SUMMARY
During cell division, genome inheritance is orchestrated by microtubule attachments formed at kinetochores of mitotic chromosomes. The primary microtubule coupler at the kinetochore, the Ndc80 complex, is regulated by Aurora kinase phosphorylation of its N-terminal tail. Dephosphorylation is proposed to stabilize kinetochore-microtubule attachments by strengthening electrostatic interactions of the tail with the microtubule lattice. Here, we show that removal of the Ndc80 tail, which compromises in vitro microtubule binding, has no effect on kinetochore-microtubule attachments in the C. elegans embryo. Despite this, preventing Aurora phosphorylation of the tail results in prematurely stable attachments that restrain spindle elongation. This premature stabilization requires the conserved microtubule-binding Ska complex, which enriches at attachment sites prior to anaphase onset to dampen chromosome motion. We propose that Ndc80 tail dephosphorylation promotes stabilization of kinetochore-microtubule attachments via the Ska complex and that this mechanism ensures accurate segregation by constraining chromosome motion following biorientation on the spindle.
Keywords: Centromere, Kinetochore, Chromosome Segregation, Mitosis, Cell Division, Ndc80 Complex, Ska Complex, Microtubule, Cell Polarity
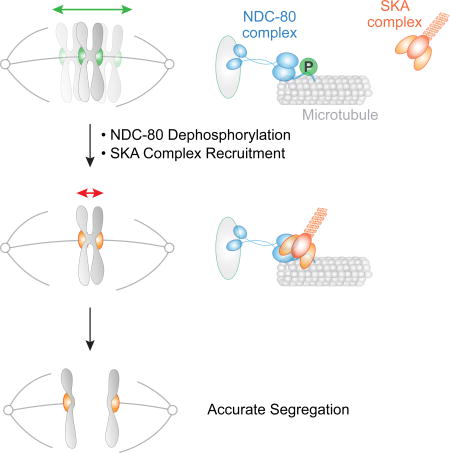
Phosphorylation by Aurora kinases regulates kinetochore-microtubule attachments during mitosis. Cheerambathur et al. show that in C. elegans one-cell embryos, Aurora phosphorylation of the Ndc80 complex, the major microtubule-binding factor at the kinetochore, controls kinetochore-microtubule attachments indirectly by regulating the recruitment of an effector, the microtubule-binding Ska complex.
INTRODUCTION
During cell division, the replicated genome is partitioned into daughter cells by interactions between the kinetochore, a multiprotein complex assembled on the centromere of mitotic chromosomes, and the spindle microtubules (Cheeseman, 2014). Accurate segregation requires bioriented kinetochore-microtubule attachments, in which sister chromatids are connected to microtubules from opposite spindle poles. Following initial capture by kinetochore-localized motors, kinetochores form end-on attachments to microtubules that are mediated by the conserved four-subunit Ndc80 complex (Cheeseman et al., 2006; Ciferri et al., 2008; Deluca et al., 2006; Wei et al., 2007). End-on kinetochore-microtubule attachments are initially dynamic to facilitate correct attachment geometry and chromosome alignment. These attachments are eventually stabilized, as best shown by reduced turnover of kinetochore microtubules in vertebrate cells (Kabeche and Compton, 2013). The suppression of kinetochore-microtubule dynamics after bi-orientation is associated with posttranslational modifications as well as alterations in kinetochore composition (Cheerambathur and Desai, 2014; Foley and Kapoor, 2012). Notably, misregulation of this step is thought to be the major source of chromosome segregation errors in cancer cells(Godek et al., 2014).
The microtubule binding activity of the Ndc80 complex resides in two distinct elements in the N-terminus of its Ndc80 subunit: a basic unstructured N-terminal tail with dispersed Aurora B kinase phosphorylation sites, and the adjacent Calponin Homology (CH) domain (Ciferri et al., 2008). High-resolution cryo-EM of Ndc80 complex-decorated microtubules indicates that the Ndc80 CH domain contacts the microtubule lattice (Alushin et al., 2012; 2010; Wilson-Kubalek et al., 2008). Mutations that disrupt this interface abrogate microtubule binding in vitro and impair attachments in vivo (Ciferri et al., 2008; Lampert et al., 2012; Sundin et al., 2011; Tooley et al., 2011). The N-terminal tail, whose deletion compromises in vitro microtubule binding, to a similar extent as CH domain mutations (Ciferri et al., 2008; Wei et al., 2007), is proposed to electrostatically interact with the negatively charged tubulin surface. However, the contribution of the Ndc80 tail to microtubule attachments in vivo differs across species. In mammalian cells, deletion of the N-tail destabilizes kinetochore-microtubule attachments, whereas, N-tail deletion does not obviously perturb chromosome segregation in one-cell C. elegans embryos and is not lethal in budding yeast (Cheerambathur et al., 2013; Guimaraes et al., 2008; Kemmler et al., 2009; Miller et al., 2008).
Phosphorylation of the Ndc80 tail, by Aurora family kinases, is implicated in regulating the dynamics of kinetochore-microtubule attachments. In mammalian cells, phosphorylation of Aurora target residues is high during early mitosis and low on bioriented chromosomes (DeLuca et al., 2011). Phosphorylation by Aurora kinase, or the introduction of phosphomimetic mutations in the Ndc80 tail, reduces microtubule binding affinity of the Ndc80 complex in vitro (Alushin et al., 2012; Cheeseman et al., 2006; Umbreit et al., 2012). In addition, phosphomimetic Ndc80 variants compromise kinetochore fiber stability and chromosome alignment in mammalian cells (Alushin et al., 2012; Guimaraes et al., 2008; Zaytsev et al., 2014). These observations have led to the proposal that N-tail phosphorylation by Aurora kinase regulates kinetochore-microtubule attachments by modulating the electrostatic affinity between the basic N-tail and the negatively-charged microtubule lattice in a graded manner (Zaytsev et al., 2015).
In addition to phosphorylation-based modulation of microtubule affinity, multiple additional factors, such as the Dam1 complex in fungi, the Ska complex in metazoans, and XMAP215 family proteins are known to enhance Ndc80’s microtubule coupling in vitro and this cooperation is proposed to be integral to proper kinetochore-microtubule attachment dynamics in vivo (Miller et al., 2016; Schmidt et al., 2012; Tien et al., 2010). Although it is known that these microtubule-associated proteins depend on Ndc80 for their kinetochore localization, how this additional machinery collaborates with the Ndc80 complex to control the properties of microtubule attachments in vivo and whether phosphoregulation of the Ndc80 complex is involved is not well understood. Notably, neither the Dam1 complex nor the Ska complex interact with the Ndc80 complex robustly in solution, highlighting that their synergy with the Ndc80 complex requires the context of the microtubule lattice (Lampert et al., 2010; Schmidt et al., 2012; Tien et al., 2010).
Here, we employ sensitive assays in the one-cell C. elegans embryo to show that dephosphorylation of the N-tail alters kinetochore-microtubule attachment dynamics via the Ska complex. We find that, while the N-tail deletion has no effect on kinetochore-microtubule attachments in vivo, abrogating N-tail phosphorylation suppresses attachment dynamics and prevents spindle elongation until the activation of a spindle-external force prior to anaphase. The suppression of attachment dynamics by the non-phosphorylatable tail is dependent on the Ska complex, indicating that N-tail phosphorylation maintains a dynamic kinetochore-microtubule interface by restraining the action of the Ska complex. Removal of the Ska complex prevents the dampening of chromosome motion that occurs prior to anaphase onset in wild-type embryos and compromises viability. These findings reveal a mechanism that acts after chromosome biorientation to suppress chromosome motion and ensure accurate segregation.
RESULTS
Subtle Kinetochore-Microtubule Attachment Defects Can be Detected in vivo by Analyzing Spindle Pole Separation Rates Prior to Anaphase
To investigate the role of NDC-80 N-tail phosphorylation, we established an in vivo assay to monitor the strength of kinetochore-microtubule attachments in the one-cell C. elegans embryo. This assay is based on a method we employed previously in which the kinetics of spindle pole separation following nuclear envelope breakdown (NEBD), measured by live imaging of embryos expressing GFP fusions to histone H2b and γ-tubulin (Fig. 1A), is used to infer the presence or absence of kinetochore-microtubule attachments (Desai et al., 2003; Oegema et al., 2001).
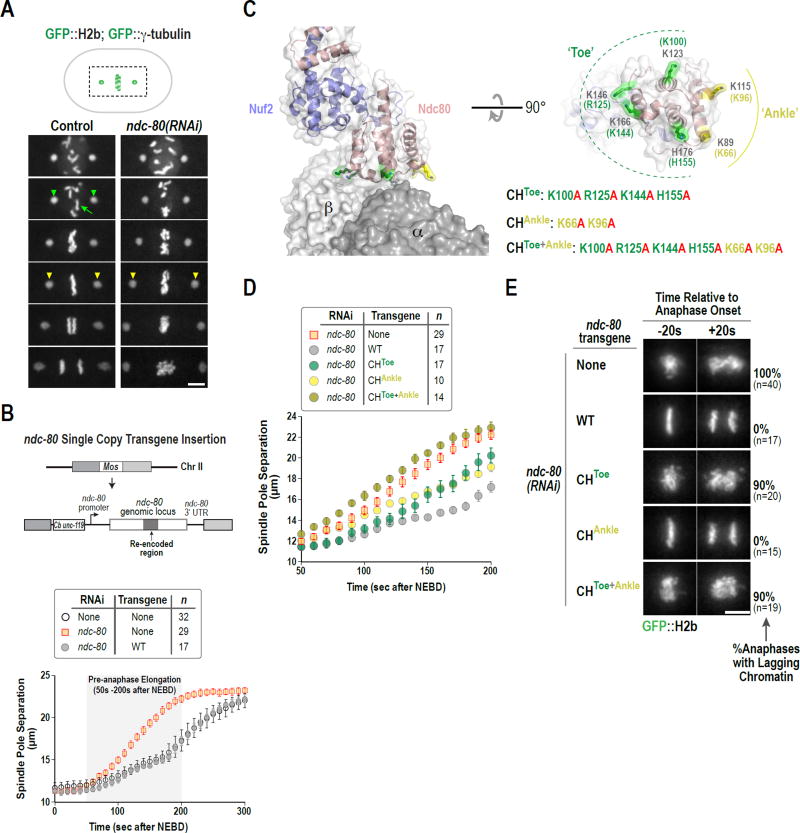
Figure 1. Quantitative analysis of spindle elongation discriminates NDC-80 mutations in vivo that alter microtubule-binding affinity.
(A) Stills from movies of control and ndc-80(RNAi) one-cell embryos expressing GFP::H2b and GFP::γ-tubulin, to label chromosomes (green arrow) and spindle poles (green arrowheads), respectively. Time is in seconds after NEBD. Yellow arrowheads highlight premature spindle pole separation in ndc-80(RNAi). Scale bar, 5 µm.
(B) Single copy transgene insertion-based NDC-80 replacement system. Spindle pole tracking analysis below shows rescue of ndc-80(RNAi) by transgene-encoded RNAi-resistant wild-type NDC-80. Error bars are the 95% confidence interval.
(C) Structure of the human Ndc80-Nuf2 head (pink/blue) bound to a tubulin dimer (gray, labeled α/β) in the microtubule lattice (PDB 3IZ0). The Toe and Ankle residues of the Ndc80 CH domain are colored in green and yellow, respectively. A rotated en face view of the microtubule-binding surface of the CH domain is on the right. Residue numbers are for human Ndc80 (black) and C. elegans NDC-80 (green/yellow).
(D) Spindle pole separation analysis for the indicated CH domain mutants following endogenous NDC-80 depletion. The 50s-200s interval after NEBD is plotted. Error bars are the 95% confidence interval. WT and No Transgene data are the same as in Fig. 1B.
(E) Chromosome segregation phenotypes for the indicated CH domain mutants highlighted by image stills from time-lapse movies 20 sec prior to and after anaphase onset; percentage of one-cell embryo anaphases with detectable lagging chromatin are indicated on the right. Scale bar, 5 µm. See also Fig. S1.
Spindle poles in embryos begin to separate ~50s after NEBD (Fig. 1B). The kinetics of spindle pole separation from this time point until anaphase onset (~200s after NEBD) is influenced by kinetochore-microtubule attachments, which resist spindle elongation forces. In control embryos, pole separation occurs at a rate of 2.0 µm/min during the pre-anaphase interval (50s-200s after NEBD). In embryos depleted of NDC-80, chromosome segregation fails and spindle poles separate rapidly (4.5 µm/min) during this interval (Fig. 1A,B & S1A, B). The 2.2-fold increase in pole separation velocity in ndc-80(RNAi) highlights the importance of NDC-80-mediated attachments in restraining pre-anaphase spindle elongation. The effect of NDC-80 depletion also indicates that kinetochore-microtubule attachments on aligning chromosomes in wild type embryos are bearing load and are likely undergoing tension-induced microtubule polymerization to allow spindle elongation.
To determine whether the spindle pole separation assay could detect a reduction in affinity of the Ndc80 complex for microtubules, rather than absence of attachments, we engineered mutants in the NDC-80 CH domain that are predicted to reduce microtubule-binding affinity. Based on prior work (Ciferri et al., 2008; Tooley et al., 2011), we targeted 2 sets of residues for mutation. The first set includes 4 residues—K100, R125, K144 and H155—which form the ‘Toe’ that contacts the inter-tubulin interface (Fig. 1C) (Alushin et al., 2010). We refer to the second set of 2 residues, K66 and K96, as the ‘Ankle’ based on their positioning relative to the Toe residues (Fig. 1C).
To assess the effect of mutating CH domain residues on kinetochore microtubule attachments in vivo, we employed a previously described NDC-80 replacement system (Cheerambathur et al., 2013) to generate mutants in which the ‘Toe’, ‘Ankle’, or ‘Toe + Ankle’ residues were mutated to alanine. In this approach, an RNA interference (RNAi)– resistant ndc-80 transgene is inserted in single copy at a fixed genomic locus under the control of endogenous ndc-80 regulatory sequences. A wildtype (WT) ndc-80 transgene (encoding NDC-80WT) fully rescued both NDC-80 depletion (Fig. 1B) and a ndc-80 null mutant, and immunoblotting has confirmed that transgene-encoded WT and various engineered NDC-80 mutants express at comparable levels to endogenous NDC-80 (Cheerambathur et al., 2013).
To monitor phenotypes associated with the ‘Toe’, ‘Ankle’, or ‘Toe + Ankle’ affinity mutants, the transgenes were introduced into a strain expressing GFP::H2b and GFP::γ-tubulin. Quantitative analysis of spindle pole separation kinetics after endogenous NDC-80 depletion revealed that the rate of pre-anaphase spindle pole separation for the CHToe+Ankle mutant (4.4 µm/min) was nearly identical to that for NDC-80 depletion (‘No Transgene’; Fig. 1D). In contrast, the rates for the CHToe and CHAnkle mutants were intermediate between the rates for NDC-80WT and NDC-80 depletion (Fig. 1D & S1A,B), which is consistent with prior work (Lampert et al., 2012; Tooley et al., 2011) suggesting that ‘Toe’ and ‘Ankle’ residues make independent contributions to the affinity of NDC-80 for microtubules. Notably, the CHToe mutation by itself resulted in a severe chromosome segregation defect, whereas the CHAnkle mutation did not (Fig. 1E), indicating that, while both sets of residues are important for attachment strength/dynamics, the Toe residues are more important for biorientation and segregation than the Ankle residues.
Based on the analysis of these three NDC-80 CH domain mutants, we conclude that the spindle pole separation assay is sufficiently sensitive to detect reduced affinity of NDC-80 for microtubules in vivo, even when the effects are mild enough to not overtly disrupt chromosome segregation.
Deletion of the NDC-80 N-Tail Does Not Affect Load-Bearing Kinetochore-Microtubule Attachment Formation in One-Cell Embryos
Deletion of the basic NDC-80 N-tail (pI=11.6) substantially reduces the in vitro microtubule binding affinity of the reconstituted C. elegans NDC-80 complex, as also observed for Ndc80 complexes from other species (Cheerambathur et al., 2013; Ciferri et al., 2008; Lampert et al., 2012; Umbreit et al., 2012; Wei et al., 2007). Surprisingly, in prior work we did not observe a significant effect of deletion of the N-Tail (NDC-80ΔTail) on kinetochore-microtubule attachments or on chromosome segregation in one-cell C. elegans embryos (Cheerambathur et al., 2013), which we confirmed in independent experiments (Fig. 2A).
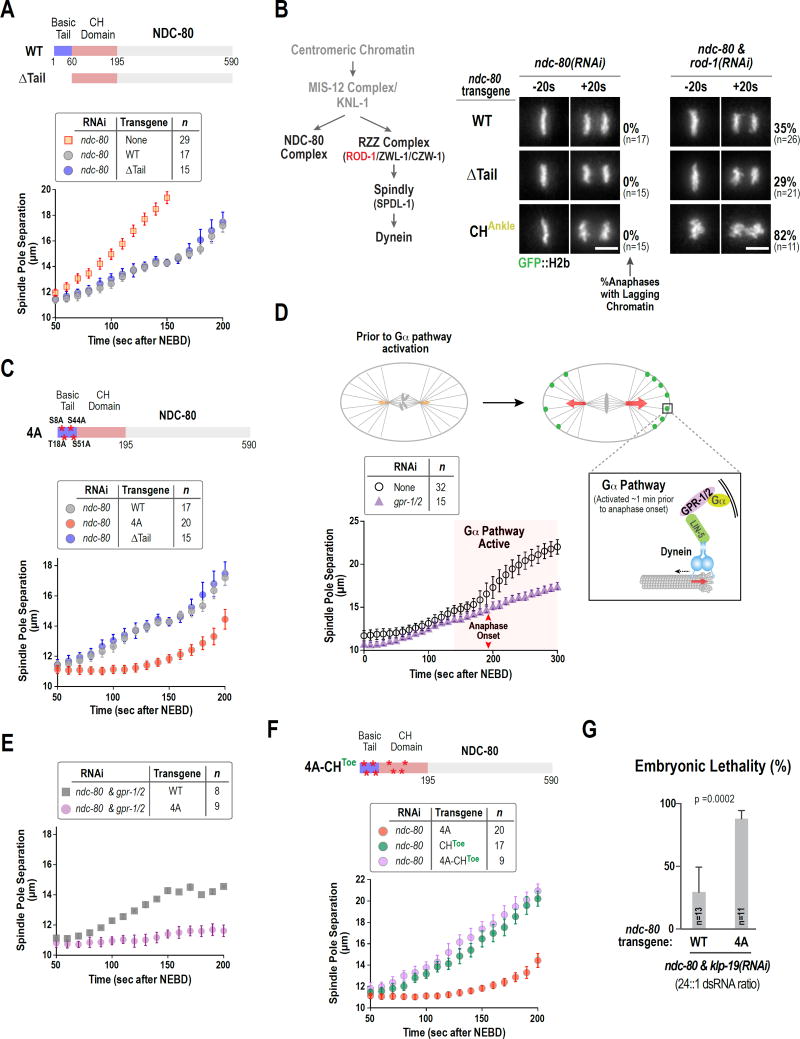
Figure 2. N-Tail deleted NDC-80 exhibits no defects in kinetochore-microtubule attachments whereas a non-phosphorylatable N-tail mutant restrains spindle elongation.
(A) Comparison of WT and ΔTail NDC-80 in the spindle pole separation assay. Error bars are the 95% confidence interval. WT data is the same as in Fig. 1B.
(B) Summary of NDC-80 complex and dynein motor recruitment to the C. elegans kinetochore (left) and chromosome segregation phenotypes for the indicated conditions (right). Scale bar, 5 µm.
(C) Spindle pole separation analysis of the indicated conditions. WT and ΔTail data are the same as in Fig. 1B and Fig. 2A, respectively. Error bars are the 95% confidence interval.
(D) Schematic of the Gα pathway that generates an asymmetric force for spindle displacement starting ~1 min prior to anaphase onset (orange & red arrows indicate forces; black dashed arrow indicates direction of motor; adapted from (Gönczy and Rose, 2005). Graph shows effect of Gα pathway inhibition by depletion of GPR-1/2 in the spindle pole separation assay. Red arrows mark anaphase onset, which is not affected by GPR-1/2 depletion. Error bars are the 95% confidence interval.
(E) & (F) Spindle pole separation analysis of the indicated conditions. Error bars are the 95% confidence interval. 4A and CHToe alone data is the same as in Fig. 2C and Fig. 1D, respectively.
(G) Embryonic lethality in WT and 4A NDC-80 following weak depletion of KLP-19. n=number of worms. See also Figs. S2 & S3.
To determine if there was a subtle kinetochore-microtubule attachment defect associated with absence of the NDC-80 N-Tail, we combined NDC-80ΔTail with removal of kinetochore-localized dynein motor. Dynein is recruited to kinetochores by the Rod/Zwilch/Zw10(RZZ)-Spindly module (ROD-1/ZWL-1/CZW-1 and SPDL-1 in C. elegans; Fig. 2B) and can be selectively removed from kinetochores by ROD-1 depletion. ROD-1 depletion leads to delayed formation of load-bearing attachments (reflected in premature pole separation that recovers prior to anaphase onset) and ~30% frequency of lagging anaphase chromatin in one-cell embryos (Fig. 2B,S2A). Even in the sensitized ROD-1 depletion, NDC-80ΔTail behaved identically to NDC-80WT (Fig. 2B,S2A). By contrast, the CHAnkle mutant of NDC-80 significantly enhanced the chromosome segregation and spindle pole separation phenotypes of ROD-1 depletion (Fig. 2B,S2A). We further assessed NDC-80ΔTail by combining it with CHToe mutations in the same transgene. The combined ΔTail-CHToe NDC-80 mutant behaved identically to the CHToe mutant in the spindle pole separation assay (Fig. S2B,C), showing no enhancement. Again, this was in contrast to the enhancement observed with the combined CHToe and CHAnkle mutations (Fig. 1D). Thus, following removal of kinetochore dynein, or in the presence of CH domain mutations, NDC-80ΔTail behaves equivalently to NDC-80WT.
We conclude that even with combinatorial inhibitions and a sensitive in vivo assay that is capable of identifying mutations that reduce the affinity of NDC-80 for microtubules, NDC-80ΔTail behaves similarly to NDC-80WT, suggesting that the N-tail does not make a significant contribution to the affinity of NDC-80 for microtubules at kinetochores in the one-cell C. elegans embryo.
Mutation of Aurora Phosphorylation Sites in the N-Tail Restrains Spindle Elongation Until Activation of the Gα Pathway
The C. elegans NDC-80 N-tail harbors four Aurora kinase phosphorylation sites that are conserved in related species (Fig S2D). Mutation of these four sites to alanine abrogates Aurora kinase-mediated phosphorylation of the reconstituted NDC-80 complex, and mutation of these sites to aspartate significantly reduces NDC-80 complex microtubule binding affinity in vitro (Cheerambathur et al., 2013; Cheeseman et al., 2006). To address the role of these Aurora target sites in vivo, we analyzed the consequences of preventing phosphorylation by changing them to alanines (NDC-804A). In contrast to NDC-80ΔTail (Fig. 2A), NDC-804A exhibited a striking phenotype in the spindle pole separation assay (Fig. 2C)—pre-anaphase spindle pole separation was significantly inhibited, with almost no spindle elongation between 50s and 130s after NEBD. To determine which Aurora kinase phosphorylates these residues in vivo, we analyzed spindle pole separation in a temperature-sensitive mutant of Aurora B/AIR-2 (Severson et al., 2000; Lewellyn et al., 2011) or following depletion of the Aurora A/AIR-1 activator TPX2/TPXL-1 (Lewellyn et al., 2011; Ozlü et al., 2005; Severson et al., 2000). This analysis suggested that the N-tail sites are likely targeted by Aurora A rather than Aurora B in vivo (Fig. S3A-E). Aurora A has also been implicated in control of kinetochore-microtubule attachments and Ndc80 tail phosphorylation in other species (Chmátal et al., 2015; Ye et al., 2015).
Although spindle elongation was inhibited by NDC-804A for a substantial portion of the NEBD-anaphase onset interval, ~1 min prior to anaphase onset the spindle poles began to separate at a rate comparable to controls (Fig. 2C). Previous work has shown that during this pre-anaphase interval, the cell polarity-regulated G-protein (Gα) pathway activates cortical dynein to generate forces pulling on the asters that position the metaphase spindle and promote its elongation (Fig. 2D;(Gönczy and Rose, 2005; Labbé et al., 2004; Nguyen-Ngoc et al., 2007). Thus, we wondered if Gα-dependent pulling forces were responsible for the late spindle elongation observed in NDC-804A. To test this, we inhibited the Gα pathway by depletion of its essential regulators GPR-1 and GPR-2 (Fig. 2D; (Colombo et al., 2003)) which does not alter the NEBD-anaphase interval, relative to control embryos, or result in chromosome segregation defects (Fig. S2E). In gpr-1/2(RNAi) embryos, the spindle elongation rate was initially similar to that in controls; however, the increase in the spindle elongation rate that occurs ~1 min prior to anaphase onset in controls, failed to occur, indicating that it is mediated by Gα pathway activation (Fig. 2D; S2E; red shaded region). In the NDC-804A mutant, inhibition of the Gα-dependent force, by depletion of GPR-1/2, abolished the spindle pole separation observed prior to anaphase onset (Fig. 2E; compare to Fig. 2D). Thus, the non-phosphorylatable NDC-80 N-tail restrains spindle elongation prior to Gα activation, during the interval when the chromosomes are aligning (50–130s after NEBD, Fig. 1A,B) but does not inhibit elongation once the Gα-dependent forces, by pulling on the asters, ramp up the tension at kinetochore-microtubule attachment sites in the spindle.
Next, to test if the suppression of spindle elongation prior to Gα activation by the non-phosphorylatable tail requires the ability of the CH domain to dock onto microtubules, we combined the non-phosphorylatable N-tail mutant with the CHToe mutant in the same ndc-80 transgene (Fig. 2F). Comparison of CHToe and 4A-CHToe forms of NDC-80 revealed identical phenotypes (Fig. 2F,S2F), indicating that the inhibition of spindle elongation observed in NDC-804A is dependent on the docking of the CH Toe to the microtubule lattice.
The NDC-804A mutant on its own exhibited normal chromosome alignment and segregation and supported embryo viability (Fig. S2F; Cheerambathur et al., 2013). To test if the NDC-804A mutant exhibited chromosome segregation phenotypes in a sensitized background, we compromised chromosome alignment by reducing the activity of the kinesin-4 family chromokinesin KLP-19. In one-cell C. elegans embryos KLP-19 provides an important non-kinetochore force for positioning and orienting chromosomes on the spindle (Powers et al., 2004). Weak inhibition of KLP-19 (generated by combining ndc-80 and klp-19 dsRNAs in 24:1 ratio) resulted in modest (~30%) embryonic lethality in the presence of NDC-80WT that was significantly elevated (to ~90%) in the presence of NDC-804A (Fig. 2G). Thus, NDC-80 N-tail phosphorylation promotes embryonic viability when the activity of the chromokinesin KLP-19 is reduced.
Suppression of Spindle Elongation by the Non-Phosphorylatable NDC-80 Tail Mutant Requires the SKA Complex
The results above suggest that the presence of the unphosphorylated NDC-80 N-tail restrains spindle elongation, likely by altering the properties of kinetochore-microtubule attachments. One possibility for how the unphosphorylated N-tail could accomplish this is by promoting the recruitment of an effector that suppresses attachment dynamics. A candidate for such an effector is the Ska complex, a kinetochore-localized microtubule-binding complex that is conserved across metazoans and is suggested to be a functional counterpart of the Dam1 complex in fungi (Gaitanos et al., 2009; Welburn et al., 2009). Inhibition of the Ska complex in cultured mammalian cells compromises kinetochore-microtubule interactions (Gaitanos et al., 2009; Raaijmakers et al., 2009; Welburn et al., 2009). Localization of the Ska complex to kinetochores is dependent on the Ndc80 complex and regulated by Aurora kinase activity (Chan et al., 2012). Although the two complexes do not interact significantly in solution, the Ska complex enhances the microtubule binding affinity and end-tracking ability of the Ndc80 complex in vitro (Schmidt et al., 2012). These observations have led to the model that the Ndc80 complex recruits the Ska complex to kinetochores and their cooperation generates robust kinetochore-microtubule attachments.
In C. elegans, the SKA complex is heterotrimeric, consisting of two SKA-1 subunits and one SKA-3 subunit (Fig. 3A; Schmidt et al., 2012). To determine if the SKA complex is required for the non-phosphorylatable NDC-80 N-tail to prevent spindle elongation, we compared the effect of SKA-3 depletion on spindle pole elongation kinetics in NDC-80WT versus NDC-804A (Fig. 3A). In the presence of NDC-80WT, SKA-3 depleted embryos exhibited spindle pole separation dynamics similar to those in controls. In contrast, SKA complex depletion in the non-phosphorylatable NDC-804A mutant eliminated the inhibition of spindle pole separation (Fig. 3A). Thus, the ability of the non-phosphorylatable NDC-80 N-tail to inhibit spindle elongation is dependent on the SKA complex.
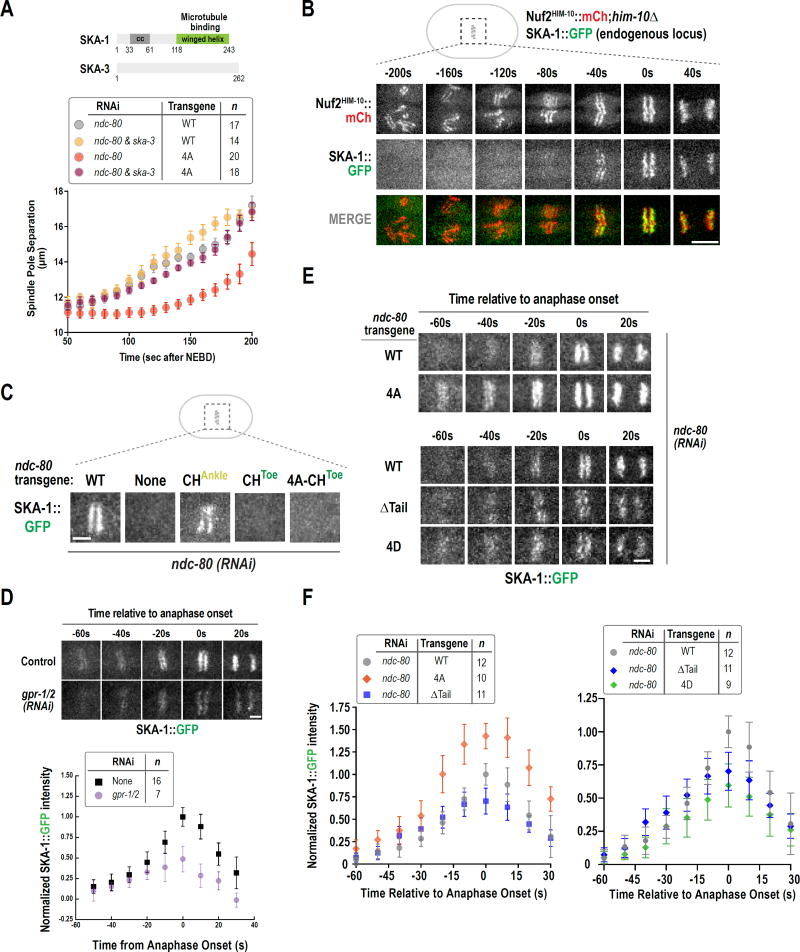
Figure 3. The non-phosphorylatable NDC-80 N-tail mutant suppresses spindle elongation via the SKA complex.
(A) Schematics of C. elegans SKA complex subunits SKA-1 and SKA-3. Spindle pole separation analysis for the indicated conditions. WT and 4A alone data are same as in Fig. 1B and Fig. 2C, respectively. Error bars are the 95% confidence interval.
(B) Stills from a time-lapse movie of a one-cell embryo co-expressing mCherry-tagged NDC-80 complex and GFP-tagged SKA complex. Scale bar, 5 µm.
(C) SKA-1::GFP localization in the indicated conditions. 10–12 embryos were imaged per condition. Scale bar, 2.5 µm.
(D) (top) Stills from time-lapse movies of SKA-1::GFP localization in control and gpr-1/2(RNAi). (bottom) Quantification of SKA-1::GFP chromosomal intensity relative to anaphase onset. Error bars are the 95% confidence interval. Measured fluorescence intensities were normalized to the averaged maximal intensity value in control.
(E) Stills from time-lapse movies of SKA-1::GFP chromosomal localization in the indicated conditions. Scale bar, 2.5 µm.
(F) Quantification of SKA-1::GFP chromosomal intensity relative to anaphase onset for the indicated conditions, performed as in Fig. 3D above. See also Fig. S4.
Dephosphorylation of the NDC-80 N-tail and Activation of the Gα Pathway Both Promote SKA Complex Recruitment
Given that the SKA complex is required for the dephosphorylated N-tail to restrain spindle elongation, we next assessed if there was a difference in SKA complex localization between NDC-80WT and NDC-804A embryos. We first analyzed a strain co-expressing SKA-1 tagged at its endogenous locus with GFP and an mCherry fusion with the NDC-80 complex subunit Nuf2HIM-10 (Fig. 3B). The NDC-80 complex localized to the diffuse kinetochores of C. elegans throughout mitosis, as expected from prior work (Desai et al., 2003). By contrast, robust SKA-1 localization was only observed in a brief time window beginning ~60s prior to anaphase onset, after chromosomes were aligned at the metaphase plate (Fig. 3B; Movie S1).
SKA-1 localization to kinetochores required NDC-80 (Fig. 3C), as expected from prior analysis in mammalian cells (Gaitanos et al., 2009; Raaijmakers et al., 2009; Welburn et al., 2009). Notably, SKA-1 recruitment was abolished by mutation of the Toe residues of the NDC-80 CH domain; by contrast, SKA-1 recruitment was not significantly affected by the CHAnkle mutant (Fig. 3C). Thus, SKA-1 localization is dependent on the NDC-80 CH domain Toe docking to the microtubule lattice.
The abrupt recruitment of the SKA complex (Fig. 3B) correlated with the activation of the Gα-dependent pulling forces that likely place bioriented kinetochore-microtubule interfaces under higher tension (Fig. 2D). Therefore, we next tested whether SKA-1 recruitment was affected by gpr-1/2 depletion. Localization of SKA-1 was significantly diminished following inhibition of Gα-dependent forces (Fig. 3D), suggesting that force applied onto NDC-80 bound to microtubules via its CH domain promotes a conformation that is recognized by the SKA complex.
In the NDC-804A mutant, spindle elongation is restrained in a SKA complex-dependent manner (Fig. 3A) suggesting that tail dephosphorylation influences the recruitment of SKA. Consistent with this idea, SKA-1 localization was significantly elevated in the presence of NDC-804A compared to NDC-80WT (Fig. 3E,F). However the presence of a non-phosphorylatable tail is not sufficient to recruit SKA-1, as SKA-1 localization was not observed in a combined non-phosphorylatable N-tail-CHToe mutant (Fig. 3C). These results suggest that the non-phosphorylatable N-tail promotes adoption of a microtubule-bound conformation of NDC-80 recognized by the SKA complex. Additional experiments suggested that tail dephosphorylation modulates but is not essential for the rapid increase in SKA-1 recruitment that occurs concomitant with Gα activation. There is a decrease in the maximum intensity of SKA-1 recruited by NDC-80ΔTail relative to NDC-80WT (Fig. 3E,F & S4B), and a phosphomimetic 4D tail mutation, which behaves analogously to NDC-80WT and NDC-80ΔTail in the spindle pole separation assay (Fig. S4A), also reduced SKA recruitment (Fig. 3E,F & S4B).
Taken together, the above data suggest that in wild-type embryos N-tail dephosphorylation and increased pulling forces resulting from Gα activation both contribute to SKA-1 recruitment after chromosomes have aligned at the metaphase plate.
The SKA Complex Dampens Chromosome Movement on the Spindle
The functional effect of the non-phosphorylatable N-tail—suppression of spindle elongation—is mediated by the SKA complex, suggesting that recruitment of the SKA complex modulates the properties of the kinetochore-microtubule interface. To address the function of the SKA complex, we used CRISPR/Cas9 to delete ska-1 (Fig. S5A) and also used RNAi to deplete SKA-3. We optimized imaging conditions to monitor chromosome dynamics at high temporal resolution (image stack acquisition every 2 seconds) and developed a method to quantify chromosome distribution over time.
In control embryos, chromosomes rapidly move (by ~100s after NEBD) to the vicinity of the metaphase plate but continue to exhibit dynamic movement. In the ~40 seconds prior to anaphase onset this motion is dampened (Fig. 4A). The tightly aligned, motion-dampened metaphase plate is then separated into two relatively evenly shaped chromosome masses in anaphase (Fig. 4A). Imaging of chromosome dynamics revealed that the dampening of movement that tightens the metaphase plate is significantly perturbed by both mutant (ska-1Δ) and RNAi (ska-3) inhibition of the SKA complex (Fig. 4A). In both conditions, chromosomes continued to exhibit dynamic movements after reaching the spindle equator, underwent large excursions from the metaphase plate and were observed to separate as irregular-shaped chromosome masses in anaphase (Fig. 4A; Movie S2). In 8 of 15 embryos imaged at high time resolution, individual chromosomes were observed to abruptly detach from the main chromosome mass and realign (green arrowhead Fig. 4A; Movie S2), a behavior that was not observed in 13 control embryos imaged under equivalent conditions.
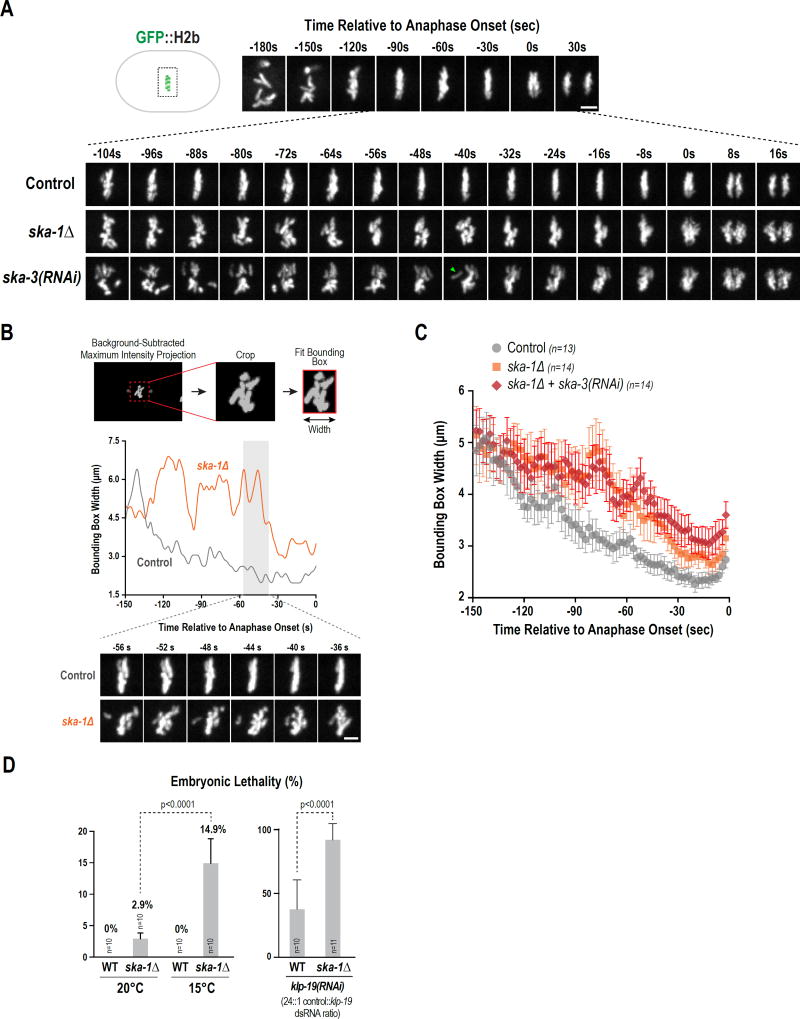
Figure 4. The SKA complex is required to dampen chromosome motion and contributes to embryonic viability.
(A) High temporal resolution analysis (6-plane z-stack at 1.5 µm spacing acquired every 2s) of chromosome dynamics. Low time resolution (30s interval) stills from early prometaphase to just after anaphase onset are shown for control on top and higher time-resolution stills for the indicated conditions during the selected interval are shown below. Scale bar, 2.5 µm.
(B) Method employed to analyze chromosome distribution along the spindle axis using a bounding box. Individual control and ska-1Δ traces are shown; stills below illustrate chromosome behavior. Scale bar, 2.5 µm.
(C) Average plot of chromosome distribution along the spindle axis for the indicated conditions. Error bars are the 95% confidence interval.
(D) (left) Comparison of embryonic lethality in WT and ska-1Δ at 20°C and 15°C. (right) Plot of embryonic lethality in control and ska-1Δ following partial depletion of KLP-19. n=number of worms. See also Fig. S5.
To quantify chromosome dynamics, we used a bounding box to encase the chromosomes in a maximal intensity projection of a background-subtracted image and plotted the width of the bounding box along the spindle axis over time. Both individual (Fig. 4B, S5B) and averaged (Fig. 4C) traces are shown. In both controls and SKA complex inhibitions, chromosomes became confined to a smaller spindle region over time. However, individual traces indicated broader chromosome distribution along the spindle axis and abrupt chromosome motions in the absence of the SKA complex (Fig. 4B), and the averaged traces indicated a significant difference in chromosome confinement on the spindle between the control and SKA complex inhibitions (Fig. 4C). We did not observe any effect of SKA complex inhibitions on the NEBD-anaphase onset interval (Fig. S5C), indicating that the role proposed for this complex in promoting anaphase onset in mammalian tissue culture cells (Sivakumar et al., 2016) is not conserved in C. elegans. ska-1Δ worms are homozygous viable at 20°C but exhibit mi ld embryonic lethality (~3%) that was elevated 5-fold at 15°C, a temperature that des tabilizes microtubules (Fig 4D). In addition, ska-1Δ worms were sensitive to reduction in activity of the chromokinesin KLP-19 (Fig. 4D), as also observed for NDC-804A (Fig 4D). Thus, while SKA complex-mediated dampening of chromosome motion after biorientation is not as essential for viability as NDC-80 complex-mediated attachments, it contributes to viability.
We conclude that the SKA complex is required for dampening chromosome motion after alignment to tightly confine bi-oriented chromosomes on the spindle and coordinate their segregation during anaphase. Based on the effect of NDC-804A, which requires the SKA complex, we suggest that this dampening is due to an alteration of the dynamic nature of NDC-80-mediated attachments. Moreover, this dampening after biorientation contributes to organismal viability, likely by ensuring accurate chromosome segregation.
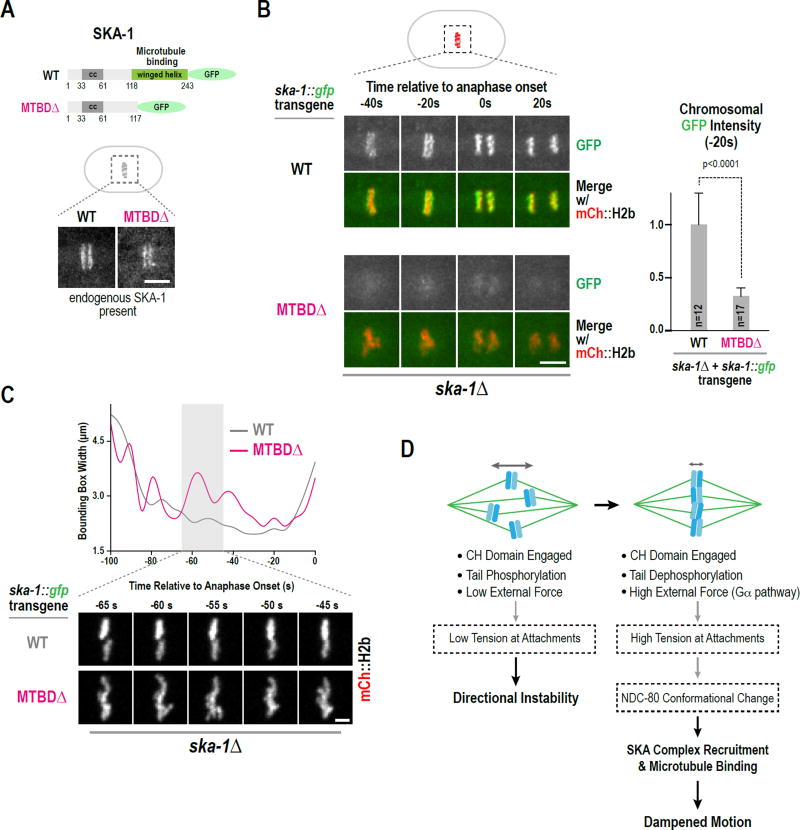
The Microtubule-Binding Domain of SKA-1 Contributes to Its Localization and Function
The microtubule-binding activity of the Ska complex has been mapped to a conserved C-terminal domain of Ska1 that adopts a winged-helix fold (Abad et al., 2014; Schmidt et al., 2012). In vitro analysis of the C. elegans SKA complex has shown that deletion of this domain inhibits microtubule binding but does not disrupt complex formation (Schmidt et al., 2012); in addition the domain and the intact complex both bind microtubules with similar weak affinity (Kd ~6–10 µM). To determine if this conserved domain contributes to SKA complex function in vivo, we employed a targeted transgene-based replacement system to generate strains expressing SKA-1::GFP (WT) and a comparable fusion lacking the microtubule-binding domain (MTBDΔ) (Fig. 5A; Fig. S5D). In the presence of endogenous SKA-1, both the wildtype and MTBDΔ SKA-1 localized to metaphase kinetochores (Fig. 5A). This observation suggests that MTBDΔ SKA-1 is expressed and associates with SKA- 3, which is in agreement with in vitro analysis (Schmidt et al., 2012). When the transgene-encoded SKA-1 variants were crossed into ska-1Δ, wild-type SKA-1 localized robustly but localization of the MTBDΔ mutant was significantly compromised (~30% relative to wild-type SKA-1 just prior to anaphase onset; Fig. 5B). This result suggests that the localization of MTBDΔ SKA-1 observed in the presence of endogenous SKA-1 is likely due to its association with wildtype SKA-1 in the trimeric SKA complex. Consistent with the reduced localization of MTBDΔ SKA-1, imaging of chromosome dynamics highlighted wider chromosome distribution with excursions away from the plate, phenotypes similar to ska-1Δ but less severe in magnitude (Fig. 5C; Fig. S5E). Thus, the microtubule-binding domain of SKA-1 contributes to NDC-80-dependent SKA complex localization on bioriented chromosomes and to the dampening of chromosome motion prior to anaphase onset.
Figure 5. The microtubule-binding domain of SKA-1 contributes to SKA complex localization and function.
(A) Schematic of transgene-expressed GFP fusions of SKA-1; the microtubule-binding domain deletion (MTBDΔ) is identical to the one characterized in vitro (Schmidt et al., 2012). Images below show metaphase stage embryos in strains where endogenous SKA-1 is present. Scale bar, 5 µm.
(B) Stills from time-lapse movies for the indicated conditions. The graph on the right plots background-subtracted chromosomal GFP fluorescence for the frame (−20s) prior to anaphase onset. Measurements were normalized to the mean value of wild-type SKA-1::GFP. Error bars are the 95% confidence interval. Scale bar, 5 µm.
(C) Examples of bounding box width analysis of mCherry::H2b timelapse sequences, performed as in Fig. 4B; stills below illustrate chromosome behavior for the two analyzed conditions. Scale bar, 2.5 µm.
(D) Model for dampening of chromosome motion after biorientation. Boxes with dashed lines contain speculations discussed in the text. See also Fig. S5.
DISCUSSION
Mechanisms that ensure chromosome biorientation on the spindle prior to anaphase onset, such as the spindle checkpoint or error correction pathways, have been the focus of many efforts to understand the high fidelity of chromosome segregation (Godek et al., 2014). Here, we describe a mechanism that acts after chromosome biorientation and contributes to organismal viability, likely by ensuring accurate segregation. Specifically, our results suggest that both N-tail dephosphorylation and spindle-external force-induced tension promote a specific conformation of microtubule-bound NDC-80 that recruits the SKA complex to dampen motion of bioriented chromosomes and spatially constrain them prior to their segregation in anaphase (Fig. 5D). This dampening is functionally important as the absence of the SKA complex compromises viability.
It has been appreciated for over a decade that kinetochore-microtubule attachments are regulated by phosphorylation of the basic N-terminal tail of the Ndc80 subunit of the Ndc80 complex (Deluca et al., 2006). The predominant view on the molecular mechanism underlying this regulation, based on in vitro studies, is that phosphorylation counteracts electrostatic interactions of the basic Ndc80 N-tail with the acidic microtubule lattice (Alushin et al., 2012; Cheeseman et al., 2006; Ciferri et al., 2008; Wei et al., 2007; Zaytsev et al., 2015). In contrast to this view, we show that the NDC-80 N-tail does not make a detectable contribution to microtubule affinity at kinetochores in the one-cell C. elegans embryo. Nevertheless, analogous to what has been reported in vertebrate cells (DeLuca et al., 2011), a non-phosphorylatable mutant of the NDC-80 N-tail prematurely stabilizes kinetochore-microtubule attachments. As this effect of non-phosphorylatable NDC-80 is entirely dependent on the SKA complex, we suggest that in metazoan species where both complexes are present, Ndc80 tail phosphorylation modulates kinetochore-microtubule attachment dynamics either exclusively or in part via the Ska complex. In vertebrate cells, Ndc80 tail deletion or alteration of tail phosphorylation is known to perturb kinetochore-microtubule interactions, and it should be feasible to test if the Ska complex contributes to these effects. Notably, work described in the accompanying manuscript (Janczyck et. al., 2017) indicates that a specific alteration of the N-tail of human Ndc80 perturbs Ska complex localization to kinetochores, revealing a link between the N-tail and the Ska complex in human cells.
In the C. elegans embryo, the action of the SKA complex on NDC-80-mediated attachments dampens chromosome motion prior to anaphase onset. Dampening of chromosome motion could be achieved by stabilizing a paused state of dynamic microtubules at the kinetochore or by promoting slow continual polymerization at bi-oriented sister kinetochores; the latter is likely the case in the C. elegans embryo, as spindle elongation continues to occur at a slow rate until anaphase onset. The abrupt detachment of individual chromosomes observed in the absence of the SKA complex also suggests that SKA helps strengthen NDC-80-mediated attachments. This idea should be testable in reconstitution experiments employing optical trapping, analogous to those conducted for budding yeast kinetochores (Tien et al., 2010).
Structural studies and in vitro microtubule-binding analysis have shown that the Ska1 subunit of the Ska complex directly binds to microtubules and synergizes with the Ndc80 complex (Abad et al., 2014; Schmidt et al., 2012). Our results show that SKA complex enrichment at the kinetochore-microtubule interface in the C. elegans embryo depends on the Toe residues in the NDC-80 CH domain and, to a significant extent, on the microtubule-binding domain of SKA-1. We propose that the SKA complex recognizes a specific microtubule-bound conformation of the NDC-80 complex and concomitant microtubule binding by the conserved winged-helix domain of SKA-1 provides additional affinity to enable SKA complex recruitment and alteration of attachment dynamics at kinetochores. We note that the microtubule-binding domain deletion does not eliminate SKA complex localization and does not fully phenocopy ska-1Δ. Thus, we suspect that the SKA complex lacking this domain is able to recognize and modulate NDC-80 complex-mediated attachments to a degree, an idea that will need to be explored in future work. The distinct microtubule lattice features recognized by the Ndc80 and Ska complexes (Abad et al., 2014; Alushin et al., 2012; Schmidt et al., 2012) are also likely to be important for their integrated action at the kinetochore-microtubule interface.
In human cells, deletion of the microtubule-binding domain of Ska1 does not prevent kinetochore localization, although it appears reduced (Redli et al., 2016; Schmidt et al., 2012). The human Ska complex has an additional subunit (Ska2), is hexameric (C. elegans SKA complex is trimeric), and the Ska3 subunits are significantly divergent. Thus, it is possible that the human Ska complex has intrinsic affinity for the Ndc80 complex in the absence of microtubule binding. While robust biochemical association of purified recombinant human Ska and Ndc80 complexes has not been reported to date, this possibility is supported by localization of Ska to kinetochores lacking microtubule attachments following Aurora B kinase inhibition in human cells (Chan et al., 2012). The microtubule-binding domain of human Ska1 has also been suggested to bind protein phosphatase 1 (Sivakumar et al., 2016) and to bind and activate Aurora B (Redli et al., 2016). We have not observed reduction in protein phosphatase 1 (GSP-2) localization at kinetochores or phenotypes associated with reduced Aurora B activity in ska-1Δ (not shown), suggesting that these proposed interactions are not conserved in C. elegans.
The abrupt timing of SKA complex recruitment observed in the C. elegans embryo is striking. We suggest that this timing is dictated primarily by the generation of a microtubule-bound NDC-80 conformation that is promoted by tension at attachment sites (Fig. 5D). In this view, the dephosphorylated N-tail or other mechanisms, including spindle-external force generation pathways such as the Gα/LGN pathway or poleward flux of kinetochore microtubules observed in other species (Matos et al., 2009), act in a unified manner by promoting generation of a microtubule-bound Ndc80 conformation that recruits Ska to trigger the synergy between these two microtubule-binding complexes. We note that a role for the N-terminal tail in Ndc80 conformation control is supported by analysis employing a conformation-sensitive sensor engineered into Ndc80 in budding yeast, which lacks Ska but harbors the functionally analogous microtubule-binding Dam complex (Suzuki et al., 2016). One important unanswered question with respect to timing of SKA recruitment is the mechanism of Ndc80 tail dephosphorylation. While specific kinetochore-localized phosphatases are suggested as being involved in dephosphorylation of Aurora substrates at the kinetochore (Liu et al., 2010; Suijkerbuijk et al., 2012), the precise mechanisms controlling Ndc80 tail dephosphorylation and its relationship to attachment and tension at the kinetochore-microtubule interface remain to be clarified.
RNAi analysis of the Ska complex in human tissue culture cells has suggested a role in kinetochore-microtubule attachments as well as in promoting the metaphase-anaphase transition (Gaitanos et al., 2009; Schmidt et al., 2012; Sivakumar et al., 2016). Our analysis of the C. elegans SKA complex using both a null ska-1 mutant and RNAi-mediated depletions of both subunits affirmed a function for this complex in the regulation of attachment dynamics after biorientation but did not reveal a role in controlling the timing of the metaphase-anaphase transition. In contrast to mutants that disrupt docking of the NDC-80 CH domain docking onto the microtubule lattice, loss of SKA complex function did not lead to penetrant lethality, indicating that SKA activity is not essential for biorientation. Nonetheless, loss of the SKA complex compromised organismal viability, especially when microtubule stability was reduced or when a different pathway that aids chromosome segregation was weakened. Thus, we propose that SKA-mediated alteration of dynamics at the kinetochore-microtubule interface after biorientation functions as a safety mechanism to ensure that daughter cells inherit the correct genome complement. Misregulation of such a mechanism may contribute to premature kinetochore microtubule stability-linked chromosome segregation errors in cancer cells, a possibility that will be interesting to address in future work.
STAR Methods
Contact for Reagent and Resource Sharing
For further information, and requests regarding reagents should be directed to and will be fulfilled by the Lead Contact, Arshad Desai (abdesai@ucsd.edu).
Experimental Model and Subject Details
The genotypes of the C. elegans strains used in this study are described in Reagents and Resources section Strains were maintained at 20°C, except for temperature-sensitive strains, which were maintained at 16°C.
Method Details
C. elegans transgenic strain construction
Strains with integrated single-copy ndc-80 transgenes, which are RNAi-resistant and ska-1::gfp transgenes were constructed using the MosSCI method (Frøkjaer-Jensen et al., 2008). The ndc-80 transgene was described previously (Cheerambathur et al., 2013). In brief, the ndc-80 genomic locus including introns, 2000 bp upstream of the start codon and 1000 bp downstream of the stop codon are present in the transgene. A 776 bp region spanning exons 3–5 was re-encoded in the transgene as described (Cheerambathur et al., 2013) to make the transgene-encoded product resistant to a dsRNA targeting this region of endogenous ndc-80. The engineering of the ska-1::gfp transgene is described in Fig. S5D. Single-copy insertions were confirmed by PCR analysis flanking the insertion site.
Endogenous GFP tagging and deletion of the ska-1 locus were done using CRISPR/Cas9 (Dickinson et al., 2013). For ska-1::gfp, the repair template included homology arms flanking the linker sequence (GGRAGSG) and GFP (1667 bp right homology arm comprised of the ska-1 coding sequence and 5’ UTR and 2050 bp left homology arm comprised of the 3’ UTR and the downstream gene locus Y106G6H.13). A Cb-unc-119 selection marker flanked by loxp sequences was inserted between the GFP and the 3’ homology arm of the repair template. For ska-1 deletion, the repair template was generated by inserting a Cb-unc-119 selection marker flanked by loxp sequences between the appropriate homology arms (2000 bp right homology arm comprised of the 5’ UTR of ska-1 coding sequence and 2050 bp left homology arm comprised of the 3’ UTR and the downstream gene locus Y106G6H.13). To modify the ska-1 locus using Cas9-triggered homologous recombination, the repair template (50ng/ µL) was co-injected with a plasmid (40ng/ µL), modified from pDD162, containing Cas9 and respective guide RNA sequences (5’-AAATTTTCATCGGTCTCTT-3’ for ska-1::gfp and 5’-AATGGAATCGTTTATTGAT-3’ for ska-1Δ) and three plasmids encoding fluorescent markers for negative selection [pCFJ90 (Pmyo-2::mCherry, 2.5 ng/µL), pCFJ104 (Pmyo-3::mCherry, 5 ng/µL) and pGH8 (Prab-3::mCherry, 10 ng/µL)] into the strain HT1593. Recombinant strains were identified by selecting moving worms without fluorescent markers and were confirmed by PCR spanning both homology regions. The Cb-unc-119 selection marker was later deleted using Cre recombinase-mediated excision. An injection mix containing pDD104 (Peft-3::Cre, 50 ng/µL) and coinjection markers [pCFJ90 (Pmyo-2::mCherry, 2.5 ng/µL) and pCFJ104 (Pmyo-3::mCherry, 5 ng/µL)] was injected into the gonads of unc-119(+) young adult ska-1::gfp or ska-1Δ animals. Cre-excised animals were identified by selecting for the Unc phenotype and were confirmed by PCR. The unc-119 mutation was subsequently removed by mating with N2 males.
RNA-mediated Interference
Double-stranded RNA synthesis was performed by first using PCR to generate DNA templates using the primers and templates listed in Table S1 in the supplement. PCR reactions were cleaned using the QIAquick PCR Purification Kit (Qiagen). Individual T3 and T7 transcription reactions using MEGAscript® T7/T3 Transcription Kit (ThermoFisher Scientific) on each template were used to generate single-stranded RNAs. The two reactions were purified using the MEGAclear™ Kit (ThermoFisher Scientific), mixed and annealed at 37°C for 30 minutes to generate double- stranded RNAs (Oegema et al., 2001). L4 hermaphrodites were injected with dsRNA and incubated at 20°C for 36–46h before dissection and imaging of embryos. For ndc-80&rod-1(RNAi) and ndc-80&gpr-1/2(RNAi), 1 mg/mL individual dsRNAs were mixed at a 1:1 ratio, except for the analysis of embryonic lethality following ndc-80&klp-19 or control hyls-1&klp-19 depletions, where a ratio of 24:1 was used to mildly reduce KLP-19 activity.
Embryonic lethality assays
For assaying embryonic lethality of ska-1Δ strains at different temperatures, L4 worms were singled onto plates and allowed to lay eggs at 15° and 20°C. After 24 hours the mother was removed and the number of eggs and hatched larvae were scored 24 hours later. N2 worms were used as controls.
For embryonic lethality assays involving partial depletions of klp-19, L4 worms were injected with dsRNA mixes containing klp-19 and ndc-80 dsRNAs or klp-19 and hyls-1 dsRNAs and incubated at 20°C. The adult worms were singled onto plates after 36 hours and viability of embryos during the 36–48 hour interval was determined.
Live Imaging, Fluorescence Intensity Quantification and Image Analysis
For imaging one-cell embryos, hermaphrodites were dissected in M9 buffer and transferred to a freshly prepared 2% agarose pad and covered with a 22×22 mm coverslip. In experiments that involved imaging of the temperature-sensitive air-2(or207ts) strain, the dissection and imaging were performed as described in (Lewellyn et al., 2011). In brief, for pre-meiotic upshift, worms were placed in a pre-heated microscope room (26.5°C) for 30–60 min prior to dissection and imaging. For post-meiotic upshift, worms were kept at ~16°C in the pre-heated room, dissected and imaged at the restrictive temperature.
For pole-tracking analysis of one cell embryos expressing GFP::Histone H2B and GFP::γ-tubulin, 5×2 µm z-stacks were collected with 2×2 binning and a 60X 1.3 NA U-planApo objective (Olympus) at 10 second intervals and 150 ms exposure at 19°C on a DeltaVision microscope (Applied Precision) equipped with a CoolSnap camera (Roper Scientific). Maximum intensity projections of z-stacks were made using Image J (Fiji) and pole tracking profiles were generated by measuring spindle length (the distance between the two centrosomes in the projections) over time using Metamorph (Molecular Devices). Spindle pole separation was plotted relative to Nuclear Envelope Breakdown (NEBD). NEBD was scored as the frame where the diffuse intranuclear histone signal equilibrated with the cytoplasm and chromosomes exhibited abrupt rapid movement indicating interaction with microtubules. Anaphase onset was scored as the frame where two distinct masses of sister chromosomes were first visible. Pre-anaphase and anaphase spindle elongation rates were calculated using linear regression analysis on the spindle pole elongation profiles within time intervals 50–200s and 200–300s, respectively.
Images of one-cell embryos expressing GFP::H2B or mCherry::H2B for analysis of chromosome dynamics, SKA1::GFP, and GFP::β-tubulin;mCherry::H2B were collected using a confocal imaging system (Revolution XD Confocal System; Andor Technology) with a spinning disk confocal scanner unit (CSU-10; Yokogawa Corporation of America) mounted on an inverted microscope (TE2000-E; Nikon) equipped with solid-state 100-mW lasers, 60X, 1.4 NA Plan Apochromat (for imaging embryos expressing SKA1::GFP or GFP::β-tubulin;mCherry::H2B) and 100X, 1.4 NA Plan Apochromat lens (for chromosome dynamics analysis and SKA1::GFP imaging in Fig. 3B), and a back-illuminated EMCCD camera (binning 1×1; iXon DV887; Andor Technology).
To quantify SKA-1::GFP fluorescence, 5×2 µm z-stacks were acquired at 20 second intervals and 500 ms exposure. Z-stacks were projected and fluorescence intensity was quantified using Image J (Fiji). A rectangular box was drawn around the SKA-1::GFP fluorescence and the integrated intensity in the box was recorded. The box was expanded by 5 pixels on each side, and the integrated intensity was measured. The signal and area difference between the expanded box and the original box were used to calculate the average background signal per pixel. The integrated chromosomal SKA-1::GFP intensity in the original box was then calculated by subtracting the background signal.
For minimum bounding box analysis used to quantify chromosome distribution, 5×1.5 µm z-stacks were acquired every 2 s in the case of GFP::H2B or 5 s in the case of mCherry::H2B. For each z-stack, a maximum intensity projection image was generated using Image J (Fiji), and rotated to position the spindle poles horizontally. Next, the intensity of all projection images of the timelapse z-series was normalized, converted to 8-bit, and both spindle poles cropped out. Subsequently, the background signal was subtracted, and to each processed projection image a minimum bounding box was fitted (only including pixels with value > 0, i.e. histone signal from the chromosomes) and the width of this box was recorded.
For generating kymographs, a maximum intensity projection of each z-stack (5×2 µm), was made using Image J (Fiji), and rotated to position the spindle poles horizontally. Next, a 30-pixel wide linescan was drawn through the spindle region and kymograph was generated using the kymograph plugin in Image J (Fiji).
Quantification and Statistical Analysis
Details of the methods employed to extract and quantify various parameters in microscopy datasets are described in the image analysis section. The statistical tests used to determine significance are described in the figure legends.
Supplementary Material
Movie S1: Localization of the SKA and NDC-80 complexes in the one-cell C. elegans embryo. SKA-1 was tagged at its endogenous locus with GFP. The NDC-80 complex subunit Nuf2HIM-10 was fused to mCherry and expressed from a single copy transgene insertion. Image z-stacks were acquired every 20 seconds and maximal intensity projections generated to visualize the dynamics of the 2 complexes. Playback rate is 100X-fold relative to real-time. Related to Figure 3.
Movie S2: Dynamics of chromosomes in control and ska-1 deletion embryos. Embryos expressed GFP::H2b, were imaged at high time resolution (z-stack acquisition every 2 seconds), and maximal intensity projections generated to visualize chromosome behavior. Related to Figure 4.
Highlights.
The Ndc80 tail is dispensable for microtubule affinity at C. elegans kinetochores
Preventing Ndc80 tail phosphorylation nonetheless stabilizes microtubule attachments
The Ska complex is required for stabilization by the dephosphorylated Ndc80 tail
Ska complex recruitment dampens chromosome motion to promote accurate segregation
Acknowledgments
We thank Paul Maddox for suggesting Gα pathway analysis, and Rebecca Green for comments on the manuscript. This work was supported by an NIH grant (GM074215) to A.D; A.D. and K.O. receive salary and other support from the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.K.C. and A.D. initiated the project; D.K.C conducted and analyzed the majority of experiments; B.P. performed SKA imaging and high-time-resolution chromosome dynamics assays; N.H. generated and analyzed the microtubule-binding domain deletion; L.L. performed the Aurora B mutant analysis; K.D.C. generated structural models and guided mutational and biochemical analysis; K.O. provided conceptual and experimental advice and helped guide the effort; the manuscript was prepared by D.K.C, K.O., and A.D., with input from all authors.
References
- Abad MA, Medina B, Santamaria A, Zou J, Plasberg-Hill C, Madhumalar A, Jayachandran U, Redli PM, Rappsilber J, Nigg EA, et al. Structural basis for microtubule recognition by the human kinetochore Ska complex. Nat Commun. 2014;5:2964. doi: 10.1038/ncomms3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat. Struct. Mol. Biol. 2012;19:1161–1167. doi: 10.1038/nsmb.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J. Cell Biol. 2012;196:563–571. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Desai A. Linked in: formation and regulation of microtubule attachments during chromosome segregation. Curr. Opin. Cell Biol. 2014;26:113–122. doi: 10.1016/j.ceb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Gassmann R, Cook B, Oegema K, Desai A. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 2013;342:1239–1242. doi: 10.1126/science.1246232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM. The kinetochore. Cold Spring Harb Perspect Biol. 2014;6:a015826. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Chmátal L, Yang K, Schultz RM, Lampson MA. Spatial regulation of kinetochore microtubule attachments by destabilization at spindle poles in meiosis I. Curr. Biol. 2015;25:1835–1841. doi: 10.1016/j.cub.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Reis, Dos G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gönczy P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- Deluca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca KF, Lens SMA, Deluca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell. Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Rybina S, Müller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2012;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen S-P, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. Embo J. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godek KM, Kabeche L, Compton DA. Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat. Rev. Mol. Cell Biol. 2014;1:57–64. doi: 10.1038/nrm3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Rose LS. Asymmetric cell division and axis formation in the embryo. WormBook. 2005:1–20. doi: 10.1895/wormbook.1.30.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janczyck PL, Skorupka KA, Tooley J, Matson DR, Kestner CA, West T, Pornillos O, Stukenberg PT. Ndc80 recruits and orients Ska to kinetochores to generate a mature microtubule attachment. 2017 doi: 10.1016/j.devcel.2017.04.020. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, Ortíz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. Embo J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé J-C, McCarthy EK, Goldstein B. The forces that position a mitotic spindle asymmetrically are tethered until after the time of spindle assembly. J. Cell Biol. 2004;167:245–256. doi: 10.1083/jcb.200406008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J. Cell Biol. 2012;200:21–30. doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn L, Carvalho A, Desai A, Maddox AS, Oegema K. The chromosomal passenger complex and centralspindlin independently contribute to contractile ring assembly. J. Cell Biol. 2011;193:155–169. doi: 10.1083/jcb.201008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos I, Pereira AJ, Lince-Faria M, Cameron LA, Salmon ED, Maiato H. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J. Cell Biol. 2009;186:11–26. doi: 10.1083/jcb.200904153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MP, Asbury CL, Biggins S. A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell. 2016;165:1428–1439. doi: 10.1016/j.cell.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr. Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gönczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozlü N, Srayko M, Kinoshita K, Habermann B, O'Toole ET, Müller-Reichert T, Schmalz N, Desai A, Hyman AA. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev. Cell. 2005;9:237–248. doi: 10.1016/j.devcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Powers J, Rose DJ, Saunders A, Dunkelbarger S, Strome S, Saxton WM. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J. Cell Biol. 2004;166:991–1001. doi: 10.1083/jcb.200403036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J. Cell. Sci. 2009;122:2436–2445. doi: 10.1242/jcs.051912. [DOI] [PubMed] [Google Scholar]
- Redli PM, Gasic I, Meraldi P, Nigg EA, Santamaria A. The Ska complex promotes Aurora B activity to ensure chromosome biorientation. J. Cell Biol. 2016;215:77–93. doi: 10.1083/jcb.201603019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. The kinetochore-bound ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 2012;23:968–980. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Hamill DR, Carter JC, Schumacher J, Bowerman B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Sivakumar S, Janczyk PŁ, Qu Q, Brautigam CA, Stukenberg PT, Yu H, Gorbsky GJ. The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. Elife. 2016;5:868. doi: 10.7554/eLife.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJE, Vleugel M, Teixeira A, Kops GJPL. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Sundin LJR, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol. Biol. Cell. 2011;22:759–768. doi: 10.1091/mbc.E10-08-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Haase J, Ohashi T, Erickson HP, Salmon ED, Bloom K. How the kinetochore couples microtubule force and centromere stretch to move chromosomes. Nat. Cell Biol. 2016;18:382–92. doi: 10.1038/ncb3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley JG, Miller SA, Stukenberg PT. The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol. Biol. Cell. 2011;22:1217–1226. doi: 10.1091/mbc.E10-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Gestaut DR, Tien JF, Vollmar BS, Gonen T, Asbury CL, Davis TN. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. U.S.a. 2012;109:16113–16118. doi: 10.1073/pnas.1209615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J. Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye AA, Deretic J, Hoel CM, Hinman AW, Cimini D, Welburn JP, Maresca TJ. Aurora A kinase contributes to a pole-based error correction pathway. Curr. Biol. 2015;25:1842–1851. doi: 10.1016/j.cub.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Mick JE, Maslennikov E, Nikashin B, Deluca JG, Grishchuk EL. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol. Biol. Cell mbc. 2015 doi: 10.1091/mbc.E14-11-1539. E14—11—1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Sundin LJR, DeLuca KF, Grishchuk EL, Deluca JG. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J. Cell Biol. 2014;206:45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: Localization of the SKA and NDC-80 complexes in the one-cell C. elegans embryo. SKA-1 was tagged at its endogenous locus with GFP. The NDC-80 complex subunit Nuf2HIM-10 was fused to mCherry and expressed from a single copy transgene insertion. Image z-stacks were acquired every 20 seconds and maximal intensity projections generated to visualize the dynamics of the 2 complexes. Playback rate is 100X-fold relative to real-time. Related to Figure 3.
Movie S2: Dynamics of chromosomes in control and ska-1 deletion embryos. Embryos expressed GFP::H2b, were imaged at high time resolution (z-stack acquisition every 2 seconds), and maximal intensity projections generated to visualize chromosome behavior. Related to Figure 4.