Abstract
Obesity and Type 2 diabetes (T2D) are associated with a variety of comorbidities that contribute to mortality around the world. Although significant effort has been expended in understanding mechanisms that mitigate the consequences of this epidemic, the field has experienced limited success thus far. The potential ability of brown adipose tissue (BAT) to counteract obesity and metabolic disease in rodents (and potentially in humans) has been a topical realization. Recently, there is also another thermogenic fat cell called beige adipocytes, which are located among white adipocytes and share similar activated responses to cyclic AMP as classical BAT. In this chapter, we review contemporary molecular strategies to investigate the role of adipose tissue depots in metabolism. In particular, we will discuss the generation of adipose tissue-specific knockout and overexpression of target genes in various mouse models. We will also discuss how to use different Cre (cyclization recombination) mouse lines to investigate diverse types of adipocytes.
Keywords: Adiponectin-cre, Ucp1-cre, Beige adipocyte, Rosa26-loxp-stop-loxp
1 Introduction
Adipocytes can be broadly classified as white or brown fat cells. While white fat cells are specialized to store energy in the form of triglyceride, brown adipocytes utilize their high mitochondrial content and uncoupling protein 1 (UCP1) to uncouple respiration and dissipate chemical energy as heat [1]. Rodents and other small mammals have copious brown fat deposits, but larger mammals often lose brown fat after infancy [2]. Recent data indicates that adult humans contain significant deposits of UCP1-positive brown fat that can be detected by PET-CT scanning methods [3–7]. Another type of adipocyte has been identified, which are “brown-like” cells that express UCP1 after cold exposure, but which appear within white depots rather than the classic interscapular BAT. These “inducible” cells have been called “beige” or “brite” cells [8–11]. The characteristics of BAT as an energy-burning tissue have led to interest in harnessing this activity to combat obesity. One might accomplish this by stimulating the activity of already existing BAT, or perhaps by inducing the appearance of new beige cells.
In order to characterize the biology among the three types of adipocytes, we will discuss the specific mouse models for investigating different fat depots.
2 Materials
2.1 Strategies for Generating Adipocyte-Specific Cre Mouse Line
Adiponectin BAC (90G21).
UCP1 BAC (148M1).
EL250 cell, Cre recombinase cassette with a Frt-flanked neomycin resistance cassette.
LA Taq DNA Polymerase (TaKaRa).
QIAGEN Plasmid Midiprep Kit.
2.2 Strategies for Generating Adipose Tissue Transgenic Mouse Line
Taq polymerase.
Mlu I and Nsi I enzymes and buffer.
T4 ligase.
2.2.4 KpnI enzyme.
1.0 % agarose gel.
Ultracentrifuge.
1.7-mL microcentrifuge tubes.
Microcapillary pipet.
DNA extraction kit (Qiagen).
Elutip-D column.
Additional reagents and equipment for checking the DNA concentration using a spectrophotometer.
Digestion buffer: 10 mM Tris–HCl (pH 7.6–8.0); 25 mM EDTA; 100 Mm NaCl; 0.5 % SDS.
Proteinase K.
3 Methods
3.1 Strategies for Generating Adipocyte-Specific Cre Mouse Line (Adiponectin-Cre as an Example)
Adiponectin promoter driving Cre expression (Adiponectin-cre) in white adipose tissue and brown adipose tissue (see Notes 1 and 2).
Electroporate adiponectin BAC (90G21) into EL250 cells.
Using PCR, amplify a linear dsDNA Cre recombinase cassette with a Frt-flanked neomycin resistance cassette that includes 70 bp of DNA with homologous flanking regions to adiponectin gene.
Electroporate the above PCR amplified Cre recombinase cassette into EL250 cells containing adiponectin BAC.
Use Luria-Bertani (LB) agar plates containing neomycin (40 μg/mL Kanamycin) to select positive targeting cassette into specific adiponectin region by homologous recombination.
Pick up the single positive colony and grow in 30 mL LB medium containing 40 μg/mL Kanamycin overnight.
Obtain positive BAC DNA using QIAGEN Plasmid Midiprep Kit, then sequence the whole Cre recombinase cassette.
Remove the Frt-flanked neomycin resistance cassette from the correct sequence of adiponectin BAC DNA containing Cre recombinase cassette [12].
Inject the adiponectin BAC DNA containing Cre recombinase cassette into fertilized mouse eggs.
Each founder line is crossed with the R26R tomato reporter line to map the activity pattern of Cre recombinase, then select the Adiponectin-cre founder line that expresses Cre recombinase-activated tomato in the greatest number of adiponectin cells. One example followed in the next part [12].
3.2 Strategies for Generating Adipose Tissue Transgenic Mouse Line (Rosa26-Transgenic Mice)
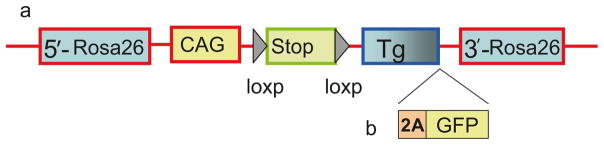
Strategies to overexpress genes in adipocytes using the aP2 (fatty acid binding protein 4, FABP4, commonly known as adipocyte Protein 2) promoter or Adiponectin promoter followed by the target genes. As previously mentioned by Kang et al., this method has its limitations, for example, variation in transgene copy number and position effects [13]. Another strategy for overexpression of genes in specific tissues is through the use of cre and loxp-stop-loxp system. Several studies have demonstrated the value of using a cre/loxp strategy; however, we are illustrating the Rosa26 targeting approach. Specifically, the transgene is cloned into the ROSA26 locus downstream of a loxP-flanked stuffer DNA sequence (STOP cassette), which abrogates transgene expression [14]. The authors modified the plasmid as shown in Fig. 1. Upon temporal and cell-type-specific induction of Cre, transcription of the transgene from the ROSA26 promoter (or from an exogenous one inserted into the ROSA26 locus) is induced as a result of the deletion of the loxP-flanked STOP cassette. To trace expression of the transgene in vivo, it can be useful to introduce downstream of the transgene an Internal Ribosome Entry Site (IRES) followed by a reporter gene coding for a fluorescence protein (such as GFP) or an enzyme (β-galactosidase) (see Note 3). However, in our case we used 2A peptide instead of IRES to increase the cleavage efficiency between genes upstream and downstream of it.
Fig. 1.
Targeting strategy to insert transgene into Rosa26 locus. Conditional transgene expressed from the CAG promoter upon Cre-mediated recombination (a). To monitor expression of the transgene in vivo a 2A-GFP cassette can be cloned downstream of the transgene of interest (b). 2A is a self-cleaving peptide, having high cleavage efficiency between genes upstream and downstream of it
3.2.1 Construct Design and Purification
Every step (from designing the transgenic construct to embryo transfer) is critical to successfully generate transgenic mice. First, preparing a clean DNA sample is a vital step because it affects the health of the embryo and the DNA integration efficiency. A DNA fragment lacking any trace of vector sequence should only be microinjected. The vector sequence, as well as any chemical residues remaining in the final DNA solution, is generally toxic to mouse zygotes and will result in death of the embryo or poor efficiency in generating transgenic mice.
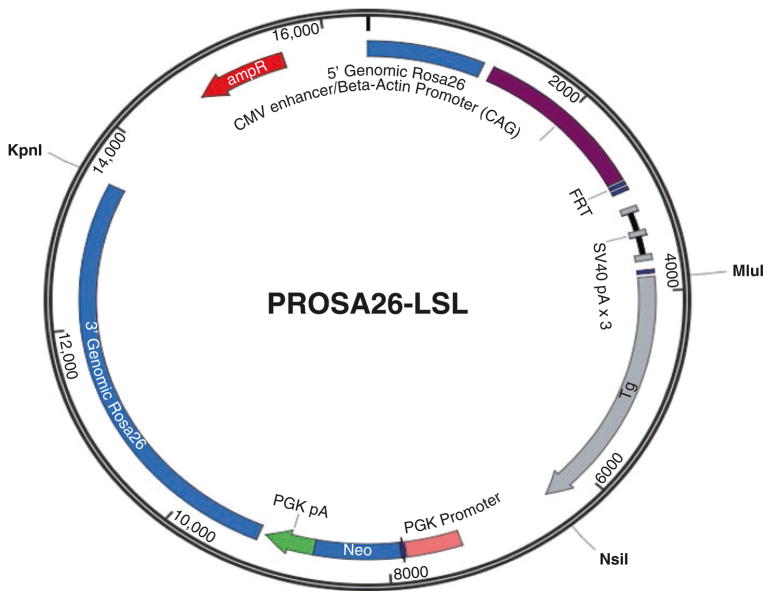
Amplify the target gene from the cDNA and use restriction enzymes and DNA ligases to cut and rejoin DNA fragments that in our case we use Mlu I and Nsi I (Fig. 2).
Linearize the cloned cassette. In our example, KpnI is used to linearize the plasmid.
DNA purification. We routinely use either of two methods to purify DNA (Elutip-D or a gel-purification method). Both methods have their own merits in that the Elutip-D method yields the purest DNA for microinjection, whereas the gel purification method is a quick and easy method to yield sufficiently DNA for microinjection. (1) Elutip-D method: use phenol: chloroform to extract and EtOH precipitate the digested DNA. Use the Elutip-D column to purify DNA. (2) Gel-purify the construct: Run the digested plasmid DNA transgenic construct on a 0.8 % agarose gel in 1× TAE buffer. Excise the transgene DNA band from the agarose gel with a clean razor blade. Use the Qiagen gel extraction kit to extract DNA.
Measure the DNA concentration. Most injection core facilities require at least 90 μg DNA to inject into ES cells.
Microinjection. DNA was injected into 129Sv embryonic stem cells (which are most commonly used) as performed by the core facility.
Fig. 2.
The construct of pRosa26-LSL
3.2.2 ES Cells Selection
Before submitting the ES cells for microinjection, collect the ES cell DNA to identify the transgene within the ES cells (Southern blot or long-range PCR). The 5′ primers are 5′-GCCAAGTGGGCAGTTTACCG-3′ and 5′-TAGGTAGGGGATCGGGACTCT-3′. The 3′ primers are 5′-GCCAGCTCATTCCTCCCACTC-3′ and 5′-GGCATGGCAATGTTCAAGCAG-3′.
Culture ES cells until confluency in a 24-well plate.
Aspirate the medium and add 400 μL of digestion buffer and 5 μL of 20 mg/mL proteinase K.
Incubate in 60 °C shaker, overnight.
Add equal volume of Phenol/Chlo, vortex (mix) well.
Extract and precipitate with 100 % cold ethanol (not add salt).
Wash twice with 70 % ethanol.
Resuspend in 30–40 μL of TE depending on the size of pellet.
Measure the concentration of DNA.
Do long-range PCR by using the primers listed above or store the samples in −20 °C.
ES cells microinjection. Intact microinjected eggs were transferred to the oviducts of pseudo-pregnant recipients by the core.
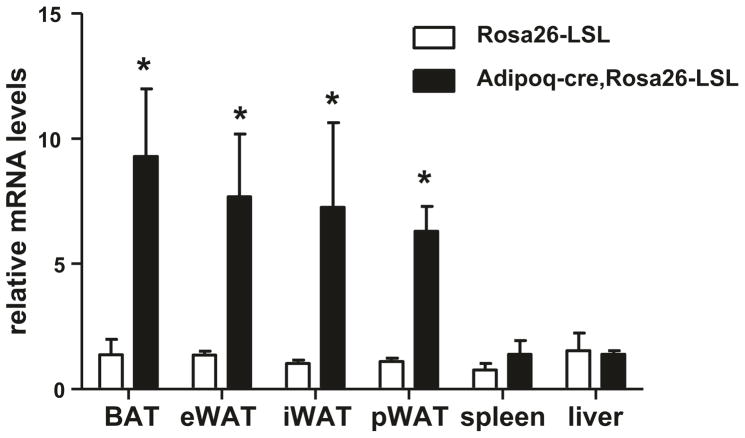
Generation of chimeric mice and identification of germ-line transmission. Ear tags or punches were applied to 2-week-old mouse pups for identification purposes and 5–10 mm tail biopsies were collected for DNA extraction. Genomic DNA was extracted and tested for the presence of Rosa26 transgene DNA with transgene specific PCR. In order to investigate the gene expression we inserted, the Rosa26 mouse was crossed with Adipoq-cre (which mentioned above) mouse, and then detect the gene expression in different fat tissues (Fig. 3) (see Note 4).
Fig. 3.
Transgene expression in different tissues. BAT brown adipose tissue, eWAT epididymal white adipose tissue, iWAT inguinal white adipose tissue, pWAT perirenal white adipose tissue
Footnotes
To achieve adipose-specific gene targeting, several Cre lines have been generated with the use of the adipose-specific gene regulatory element [15–23]. Tissue specificity and efficacy of recombination using the Cre/LoxP (cyclization recombination-locus of X over P1) system are affected by many parameters, including the genomic location and distance between the loxP sites of the target locus as well as insertion site and copy number of Cre transgene [24], reviewed in detail by Kang S. [13].
So far, the field currently lacks a Cre line that targets WAT-specific or beige-specific recombination. The Adiponectin-cre targets all fat depots while the Ucp1-cre is restricted to BAT. However, given that beige adipocytes can be driven to upregulate the expression of Ucp1 by cold and other stimuli, Ucp1-cre recombination would occur in depots outside classical BAT. Importantly, Wu and Spiegelman have identified markers selectively for beige cells, such as CD137, CD40, and TMEM26 [2]. Thus, it will be at least theoretically possible to generate a beige-specific Cre mouse line.
The traditional transgenic mice are usually generated by direct microinjection of DNA fragments into fertilized one-cell mouse embryos, followed by transfer of these embryos into recipient mothers that can carry the pregnancy to term. Our strategy is gene targeting approach requires embryonic stem cells (ES cells), which are currently only available for a very few strains of mice (i.e., 129Sv and C57/bL6J). However, the major advantage of using Rosa26 for transgenic studies is that it can overcome positional effects to produce integration-site independent, copy-number dependent, and accurate transgene expression in vivo. Importantly, the Rosa26 locus itself can be targeted with relative ease and shows broad expression across most cell types. Furthermore, contribute to the loxp-stop-loxp site, the target gene can be expressed in a time- and cell-type specific fashion.
Finally, the Cre/loxP system can control transgene expression in a time- and cell-type specific fashion. However, once induced, the transgene cannot be silenced any longer. To overcome this limitation, the tetracycline (Tet)-controlled system can be applied to generate inducible ROSA26 transgenes [25].
References
- 1.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. doi: 10.1016/j.cell.2013.12.012. S0092-8674(13)01546-8 [pii]. Epub 2014/01/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. S0092-8674(12)00595-8 [pii]. Epub 2012/07/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. 360/15/1509 [pii]. Epub 2009/04/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirbolooki MR, Constantinescu CC, Pan ML, Mukherjee J. Quantitative assessment of brown adipose tissue metabolic activity and volume using 18F-FDG PET/CT and β3-adrenergic receptor activation. EJNMMI Res. 2011;1(1):30. doi: 10.1186/2191-219X-1-30. 2191-219X-1-30 [pii]. Epub 2012/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. 360/15/1518 [pii]. Epub 2009/04/10. [DOI] [PubMed] [Google Scholar]
- 6.von Heydebreck A, Huber W, Poustka A, Vingron M. Identifying splits with clear separation: a new class discovery method for gene expression data. Bioinformatics. 2001;17(Suppl 1):S107–S114. doi: 10.1093/bioinformatics/17.suppl_1.s107. Epub 2011/08/02. [DOI] [PubMed] [Google Scholar]
- 7.Orava J, Nuutila P, Lidell ME, Oikonen V, Noponen T, Viljanen T, et al. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metab. 2011;14(2):272–279. doi: 10.1016/j.cmet.2011.06.012. S1550-4131(11)00261-0 [pii]. Epub 2011/08/02. [DOI] [PubMed] [Google Scholar]
- 8.Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121(1):96–105. doi: 10.1172/JCI44271. 44271 [pii]. Epub 2010/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285(10):7153–7164. doi: 10.1074/jbc.M109.053942. M109.053942 [pii]. Epub 2009/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishibashi J, Seale P. Medicine. Beige can be slimming. Science. 2010;328(5982):1113–1114. doi: 10.1126/science.1190816. science.1190816 [pii]. Epub 2010/05/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. nature07182 [pii]. Epub 2008/08/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhillon H, Zigman JM, Ye C, Lee CE, McGovern RA, Tang V, et al. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron. 2006;49(2):191–203. doi: 10.1016/j.neuron.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 13.Kang S, Kong X, Rosen ED. Adipocyte-specific transgenic and knockout models. Methods Enzymol. 2014;537:1–16. doi: 10.1016/B978-0-12-411619-1.00001-X. B978-0-12-411619-1.00001-X [pii]. Epub 2014/02/01. [DOI] [PubMed] [Google Scholar]
- 14.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. Epub 1999/01/23. [DOI] [PubMed] [Google Scholar]
- 15.Graves RA, Tontonoz P, Platt KA, Ross SR, Spiegelman BM. Identification of a fat cell enhancer: analysis of requirements for adipose tissue-specific gene expression. J Cell Biochem. 1992;49(3):219–224. doi: 10.1002/jcb.240490303. Epub 1992/07/01. [DOI] [PubMed] [Google Scholar]
- 16.Ross SR, Graves RA, Greenstein A, Platt KA, Shyu HL, Mellovitz B, et al. A fat-specific enhancer is the primary determinant of gene expression for adipocyte P2 in vivo. Proc Natl Acad Sci U S A. 1990;87(24):9590–9594. doi: 10.1073/pnas.87.24.9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barlow C, Schroeder M, Lekstrom-Himes J, Kylefjord H, Deng CX, Wynshaw-Boris A, et al. Targeted expression of Cre recombinase to adipose tissue of transgenic mice directs adipose-specific excision of loxP-flanked gene segments. Nucleic Acids Res. 1997;25(12):2543–2545. doi: 10.1093/nar/25.12.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409(6821):729–733. doi: 10.1038/35055575. Epub 2001/02/24. [DOI] [PubMed] [Google Scholar]
- 19.He W, Barak Y, Hevener A, Olson P, Liao D, Le J, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. 2536828100 [pii]. Epub 2003/12/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13(3):249–259. doi: 10.1016/j.cmet.2011.02.005. Epub 2011/03/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai T, Jiang M, Chambon P, Metzger D. Impaired adipogenesis and lipolysis in the mouse upon selective ablation of the retinoid X receptor alpha mediated by a tamoxifen-inducible chimeric Cre recombinase (Cre-ERT2) in adipocytes. Proc Natl Acad Sci U S A. 2001;98(1):224–228. doi: 10.1073/pnas.011528898. 011528898 [pii]. Epub 2001/01/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ZV, Deng Y, Wang QA, Sun K, Scherer PE. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology. 2010;151(6):2933–2939. doi: 10.1210/en.2010-0136. en.2010-0136 [pii]. Epub 2010/04/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell. 2014;158(1):69–83. doi: 10.1016/j.cell.2014.04.049. S0092-8674(14)00723-5 [pii]. Epub 2014/07/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kos CH. Cre/loxP system for generating tissue-specific knockout mouse models. Nutr Rev. 2004;62(6 Pt 1):243–246. doi: 10.1301/nr2004.jun243-246. [DOI] [PubMed] [Google Scholar]
- 25.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44(1):23–28. doi: 10.1002/gene.20180. Epub 2006/01/10. [DOI] [PubMed] [Google Scholar]