Summary
Antibodies play a crucial role in host defense and are indispensable research tools, diagnostics, and therapeutics. Antibody generation involves binding of genomically encoded germline antibodies followed by somatic hypermutation and in vivo selection to obtain antibodies with high affinity and selectivity. Understanding this process is critical for developing monoclonal antibodies, designing effective vaccines, and understanding autoantibody formation. Prior studies have found that antibodies to haptens, peptides, and proteins evolve from polyspecific germline antibodies. The immunological evolution of antibodies to mammalian glycans has not been studied. Using glycan microarrays, protein microarrays, cell binding studies, and molecular modeling, we demonstrate that therapeutic antibodies to the tumor-associated ganglioside GD2 evolved from highly specific germline precursors. The results have important implications for developing vaccines and monoclonal antibodies that target carbohydrate antigens. In addition, they demonstrate an alternative pathway for antibody evolution within the immune system that is distinct from the polyspecific germline pathway.
Keywords: Antibody Evolution, Ganglioside, GD2, Glycan Microarray, Germline Antibody
Graphical abstract
Introduction
Antibodies, immunoglobulins produced by B cells, play a crucial role in host defense and homeostasis (Schroeder and Cavacini, 2010). Antibodies protect us from infections by binding antigens on pathogens and neutralizing them, causing agglutination and/or tagging them for destruction by the immune system. Improper control of antibody production can be harmful. For example, generation of antibodies to certain self-antigens can cause autoimmune diseases, such as Guillain-Barré syndrome. In addition to natural physiological roles of antibodies, monoclonal antibodies are widely used for basic research, disease diagnosis, and disease therapy (Chan and Carter, 2010; Oldham and Dillman, 2008; Weiner et al., 2010).
A detailed comprehension of how the immune system generates antibodies is critical for producing monoclonal antibodies, designing suitable antigens for vaccines, and for understanding and controlling the development of autoantibodies (Mauri and Bosma, 2012). Each B cell encodes a unique antibody sequence, which can be produced as a soluble secreted protein or as a membrane bound B cell receptor. When the B cell receptor binds an antigen and is activated, the cell begins to proliferate. As activation and proliferation continues, mutations are introduced into the antibody gene via somatic hypermutation. B cells with mutations that confer improved affinity will outcompete other B cells, leading to selection and enrichment of improved antibodies. Since the entire process is only initiated if a germline B cell receptor is activated, the binding properties of germline antibodies are of fundamental importance for the development of all antibodies. Unfortunately, little is known about the binding properties of most germline antibodies/B cell receptors.
Prior studies have suggested that germline encoded antibodies are polyspecific; these studies include antibodies to proteins/peptides (Manivel et al., 2000; Sethi et al., 2006), haptens (Adhikary et al., 2015; James et al., 2003; Jimenez et al., 2004; Romesberg et al., 1998; Wedemayer et al., 1997a; Yin et al., 2003), bacterial polysaccharides (Evans et al., 2011; Nguyen et al., 2003), and HIV (Finton et al., 2014; Hoot et al., 2013; Scharf et al., 2016; Xiao et al., 2009). According to the polyspecificity hypothesis, B cells initially produce a surface bound germline antibody with a very malleable antigen binding site that is capable of recognizing numerous antigens with low affinity. By producing polyspecific germline antibodies, the immune system has the potential to respond to a much larger variety of foreign antigens while still focusing resources on the subset of antigens that are actually encountered.
Once activated, the affinity maturation process leads to antibodies with much higher affinity and selectivity. This improvement is hypothesized to be the result of a rigidification/pre-organization of the antigen binding pocket biased towards the target antigen/immunogen. Resultantly, the immune system is now primed to produce large amounts of antibody specific to the target antigen without the potential hazards of polyspecific binding.
Although polyspecificity is a powerful and appealing concept, experimental evidence of polyspecificity is only available for a relatively small number of germline antibodies. Rao et al. evaluated three germline antibodies against two different cDNA libraries and found extensive cross-reactivity with many proteins (Manivel et al., 2000). In contrast, affinity matured antibodies derived from these germlines did not cross-react at all with any proteins in either cDNA library. Schultz et al. found that the germline antibody of 39-A11 bound 9 structurally distinct small molecules with affinities within about 10 fold of the actual hapten (Romesberg et al., 1998). Evans et al. has examined binding of germline antibodies to a small panel of bacterial oligosaccharides, showing polyspecificity for the germline antibody (Brooks et al., 2008; Evans et al., 2011; Gerstenbruch et al., 2010). A number of other studies have evaluated binding of germline antibodies to small panels of potential ligands (Adhikary et al., 2015; Finton et al., 2014; Hoot et al., 2013). While not exhaustive, these studies collectively support the concept of germline polyspecificity. From a mechanistic point of view, there are a number of elegant structural studies demonstrating that CDR loops of germline antibodies have a higher degree of conformational flexibility than affinity matured antibodies derived from them (Jimenez et al., 2004; Nguyen et al., 2003; Patten et al., 1996; Wedemayer et al., 1997a; Wedemayer et al., 1997b; Zimmermann et al., 2006). For example, Schultz and colleagues generated crystal structures of antigen binding fragments (Fab) of 48G7 and its germline precursor in both liganded and unliganded forms (Wedemayer et al., 1997a). They found that the binding site of the germline antibody underwent significant conformational changes upon binding the ligand, whereas the binding site of the affinity matured antibody, 48G7, was pre-organized for binding, consistent with a lock and key mechanism. These findings have been reproduced with molecular dynamics simulations as well. For example, a study of four pairs of germline and mature antibodies (7G12, AZ28, 28B4, and 48G7) showed clear decreases in flexibility in the affinity mature structures, especially in CDR H3, and offered structural explanations for how individual mutations are restricting loop mobility (Wong et al., 2011). Although these excellent studies provide strong evidence for polyspecificity in some cases, the scope and generality of germline polyspecificity is not clear. For example, almost nothing is known about the binding properties of germline antibodies to mammalian glycans.
In this study, we used a combination of glycan and protein microarray technologies, cell based assays, and molecular dynamics to study the immunological evolution of two clinical relevant antibodies, ch14.18 (Unituxin) and 3F8. Both antibodies target GD2, a ganglioside that is highly overexpressed on a variety of cancers, including neuroblastoma and melanoma. Antibodies ch14.18 and 3F8 have demonstrated exceptional clinical results for the treatment of neuroblastoma, and ch14.18 (Unituxin) has received FDA approval (Cheung et al., 1998; Cheung et al., 2014; Yu et al., 2010). We compared the binding properties of ch14.18 and 3F8 with their corresponding germline antibodies. Surprisingly, we found that the germline antibodies are highly selective for GD2, demonstrating no observable polyspecificity. These results allude to the native development of germline antibodies specific for a single antigen.
Results
Design of the system
A key consideration of antibody evaluation is the antibody format. Prior studies on affinity maturation have primarily used fragment antigen binding (Fab) structures, which contain a single binding site. This monovalent format is often sufficient for the study of interactions with proteins, peptides, or small molecule haptens, as these monovalent binding events frequently have low nanomolar or better affinities. However, monovalent interactions with carbohydrate antigens are typically weak, with dissociation constants greater than 10 µM. To achieve physiologically relevant affinity, carbohydrate-binding antibodies typically rely on the formation of multivalent complexes. In addition to affinity, the selectivity of a multivalent binding event can be quite different than a comparable monovalent binding event. For example, Kiessling et al. demonstrated that the specificity observed in a monovalent binding event can be amplified in a multivalent binding event (Gordon et al., 1998). Additionally, Kahne et al. showed that the preferred ligands in a multivalent binding event can be different from those observed in a monovalent event (Liang et al., 1997). Given the importance of multivalent binding for carbohydrate recognition and that a whole IgG is similar in structure to the B cell receptor, we selected a divalent, whole IgG format for our studies. The use of a whole IgG would also allow us to directly compare binding properties of ch14.18 and 3F8 measured in our assay with previously published data.
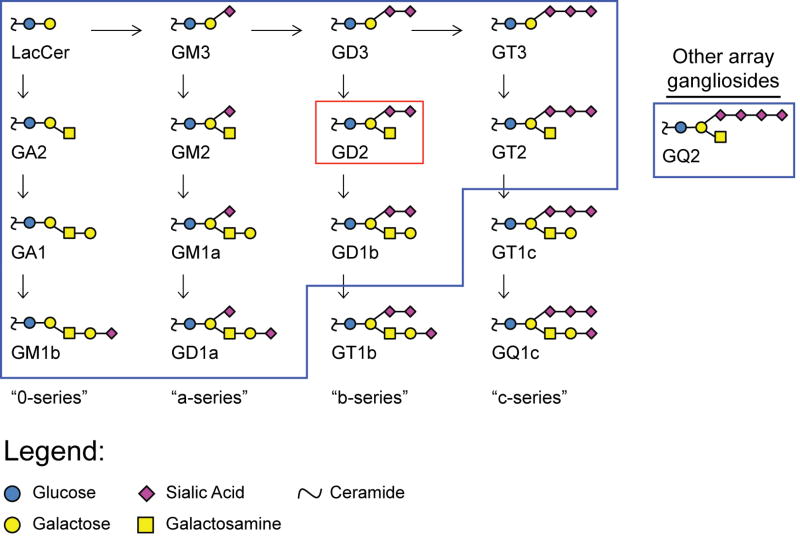
A second consideration was the approach for evaluating selectivity. The approach must assess binding in a multivalent context and have sufficient throughput to allow evaluation of many variations of structure and presentation. To evaluate potential binding to a large and diverse panel of carbohydrates, we evaluated each recombinantly designed antibody on our glyco-antigen microarray. The array contains over 500 components, including a diverse set of glycopeptides, N- and O- linked glycans, Lewis and blood group antigens, glycolipids and glycosphingolipids, and glycopeptides. Many of these array components are printed at both “high” and “low” density to provide variations in glycan spacing/presentation (Figure S1). Of particular relevance to this study, many of the array components contain glycan sequences that are closely related to GD2 such as GD1a, GD1b, GD3, GM1a, GM1b, GM2, GM3, GT1a, GT2, GT3, GQ2, and GA2 (Figure 1). We (Gildersleeve et al., 2008; Gildersleeve and Wright, 2016; Oyelaran et al., 2009; Shoreibah et al., 2011; Zhang et al., 2010) and others (Chang et al., 2010; Dupin et al., 2015; Godula and Bertozzi, 2012; Goudot et al., 2013; Hung et al., 2013; Jaipuri et al., 2008; Karamanska et al., 2008; Ligeour et al., 2015; Park and Shin, 2007; Shivatare et al., 2013; Song et al., 2008; Wang et al., 2008) have used glycan microarrays to evaluate apparent KD values for carbohydrate-protein interactions and have found good agreement with previously published values measured by surface plasmon resonance.
Figure 1.
Biosynthetic pathway for the synthesis of gangliosides. Structural diversity is generated through the addition of sialic acid to form α3 branching with galactose and the addition of glucosamine to form β4 branching. The target antigen, GD2, is boxed in red. Other gangliosides present on the glycan array are contained within blue boxes.
Comparison of antibody sequences with putative germlines
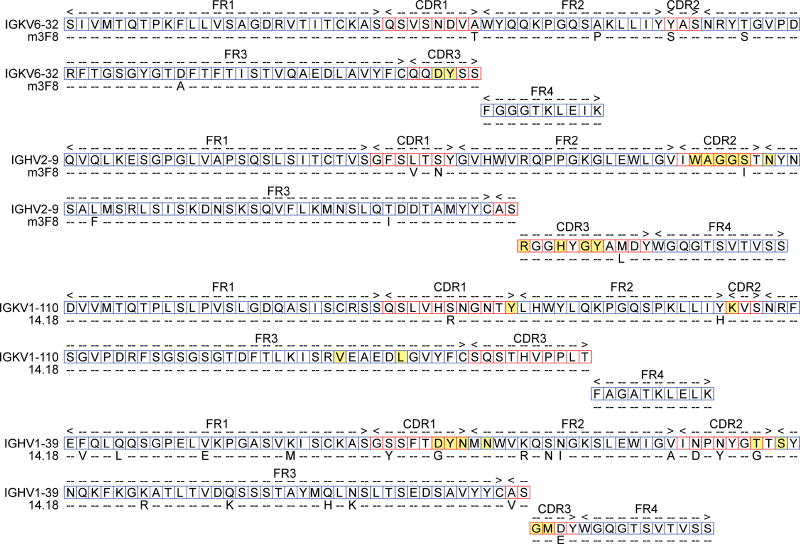
To facilitate comparisons of our work with previous work on immunological evolution of antibodies, we used the same methodology as prior studies to assign the putative germline genes. A brief description is below, and a more detailed description can be found in the Supporting Information. Using NCBI-BLAST and IMGT/V-Quest alignment algorithms in tandem, nucleotides sequences of 3F8 and ch14.18 that had been previously published in patents and literature were aligned with putative germline sequences. The amino acid sequences of the affinity mature and putative germline sequences were aligned and mutations from the putative germline genes were determined (Figure 2). We found that for all of the aligned anti-GD2 sequences, nucleotide alignment was greater than 92% similarity in all cases, with most of the cases being greater than 96% similarity. This degree of homology is high and on par with other studies on immunological evolution of antibodies.
Figure 2.
Alignment of anti-GD2 antibodies with their putative germline sequences. Top rows representative of germline amino acid structure. Bottom rows represent mutations from the germline antibody (dashed = no mutation). Amino acids relevant in antigen binding, as proposed by Ahmed et al. (2013), are highlighted in yellow. Complementary determining regions (CDRs) are labeled as designated by IMGT analyses.
Ch14.18 had 97% nucleotide similarity for the light chain and 92% similarity for the heavy chain nucleotide sequences. These alignments resulted in 2 mutations of the light chain variable region and 19 within the heavy chain. Interestingly, the majority of these heavy chain mutations (12 mutations) fall within the framework regions (FRs) of the variable gene, as opposed to CDRs.
Antibody 3F8 demonstrated a greater degree of similarity between germline and affinity mature sequences. Nucleotide alignments were 97% similar for both the light and heavy chain nucleotide sequences. These alignments indicated the presence of 5 germline mutations of the light chain variable region, as well as 6 mutations of the heavy chain germline region. Interestingly, only one of these mutations (HC: S56I) was hypothesized, by molecular modeling, to be a residue implicated in antigen binding (Ahmed et al., 2013).
Evaluation of ch14.18 affinity mature and germline antibodies
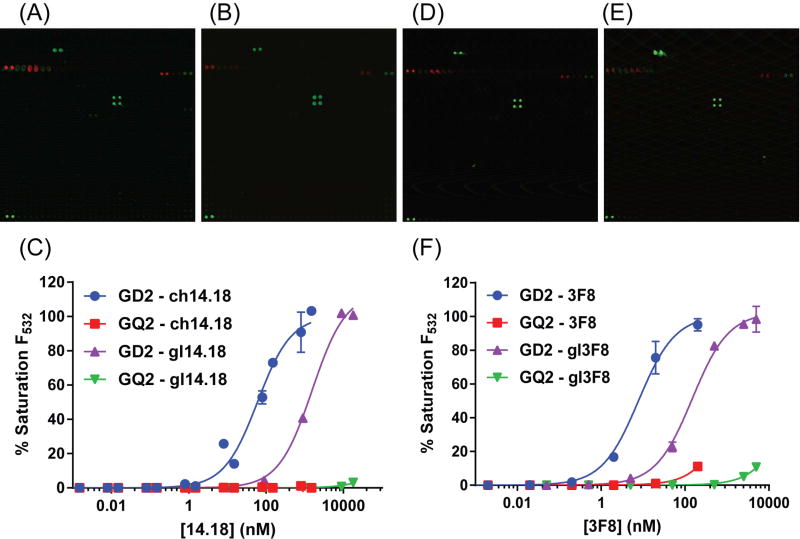
Ch14.18 was the first of the anti-GD2 constructs to be studied on the glycan microarray. Ch14.18 is a standout in the field of anti-carbohydrate immunotherapy, with incredible success in a Phase III clinical trial and recent FDA approval. Following the recombinant expression and purification of ch14.18, the antibody was profiled on the glycan microarray and found to have an apparent KD,GD2 of approximately 60 nM (Figure 3 & S3A; Tables 1 & S2). This value is in good agreement with the previously reported value of 77 nM. Additionally, the ch14.18 construct demonstrated remarkably high selectivity for GD2 on the glycan microarray. Only GT2 [Neu5Acα2-8Neu5Acα2-8Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glc-] and GQ2 [Neu5Acα2-8Neu5Acα2-8Neu5Acα2-8Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glc-], GD2-like structures with elongated sialic acid side chains, had measurable levels of interaction with the expressed ch14.18 antibody. Although the binding to these glycans was too weak to accurately measure an apparent KD, we could estimate at least 1000-fold higher affinity for GD2 than any other glycan (Table 1). Interestingly, the structurally similar gangliosides GD1b [Neu5Acα2-8Neu5Acα2-3(Galβ1-3GalNAcβ1-4)Galβ1-4Glc-], GD3 [Neu5Acα2-8Neu5Acα2-3Galβ1-4Glc-], and GM2 [Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glc-] demonstrated no identifiable levels of interaction with ch14.18, even at the highest antibody concentration tested, suggesting a high preference for branched structures with a minimum of di-sialylation and GalNAcβ1-4 termination.
Figure 3.
High specificity of anti-GD2 antibodies for antigen GD2. (A) Affinity mature 14.18 and (B) germline 14.18 antibody react with high specificity towards antigen GD2. (C) The non-target antigen with the highest observable binding was GQ2, at greater than 500-fold reduced levels. (D) Affinity mature 3F8 and (E) germline 3F8 were also extremely specific for GD2. (F) GQ2 binding was observed in 3F8 at levels greater than 250-fold reduced versus that of GD2. Linear formats of these same data are demonstrated in Figure S3 to fully illustrate antibody dose saturation. Data are represented as mean ± standard deviation (minimum of 2 independent array experiments, duplicate spots per array). See also Figures S1–S5 and Table S2.
Table 1.
Apparent KD values of recombinant anti-GD2 antibodies. Apparent KD values were calculated using glycan microarray analysis.
| Antibody | KD,App (nM)¶ | Literature† KD (nM) | Fold Preference‡ GD2 vs. GT2 |
Fold Preference‡ GD2 vs. GQ2 |
|---|---|---|---|---|
| 3F8 | 8.5 | 13 ± 3 | 4000 | 250 |
| Germline 3F8 | 146 | -- | > 5000 | 1000 |
| Humanized 3F8 | 7.7 | 11 ± 1 | 1500 | 200 |
| 3F8_mut (HC:I56S) | 128 | -- | 5000 | 500 |
| Dinutuximab (ch14.18) | 60 | 77 ± 8 | > 5000 | 1000 |
| Germline 14.18 | 1600 | -- | > 5000 | 250 |
All tested antibodies were of human IgG1 isotype.
Literature values, for comparative purposes, from Cheung et al. (2012)
Fold preferences indicate relative binding of GD2 over GT2 or GQ2 as estimated from the array data.
Next, the germline antibody of ch14.18 was evaluated. No previously published literature exists discussing the expression of germline antibodies to gangliosides. Our expectations were that these antibodies would follow the polyspecificity hypothesis; a significant decrease (>1000-fold) in affinity and wide-spread binding events. Following expression, purification, and dose-response glycan microarray analysis, we found that this germline had modest affinity but remarkable selectivity. The apparent KD,GD2 of the germline 14.18 structure was ~1.6 µM (Figure 3 & S3B; Tables 1 & S2). Though a ~30-fold decrease in affinity is significant, this change is small compared to the >1000-fold changes in affinity during maturation commonly seen with antibodies to haptens, proteins, and peptides. Like ch14.18, the germline antibody only bound GD2, with at least 250-fold selectivity over the next best glycan on the array (Table 1).
Screening chimeric and germline 3F8
3F8 was the next anti-GD2 antibody to be expressed and profiled. 3F8 is noteworthy for its low nanomolar affinity (KD,GD2 = 5–15 nM) and demonstrated success in Phase I and Phase II clinical trials. In addition to the affinity mature and germline antibodies, we opted to additionally express the previously described humanized construct as an additional validation structure.
Consistent with prior publications, the chimeric 3F8 (ch3F8) and humanized 3F8 (hu3F8) constructs demonstrated high avidity and selectivity on our array. Ch3F8 demonstrated an apparent KD,GD2 value of approximately 8.5 nM (Figures 3, S3C; Tables 1, S2), very similar to the previously reported value of 13 nM (Cheung et al., 2012). Similarly to ch14.18, the ch3F8 structure was highly specific for GD2. GT2 and GQ2 were nominal binders of ch3F8, with >250-fold reduced affinity as compared to GD2. No other binding events were observed with ch3F8. Hu3F8 was found to have an apparent KD,GD2 of 7.7 nM (Figure S3I–J; Tables 1, S2), which is comparable with the literature reported value of 11nM (Cheung et al., 2012). In agreement with ch3F8, gangliosides GT2 and GQ2 were the only other glycan structures with observable levels of interaction with the humanized construct, at >250-fold reduced affinity for GQ2 and >4000-fold reduced affinity for GT2 (Table 1). No other glycans demonstrated significant binding levels at tested antibody concentrations.
Next, 3F8 germline antibodies were expressed and assayed. The apparent KD,GD2 of the 3F8 germline antibody was approximately 146 nM (Figures 3, S3D; Tables 1, S2), only a 18-fold decrease versus that of the affinity mature structure and nearly as good as the FDA approved antibody ch14.18. Remarkably, the specificity profile of the 3F8 germline antibody was nearly identical to that of the affinity mature structure, with high selectivity for GD2 over all other array components.
Mutational Intermediates of 3F8
To test the significance of individual mutations in affinity maturation, we constructed a set of antibodies wherein mutations in the CDR loops of 3F8 were reverted back to the germline sequence. Six mutant structures were generated corresponding to the six mutations to CDR amino acids (LC: T34A and S50Y, HC: V29L, N31S, I56S, and L106M). None of these structures deviated significantly in specificity from that of affinity mature 3F8 (Figures S4A–G). One of these structures, ch3F8_mut (HC:I56S), was selected for a full dose-response characterization. We chose this structure due to it being both a mutation within a CDR and a mutation into a hypothesized antigen binding amino acid, as per literature (Ahmed et al., 2013). The apparent KD,GD2 of this structure was approximately 128 nM (Figures S4H–K; Tables 1, S2), suggesting that a majority of the improvement in affinity from germline to mature structures is dependent on this particular mutation. This ch3F8 mutant demonstrated small levels of interaction with GQ2 (Table 1), but non-identifiable interactions with other glycan antigens.
Screening 3F8 structures against protein microarrays
Given the unexpectedly high selectivity for the germline antibodies, we considered the possibility that the germline antibodies could bind non-glycan structures. Others have demonstrated evidence that anti-carbohydrate antibodies may bind peptide and protein targets by molecular mimicry. As such, we sought to evaluate the binding events of the affinity mature and germline 3F8 antibodies on a platform separate from our glycan microarray.
The HuProt v2.0 human proteome slides are a microarray platform with greater than 19,000 human proteins with high diversity of functional families. As a positive control, we manually spotted a GD2 neoglycoprotein on the array surface.
Germline and affinity mature ch3F8 were assayed on the HuProt v2.0 human proteome microarray. Both antibodies were assayed at concentrations 2-times greater than the calculated KD,GD2 in an effort to promote as many binding events as possible. The ch3F8 antibodies successfully bound the GD2-positve control spots, but no specific antibody-protein interactions were detected (Figure S5). These results further support the remarkable selectivity of affinity mature and germline 3F8 antibodies for GD2.
Cell-based Assays – Live Cell Imaging and Direct Cytotoxicity
The glyco-antigen microarray provides a rapid tool to assess potential binding to a large number of glycan structures, but it does not encompass the full mammalian glycome. As additional evidence of selectivity, we assayed binding of the affinity mature and germline ch3F8 antibodies in cell binding assays. The melanoma cell line M14 was selected as the GD2-positive cell line. The colorectal cancer cell line SW620 was included as a GD2-negative cell line.
Ch3F8 antibodies were first tested for their binding capacity in a live cell environment. M14 and SW620, plated and incubated overnight, were treated with affinity mature and germline antibodies at concentrations 2-times greater than the calculated KD,GD2. Antibody binding was detected by addition of fluorescently-label secondary reagent. Corresponding secondary-only control experiments were also considered. Consistent with our previously observed data, the germline and affinity mature 3F8 structures were both capable of binding the GD2-positive M14 cell line, but were incapable of binding the GD2-negative SW620 cell line. No secondary-reagent binding was observed in the negative control experiments (Figure S6). These results were consistent with cells studied under fixed, suspension conditions (Figures S6, Table S3).
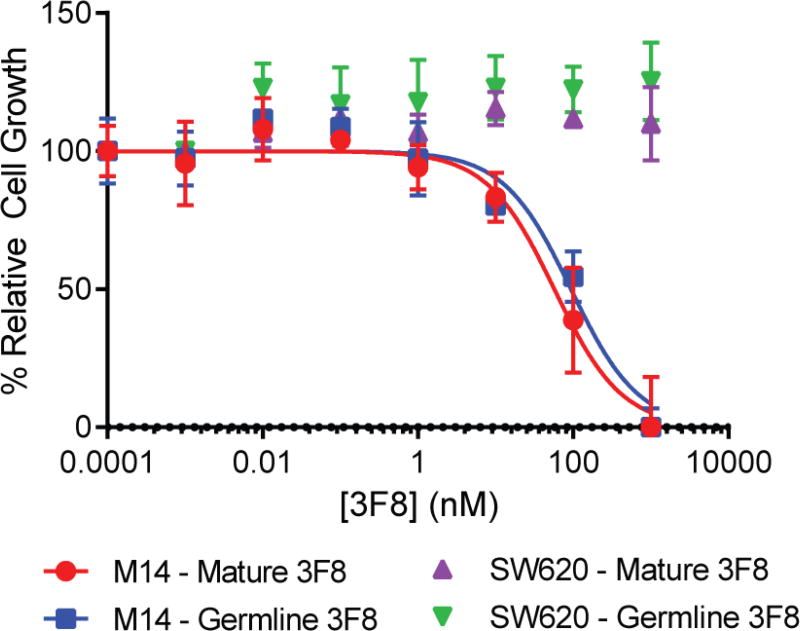
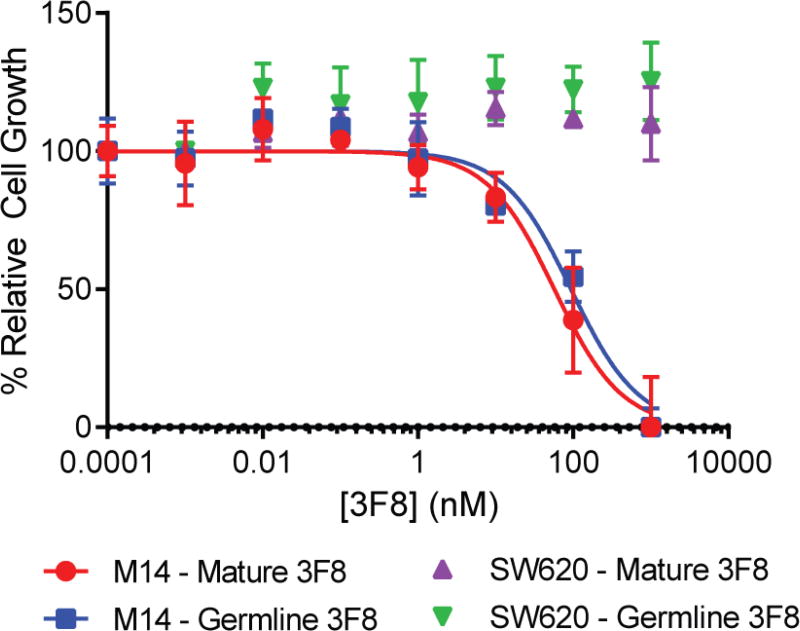
Ch3F8 antibodies were subsequently considered for their direct cytotoxicity. The GD2-postive melanoma cell line M14 and GD2-negative colorectal cancer cell lines SW620 were treated with increasing concentrations of antibody for 72 hours. Upon assay, the affinity mature and germline ch3F8 antibodies had similar potencies towards M14 cells (Figure 4; EC50 for ch3F8 = 8.6 ± 1.7 µg/mL; gl_3F8 = 14.1 ± 3.1 µg/mL). These potencies were also similar to that of affinity mature 3F8 for the GD2-positive neuroblastoma cell line LAN-1 (Cheung et al., 2012). Neither antibody was capable of directing cell death/slowing growth of the SW620 cell line.
Figure 4.
3F8 antibody direct cytotoxicity. The 3F8 antibodies (mature and germline) were capable of promoting direct cytotoxicity against the GD2-presenting M14 melanoma cell lines. These antibodies were incapable of inducing direct cytotoxicity against the colorectal cancer cell line SW620. Data are represented as mean ± standard deviation of quadruplicate wells. See also Figure S6 and Table S3.
Molecular Dynamics
As discussed earlier, polyspecific binding properties of germline antibodies are thought to derive from highly flexible and adaptable binding pockets. In contrast, the affinity maturation process produces binding pockets that are rigidified and either stabilized into a binding conformation for a single antigen (lock and key model) or energetically favored to adopt that binding conformation a higher percentage of the time (conformational selection model). Given that our experimental results had shown an unexpectedly high degree of selectivity for GD2 already in its germline antibody, and a relatively modest gain in binding affinity upon maturation, we decided to investigate whether the differences in structural flexibility observed in other antibodies between germline and mature structures are present and visible in anti-GD2 antibodies using molecular dynamics simulations.
We began with the high-resolution crystal structure for the Fab fragment of 3F8 (PDB ID: 3VFG, resolution 1.6Å), published previously by Ahmed et al., and generated a homology model for its putative germline precursor. There are only 11 residue differences, so the starting structures for germline and affinity mature 3F8 were very similar. While it is possible to use a variety of enhanced sampling simulation methods to drive exploration of conformational space, we wanted rather to evaluate the intrinsic flexibility of each structure as accurately and realistically as possible and therefore performed molecular dynamics at constant pressure and temperature in a periodic box of explicit water. This method of molecular modeling is consistent with previously published methods of protein/antibody molecular modeling simulations. Sampling was increased by running four independent simulations for each structure, for a total of 100 ns.
The equilibration and overall stability of each simulation was evaluated by calculating the RMS deviation of the backbone atoms in each domain from the starting conformation over time. All simulations reached a stable conformation within 0.8 – 1.2 Å after approximately 5 ns of simulation time (Figure S7A–D). RMS fluctuations away from the average for each residue were also calculated and plotted. These were compared to the crystallographic B-factors from the affinity mature 3F8 structure (Ahemd et al., 2013) as a check on whether the simulations are reproducing experimentally observed flexibilities. B-factors include many other sources of disorder in the crystal along with thermal vibrations, so they are larger in scale than the calculated atomic RMS fluctuations, but regions of increased and decreased flexibility are qualitatively reproduced (Figure S7E–H).
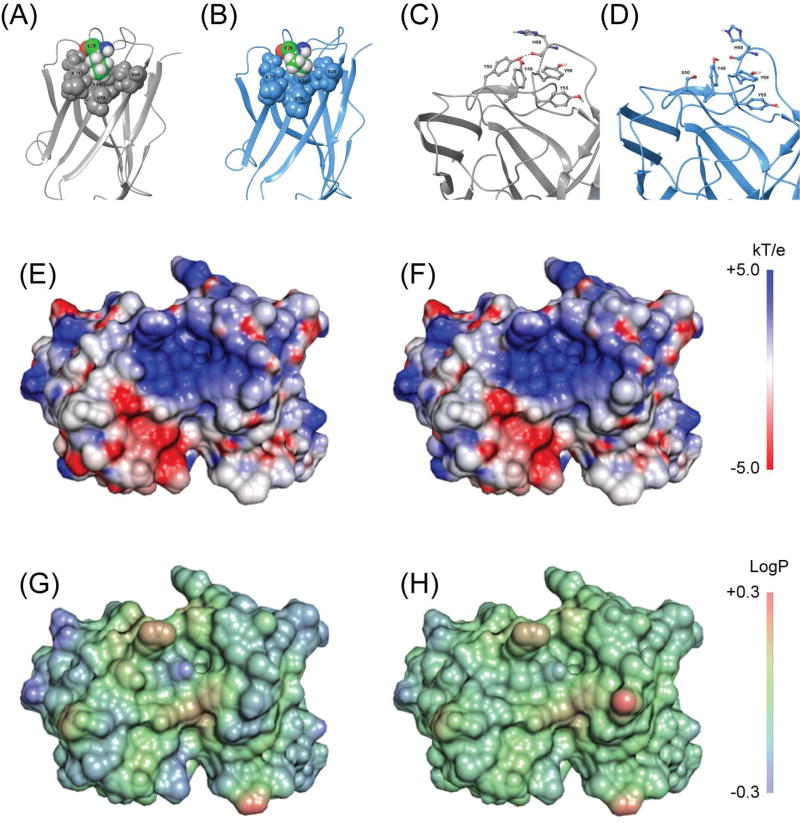
We found that backbone fluctuations of the mature Fab were slightly but significantly greater than those of the germline antibody, particularly within CDRs H1 and H3 (Figures S7G–H; Table S4). This observation suggests that rather than becoming more rigid, the affinity mature structure of 3F8 shows an increase in flexibility relative to the germline structure. This feature is illustrated in Figure 5 via a worm plot comparison of the two structures. Here the RMSF average for the backbone atoms of each residue is mapped to the radius of the backbone trace, so that rigid regions of the structure are narrow, and more flexible regions appear fatter. The largest increases in flexibility are seen in CDR loops H1 and H3 (Figure 5F, 5H), and to a lesser extent portions of L1 and L3 (Figure 5C, 5E).
Figure 5.
Molecular dynamics analyses of unliganded 3F8. Worm plots of (A) germline 3F8 and (B) affinity mature 3F8 illustrate a relatively rigid germline structure that loosens slightly during affinity maturation. CDR loops are labeled and residue mutations between germline and affinity mature are shown in stick representation. Histograms of RMSD distribution from the starting conformation for CDR loops during the simulations. Light chain CDRs 1–3 (Figures C–E respectively) and heavy chain CDRs 1–3 (Figures F–H respectively) illustrate the propensity of mature CDRs to explore conformational space more liberally. See also Figure S7 and Table S4.
We further analyzed the conformational space explored by the CDRs by calculating the RMS distance from the starting structure that is reached by the residues in each loop over time. These distances were binned and normalized and the resulting histograms are shown in Figures 5C–H. Strikingly, the CDRs in the germline structure tend to explore a relatively limited region of conformational space (narrow peaks in the histogram), at or less than 2 Å away from their starting conformation. In contrast, apart from L2, the affinity mature CDRs cover a much wider range of spatial distances.
We extracted a set of 20 diverse frames from the simulations of both the germline and mature structures to visualize and analyze how changes in sequence upon maturation might be driving the observed increases in flexibility. In the case of CDR H1, the L29V mutation appears to destabilize this loop by a decrease in van der Waals packing interactions with surrounding hydrophobic residues HC V34, HC V78 and the acyl chain of HC K71 (Figure 6A–B). The Y50S mutation located in CDR L2 does not affect the L2 loop itself, which remains rigid, however Tyr at this position hydrogen bonds to the backbone carbonyl of HC H98 on CDR loop H3, and forms a well-organized multiple ring-stacking interaction with LC Y49, LC Y55, and HC Y99 (Figure 6C–D). These interactions are diminished upon mutation to Ser, and this appears at least partially responsible for the increase in flexibility of the H3 loop.
Figure 6.
(A–D) Snapshots of residues identified as contributing to antibody flexibility. (A) In the germline structure, L29 of the heavy chain (shown in CPK representation with green carbons) is involved in van der Waals packing with heavy chain residues V34, V78, and K71. (B) In the mature structure, L29 is mutated to V29. The result is a destabilization of the HC loop by a decrease in van der Waals packing. (C) In the germline light chain, Y50 hydrogen bonds to the backbone carbonyl of HC H98, and forms a multiple ring-stacking interaction with LC Y49, LC Y55, and HC Y99. (D) The mutation in the mature structure to LC S50 weakens these interactions. (E–H) Top-down view of the antigen binding site surface of 3F8. Electrostatic surface of (E) germline and (F) affinity mature 3F8 structures. Surface hydrophobicity of (G) germline and (H) affinity mature 3F8 structures. Surfaces were calculated using VASCo and DelPhi and rendered in PyMOL.
Interestingly, HC S56I, which produces the majority of the gain in GD2 binding affinity based on our individual mutation studies, is located in CDR H2 and does not appear to cause any changes in conformation or flexibility in that loop or in the binding site as a whole. Conversely, neither HC L29V nor LC Y50S affects GD2 binding affinity. This suggests that flexibility and affinity are not directly linked in the 3F8 antibody. Other mechanisms proposed for increased antigen affinity include changes in electrostatic surface potential (Zhao et al., 2015) or increased buried hydrophobic surface area with antigen binding. We calculated the molecular hydrophobic potential based on the surface projection of atomic logP values (Ghose et al., 1998; Steinkellner et al., 2009), along with the surface electrostatic potential and saw no large charges in 3F8 between the germline and affinity mature structures (Figure 6E–H).
The crystal structure of the related mouse 14G2a antibody (Horwacik et al., 2015) showed that GD2 binding in 14G2a/ch14.18 does not result in a large conformational change but rather an overall closing of the binding site loops, and that binding is mediated by an extensive network of water molecules. Although the glycan binding site in 14G2a is shaped quite differently from that in 3F8, this structural information allows us to speculate that in GD2-specific antibodies, the increase in binding affinity with maturation might be enthalpically driven via increased direct and water-mediated hydrogen-bonding in the binding site, rather than entropically driven via a decrease in flexibility (Klebe, 2015). The flexibility increase with maturation, as observed in the molecular dynamics simulations, might allow better accommodation and reorganization of the water network in the antigen binding pocket upon GD2 binding.
Discussion
An in depth understanding of the binding properties of germline antibodies and the effects of affinity maturation are critical for a wide range of basic research and clinical applications. For example, information about germline specificity can help with the design of vaccine antigens. Studies from the HIV field exemplify this concept. Certain broadly HIV neutralizing monoclonal antibodies recognize the HIV envelope with high affinity. Interestingly, the corresponding germline antibodies do not bind the HIV envelope at all (Doores et al., 2010; Xiao et al., 2009). Thus, HIV envelope would not be a suitable immunogen to activate the B cells that give rise to those same broadly neutralizing antibodies. Information about germline specificity can also be instructive for generating high quality monoclonal antibodies or for understanding the origins and development of autoantibodies.
The binding properties of germline antibodies are not easily predicted. Prior studies on small molecule haptens, peptides, proteins, and bacterial oligosaccharides have shown that many germline antibodies have flexible binding sites and are capable of recognizing multiple antigens. Thus, the exact structures recognized by a germline antibody can be quite difficult to anticipate. For example, the germline precursor to the Diels-Alderase antibody 39-A11 was shown to bind a highly diverse set of molecules, including a bicyclo[2.2.2]octene hapten, a nucleoside analog, a steroid, and a peptide.(Romersberg et al., 1998) The binding properties of germline antibodies can also be distinct from the corresponding affinity matured antibody, as the HIV examples discussed above illustrate. For these reasons, germline antibody specificity and affinity must be determined experimentally.
Using a combination of glycan microarrays, proteins microarrays, cell binding, and molecular dynamics simulations, we studied the immunological evolution of two clinically useful antibodies, 3F8 and ch14.18 (Unituxin). Both antibodies target the ganglioside GD2, a tumor associated carbohydrate antigen highly overexpressed on a variety of cancers. Potential recognition of many different structurally-defined antigen families was investigated, including 19,000 proteins, and hundreds of N-linked glycans, O-linked glycans, glycolipids, non-human glycans, and glycopeptides. In addition, binding to GD2 positive and negative cell lines was tested. Surprisingly, the putative germline antibodies demonstrated remarkable selectivity for GD2, with only very weak binding (~250–1000 fold worse; Table 1) to GQ2 and GT2 observed among the thousands of potential antigens tested. Based on molecular dynamics simulations, the binding pockets of the germline antibodies appear to be relatively rigid and pre-organized for recognition of GD2. In fact, 3F8 was found to be more flexible than its germline precursor.
The high degree of selectivity displayed by the GD2 antibodies and their germline precursors is impressive and surprising. Many monoclonal antibodies to mammalian glycans have modest selectivity, even after the affinity maturation process. For example, in a prior study of commercially-available antibodies, about half were found to bind glycans other than the listed target glycan (Manimala et al., 2007). Thus, high selectivity is uncommon, even among monoclonal antibodies chosen for commercial production. Given that most germline antibodies to other antigen families are polyspecific and that many affinity matured antibodies to glycans have modest selectivity, the nearly exclusive binding of the germline antibodies to GD2 is especially noteworthy.
Our results have a number of important implications. First, high quality antibodies to glycans are often difficult to obtain (Sterner et al., 2016). Some glycan targets yield good antibodies across multiple experiments whereas other seemingly similar glycan antigens consistently yield antibodies with poor selectivity. The factors that determine whether one will obtain a high quality monoclonal antibody are not well understood. Self vs non-self and experimental parameters (e.g. adjuvant, number of boosts, antigen source) certainly influence the outcome, but these elements do not fully explain the variable results. Our results suggest that mammals already possess B cell receptors with modest to good affinity and high selectivity for certain glycans. Thus, mammals are predisposed to produce high quality antibodies to those glycans. In addition to helping explain the variable outcomes, our results also suggest that the germline repertoire may be a better source of carbohydrate-binding antibodies than previously thought, at least for some targets.
Second, our results have implications for vaccine design, especially in the areas of cancer and HIV. There are considerable efforts to develop cancer vaccines that induce antibodies to GD2 and other tumor-associated carbohydrates, such as GM2, GD3, and fucosyl-GM1 (Dube and Bertozzi, 2005). A number of strategies utilize either partial structures or unnaturally modified structures to break tolerance (Astronomo and Burton, 2010; Huang et al., 2013). Our results indicate that germline B cells that give rise to antibodies like 3F8 and ch14.18 are only activated by a very narrow set of glycan structures. Therefore, design of a suitable vaccine antigen may be especially critical for these vaccines. Evaluation of binding with relevant germline antibodies might facilitate selection of appropriate vaccine antigens. Induction of antibodies to glycans is also a prominent strategy in the development of HIV vaccines (Scanlan et al., 2007). Many broadly neutralizing antibodies target high mannose and other N-linked glycans on gp120. Like the cancer vaccines, the HIV vaccines must overcome self-tolerance. One general mechanism for overcoming tolerance involves engaging a polyspecific B cell receptor with weak affinity for the self-antigen and then evolving enhanced affinity. This approach may be most relevant for B cells expressing a polysepcific germline receptor such as those recognizing proteins, peptides, and small molecule haptens. When targeting mammalian self-glycans, overcoming tolerance may involve activating a B cell with selectivity for a related but distinct glycan and then evolving altered selectivity for the self-glycan.
Our results also raise several important questions. First, why would a mammal possess germline encoded antibodies to GD2? Second, why would germline antibodies to GD2 have high selectivity, while germline antibodies to other antigens are polyspecific? Third, are there other germline antibodies with very high selectivity? While these questions are difficult to answer, some insight can be gained from considering the advantages and disadvantages of selectivity.
The immune system has a limited set of resources to defend the host. Polyspecificity provides a mechanism to maximize reactivity with a finite number of B cells. The main disadvantage is that polyspecificity increases the chances of generating autoantibodies. Therefore, the immune system may encode polyspecific antibodies to families of antigens that are typically not found on cells, such as bacterial polysaccharides and small molecule haptens. For antigen families where there is a high potential for generating self-reactivity, the immune system may encode germline antibodies with high selectivity, such that it can bind important antigens while minimizing risks of autoimmunity.
In the case of GD2, GD2-like structures are found on certain pathogens, such a C. jejuni. In addition, antibodies to GD2 have anti-cancer activity. Therefore, it is possible that these antibodies impart survival advantages to the host. At the same time, there are many closely related glycans that are found normally on mammalian cells. Polyspecificity within the ganglioside family of antigens could increase susceptibility to autoimmune diseases. In fact, antibodies to a several structurally-related gangliosides are implicated in autoimmune diseases, such as antibodies to GM2 in Guillain-Barre syndrome and antibodies to GQ1b in Miller-Fisher syndrome. While additional experiments would be needed to more fully evaluate this model, it provides one potential explanation for the variations in germline specificity.
The study of germline antibody maturation is critical to our understanding of immune response. As the available toolkit of microarray technologies has expanded rapidly, so too has our capacity to probe germline antibody binding. While previous studies have demonstrated germline polyspecificity with a small library of antigens, our studies demonstrate a unique case in which the germline immune system has a pre-set capacity to specifically target GD2. These microarray results, accompanied with molecular dynamics simulations and cell binding/cytotoxicity assays, demonstrate an alternative pathway for antibody evolution within the immune system that is distinct from the polyspecific germline pathway.
Experimental Procedures
Anti-glycan antibody expression, purification, and array profiling
Details for the preparation of the plasmids (pFuse vector), transfection, and expression can be found in the Supporting information and Table S1. Secreted antibody was isolated from the supernatant by protein L affinity chromatography. Antibody purity was validated by polyacrylamide gel electrophoresis and size exclusion chromatography (Figures S1 and S2). Antibody concentration was calculated by NanoDrop and Easy-Titer Human IgG Assay Kit (Thermo Scientific, Rockford, IL).
Glycan microarray slides were prepared and assayed in laboratory as previously described (Campbell et al., 2010). Detailed information about the array quality assessment and full experimental details can be found in the Supplemental Information. Briefly, the microarrays used in this experiment had 503 components. Serial dilutions of recombinantly expressed anti-GD2 antibodies in 3% w/v bovine serum albumin (BSA)(Sigma; St. Louis, MO) and 1% w/v human serum albumin (HSA)(Sigma; St. Louis, MO) PBST buffer were assayed on slide at 37°C with gentle shaking (100 RPM) for 4 hours. Following incubation, the slides were washed with PBST and appropriate, fluorescently-labeled secondaries, diluted 1:500 in 1% BSA + 3% HSA in PBS solution, were incubated on slide at 37°C with gentle shaking (100 RPM) for 2 hours in the dark. Finally, slides were washed in PBST, dried by centrifugation at 200 rcf for 5 minutes, and then immediately scanned. Example pre-scan and data images can be found in Figure S1.
Slide Scanning and Data Analysis
Glycan-antibody binding on the microarray was quantified using an InnoScan 1100 AL fluorescence scanner (Innopsys; Chicago, IL). Images were analyzed using GenePix Pro 7.0. Fluorescent intensity values calculated in GenePix Pro 7.0 were plotted using GraphPad Prism 6.0. Each neoglycoprotein was printed in duplicate in each well, and each antibody was assayed in a minimum of 2 independent array experiments for each concentration tested. The final intensity values for each antibody-neoglycoprotein interaction (Figures 3, S3 and S4) were calculated from the average of corresponding spots in each of the two (or more) wells (duplicate spots in each of two or more wells; total of 4 or more spots). Apparent KD values were calculated using a non-linear regression, one site binding (hyperbola) fitting model following the method of MacBeath (Gordus and MacBeath, 2006). The list of array components and full microarray data will be publicly available in the National Center for Biotechnology Information’s (NCBI) Gene Expression Omnibus (GEO) (Edgar et al., 2002) via GEO Series accession number (GSE100438).
3F8 Interactions with the Human Proteome
HuProtv2.0 human proteome microarray slides were purchased from CDI-Labs (Baltimore, MD). The HuProt v2.0 array contains 19,000 proteins, each printed in duplicate on an epoxide-coated glass slide. Prior to experimentation, GD2 variants from our array were manually spotted to the proteome array to serve as an experimental positive control. GD2 neoglycoproteins will adhere to the epoxide surface. Briefly, a microarray pin was dipped into a 1mg/mL solution of GD2-BSA and manually spotted on the slide 4 times in an unprinted region near the top of the slide. HuProt v2.0 microarray slides were assayed using the same protocol as the glycan microarray slides. The final intensity value for each antibody-protein interaction was calculated as the average of duplicate spots on 2 independent HuProt2.0 array slides (4 spots total).
Direct Cytotoxicity
The melanoma cell line M14 and the colon cancer cell line SW620 were obtained from the National Cancer Institute Developmental Therapeutics Program (NCI DTP, Frederick, MD) tumor cell line repository. These cell lines were maintained in RPMI-1640 supplemented with 10% fetal bovine serum and 2 mM L-glutamine and passaged at regular intervals.
M14 and SW620 cells were plated into flat bottom 96-well plates at a density of 2.0 × 104 cell per well. After 24 hour incubation at 37°C and 5% CO2, increasing concentrations of germline or mature 3F8 antibodies were added to each well. The control wells received RPMI1640 media alone. Four replicates were performed for each concentration. Plates were incubated at 37°C and 5% CO2 for 72 hours. 10 uL of Cell Counting Kit-8 (CCK-8; Sigma Aldrich) reagent was added to each well and the plate was incubated an additional 4 hours. Cell number was measured by OD readings at 450 nm using a BioTek Synergy2 spectrophotomer.
Molecular Dynamics
A full description of the molecular dynamics simulation systems and parameters is available in the Supplemental Information. Briefly, a germline homology model of the 3F8 Fab was generated based on the previously published affinity mature 3F8 structure (Ahmed et al., 2013). Multi-copy simulations were run using Amber 15 in the NPT ensemble with explicit water, for a total simulation time of 100 ns for each Fab.
Supplementary Material
Highlights.
Study of immunological evolution of antibodies to a mammalian carbohydrate
Germline antibodies to ganglioside GD2 have unexpectedly high selectivity
Germline cross-reactivity not observed on glycan or human proteome microarrays
Mature and germline GD2 antibodies demonstrate similar levels of rigidity
Significance.
Sterner et al. demonstrate that germlines antibodies to the mammalian glycan GD2 have unexpectedly high selectivity. No cross reactivity was observed on a glycan microarray with 500 components or a human proteome array with 19,000 proteins. Molecular dynamics reveal pre-organized and relatively rigid binding pockets for the germline antibodies.
Acknowledgments
We thank the Consortium for Functional Glycomics (GM62116; The Scripps Research Institute), Professors Tom Tolbert (University of Kansas), Lai-Xi Wang (University of Maryland), Xuefei Huang (Michigan State University), Todd Lowary (University of Alberta) Dr. Joseph Barchi (National Cancer Institute), and Omicron Biochemicals Inc. for contributing glycans for the array. The molecular dynamics simulations utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). This work was supported in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, and in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ Contributions: E.S., M.L.P., and J.C.G. designed the study; E.S. performed biological studies and analyzed microarray data under the supervision of J.C.G.; M.L.P. carried out molecular dynamics simulations under the supervision of M.C.N.; all authors contributed to writing the paper.
Data and materials availability: The full microarray data will be publicly available in the National Center for Biotechnology Information’s (NCBI) Gene Expression Omnibus (GEO) (Edgar et al., 2002) via GEO Series accession number (GSE100438).
Disclosure of Potential Conflicts of Interest: The authors declare no conflicts of interest.
References
- Adhikary R, Yu W, Oda M, Walker RC, Chen T, Stanfield RL, Wilson IA, Zimmermann J, Romesberg FE. Adaptive mutations alter antibody structure and dynamics during affinity maturation. Biochemistry. 2015;54:2085–2093. doi: 10.1021/bi501417q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Goldgur Y, Hu J, Guo HF, Cheung NK. In silico driven redesign of a clinically relevant antibody for the treatment of GD2 positive tumors. PLoS One. 2013;8:e63359. doi: 10.1371/journal.pone.0063359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astronomo RD, Burton DR. Carbohydrate vaccines: developing sweet solutions to sticky situations? Nat. Rev. Drug Discov. 2010;9:308–324. doi: 10.1038/nrd3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Muller-Loennies S, Brade L, Kosma P, Hirama T, MacKenzie CR, Brade H, Evans SV. Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes. J. Mol. Biol. 2008;377:450–468. doi: 10.1016/j.jmb.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Campbell CT, Zhang Y, Gildersleeve JC. Construction and use of glycan microarrays. Curr. Protoc. Chem. Biol. 2010;2:37–53. doi: 10.1002/9780470559277.ch090228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 2010;10:301–316. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- Chang SH, Han JL, Tseng SY, Lee HY, Lin CW, Lin YC, Jeng WY, Wang AH, Wu CY, Wong CH. Glycan array on aluminum oxide-coated glass slides through phosphonate chemistry. J. Am. Chem. Soc. 2010;132:13371–13380. doi: 10.1021/ja1046523. [DOI] [PubMed] [Google Scholar]
- Cheung NK, Guo H, Hu J, Tassev DV, Cheung IY. Humanizing murine IgG3 anti-GD2 antibody m3F8 substantially improves antibody-dependent cell-mediated cytotoxicity while retaining targeting in vivo. Oncoimmunology. 2012;1:477–486. doi: 10.4161/onci.19864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung NK, Kushner BH, Yeh SD, Larson SM. 3F8 monoclonal antibody treatment of patients with stage 4 neuroblastoma: a phase II study. Int. J. Oncol. 1998;12:1299–1306. doi: 10.3892/ijo.12.6.1299. [DOI] [PubMed] [Google Scholar]
- Cheung NKV, Cheung IY, Kramer K, Modak S, Kuk D, Pandit-Taskar N, Chamberlain E, Ostrovnaya I, Kushner BH. Key role for myeloid cells: Phase II results of anti-GD2 antibody 3F8 plus granulocyte-macrophage colony-stimulating factor for chemoresistant osteomedullary neuroblastoma. Int. J. Cancer. 2014;135:2199–2205. doi: 10.1002/ijc.28851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Fulton Z, Huber M, Wilson IA, Burton DR. Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged. J. Virol. 2010;84:10690–10699. doi: 10.1128/JVI.01110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- Dupin L, Zuttion F, Gehin T, Meyer A, Phaner-Goutorbe M, Vasseur JJ, Souteyrand E, Morvan F, Chevolot Y. Effects of the surface densities of glycoclusters on the determination of their IC50 and KD value determination by using a microarray. Chem Bio Chem. 2015;16:2329–2336. doi: 10.1002/cbic.201500371. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acid Res. 2002;14:657–665. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DW, Muller-Loennies S, Brooks CL, Brade L, Kosma P, Brade H, Evans SV. Structural insights into parallel strategies for germline antibody recognition of lipopolysaccharide from Chlamydia. Glycobiology. 2011;21:1049–1059. doi: 10.1093/glycob/cwr041. [DOI] [PubMed] [Google Scholar]
- Finton KA, Friend D, Jaffe J, Gewe M, Holmes MA, Larman HB, Stuart A, Larimore K, Greenberg PD, Elledge SJ, et al. Ontogeny of recognition specificity and functionality for the broadly neutralizing anti-HIV antibody 4E10. PLoS Pathog. 2014;10:e1004403. doi: 10.1371/journal.ppat.1004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstenbruch S, Brooks CL, Kosma P, Brade L, Mackenzie CR, Evans SV, Brade H, Muller-Loennies S. Analysis of cross-reactive and specific anti-carbohydrate antibodies against lipopolysaccharide from Chlamydophila psittaci. Glycobiology. 2010;20:461–472. doi: 10.1093/glycob/cwp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose A, Viswanadhan V, Wendoloski J. Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: An analysis of ALOGP and CLOGP methods. J. Phys. Chem. A. 1998;102:3762–3772. [Google Scholar]
- Gildersleeve JC, Oyelaran O, Simpson JT, Allred B. Improved procedure for direct coupling of carbohydrates to proteins via reductive amination. Bioconjug. Chem. 2008;19:1485–1490. doi: 10.1021/bc800153t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildersleeve JC, Wright WS. Diverse molecular recognition properties of blood group A binding monoclonal antibodies. Glycobiology. 2016;26:443–448. doi: 10.1093/glycob/cwv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godula K, Bertozzi CR. Density variant glycan microarray for evaluating cross-linking of mucin-like glycoconjugates by lectins. J. Am. Chem. Soc. 2012;134:15732–15742. doi: 10.1021/ja302193u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EJ, Sanders WJ, Kiessling LL. Synthetic ligands point to cell surface strategies. Nature. 1998;392:30–31. doi: 10.1038/32073. [DOI] [PubMed] [Google Scholar]
- Gordus A, MacBeath G. Circumventing the problems caused by protein diversity in microarrays: implications for protein interaction networks. J. Am. Chem. Soc. 2006;128:13668–13669. doi: 10.1021/ja065381g. [DOI] [PubMed] [Google Scholar]
- Goudot A, Pourceau G, Meyer A, Gehin T, Vidal S, Vasseur JJ, Morvan F, Souteyrand E, Chevolot Y. Quantitative analysis (KD and IC50) of glycoconjugates interactions with a bacterial lectin on a carbohydrate microarray with DNA Direct Immobilization (DDI) Biosens. Bioelectron. 2013;40:153–160. doi: 10.1016/j.bios.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, Diskin R, Scheid JF, Sather DN, Burton DR, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 2013;9:e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwacik I, Golik P, Grudnik P, Kolinski M, Zdzalik M, Rokita H, Dubin G. Structural basis of GD2 ganglioside and mimetic peptide recognition by 14G2a antibody. Mol. Cell. Proteomics. 2015;14:2577–2590. doi: 10.1074/mcp.M115.052720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Hung JT, Cheung SK, Lee HY, Chu KC, Li ST, Lin YC, Ren CT, Cheng TJ, Hsu TL, et al. Carbohydrate-based vaccines with a glycolipid adjuvant for breast cancer. Proc. Natl. Acad. Sci. USA. 2013;110:2517–2522. doi: 10.1073/pnas.1222649110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung TC, Lin CW, Hsu TL, Wu CY, Wong CH. Investigation of SSEA-4 binding protein in breast cancer cells. J. Am. Chem. Soc. 2013;135:5934–5937. doi: 10.1021/ja312210c. [DOI] [PubMed] [Google Scholar]
- Jaipuri FA, Collet BY, Pohl NL. Synthesis and quantitative evaluation of Glycero-D-manno-heptose binding to concanavalin A by fluorous-tag assistance. Angew. Chem. Int. Ed. 2008;47:1707–1710. doi: 10.1002/anie.200704262. [DOI] [PubMed] [Google Scholar]
- James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- Jimenez R, Salazar G, Yin J, Joo T, Romesberg FE. Protein dynamics and the immunological evolution of molecular recognition. Proc. Natl. Acad. Sci. USA. 2004;101:3803–3808. doi: 10.1073/pnas.0305745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanska R, Clarke J, Blixt O, Macrae JI, Zhang JQ, Crocker PR, Laurent N, Wright A, Flitsch SL, Russell DA, et al. Surface plasmon resonance imaging for real-time, label-free analysis of protein interactions with carbohydrate microarrays. Glycoconjugate J. 2008;25:69–74. doi: 10.1007/s10719-007-9047-y. [DOI] [PubMed] [Google Scholar]
- Klebe G. Applying thermodynamic profiling in lead finding and optimization. Nat. Rev. Drug Discov. 2015;14:95–110. doi: 10.1038/nrd4486. [DOI] [PubMed] [Google Scholar]
- Liang R, Loebach J, Horan N, Ge M, Thompson C, Yan L, Kahne D. Polyvalent binding to carbohydrates immobilized on an insoluble resin. Proc. Natl. Acad. Sci. USA. 1997;94:10554–10559. doi: 10.1073/pnas.94.20.10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligeour C, Dupin L, Angeli A, Vergoten G, Vidal S, Meyer A, Souteyrand E, Vasseur JJ, Chevolot Y, Morvan F. Importance of topology for glycocluster binding to Pseudomonas aeruginosa and Burkholderia ambifaria bacterial lectins. Org. Biomol. Chem. 2015;13:11244–11254. doi: 10.1039/c5ob01445j. [DOI] [PubMed] [Google Scholar]
- Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- Manivel V, Sahoo NC, Salunke DM, Rao KV. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity. 2000;13:611–620. doi: 10.1016/s1074-7613(00)00061-3. [DOI] [PubMed] [Google Scholar]
- Mauri C, Bosma A. Immune regulatory function of B cells. Annu. Rev. Immunol. 2012;30:221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Seto NO, MacKenzie CR, Brade L, Kosma P, Brade H, Evans SV. Germline antibody recognition of distinct carbohydrate epitopes. Nat. Struct. Biol. 2003;10:1019–1025. doi: 10.1038/nsb1014. [DOI] [PubMed] [Google Scholar]
- Oldham RK, Dillman RO. Monoclonal antibodies in cancer therapy: 25 years of progress. J. Clin. Oncol. 2008;26:1774–1777. doi: 10.1200/JCO.2007.15.7438. [DOI] [PubMed] [Google Scholar]
- Oyelaran O, Li Q, Farnsworth D, Gildersleeve JC. Microarrays with varying carbohydrate density reveal distinct subpopulations of serum antibodies. J. Proteome Res. 2009;8:3529–3538. doi: 10.1021/pr9002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Shin I. Carbohydrate microarrays for assaying galactosyltransferase activity. Org. Lett. 2007;9:1675–1678. doi: 10.1021/ol070250l. [DOI] [PubMed] [Google Scholar]
- Patten PA, Gray NS, Yang PL, Marks CB, Wedemayer GJ, Boniface JJ, Stevens RC, Schultz PG. The immunological evolution of catalysis. Science. 1996;271:1086–1091. doi: 10.1126/science.271.5252.1086. [DOI] [PubMed] [Google Scholar]
- Romesberg FE, Spiller B, Schultz PG, Stevens RC. Immunological origins of binding and catalysis in a Diels-Alderase antibody. Science. 1998;279:1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- Scharf L, West AP, Sievers SA, Chen C, Jiang S, Gao H, Gray MD, McGuire AT, Scheid JF, Nussenzweig MC, et al. Structural basis for germline antibody recognition of HIV-1 immunogens. eLife. 2016;5:e13783. doi: 10.7554/eLife.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Cavacini L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010;125:S41–52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi DK, Agarwal A, Manivel V, Rao KV, Salunke DM. Differential epitope positioning within the germline antibody paratope enhances promiscuity in the primary immune response. Immunity. 2006;24:429–438. doi: 10.1016/j.immuni.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Shivatare SS, Chang SH, Tsai TI, Ren CT, Chuang HY, Hsu L, Lin CW, Li ST, Wu CY, Wong CH. Efficient convergent synthesis of bi-, tri-, and tetra-antennary complex type N-glycans and their HIV-1 antigenicity. J. Am. Chem. Soc. 2013;135:15382–15391. doi: 10.1021/ja409097c. [DOI] [PubMed] [Google Scholar]
- Shoreibah MG, Jackson CL, Price PW, Meagher R, Godwin AK, Cai Q, Gildersleeve JC. Anti-human embryonic stem cell monoclonal antibody Hesca-2 binds to a glycan epitope commonly found on carcinomas. Stem Cells Dev. 2011;20:515–525. doi: 10.1089/scd.2010.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xia B, Lasanajak Y, Smith DF, Cummings RD. Quantifiable fluorescent glycan microarrays. Glycoconjugate J. 2008;25:15–25. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- Steinkellner G, Rader R, Thallinger GG, Kratky C, Gruber K. VASCo: computation and visualization of annotated protein surface contacts. BMC Bioinformatics. 2009;10:32. doi: 10.1186/1471-2105-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner E, Flanagan N, Gildersleeve JC. Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database. ACS Chem. Biol. 2016;11:1773–1783. doi: 10.1021/acschembio.6b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SK, Liang PH, Astronomo RD, Hsu TL, Hsieh SL, Burton DR, Wong CH. Targeting the carbohydrates on HIV-1: Interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc. Natl. Acad. Sci. USA. 2008;105:3690–3695. doi: 10.1073/pnas.0712326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997a;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- Wedemayer GJ, Wang LH, Patten PA, Schultz PG, Stevens RC. Crystal structures of the free and liganded form of an esterolytic catalytic antibody. J. Mol. Biol. 1997b;268:390–400. doi: 10.1006/jmbi.1997.0974. [DOI] [PubMed] [Google Scholar]
- Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat. Rev. Immunol. 2010;10:317–327. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SE, Sellers BD, Jacobson MP. Effects of somatic mutations on CDR loop flexibility during affinity maturation. Proteins. 2011;79:821–829. doi: 10.1002/prot.22920. [DOI] [PubMed] [Google Scholar]
- Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, Dimitrov DS. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Comm. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Andryski SE, Beuscher AEt, Stevens RC, Schultz PG. Structural evidence for substrate strain in antibody catalysis. Proc. Natl. Acad. Sci. USA. 2003;100:856–861. doi: 10.1073/pnas.0235873100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman SG, Chen HX, Smith M, Anderson B, Villablanca JG, Matthay KK, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med. 2010;363:1324–1334. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Campbell C, Li QA, Gildersleeve JC. Multidimensional glycan arrays for enhanced antibody profiling. Mol. Biosyst. 2010;6:1583–1591. doi: 10.1039/c002259d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Ahmed M, Guo H, Cheung IY, Cheung N-KV. Alteration of electrostatic surface potential enhances affinity and tumor killing properties of anti-ganglioside GD2 monoclonal antibody hu3F8. J. Biol. Chem. 2015;290:13017–13027. doi: 10.1074/jbc.M115.650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J, Oakman EL, Thorpe IF, Shi X, Abbyad P, Brooks CL, 3rd, Boxer SG, Romesberg FE. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc. Natl. Acad. Sci. USA. 2006;103:13722–13727. doi: 10.1073/pnas.0603282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.