Abstract
The binding of oligodeoxynucleotides modified with adenine 2′-O-methyl riboside, 2,6-diaminopurine 2′-O-methyl riboside, cytosine 2′-O-methyl riboside, 2,6-diaminopurine deoxyriboside or 5-bromodeoxyuridine was studied with a microarray containing all possible (4096) polyacrylamide-bound hexadeoxynucleotides (a generic microchip). The generic microchip was manufactured by using reductive immobilization of aminooligonucleotides in the activated copolymer of acrylamide, bis-acrylamide and N-(2,2-dimethoxyethyl) acrylamide. The binding of the fluorescently labeled modified octanucleotides to the array was analyzed with the use of both melting profiles and the fluorescence distribution at selected temperatures. Up to three substitutions of adenosines in the octamer sequence by adenine 2′-O-methyl ribosides (Am), 2,6-diaminopurine 2′-O-methyl ribosides (Dm) or 2,6-diaminopurine deoxyribosides (D) resulted in increased mismatch discrimination measured at the melting temperature of the corresponding perfect duplex. The stability of complexes formed by 2′-O-methyl-adenosine-modified oligodeoxynucleotides was slightly decreased with every additional substitution, yielding ∼4°C of total loss in melting temperature for three modifications, as followed from microchip thermal denaturation experiments. 2,6-Diaminopurine 2′-O-methyl riboside modifications led to considerable duplex stabilization. The cytosine 2′-O-methyl riboside and 5-bromodeoxyuridine modifications generally did not change either duplex stability or mismatch resolution. Denaturation experiments conducted with selected perfect duplexes on microchips and in solution showed similar results on thermal stabilities. Some hybridization artifacts were observed that might indicate the formation of parallel DNA.
INTRODUCTION
In the last few years, high density DNA probe arrays or oligonucleotide microchips have become an efficient instrument in DNA and RNA sequence analysis. Probe arrays or microchips contain a large number of short single-stranded oligodeoxynucleotides immobilized or directly synthesized on a durable solid support (1–3). To date, the main attention in microchip-based experiments has been paid to biological applications, such as genetic mutation analysis, polymorphism screening or sequencing by hybridization of unknown DNA segments (4–7). Only a few studies have taken advantage of the microchip approach in physicochemical analysis of nucleic acids. Those that have used this technique include thermodynamic analysis of octamer duplexes formed with a small hydrazide-polyacrylamide gel oligonucleotide array (8), the comparative hybridizations of modified and unmodified RNA with high density probe arrays (9) and the studies of the DNA binding to membrane-type PNA grids (10). One of the important applications of microarrays that could considerably benefit from their diversity is the analysis of interactions between natural sequences and synthetic DNA or RNA analogs. The increasing utilization of modified oligonucleotides as diagnostic and therapeutic resources requires the detailed studies. The search for nucleotide analogs that stabilize A–T base pairs and do not compromise the discrimination of perfect and mismatched duplexes is of particular interest. In this paper we report the use of the generic microchip, i.e. the microchip containing all possible (4096) polyacrylamide-bound hexanucleotides (11) for the analysis of the stability and the discrimination efficiency of oligonucleotides modified with adenine 2′-O-methyl riboside (Am), 2,6-diaminopurine 2′-O-methyl riboside (Dm), cytosine 2′-O-methyl riboside (Cm), 2,6-diaminopurine deoxyriboside (D) and 5-bromodeoxyuridine (B). These modifications have been shown to be useful in antisense drug design, constructing hybridization probes with high affinity, structural studies and studies of protein–DNA interactions (12–15). The most intensively studied analog among these non-natural nucleotides is D. The reported effect of this base analog on the stability of oligo- and polynucleotide duplexes was, on average, ∼1.5° per modification (16–18). However in some cases the stability of short modified double-stranded DNA appeared to be lower when compared with unmodified duplexes (19). The specificity data on 2,6-diaminopurine-containing duplexes are limited by the study of duplexes with only few substitutions in complementary strands opposite replaced deoxyadenosines, indicating considerably lower stability for mismatches (18,19). Polyribonucleotides containing 2-aminoA replacements for adenosines showed a much higher stabilization effect in RNA duplexes (∼30°C) as compared to corresponding poly(2-amino-dA):poly(dT) replacements, which showed only 12°C increase in Tm (20). Higher priming efficiency of (2-amino-dA)-containing oligodeoxynucleotides was succesively used in PCR experiments (21). Other non-natural nucleotides selected for current on-chip experiments have not been the subject of intensive DNA duplex studies. Modifications of ribose 2′-hydroxyl units with methyl groups have found a wide application mostly in antisense technology due to their expressed ability to stabilize duplexes with complementary RNA (22). Some stabilization effect was reported for 5Br-dU modified DNA obtained from 5Br-dU substituted cells (23).
MATERIALS AND METHODS
Oligonucleotide synthesis
Oligonucleotides were synthesized on a 1 µmol scale with a 394ABI synthesizer (Applied Biosystems). Amino-oligonucleotides were labeled at the 3′ end by tetramethylrhodamine succinimide ester (Molecular Probes) according to the manufacturer’s protocol. All labeled oligonucleotides were purified by reverse phase HPLC.
Generic microchip
The microchip was manufactured using previously described techniques (24–26) with a library of 4096 amino-oligonucleotides. A grid of gel elements on a glass surface was generated by photopolymerization (24) using a stock solution containing 5% acrylamide–bisacrylamide (19:1), 0.5% N-(2,2-dimethoxyethyl) acrylamide, 40% glycerol in 0.1 M sodium phosphate buffer. Aldehydes in the gel were generated by treatment with 2% aqueous trifluoroacetic acid for 10 min. Droplets of individual oligonucleotide solutions (∼0.2 nl, 2.5 mM) were transferred from stock plates to gel pads of the microarray using homemade robotic equipment (2). Phase–transfer reduction on the microchip was carried out for 8 h in 0.1 M solution of pyridine–borane complex in chloroform covered with water (25). The final loading density of oligonucleotides was in the range of 0.1–0.2 pmol per gel pad.
Hybridization and denaturation experiments
The generic microchip was hybridized with 1 µM solution of labeled octanucleotide at 4°C in 6× SSPE buffer (1 M NaCl, 0.05 M phosphate, 5 mM EDTA pH 7.0) containing 1% Tween-20. Equilibrium melting profiles were obtained with a fluorescent microscope equipped with a CCD camera, Peltier thermotable and XY stages. Data acquisition was controlled by home-written software running under Labview‘ (8).
UV denaturation experiments
Solution melting experiments were performed in 6× SSPE buffer at the total oligonucleotide concentration of 2 µM using HP1852 diode array spectrophotometer (Hewlett Packard).
RESULTS
Immobilization of oligonucleotides
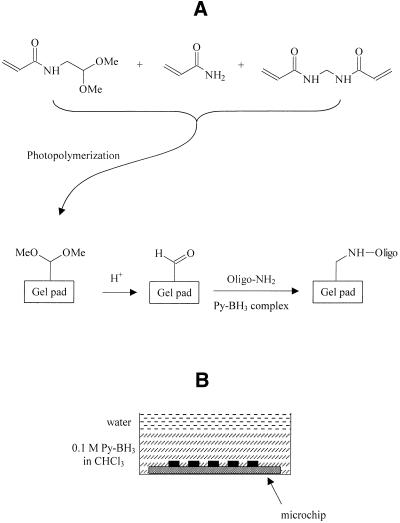
Chemistry of oligonucleotide immobilization in elements of the photochemically generated gel matrix was essentially the same as previously described (26) except for minor modifications. We used a copolymer of acrylamide, bis-acrylamide and N-(2,2-dimethoxyethyl) acrylamide to introduce the latent aldehyde function into the gel (Fig. 1). Aldehydes in the gel were activated by brief acid treatment. Loading all the hexadeoxynucleotides containing the 5′-aminolink was followed by Schiff-base formation and reduction with phase–transfer of reducing agent. In the latter procedure, shown schematically in Figure 1, the intermediate organic phase covered the whole microchip, thus preventing any cross-contamination between gel pads. Reduction of oligonucleotides captured in the form of imines inside dry gel pads may be induced only after swelling of the gel. Both water, the most suitable solvent for this purpose, and the reducing agent (pyridine borane complex) were delivered simultaneously to the gel pads from organic phase (Fig. 1B).
Figure 1.
Scheme of amino-oligonucleotides immobilization on the aldehyde-functionalized polyacrylamide copolymer (A) and phase–transfer reduction (B).
Each oligonucleotide immobilized on the generic microchip was actually eight bases long and consisted of the variable hexanucleotide part flanked from both the 3′ and 5′ ends with a degenerated nucleotide (mixture of dA, dG, dC and dT). As a result, octanucleotides formed more stable duplexes than plain hexanucleotides. Taking into account the relatively low stability of hexamer duplexes, this technique was very helpful and allowed us to increase the range of working temperatures. During the thermal denaturation experiments, the 3′- and 5′-extended probes also provided an advantage of observing the low temperature plateau for many duplexes with medium and high AT content.
Specificity of interactions
Five commercially available non-natural nucleotides were selected for microchip analysis: Am, Dm, Cm, D and B. We used octanucleotides (Table 1) containing one to three modified units and fluorescently labeled at the 3′ end by tetramethyl rhodamine (TMR). The hybridization of the labeled octanucleotides with the generic microchip was monitored in real time with a fluorescent microscope in the temperature range of 5–50°C. Scanning the hybridization patterns at different temperatures under equilibrium conditions resulted in 4096 denaturation profiles. Analysis of the binding of modified oligonucleotides was performed using both melting profiles and fluorescence distribution at selected temperatures. Raw hybridization data were treated using home-written software running under a Matlab‘ environment. The microchip layout used in analysis differed from the physical array map and was based on so-called mismatch topology. In this layout the square 64×64 array was divided into 16 fields differing from each other by variations in two central nucleotides, i.e. third and fourth positions of hexamer, as shown in Figure 2. Every field consisted of 16 clusters differing from each other by variations in the second and fifth positions. Finally, every cluster combined 16 hexanucleotides with variations in terminal positions (11,27). In this description we do not take into account base mixtures on the ends of each oligonucleotide. Such a presentation allowed an easy visual observation of all mismatches for every perfect duplex. Thus, any perfect duplex may be found as a spot on a crossing of mismatches. Another advantage of the described array layout is a convenient description of the array. Every row of the array contained sequences with the same 5′-trinucleotide part, while every column contained sequences with the identical 3′-trinucleotide parts (Fig. 2).
Table 1. Structure, stability and thermodynamics of modified and unmodified oligonucleotides.
| Oligo # |
Sequencea |
Tmchip (°C)b |
Tmsol (°C)c |
ΔH (kcal
mol-1)b |
| 1 | AAGTAACA | 18.6 | 20.6 | 30.4; 31.4c |
| 2 | AAmGTAACA | 18.7 | – | 32.9 |
| 3 | AAmGTAmACA | 15.0 | 13.1 | 33.7; 28.6c |
| 4 | AAmGTAmAmCA | 14.5 | – | 35.8 |
| 5 | ADGTAACA | 21.3 | – | 31.3 |
| 6 | ADGTDACA | 26.4 | 27.2 | 32.9; 52.2c |
| 7 | ADmGTAACA | 20.6 | – | 32.0 |
| 8 | ADmGTDmACA | 23.3 | – | 40.9 |
| 7–9 | ACGACCTA | 28.3 | – | 26.8 |
| 8–10 | ACGACCmTA | 28.4 | – | 31.3 |
| 9–11 | ACmGACmCmTA | 27.3 | – | 43.4 |
| 10–12 | ATGATTCA | 20.7 | – | 36.8 |
| 11–13 | ABGATTCA | 20.4 | – | 35.9 |
| 12–14 | ABGABTCA | 21.5 | – | 37.0 |
aThe oligonucleotides were labeled at the 3′ end with. Unmodified oligonucleotides are in bold.
bTm and enthalpy values obtained in microchip experiments were determined as previously described (6). Concentration of the labeled target in hybridization buffer was 10–6 M. Errors did not exceed ±1°C for Tm and ±8 kcal/mol for ΔH.
cData obtained from UV denaturation studies in solution. Total strand concentration used was 2 × 10–6 M. Errors were within ±0.5°C for Tm and ±5 kcal/mol for ΔH.
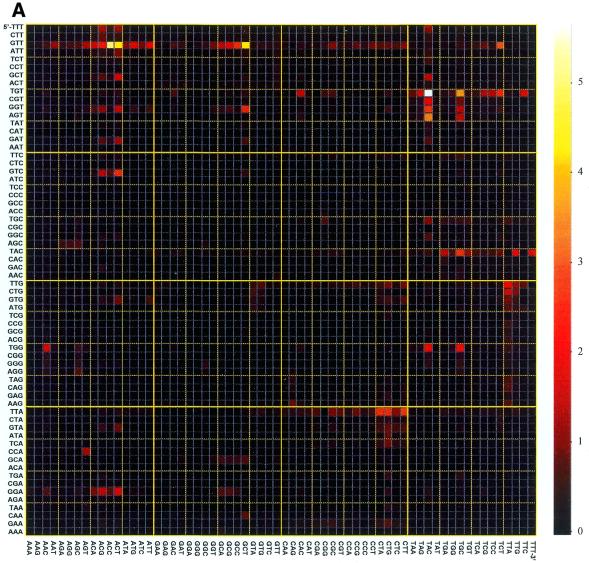
Figure 2.
Hybridization image for the labeled oligonucleotide 5′-ADGTDACA at 7°C (A) and 26°C (B). Color bars on the right of hybridization images represent the relative scale of fluorescence intensity.
Although there are three perfect duplexes formed by the labeled 8mer oligonucleotides and hexamer probes on the chip, we focused our attention on the perfect duplex in the middle of the hybridized sequence and its mismatches. In this case interactions between the terminal bases of the hybridized 8mer and mixtures of bases around hexamer core sequence are considered to be more or less uniform and having no undesirable specific 3′- or 5′-terminal effects.
For all modifications of dA we used the same basic sequence 5′-AAGTAACA. It allowed the direct comparison of the effect of different nucleotide analogs on the specificity of interaction. Modified bases were introduced into the second, fifth and sixth positions from the 5′ end of the octamer (Table 1).
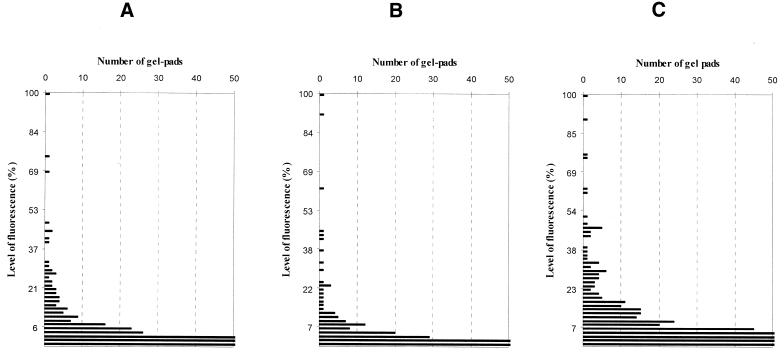
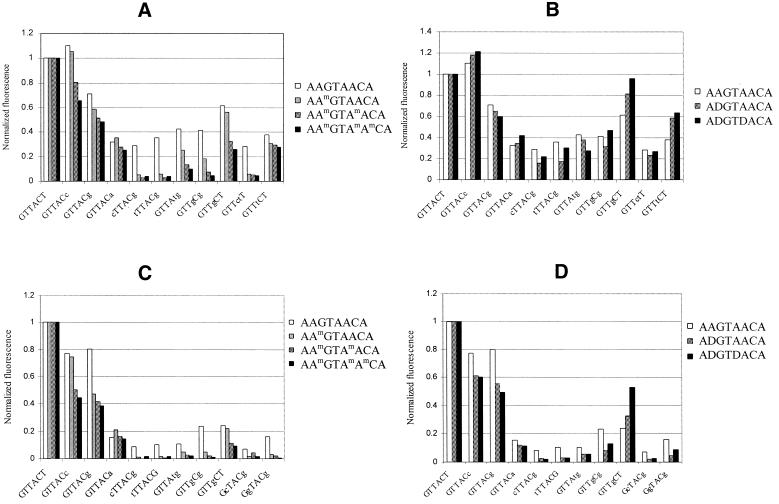
First, we analyzed the hybridization behavior of Am-, D-modified and unmodified oligonucleotides. At a low temperature (7°C), where the fluorescence level was highest, the fluorescence distribution was the narrowest for the Am-modified oligonucleotides and broader for the unmodified and D-substituted octamers (Fig. 3A–C). Such behavior was associated with the specificity of duplex formation or mismatch discrimination. We determined this parameter, i.e. discrimination between perfect and mismatched duplexes, as a ratio of fluorescence intensities from appropriate gel pads. The quantitative data on fluorescence registered at 7°C for perfect duplexes AXGTXXCA-NGTTACTN, where X = dA, Am or D and their stable mismatches are shown in Figure 4A and B. To restrict the number of analyzed spots we selected only those gel pads on the generic microchip that provided ≥10% of the perfect duplex fluorescence upon hybridization with the unmodified octanucleotide. The evident improvement in mismatch discrimination was observed for all the Am substitutions (Fig. 4A). The higher the content of modified units, the better the discrimination of mismatches. In the case of the D-modified oligonucleotides (Fig. 4B), the mismatch discrimination was nearly the same or sometimes worse, when compared with the unmodified reference sequence.
Figure 3.
Distribution of fluorescence on the generic microchip at 7°C for the hybridized labeled oligonucleotides 5′-AAGTAACA (A), 5′-AAmGTAmACA (B) and 5′-ADGTDACA (C).
Figure 4.
Comparative fluorescent signals from the duplexes formed by selected gel-immobilized hexamers with labeled Am- (A) and D-modified (B) oligonucleotides at 7°C and with the same Am- (C) and D-modified (D) oligonucleotides at Tm of the corresponding perfect duplexes. Fluorescence intensities of the perfect duplexes were assigned to 1. Mismatched nucleotides shown in lowercase.
However, the data on mismatch discrimination obtained at the selected temperature did not seem to be adequate since the stabilization or destabilization effect of modifications was not taken into account. Indeed, the stabilization effect of 2,6-diaminopurine substitutions shifted melting profiles to the high temperature area for both perfect and mispaired duplexes. As a result, the selected temperature (7°C) appeared to be closer to the low temperature plateau for the D-modified oligonucleotides. In contrast, a destabilizing action of the Am modifications (Table 1) caused the opposite effect that might be misinterpreted as an increase in mismatch discrimination. Evidently, more appropriate discrimination data could be obtained at the temperatures that had been selected specifically for particular duplexes, for example melting temperatures. Moreover, at the experimental conditions we routinely use in hybridization experiments (large excess of the hybridized oligonucleotide), the discrimination data estimated as a ratio of the hybridization signals at low temperatures (equivalent to the concentration of duplexes) may considerably differ from the ratio of corresponding equilibrium constants and would not reflect true discriminative ability.
To obtain the correct estimation of the binding specificity we measured melting profiles for all the microchip-bound duplexes and analyzed the fluorescence distribution on the generic microchip at the melting temperature of the corresponding perfect duplex. Figure 4C and D shows the mismatch discrimination data obtained for the same octanucleotides as in Figure 4A and B at the Tm of the corresponding perfect duplex. Due to the lower general level of fluorescence, the threshold of mismatch selection was reduced to 5% of the fluorescence intensity of the perfect duplex registered on the hybridization pattern of the unmodified octamer. As follows from Figure 4C, the mismatch discrimination for the Am analogs was further increased, leaving only one probe with the same level of normalized fluorescence (GTTACA) and two more probes with relatively high fluorescence (GTTACG and GTTACC), i.e., ‘terminal’ AA, AG and AC mismatches, respectively.
For the D-substituted analogs the discrimination data registered at the melting temperature of the corresponding perfect duplexes (Figs 2B and 4D) considerably differed from those obtained at 7°C. From all the imperfect duplexes, only one (GTTGCT) gained fluorescence upon modification of the octanucleotide target. The origin of the stabilization effect of this internal GT mismatch was most likely related to stacking interaction with 2,6-diaminopurine in the neighboring base pair of the duplex. Two other probes showing the relatively high signals, GTTACC and GTTACG, represented the AC and AG ‘terminal’ mismatches, similarly to Am-modified octanucleotides. Formation of non-centered D-modified duplex with immobilized probe TGTTAC was accompanied by the formation of relatively stable duplex with internal GT mismatch (probe TGTTGC, Fig. 2). Neighboring DT base pair and pairing of 2,6-diaminopurine with different bases of the terminal base mixture may explain the observed mismatch stability. Low intensity of the other non-centered perfect duplex (probe TTACTT) as compared to its mismatches is apparently related to the effect of dangling base mixture.
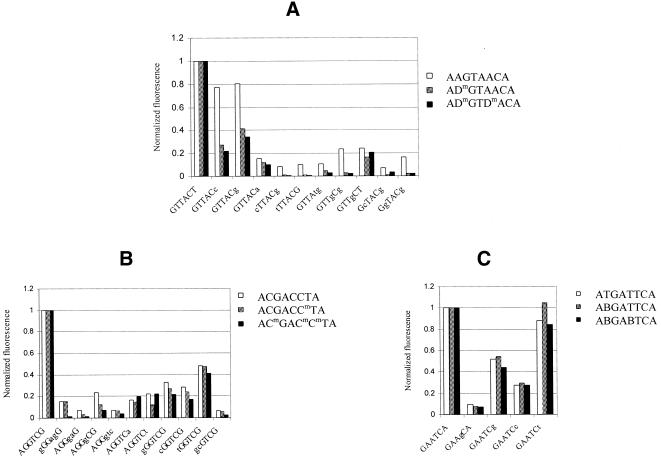
Oligonucleotides containing Dm substitutions represented mixed (i.e. base and sugar) modification. Discrimination data obtained for these modified oligonucleotides are shown in Figure 5A. The effect of Dm nucleotides in the labeled octamers on the specificity of binding appeared to be superior over Am and D analogs. Taking into account the expressed stabilization effect of Dm analogs (Table 1) this type of modification may be considered the best among all studied substitutions of dA.
Figure 5.
Comparative fluorescent signals from the duplexes formed by selected gel-immobilized hexamers with labeled and Dm- (A), Cm- (B) and B-modified (C) oligonucleotides at the Tm of the corresponding perfect duplexes. See also the legend to Figure 4.
Finally, we characterized the Cm- and B-substituted oligonucleotides (Fig. 5B and C). In the case of labeled octamers #8, #9, #11 and #12 (Table 1) no noticeable effect of a base analog on the specificity of binding was found, as compared with their unmodified counterparts #7 and #10, respectively.
Raising the temperature changed mismatch discrimination. The calculated temperature dependence of the perfect-to-mismatch ratio for model duplexes with fixed ΔS and ΔH (assuming the lower ΔH value for mismatched duplex) had a maximum that shifts to the low temperature region for lower concentrations of the hybridized oligonucleotide. We should note here that the temperature dependence of mismatch discrimination calculated as the ratio of duplex stabilities, i.e. ratio of equilibrium constants of perfect and mismatched duplexes, does not have a maximum and is highest at low temperatures. In the chosen experimental conditions the best discrimination between particular mismatch and given perfect duplex would be observed at some specific temperature, which could be easily determined from the selected pair of denaturation profiles. Thus, we observed the maximum discrimination between the mentioned above internal GT mismatch and the corresponding perfect duplex at 27°C for the unmodified octamer AAGTAACA and at 33°C for oligonucleotide ADGTDACA.
These data clearly demonstrate the benefits of assessing all the equilibrium melting profiles, thus allowing both the correct estimation of specificity and selection of optimized conditions for better discrimination of mismatches.
Duplex stability and thermodynamics
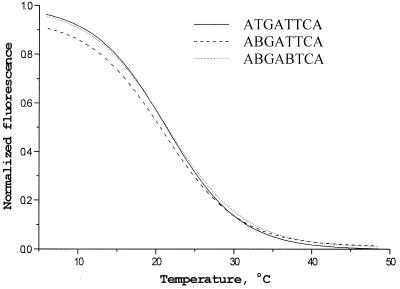
The melting profiles obtained from the generic microchip for perfect duplexes with different extents of substitution indicated the expected stabilization effect of the 2,6-diaminopurine analogs (16). An average gain of ∼4°C in Tm value was observed for every additional replacement. Remarkable stabilization was observed also for Dm-modified oligonucleotides. Substitutions of dAs with the Am residues resulted in a slight decrease of stability, while Cm and B modifications practically did not change stability, as compared with their unmodified precursors (Table 1 and Fig. 6).
Figure 6.
Melting profiles of the duplexes formed by B-substituted analogs and the gel-immobilized oligonucleotide 5′-GAATCA.
We compared denaturation profiles of three selected duplexes on the chip with UV melting curves in solution (Table 1). The duplexes consisted of labeled octamers and corresponding hexadeoxynucleotides flanked by nucleotide mixtures and containing 5′-aminolink. The difference in melting temperatures derived from the chip and solution experiments fell into the range of ±2°C (Table 1). These comparative data on duplex stability on-chip and in solution differ from those obtained previously on hydrazide–polyacrylamide gel supports (8). We believe that difference in the type of gel and nature of linker group may be responsible for such an effect.
The thermodynamics parameters, which we were able to obtain from this type of gel microarray, in some cases appeared to be very close to the results derived from the solution studies of the same duplexes. Interestingly, the decrease of stability for the Am analogs was accompanied by some growth of the ΔH values. Increasing the number of the Cm units also induced the similar enthalpy changes. It should be noted, however, that the data presented in Table 1 characterize no individual duplexes. Instead, we dealt with a mixture of 16 different duplexes and this has influenced the shape of the resulting curve, the cooperativity of transition and the ΔH values. Moreover, both the fluorescent dye covalently bound to the octanucleotide sequence and aminolink on the immobilized oligonucleotide may be responsible for some specific effects.
Unexpected hybridization signals
In hybridization patterns of the D-modified oligonucleotide ADGTDACA, we found a few relatively bright signals that were not related to either possible mismatches or shorter duplexes. In particular, three probes and their accompanying mismatches attracted our attention: NAGCAGCN, NAAGCAGN and NGCAGCTN (Fig. 2). Comparing the selected probes with the hybridized sequence AAGTAACA raises the suggestion of a parallel duplex with DA, GG, TC, AG and CC base pairs. We did not confirm the presumed motif by any other experiments. Nevertheless, some of these base pairs have been observed in parallel DNA structures (28,29).
DISCUSSION
To follow all the possible interactions of the selected single-stranded DNA target with oligodeoxynucleotide probes, it seemed reasonable to use an array containing a complete set of probes of given length. The apparent advantage of such an approach is that it allows the analysis of any mispaired duplex and thus to assess the detailed information about the specific recognition of modified DNAs or RNAs. Recently, we described the use of a microchip containing all possible hexanucleotides immobilized in polyacrylamide gel pads, a so-called generic microchip, to analyze the interaction of Hoechst dye with short DNA duplexes (11). In the present paper, we demonstrate the application of the generic microchip for studies of the modified DNA duplexes.
We have introduced some additional details to immobilization technique used in microchip manufacturing. The immobilization technique we originally developed for stable attachment of amino-oligodeoxynucleotides to aldehyde-functionalized polyacrylamide (26) was based on the use of N-(5,6-di-O-isopropylidene) hexyl acrylamide as a source of aldehydes. Both the two-step activation procedure and the relatively expensive synthesis were evident drawbacks to this method. The acrylic monomer, N-(2,2-dimethoxyethyl) acrylamide, was chosen as an expedient alternative providing easy synthesis and convenient acid activation of the gel. In combination with the phase–transfer reduction, this technique has been shown to be adequate to produce the generic microchip.
Studies of the effect of nucleotide substitutions on stability and specificity of duplex formation revealed some essential details of the selected method. The short length of the core sequence of probes considerably simplifies hybridization patterns for short model oligonucleotides and allows carrying out the detailed analysis of base pairing. In addition, the rearrangement of probes during analysis makes interactions visually easy to follow. A disadvantage, however, is the low stability of the formed short duplexes. Extension of probes by the mixtures of four bases at both ends improved this parameter. In turn, such a methodology affects the terminal mismatch discrimination. The additional degenerated base (or base mixture) converts the terminal base pair into the internal one, providing better mismatch discrimination. This should be taken into account when considering the data shown in Figures 4 and 5. Despite the general conclusion that mismatch discrimination should remain the same for plain hexamers, the difference between perfect and true terminal mismatches is expected to be lower.
In the current study we compared different nucleotide analogs in terms of mismatch discrimination and used the ratio of fluorescence intensities as the suitable experimental criteria to characterize modifications in this respect. In fact, the true measure of discriminative ability is the ratio of equilibrium constants of perfect and mismatched duplex. This parameter could be obtained directly from the experiment as a ratio of fluorescence intensities if the hybridized oligonucleotide was at very low concentration. This latter approach, however, would considerably complicate the experiment and would not generally change the conclusions.
The Tm of the perfect duplexes was selected as the reasonable criteria to characterize mismatch discrimination. This approach provides data that do not need to be corrected for differences in thermal stabilities. The increase of specificity for the D-substituted targets we observed at Tm and the apparent loss of mismatch discrimination observed at low temperature for the same oligonucleotides indicate that monitoring of equilibrium in a wide temperature range is the most appropriate method for such studies. This technique performed in parallel for the large library of immobilized oligonucleotides provides the possibility to find the optimum hybridization conditions as well.
On-chip studies of duplex stabilities gave Tm values very close to those obtained in solution experiments. This feature of the generic microchip offers the possibility of correct estimation of thermal stability for modified oligonucleotides on the array. As to the thermodynamics obtained from the microchip experiments, the data obtained do not have absolute value until reliable correlation between gel-based and solution methods has been established, still allowing, however, the comparative studies of different modifications.
Another significant aspect of the parallel analysis on arrays that came out in the current study is the detection of non-canonical DNA–DNA or DNA–RNA complexes that modified nucleic acids may presumably form.
CONCLUSIONS
We prepared a 4096-oligonucleotide microchip and used it to the analyze the stability and specificity of binding of short modified DNA duplexes. In terms of binding specificity the highest benefit from DNA modification was observed for Dm substituted oligonucleotides. These analogs also provided a significant stabilization effect. Other modified nucleotides induced less or no stabilization/specificity effect. Parallel studies on the generic microchip provided comparative thermodynamic, stability and specificity data for selected perfect and mismatched modified hexamer duplexes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Gemmell for the preparation of photopolymerized polyacrylamide arrays, and G. Yershov and A. Belgovsky for loading microarrays with oligonucleotide solutions. We are grateful to Dr M. A. Livshits and Dr Y. Lysov for their critical reading of this manuscript and I. Rudyh and A. Kolchinsky for editorial assistance. This work was supported by the US Department of Energy, Office of Health and Environmental Research, under Contract No. W-31-109-Eng-38, the Russian Human Genome Program under Grant 123/97 and the Russian Foundation of Fundamental Research under Grant 96-04-49858.
References
- 1.McGall G., Labadie,J., Brock,P., Wallraff,G., Nguyen,T. and Hinsberg,W. (1996) Light-directed synthesis of high-density oligonucleotide arrays using semiconductor photoresists. Proc. Natl Acad. Sci. USA, 93, 13555–13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yershov G., Barsky,V., Belgovskiy,A., Kirillov,E., Kreindlin,E., Ivanov,I., Parinov,S., Guschin,D., Drobishev,A., Dubiley,S. and Mirzabekov,A. (1996) DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl Acad. Sci. USA, 93, 4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Southern E.M., Maskos,U. and Elder,J.K. (1992) Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics, 13, 1008–1017. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 5.Schena M., Shalon,D., Heller,R., Chai,A., Brown,P.O. and Davis,R.W. (1996) Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl Acad. Sci. USA, 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D.G., Fan,J.-B., Siao,C.-J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J. et al. (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 7.Drobyshev A., Mologina,N., Shik,V., Pobedimskaya,D., Yershov,G. and Mirzabekov,A. (1997) Sequence analysis by hybridization with oligonucleotide microchip: identification of β-thalassemia mutations. Gene, 188, 45–52. [DOI] [PubMed] [Google Scholar]
- 8.Fotin A.V., Drobyshev,A.L., Proudnikov,D.Y., Perov,A.N. and Mirzabekov,A.D. (1998) Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res., 26, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hacia J.G., Woski,S.A., Fidanza,J., Edgemon,K., Hunt,N., McGall,G., Fodor,S.P.A. and Collins,F.S. (1998) Enhanced high density oligonucleotide array-based sequence analysis using modified nucleoside triphosphates. Nucleic Acids Res., 26, 4975–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiler J., Gausepohl,H., Hauser,N., Jensen,O.N. and Hoheisel,J.D. (1997) Hybridisation based DNA screening on peptide nucleic acid (PNA) oligomer arrays. Nucleic Acids Res., 25, 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drobyshev A.L., Zasedatelev,A.S., Yershov,G.M. and Mirzabekov,A.D. (1999) Massive parallel analysis of DNA-Hoechst 33258 binding specificity with a generic oligodeoxyribonucleotide microchip. Nucleic Acids Res., 27, 4100–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altmann K.-H., Fabbro,D., Dean,N.M., Geiger,T., Monia,B.P., Muller,M. and Nicklin,P. (1996) Second-generation antisense oligonucleotides: structure-activity relationships and the design of improved signal-transduction inhibitors. Biochem. Soc. Trans, 24, 630–637. [DOI] [PubMed] [Google Scholar]
- 13.Kutyavin I.V., Rhinehart,R.L., Lukhtanov,E.A., Gorn,V.V., Meyer,R.B. and Gamper,H.B. (1996) Oligonucleotides containing 2-aminoadenine and 2-thiothymine act as selectively binding complementary agents. Biochemistry, 35, 11170–11176. [DOI] [PubMed] [Google Scholar]
- 14.Bailly C. and Waring,M.J. (1998) The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA. Nucleic Acids Res., 26, 4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen A., Powell,L.M., Dryden,D.T.F., Murray,N.E. and Brown,T. (1995) Tyrosine 27 of the specificity polypeptide of EcoKI can be UV crosslinked to a bromodeoxyuridine-substituted DNA target sequence. Nucleic Acids Res., 23, 1177–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chazin W.J., Rance,H., Chollet,A. and Leupin,W. (1991) Comparative NMR analysis of the decadeoxynucleotide d(GCATTAATGC)2 and an analogue containing 2-aminoadenine. Nucleic Acids Res., 19, 5507–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoheisel J.D. and Lehrah,H. (1990) Quantitative measurements on the duplex stability of 2,6-diaminopurine and 5-chlorouracil nucleotides using enzymatically synthesized oligomers. FEBS Lett., 27, 103–106. [DOI] [PubMed] [Google Scholar]
- 18.Chollet A. and Kawashima,E. (1988) DNA containing base analogue 2-aminoadenine: preparation, use as hybridization probes and cleavage by restriction endonucleases. Nucleic Acids Res., 16, 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheong C., Tinoco,I. and Chollet,A. (1988) Thermodynamic studies of base pairing involving 2,6-diaminopurine. Nucleic Acids Res., 16, 5115–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard F.B. and Miles,H.T. (1983) Poly(2-aminodeoxyadenylic acid): circular dichroism and thermal stability of its complexes and their relevance to phage DNA in which A is replaced by 2NH2A. Biopolymers, 22, 597–600. [DOI] [PubMed] [Google Scholar]
- 21.Lebedev Y., Akopyants,N., Azhikina,T., Shevchenko,Y., Potapov,V., Stecenko,D., Berg,D. and Sverdlov,E. (1996) Oligonucleotides containing 2-aminoadenine and 5-methylcytosine are more effective as primers for PCR amplification than their unmodified counterparts. Genet. Anal., 13, 15–21. [DOI] [PubMed] [Google Scholar]
- 22.Inoue H., Hayase,Y., Imura,A., Iwai,S., Miura,K. and Ohtsuka,E. (1987) Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res., 15, 6131–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cullen B.R. and Bick,M.D. (1976) Thermal denaturation of DNA from bromodeoxyuridine substituted cells. Nucleic Acids Res., 3, 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guschin D., Yershov,G., Zaslavsky,A., Gemmell,A., Shick,V., Proudnikov,D., Arenkov,P. and Mirzabekov,A. (1997) Manual manufacturing of oligonucleotide, DNA and protein microchips. Anal. Biochem. 250, 203–211. [DOI] [PubMed] [Google Scholar]
- 25.Proudnikov D., Timofeev,E. and Mirzabekov,A. (1998) Immobilization of DNA in polyacrylamide gel for the manufacture of DNA and DNA–oligonucleotide microchips. Anal. Biochem., 259, 34–41. [DOI] [PubMed] [Google Scholar]
- 26.Timofeev E., Kochetkova,S., Mirzabekov,A. and Florentiev,V. (1996) Regioselective immobilization of short oligonucleotides to acrylic copolymer gels. Nucleic Acids Res., 24, 3142–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chechetkin V.R., Turygin,A.Y., Proudnikov,D.Y., Prokopenko,D.V., Kirillov,E.V. and Mirzabekov,A.D. (2000) Sequencing by hybridization with the generic 6mer oligonucleotide microarray: an advanced scheme for data processing. J. Biomol. Struct. Dyn., 18, 83–101. [DOI] [PubMed] [Google Scholar]
- 28.Mergny J.-L. (1999) Fluorescence energy transfer as a probe for tetraplex formation: the i-motif. Biochemistry, 38, 1537–1581. [DOI] [PubMed] [Google Scholar]
- 29.Germann M.W., Kalisch,B.W. and van de Sande,J.H. (1998) Structure of d(GT)n·d(GA)n sequences: formation of parallel stranded duplex DNA. Biochemistry, 37, 12962–12970. [DOI] [PubMed] [Google Scholar]