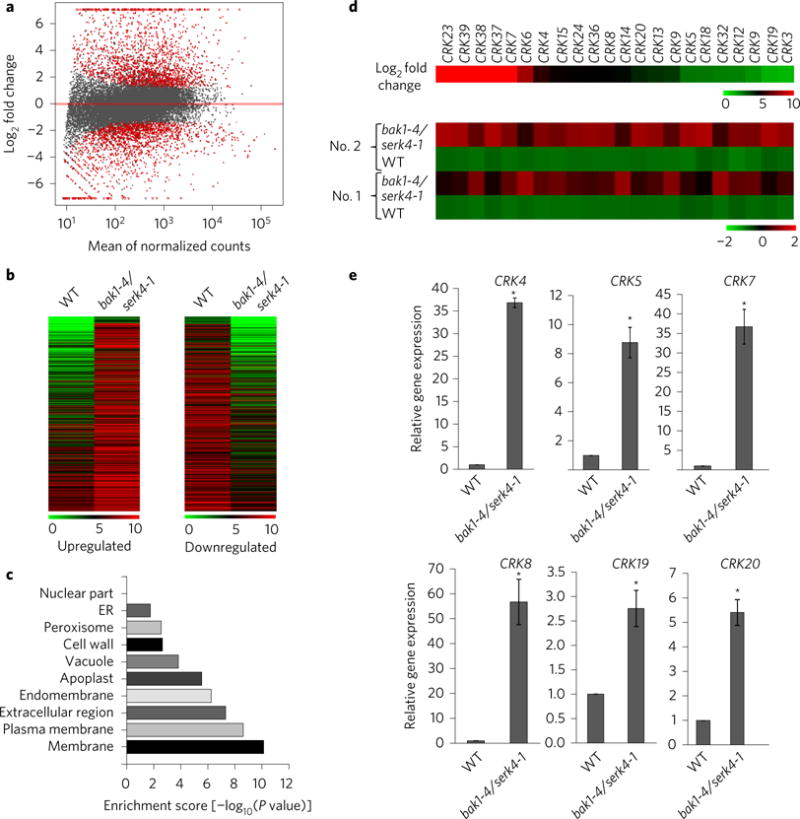
a, DESeq’s plotMA displays differential expression (log2 fold changes between bak1-4/serk4-1 and WT on the y axis) versus expression strength (the mean of normalized counts on the x axis). Genes with significantly differential expression with FDR < 0.10 and fold change ≥2 are in red. b, Heatmaps of upregulated and downregulated genes in the bak1/serk4 mutant compared with WT. The original means of read counts were subjected to data adjustment by log2 transformation using MeV4.0 for the heatmaps. The genes are plotted in the order of fold changes. c, Gene ontology (GO) enrichment analysis based on the cellular component. The x axis indicates the enrichment scores [−log10 (P value)] (the P value indicates the possibility of significant enrichment) for each cellular component GO item on the y axis. d, Members of CRKs are induced in bak1/serk4. The upper panel indicates log2fold change of different CRK genes (bak1-4/serk4-1 vs. WT) and the bottom panel shows normalized expression level from two independent repeats (No. 1 and No. 2). e, Expression of some CRKs by qRT-PCR analysis. Ten-day-old seedlings grown on ½MS plates were subjected to qRT-PCR analysis. The data are shown as means ± s.e.m. from three biological replicates. *P < 0.05 significant difference compared with WT using Student’s t-test.
It has been reported that overexpression of certain CRKs, such as CRK4, CRK5, CRK13, CRK19 and CRK20, was able to induce cell death in Arabidopsis transgenic plants44–46. When transiently expressed in Nicotiana benthamiana, CRK4 and CRK5 elicited water-soaking and subsequent cell death (Fig. 6a and Supplementary Fig. 5a). We tested whether CRK proteins were glycosylated and whether STT3a-mediated protein glycosylation was required for CRK-mediated cell death. Glycosylated proteins often display slower migration than non-glycosylated proteins in an immunoblot. When treated with tunicamycin, an inhibitor of protein N-glycosylation, both CRK4 and CRK5 proteins exhibited faster migration than proteins without treatment, suggesting that CRK4 and CRK5 proteins are likely to be N-glycosylated in Arabidopsis cells (Fig. 6b). Consistently, Endoglycosidase H treatment released CRK4 proteins with reduced molecular weight (Supplementary Fig. 5b). CRK4 proteins when expressed in stt3a-2 migrated faster than that in WT (Fig. 6c), indicating that STT3a is required for CRK4 glycosylation. Notably, we have consistently observed that the accumulation of CRK4 proteins after tunicamycin treatment or in stt3a appeared to be reduced when compared with proteins without treatment or in WT (Fig. 6b,c), suggesting that N-glycosylation may regulate CRK4 protein stability.
Figure 6. CRK4-induced cell death requires STT3a-mediated N-glycosylation.
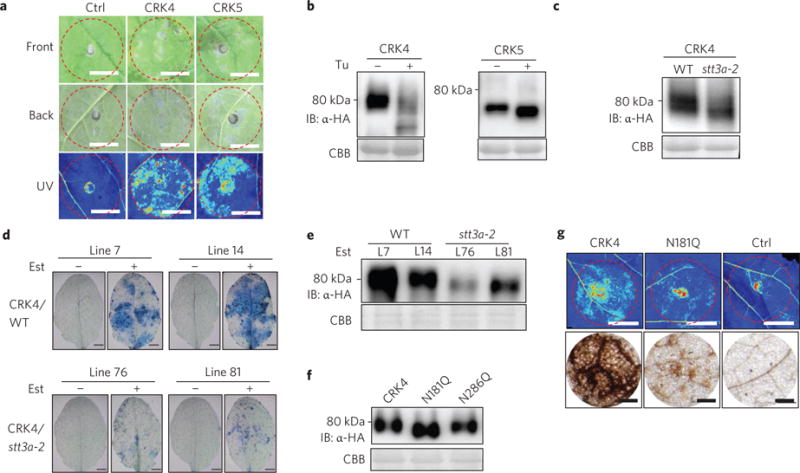
a, Expression of CRK4 or CRK5 in N. benthamiana induces cell death. Cell death was visualized on the front (top panel) or back (middle panel) side of different leaves or under UV light with the ChemiDoc Imaging System (bottom panel) 4 days after infiltration. The infiltrated areas are labelled with red dashed circles. Scale bar, 1 cm. b, Tunicamycin treatment modifies protein migration of CRK4 and CRK5. Protoplasts from Col-0 WT were transfected with CRK4-HA or CRK5-HA and treated without or with tunicamycin (1 μM). The proteins were detected by immunoblotting with α-HA antibody (upper panel). Coomassie Brilliant Blue (CBB) staining is a loading control (lower panel). c, CRK4 exhibits elevated protein migration rate in stt3a-2. Protoplasts from Col-0 WT or the stt3a-2 mutant were transfected with CRK4-HA. d, STT3a is required for CRK4-induced cell death in Arabidopsis. Leaves of four-week-old transgenic plants of oestradiol-inducible CRK4-HA in WT and stt3a background were infiltrated with oestradiol (10 μM) or mock. Cell death staining was performed 4 days after infiltration. Two independent lines for each background are shown. Scale bar, 2 mm. e, CRK4 protein expression in transgenic plants. Detached leaves from 4-week-old transgenic plants were soaked in oestradiol solution (10 μM) for 24 h. The HA-tagged CRK4 protein was detected by immunoblotting with α-HA antibody. f, Expression of CRK4 and its putative N-glycosylation mutants in Arabidopsis protoplasts. g, CRK4-induced cell death is partially blocked by N181Q mutation in N. benthamiana. Cell death was documented 4 days after inoculation of Agrobacterium carrying different constructs in N. benthamiana. Upper panel, cell death was visualized as autofluorescence under UV light with the ChemiDoc Imaging System. Scale bar, 1 cm. Bottom panel, H2O2 accumulation by DAB staining. Scale bar, 100 μm.
We further introduced CRK4 under the control of an oestrogen-inducible promoter into WT and stt3a-2. The cell death caused by ectopic expression of CRK4 in WT plants was largely alleviated in stt3a-2 when oestradiol was applied (Fig. 6d). Similar to the above transient assays, the CRK4 protein level was lower in stt3a-2 than that in WT in multiple transgenic lines (Fig. 6e). We aligned the extracellular domain of CRK4 with its closest homologue CRK5. Out of five potential N-glycosylation sites (Asn in Asn-X-Ser/Thr, where X is any amino acid except Pro), we identified two sites that are conserved between CRK4 and CRK5 (Supplementary Fig. 5c). We mutated them in CRK4 to Gln (CRK4N181Q and CRK4N286Q) and found that CRK4N181Q showed faster migration than WT CRK4 when expressed in Arabidopsis protoplasts (Fig. 6f) or N. benthamiana (Supplementary Fig. 5d), resembling the CRK4 expression in stt3a (Fig. 6c). The protein level of CRK4N181Q was similar to that of WT CRK4, suggesting that mutation of one glycosylation site may not affect CRK4 protein stability. Significantly, the CRK4N181Q mutant reduced cell death intensity and H2O2 accumulation compared with WT CRK4 when transiently expressed in N. benthamiana (Fig. 6g). Since CRK4N181 is a conserved site between CRK4 and CRK5, it is likely to be that the corresponding site in CRK5 is required for its cell death-inducing ability. The results suggest that N-glycosylation is essential for CRK4-mediated cell death, and CRK4 is one of the substrates of STT3a in bak1/serk4 cell death. To test whether loss of CRK4 suppresses bak1/serk4 cell death, we silenced BAK1/SERK4 in the crk4 mutant. However, the crk4 mutant did not affect cell death by VIGS of BAK1/SERK4 (Supplementary Fig. 5e,f), indicating that mutation of CRK4 is not sufficient to suppress bak1/serk4 cell death.
Discussion
The mechanisms of cell death control are poorly understood in plants. To uncover the pathways and mechanisms regulating bak1/serk4 cell death, we have developed an unbiased and highly efficient genetic screen which combines the features of both forward and reverse genetics. We have identified stt3a as a suppressor of bak1/serk4 cell death. Systematic investigation of components in protein N-glycosylation pathways and ERQC indicates that N-glycan production in ER and specific ERQC components involved in protein folding are essential in the activation of bak1/ serk4 cell death. By combining RNA-seq, genetic and biochemical analyses, we provide evidence that CRK4 serves as one of the client proteins for STT3a-mediated protein N-glycosylation and ERQC in bak1/serk4 cell death.
Although BAK1 and BIR1 are in a complex, the mechanisms regulating their functions in cell death control are distinct. The bir1-mediated cell death depends on SOBIR1 and the R proteinmediated signalling20. However, sobir1 and R protein signalling mutants did not affect bak1/serk4 cell death (Fig. 1f–h). Both bak1/serk4 and bir1 cell death require STT3a (Fig. 3), although the SOBIR1 protein level was not affected in stt3a (ref. 23). It has been shown that SOBIR1 protein accumulation was reduced in erdj3b, suggesting that ERQC is important for biogenesis of SOBIR1 (ref. 32). It is likely that activation of SOBIR1 also requires N-glycosylation modification in ER. Thus, protein glycosylation and ERQC are common features in BAK1/SERK4- and BIR1-regulated cell death, but different client proteins are likely to be deployed for N-glycosylation modification.
BAK1 family RLKs serve as a shared signalling node that modulates the interconnected architecture of the complex cellular signalling networks yet disseminates different biological outcomes6,47. Mounting evidence suggests that BAK1 family RLKs function independently in different signalling pathways. For instance, the involvement of BAK1 and SERK4 in cell death control can be separated from their involvement in brassinosteroid signalling18,19. Similarly, their function in stomatal patterning is uncoupled from function in brassinosteroid signalling15. The bak1-5 mutant had severe defects in plant immunity, but did not affect brassinosteroid signalling and cell death control48. These results are consistent with our observation that stt3a did not suppress bak1 or bak1/serk4 deficiency in flg22 and brassinosteroid signalling, reinforcing that the signalling pathway of BAK1/SERK4-regualted cell death is uncoupled from their functions in flg22-triggered immunity and brassinosteroid-mediated development.
Many CRK genes are induced by defence hormone salicylic acid treatment and bacterial pathogen infections44,45. Constitutive expression of CRK4 or CRK5 at a modest level enhanced plant resistance to virulent bacteria Pseudomonas syringae45,49. In addition, chemical-induced expression of CRK4 or CRK5 triggered cell death in Arabidopsis44,45. These observations point to the potential role of CRKs in plant defence responses. We observed an enrichment of CRK genes among upregulated genes in bak1/serk4 (Fig. 5d,e). In addition, CRK4-induced cell death depends on STT3a-mediated protein N-glycosylation (Fig. 6d). The CRK4 N-glycosylation mutant reduced the ability to trigger cell death (Fig. 6g). It is likely to be that the non-glycosylated or under-glycosylated CRK4 protein is misfolded in ER and finally removed by ER-associated degradation, thereby resulting in the reduced protein level in stt3a (Fig. 6e). However, crk4 did not suppress bak1/serk4 cell death (Supplementary Fig. 5e,f). This may be due to the redundant function of several CRKs, which are able to induce cell death. We also could not exclude the possible contribution of other genes. Notably, SOBIR1 is also moderately induced in bak1/serk4. Thus, CRK4 is one of the client proteins of N-glycosylation and ERQC involved in BAK1/SERK4-regualted cell death.
Methods
Plant materials and growth conditions
Arabidopsis accessions Col-0 and C24 (WT), various mutants and transgenic plants used in this study were grown in soil (Metro Mix 366) in a growth room at 23 °C, 60% relative humidity, 70 μE m−2 s−1 light with a 12-h light/12-h dark photoperiod for 2 weeks before VIGS assay or 30 days for protoplast isolation. The bak1-4, serk4-1, sobir1-12, stt3a-1, hgl1-1, rsw3-1, fucTa/fucTb/xylT, erdjb3b-1, sdf2-2 and crt3-1 mutants and NahG transgenic plants have been reported previously15,25,32,41. The ire1a/ire1b and bzip60/bzip28 double mutants were obtained from Stephen H. Howell42. The sets of confirmed Arabidopsis T-DNA insertion lines (CS27941, CS27943 and 0827944), stt3a-2, ost3/6 (SALK_067271C), stt3b-2 (SALK_078498C), stt3b-3 (Salk_134449C)’ alg3 (sALK_040296C), alg10-1 (SAIL_515_F10), alg12 (SALK_200867C) and crk4 (CS859967) were obtained from the Arabidopsis Biological Resource Center (ABRC) and confirmed by PCR using primers listed in Supplementary Table 5. Seedlings were grown on agar plates containing ½ Murashige and Skoog medium (½MS) with 0.5% sucrose, 0.8% agar and 2.5 mM MES at pH 5.7, in a growth chamber at 23 °C or 30 °C, 70 μE m−2 s−1 light with a 12-h light/12-h dark photoperiod.
Plasmid construction and generation of transgenic plants
To generate the VIGS BAK1/SERK4 construct, fragments of BAK1 (319 bp) and SERK4 (310 bp) were PCR amplified from Arabidopsis Col-0 cDNA with primers listed in Supplementary Table 5, digested with EcoRI and Ncol for the SERK4 fragment, and Ncol and KpnI for the BAK1 fragment, and ligated with the pYL156 (pTRV-RNA2) vector pre-cut with EcoRI and KpnI21. To generate VIGS constructs for individual genes, fragments of SERK4 (310 bp), CLAI (541 bp), MEKK1 (520 bp) and BIR1 (491 bp) were PCR amplified from Arabidopsis Col-0 cDNA using primers listed in Supplementary Table 5, digested with EcoRI and KpnI, and ligated into the pYL156 (pTRV-RNA2) vector pre-cut with EcoRI and KpnI. All the clones were confirmed by sequencing.
The CRK4 and CRK5 genes were amplified from Col-0 cDNA with primers containing BamHI or NcoI at the amino (N) terminus and StuI at the carboxy (C) terminus (Supplementary Table 5), and ligated into a plant protoplast expression vector pHBT with a CaMV 35S promoter at the N terminus and haemagglutinin (HA) epitope tag at the C-terminus. The point mutations of CRK4N181Q and CRK4N286Q were generated by site-directed mutagenesis with primers listed in Supplementary Table 5. To construct the pCB302 binary vector containing CRKs for Agrobacterium-mediated transient expression assay in N. benthamiana, the CRKs fragment was released from the pHBT vector digested with BamHI or NcoI and StuI and ligated into the pCB302 binary vector. The Est::CRK4 binary vector construct was generated by inserting the PCR products of the CRK4 open reading frame with an HA epitope tag at its C terminus from the pHBT-CRK4-HA vector into the pER8 vector using the XhoI and SpeI sites15. All the clones were confirmed by sequencing.
STT3a complementation transgenic plants in the stt3a-2 background have been reported previously25. Arabidopsis transgenic plants were generated using Agrobacterium-mediated transformation by the floral-dip method. For oestradiol induction of CRK4 expression, the detached leaves of Est::CRK4 T1 transgenic plants were treated with 10 μM oestradiol for 24 h and transgene expression was detected using immunoblotting with α-HA antibody.
Arabidopsis protoplast and Nicotiana benthamiana transient assays
For Arabidopsis protoplast transient expression, protoplasts from WT or stt3a mutant were transfected with HA-tagged CRKs in the pHBTvector and incubated for 12 h. Proteins were isolated with 2×SDS loading buffer and subjected to immunoblot analysis with anti-HA antibody.
For N. benthamiana transient expression, Agrobacterium tumefaciens strain GV3101 containing pCB302 vector was cultured overnight in LB medium at 28 °C. Bacteria were harvested by centrifugation and resuspended with buffer (10 mM MES, pH 5.7, 10 mM MgCl2, 200 μM acetosyringone) at A600 = 0.75. Leaves of 4-week-old soil-grown N. benthamiana were hand-infiltrated using a needleless syringe with Agrobacterium cultures. Leaf samples were collected 36 h after infiltration for protein isolation and immunoblot analysis. The cell death phenotype was observed and leaf pictures were taken 4 days after infiltration under UV light with a ChemiDoc system.
Trypan blue and DAB staining
Trypan blue staining and 3,3′-diaminobenzidine (DAB) staining were performed according to procedures described previously with modifications50. Briefly, the excised plant tissues were immersed in trypan blue staining solution (2.5 mg ml−1 trypan blue in lactophenol (lactic acid, glycerol, liquid phenol and H2O in a ratio of 1:1:1:1)) or DAB solution (1 mg ml−1 DAB in 10 mM Na2HPO4 and 0.05% Tween 20). Samples were vacuum-in filtrated for 30 min and then incubated for 8 h at 25 °C with gentle shaking at 75 rpm. Subsequently, samples were transferred to trypan blue destaining solution (ethanol and lactophenol in a ratio of 2:1) or DAB destaining solution (ethanol, acetic acid and glycerol in a ration of 3:1:1) and incubated at 65 °C for 30 min. The samples were then incubated in fresh destaining solution at room temperature until complete destaining. Pictures were taken under a dissecting microscope with samples in 10% glycerol.
Agrobacterium-mediated virus-induced gene silencing assay
Plasmids containing binary TRV vectors pTRV-RNA1 and pTRV-RNA2 derivatives, pYL156-BAK1/SERK4, pYL156-SERK4, pYL156-MEKK1, pYL156-BIR1, pYL156-GFP (the vector control) or pYL156-CLA1 were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Agrobacterium cultures were first grown overnight in LB medium containing 50 pg ml−1 kanamycin and 25 pg ml−1 gentamicin and then subcultured in fresh LB medium containing 50 μg ml−1 kanamycin and 25 μg ml−1 gentamicin supplemented with 10 mM MES and 20 μM acetosyringone overnight at 28 °C in a roller drum. Cells were pelleted by 4,200 r.p.m. centrifugation, resuspended in a solution containing 10 mM MgCl2, 10 mM MES and 200 μM acetosyringone, adjusted to A600 of 1.5 and incubated at 25 °C for at least 3 h. Agrobacterium cultures containing pTRV-RNA1 and pTRV-RNA2 derivatives were mixed in a 1:1 ratio and inoculated into the first pair of true leaves of 2-week-old soil-grown plants using a needleless syringe. The pYL156-CLA1 construct, which leads to plant albino phenotype 2 weeks after Agrobacterium infiltration, was included as a visual marker for VIGS efficiency.
Supplementary Material
Acknowledgments
We thank S. H. Howell and the Arabidopsis Biological Resource Center for various Arabidopsis mutant seeds and transgenic plants, and members of the laboratories of L.S. and P.H. for discussions and comments about the experiments. The work was supported by grants from the National Institutes of Health (NIH) (R01GM092893) and the National Science Foundation (NSF) (IOS-1252539) to P.H., the NIH (R01GM097247) and the Robert A. Welch Foundation (A-1795) to L.S. and the NSF (IOS-1547551) to H.K. The Next-Generation Sequencing (NGS) was supported by a Texas AgriLife Genomics[JS(S6] Seed Grant. G.X. was partially supported by the China Scholarship Council (CSC). L.S.V. and S.A.S. were partially supported by the CAPES Foundation (Coordination for the Improvement of Higher Education Personnel), Brazil. A.C.I. was partially supported by the Rio de Janeiro State Research Foundation (FAPERJ), Brazil. A.B. was an undergraduate student supported by a NSF Research Experiences for Undergraduates (REU) programme.
Footnotes
Author contributions
M.V.V.O., G.X., B.L., L.S. and P.H. conceived and designed the experiments and wrote the manuscript with input from all co-authors. M.V.V.O., G.X., B.L., L.S.V., X.M., X.C., X.Y., S.A.S., A.C.I., A.M.M. and A.L.B performed the experiments; M.V.V.O., G.X., B.L., L.S.V., G.A.S.F., L.S. and P.H. analysed data; H.K. provided reagents.
Additional information
Supplementary information is available online.
Competing interests
The authors declare no competing financial interests.
References
- 1.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J. The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem Sci. 2014;39:447–456. doi: 10.1016/j.tibs.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 5.Gou X, et al. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012;8:e1002452. doi: 10.1371/journal.pgen.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aan den Toorn M, Albrecht C, de Vries S. On the origin of SERKs: bioinformatics analysis of the somatic embryogenesis receptor kinases. Mol Plant. 2015;8:762–782. doi: 10.1016/j.molp.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES 1 and 2 control male sporogenesis. Plant Cell. 2005;17:3337–3349. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES 1 and 2 are essential for tapetum development and microspore maturation. Plant Cell. 2005;17:3350–3361. doi: 10.1105/tpc.105.036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–U412. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 10.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postel S, et al. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol. 2010;89:169–174. doi: 10.1016/j.ejcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Roux M, et al. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/ SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23:2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 14.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 15.Meng X, et al. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol. 2015;25:2361–2372. doi: 10.1016/j.cub.2015.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature. 2015;525:265–268. doi: 10.1038/nature14858. [DOI] [PubMed] [Google Scholar]
- 17.Ladwig F, et al. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell. 2015;27:1718–1729. doi: 10.1105/tpc.15.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He K, et al. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol. 2007;17:1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 19.Kemmerling B, et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 20.Gao M, et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe. 2009;6:34–44. doi: 10.1016/j.chom.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 21.Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 2006;142:21–27. doi: 10.1104/pp.106.084624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao MH, et al. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18:1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Sun T, Zhang Y. ER quality control components UGGT and STT3a are required for activation of defense responses in Bir1-1. PLoS ONE. 2015;10:e0120245. doi: 10.1371/journal.pone.0120245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua J. Modulation of plant immunity by light, circadian rhythm, and temperature. Curr Opin Plant Biol. 2013;16:406–413. doi: 10.1016/j.pbi.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 25.Koiwa H, et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003;15:2273–2284. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farid A, et al. Specialized roles of the conserved subunit OST3/6 of the oligosaccharyltransferase complex in innate immunity and tolerance to abiotic stresses. Plant Physiol. 2013;162:24–38. doi: 10.1104/pp.113.215509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Li J. Endoplasmic reticulum-mediated protein quality control in Arabidopsis. Front Plant Sci. 2014;5:162. doi: 10.3389/fpls.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pattison RJ, Amtmann A. N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 2009;14:92–99. doi: 10.1016/j.tplants.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Farid A, et al. Arabidopsis thaliana alpha1,2-glucosyltransferase (ALG10) is required for efficient N-glycosylation and leaf growth. Plant J. 2011;68:314–325. doi: 10.1111/j.1365-313X.2011.04688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong Z, et al. Evolutionary conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:11437–11442. doi: 10.1073/pnas.1119173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howell SH. Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol. 2013;64:477–499. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- 32.Sun T, Zhang Q, Gao M, Zhang Y. Regulation of SOBIR1 accumulation and activation of defense responses in bir1-1 by specific components of ER quality control. Plant J. 2014;77:748–756. doi: 10.1111/tpj.12425. [DOI] [PubMed] [Google Scholar]
- 33.Nekrasov V, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saijo Y, et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu X, et al. Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc Natl Acad Sci USA. 2009;106:22522–22527. doi: 10.1073/pnas.0907711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA. 2009;106:15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haweker H, et al. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J Biol Chem. 2010;285:4629–4636. doi: 10.1074/jbc.M109.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Numers N, et al. Requirement of a homolog of glucosidase II beta-subunit for EFR-mediated defense signaling in Arabidopsis thaliana. Mol Plant. 2010;3:740–750. doi: 10.1093/mp/ssq017. [DOI] [PubMed] [Google Scholar]
- 39.Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin H, Hong Z, Su W, Li JM. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:13612–13617. doi: 10.1073/pnas.0906144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang JS, et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA. 2008;105:5933–5938. doi: 10.1073/pnas.0800237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng Y, Srivastava R, Howell SH. Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci USA. 2013;110:19633–19638. doi: 10.1073/pnas.1314749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S, Shan L, He P. Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci. 2014;228C:118–126. doi: 10.1016/j.plantsci.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen K, Fan B, Du L, Chen Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol Biol. 2004;56:271–283. doi: 10.1007/s11103-004-3381-2. [DOI] [PubMed] [Google Scholar]
- 45.Chen K, Du L, Chen Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol Biol. 2003;53:61–74. doi: 10.1023/B:PLAN.0000009265.72567.58. [DOI] [PubMed] [Google Scholar]
- 46.Acharya BR, et al. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 2007;50:488–499. doi: 10.1111/j.1365-313X.2007.03064.x. [DOI] [PubMed] [Google Scholar]
- 47.Liebrand TWH, van den Burg HA, Joosten MHAJ. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014;19:123–132. doi: 10.1016/j.tplants.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Schwessinger B, et al. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7:e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeh YH, Chang YH, Huang PY, Huang JB, Zimmerli L. Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Frontiers in plant science. 2015;6:322. doi: 10.3389/fpls.2015.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao XQ, et al. Bifurcation of Arabidopsis NLR immune signaling via Ca2+− dependent protein kinases. PLoS Pathog. 2013;9:14. doi: 10.1371/journal.ppat.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


