Abstract
Nonalcoholic fatty liver disease (steatosis) is the most prevalent liver disease in the Western world. One of the advanced pathologies is nonalcoholic steatohepatitis (NASH), which is associated with induction of the unfolded protein response (UPR) and disruption of autophagic flux. However, the mechanisms by which these processes contribute to the pathogenesis of human diseases are unclear. Herein, we identify the α isoform of the inhibitor of Bruton's tyrosine kinase (IBTKα) as a member of the UPR, whose expression is preferentially translated during endoplasmic reticulum (ER) stress. We found that IBTKα is located in the ER and associates with proteins LC3b, SEC16A, and SEC31A and plays a previously unrecognized role in phagophore initiation from ER exit sites. Depletion of IBTKα helps prevent accumulation of autophagosome intermediates stemming from exposure to saturated free fatty acids and rescues hepatocytes from death. Of note, induction of IBTKα and the UPR, along with inhibition of autophagic flux, was associated with progression from steatosis to NASH in liver biopsies. These results indicate a function for IBTKα in NASH that links autophagy with activation of the UPR.
Keywords: autophagy, eukaryotic initiation factor 2 (eIF2), eukaryotic translation initiation, translation control, unfolded protein response (UPR)
Introduction
Nonalcoholic fatty liver disease (NAFLD)3 is the most prevalent liver disease in the Western world, and its associated pathologies range from simple steatosis to nonalcoholic steatohepatitis (NASH); hallmarks of NASH include hepatic inflammation, increased liver enzymes, and fibrosis that increases the potential to progress to cirrhosis, liver failure, and hepatocellular carcinoma (1–3). The liver plays a central role in metabolizing and processing fatty acids for export as complex lipids to the adipose tissue for storage; however, metabolic diseases such as obesity and insulin resistance trigger adipocytes to release excess levels of circulating free fatty acids (FFAs) into the serum. Elevated levels of saturated FFAs are suggested to cause lipotoxicity in peripheral tissues, including liver, activating cellular stress pathways, which can in turn lead to increased inflammation and hepatocyte death (4, 5).
Saturated FFAs can induce the unfolded protein response (UPR), allowing for changes in both transcriptional and translational modes of gene expression that can serve to alleviate stresses afflicting the endoplasmic reticulum (ER) (6, 7). The UPR is composed of sensor proteins for ER stress, including PERK (EIF2AK3/PEK) that phosphorylates the α subunit of eIF2 (eIF2α-P), which represses global translation initiation and lowers the influx of nascent polypeptides into the stressed ER (8). Additionally, eIF2α-P leads to preferential translation of key UPR genes that function in stress adaptation (9). Recently, we showed that eIF2α-P by PERK results in preferential translation of the transcription factor CHOP (GADD153/DDIT3), which plays a central role in saturated FFA-induced lipotoxicity and hepatic inflammation (10). Increased CHOP expression triggers a signaling pathway that induces NF-κB-directed transcriptional expression and consequent secretion of pro-inflammatory cytokines, such IL-8 and TNFα, providing a linkage between the UPR and inflammation to disease progression (6, 7, 10). Although activation of CHOP by saturated FFAs is suggested to play a direct role in both the hepatotoxicity and inflammation observed during the progression of NASH, the underlying mechanisms of hepatocellular death remain unclear.
Recent literature has shown that autophagy is associated with the progression of NASH (11–14). Autophagy requires formation of double membrane vesicles that carry cytoplasmic cargo to the lysosome for degradation (15). Hepatic autophagy was reported to be impaired in NASH patients, as well as mice that displayed liver steatosis due to a high fat diet (11, 12). Of interest, although shorter term treatment of cultured human hepatocytes with the saturated FFA palmitate triggered autophagy, prolonged exposure led to a block in autophagic flux and cell death (11). It was reasoned that the block in autophagic flux contributes to the sensitivity of hepatocytes to prolonged exposure to saturated FFA, and it was reported that loss of CHOP function lowered the blockage of autophagic flux triggered by saturated FFA and protected hepatocytes from cell death (11). CHOP as well as its upstream effector–transcription factor ATF4 are suggested to contribute to the expression of select genes implicated in the formation and function of autophagosomes (16–18), which suggests that a blockage in autophagic flux rather than lowering of autophagy per se is a critical contributor to hepatoxicity by saturated FFAs. A role for the UPR in the initiation of autophagy is also supported by the finding that substitutions in eIF2α that block phosphorylation by PERK result in defective lipidation of LC3b and a consequent block in phagophore initiation (19). This finding suggests that there may be additional unknown direct translational target(s) of eIF2α-P that are essential for phagophore initiation during stress. The underlying mechanisms by which PERK and the UPR contribute to induction of autophagy will be a central focus of this study.
We recently identified the α isoform of inhibitor of Bruton's tyrosine kinase (IBTKα) as being preferentially translated in response to eIF2α-P and ER stress (20). Although the biological functions of IBTKα are not yet understood, it is noted that IBTKα contains protein–protein interaction domains, including ankyrin repeats and the BTB/POZ domain, which is suggested to enable IBTKα to serve as a substrate adapter for the E3 ubiquitin ligase CUL3 (21, 22). In this study, we show that IBTKα expression is rapidly induced by PERK upon exposure to saturated FFA and that the ensuing activation of IBTKα plays an essential role in phagophore initiation by IBTKα assembly into a multisubunit complex with LC3b, SEC16A, and SEC31A at the endoplasmic reticulum exit site (ERES). We also show that depletion of IBTKα results in impaired autophagy, along with reduced protein secretion, resulting in suppression of FFA-induced hepatocyte cell death. Finally, we use human liver biopsy samples to demonstrate that the UPR-inflammation signaling axis and increased IBTKα expression are associated with progression from steatosis to NASH, thereby linking the UPR, inflammation, and inhibition of autophagic flux to disease pathogenesis in humans.
Results
IBTKα is a novel UPR member induced by saturated FFAs
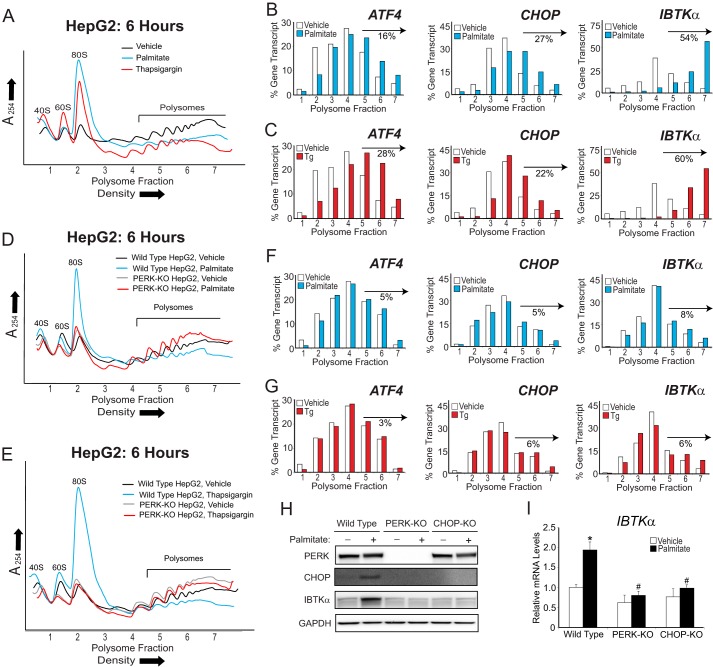
To determine whether IBTKα is preferentially translated in human hepatocytes following metabolic stress, we treated human hepatoma HepG2 cells with palmitate or thapsigargin, a pharmacological agent that potently induces ER stress. Following 6 h of treatment, we performed polysome profiling (Fig. 1A). Both thapsigargin and palmitate resulted in a reduction of heavy polysomes coincident with accumulation of monosomes, indicative of lowered global translation initiation compared with vehicle treatment. IBTKα mRNA, as well as those encoding preferentially translated controls ATF4 and CHOP, was then measured by comparing the percent of each gene transcript in the gradient fractions (Fig. 1, B and C). After either stress treatment, there was a significant shift of IBTKα mRNA toward large polysomes compared with vehicle, similar to the expected increase in the ATF4 and CHOP transcripts. Interestingly, IBTKα was present in the heaviest polysome fractions 6 and 7 after either thapsigargin or palmitate treatment, whereas ATF4 and CHOP were predominantly in polysome fractions 4–6. This shift of the IBTKα mRNA to the heaviest polysome fractions is consistent with the fact that IBTKα has a longer coding sequence that can accommodate more translating ribosomes compared with ATF4 and CHOP. HepG2 cells deleted for PERK (PERK-KO) by using CRISPR/Cas9 retained high levels of translation as viewed by heavy polysomes independent of stress (Fig. 1, D and E) and showed only modest changes in fraction distributions of IBTKα, ATF4, or CHOP mRNAs (Fig. 1, F and G). We conclude that PERK is required for repression of global protein synthesis, coincident with preferential translation of IBTKα and UPR members in human hepatocytes in response to ER stress triggered by lipotoxicity.
Figure 1.
PERK is required for preferential translation of IBTKα during palmitate exposure. A, polysome profiles of lysates prepared from HepG2 cells treated with palmitate, thapsigargin, or vehicle for 6 h. B and C, following polysome analysis, fractions 1–7 were collected, and the percentage of ATF4, CHOP, and IBTKα mRNA in each was quantified by qPCR and shown as a histogram. The percentage of the total gene transcripts in the heavy polysomes (fractions 5–7) for WT HepG2 treated with either palmitate or thapsigargin versus vehicle is indicated for each polysome profile. D and E, polysome profiles of lysates prepared from either WT or PERK-KO HepG2 cells treated for 6 h with palmitate, thapsigargin, or vehicle, as indicated. F and G, following polysome analysis of lysates prepared from PERK-KO HepG2 cells, the percentage of ATF4, CHOP, and IBTKα mRNA distributed in fractions 1–7 was quantified by qPCR and illustrated as histograms. Changes in the percentage of each gene transcript in the heavy polysomes (fractions 5–7) are indicated for the PERK-KO cells that were treated with either palmitate or thapsigargin versus vehicle. H, WT, PERK-KO, or CHOP-KO HepG2 cells were treated with either vehicle (−) or palmitate (+) for 12 h, followed by immunoblot analyses for the indicated proteins. I, WT, PERK-KO, or CHOP-KO HepG2 cells were treated with either vehicle (−) or palmitate (+) for 12 h, followed by qPCR measurements of IBTKα mRNA.
PERK and its downstream effector CHOP also trigger transcriptional expression of UPR target genes to alleviate stress or activate inflammation (8, 23). To determine whether IBTKα expression was also regulated at the transcriptional level during treatment with palmitate, we generated CHOP knock-out (CHOP-KO) HepG2 cells and exposed these cells along with their wild-type (WT) counterparts to saturated FFAs or vehicle (Fig. 1, H and I). IBTKα mRNA and protein were induced only in WT HepG2 cells treated with palmitate, whereas basal levels remained unchanged between WT and CHOP-KO cells. These results indicate that PERK activation and its downstream effector CHOP are also required for induced IBTKα mRNA expression in the UPR.
Saturated FFAs induce cell death through inhibition of autophagic flux
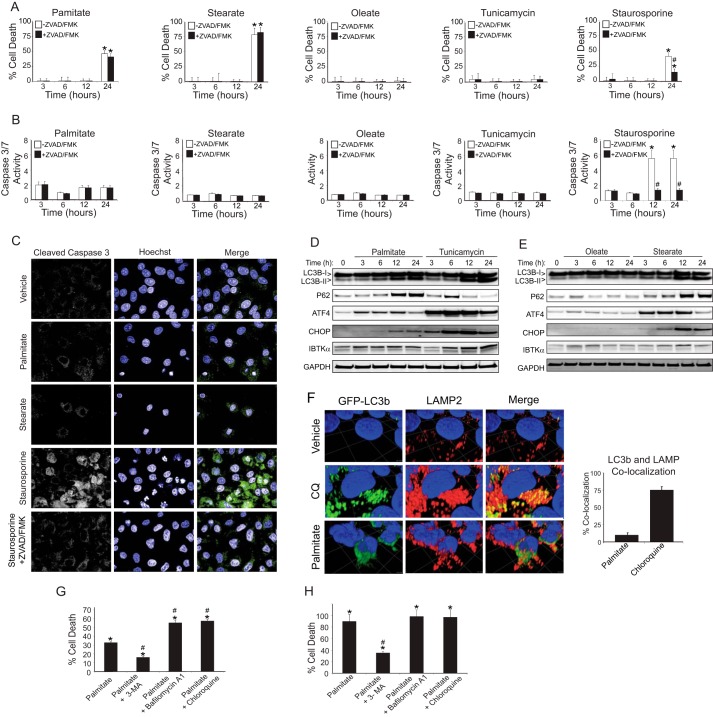
Although it was suggested that both apoptosis and autophagy are associated with lipotoxicity during NASH (5, 13, 24), it is unclear whether either of these pathways play a direct role in hepatocyte death. To determine whether apoptotic pathways are involved, we investigated the role of caspases in hepatocyte death during lipotoxicity by treating HepG2 cells with saturated and unsaturated FFAs, either alone or in the presence of pan-caspase inhibitor ZVAD/FMK (Fig. 2, A and B). In parallel, we also used controls, including staurosporine, that induce apoptosis and the potent ER stress agent tunicamycin. Treatment with saturated FFAs palmitate and stearate produced a modest increase in caspase activity, but the addition of ZVAD/FMK did not rescue cell death. By contrast, ZVAD/FMK blocked both staurosporine-induced cell death and caspase 3/7 activation. Neither the unsaturated FFA oleate nor the canonical UPR activator tunicamycin resulted in appreciable cell death after 24 h. During apoptosis, nuclear localization of cleaved caspase 3 is essential for breakdown of the nuclear lamina and DNA fragmentation (25). Although cleaved caspase 3 was localized predominantly to the nucleus after staurosporine treatment as judged by immunocytochemistry, total cleaved caspase 3 was reduced and retained in the cytoplasm with the addition of ZVAD/FMK (Fig. 2C). By contrast, the low levels of caspase 3 activation determined during treatment with saturated FFAs were coincident with caspase 3 being retained in the cytoplasm. These results indicate that apoptosis is not the predominant mode of hepatocyte death following exposure to saturated FFAs.
Figure 2.
Saturated FFAs induce hepatocyte cell death through inhibition of autophagic flux. HepG2 cells were treated with palmitate, stearate, oleate, tunicamycin, or staurosporine in the presence or absence of 20 μm ZVAD/FMK for up to 24 h, and cell viability was determined by LDH release (A), and fold change of caspase 3/7 activity relative to vehicle was measured using Apo-ONE biochemical assay (B). C, subcellular localization of cleaved caspase 3 and DNA integrity was visualized using immunofluorescence microscopy in HepG2 cells treated for 24 h. D and E, HepG2 cells were treated for up to 24 h with palmitate, tunicamycin, oleate, or stearate, and the indicated proteins were measured by immunoblot analyses. F, HepG2 cells stably transduced with GFP-LC3b were treated with vehicle, chloroquine (CQ), or palmitate for 12 h, and co-localization with LAMP2 was visualized using immunofluorescence using spinning disk confocal microscopy on the Opera. The three-dimensional images were deconvolved and quantified at the pixel level by using the program Volocity. G and H, HepG2 and primary human hepatocytes were treated for 24 h, and viability was assessed using LDH release.
Inhibition of autophagic flux is linked with NASH in human patients (11, 26). To determine whether autophagy was associated with activation of the UPR, induction of IBTKα, and cell death, we treated HepG2 cells with saturated or unsaturated FFAs for up to 24 h and measured key markers of UPR activation as well as LC3b and P62/SQSTM1 to assess changes in autophagy (Fig. 2, D and E). Increased levels of ATF4 and IBTKα proteins were observed by 3 h after treatment with palmitate and 6 h of tunicamycin, prior to induction of CHOP. Furthermore, palmitate and stearate triggered accumulation of lipidated LC3b-II and P62, suggesting that saturated FFAs either induced autophagy or, alternatively, blocked autophagic flux. Oleate had only a modest effect on the UPR and did not alter the autophagic markers, whereas treatment with tunicamycin led to potent induction of the UPR and increased LC3b-II but produced only a transient increase in P62.
To confirm whether prolonged exposure to saturated FFAs interferes with autophagic flux as reported (11), we expressed N-terminal GFP-LC3b in HepG2 cells and assessed its co-localization with lysosomes by staining with LAMP2 (Fig. 2F). First, we treated the HepG2 cells with chloroquine, which inhibits proteolysis in the lysosome and hence stabilizes LC3b; 75% of GFP-LC3b was co-localized with the lysosomal marker LAMP2, indicating that autophagosomes were properly trafficked to the lysosomes. By contrast, treatment with palmitate disrupted autophagosome trafficking to the lysosome, with only 10% of GFP-LC3b being co-localized with LAMP2 despite a robust induction of autophagy. These results are consistent with prior reports that prolonged treatment with palmitate leads to a block in autophagic flux, which would impair proper trafficking of damaged macromolecules and organelles to the lysosome for degradation.
Does disruption in the autophagic process play an active role in hepatocyte death upon exposure to saturated FFAs? We treated HepG2 cells with palmitate alone or in combination with agents that disrupt either autophagy initiation (3-methyladenine) or lysosomal function (chloroquine or bafilomycin A1) for 24 h and measured cell death (Fig. 2, G and H). To determine whether this mechanism was relevant to human hepatocytes, we also repeated the experiment with primary human hepatocytes. Addition of 3-methyladenine, which prevents induction of autophagy by inhibiting PI3K, increased the survival of both HepG2 cells and primary human hepatocytes, whereas the lysosome inhibitors resulted in increased death of HepG2 cells. Human primary hepatocytes were more sensitive to palmitate-induced cell death (>80% compared with 30% in HepG2 cells); therefore, adding chloroquine and bafilomycin A1 did not increase cell death further. These results suggest that inhibition of autophagic flux, rather than inhibition of phagophore formation per se, plays an important role in palmitate-induced hepatotoxicity.
IBTKα is required for autophagosome formation and sensitizes hepatocytes to lipotoxicity
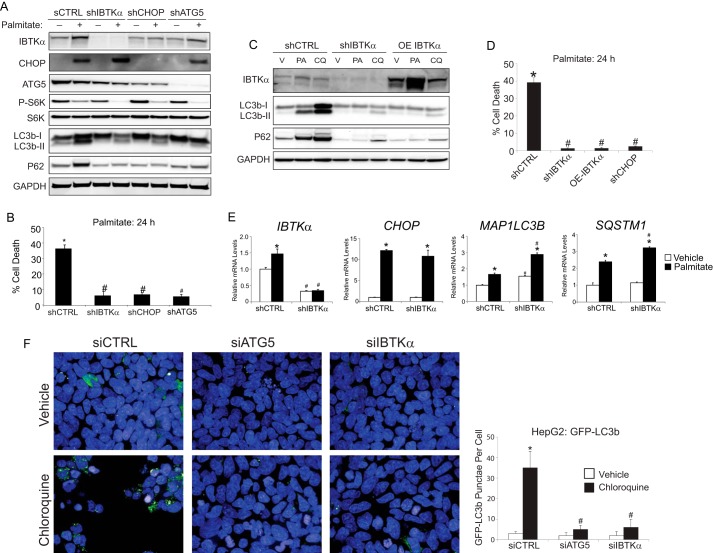
To explore the hypothesis that IBTKα regulates cell survival by induction of autophagy, we generated IBTKα knockdown (shIBTKα) HepG2 cells using shRNA and measured key protein markers of stress and autophagy, in addition to cell death following palmitate treatment (Fig. 3A). As controls, we also knocked down ATG5, which is important for formation of the autophagosome (27), as well as CHOP that is suggested to contribute to expression of select autophagy genes. Following palmitate exposure, each of the three gene knockdowns resulted in decreased conversion of LC3b-I to LC3b-II and levels of P62 coincident with lowered cell death (Fig. 3, A and B). Of interest, either knockdown of IBTKα or overexpression of IBTKα similarly reduced LC3b lipidation and P62 levels, suggesting that the appropriate amount of IBTKα expressed in cells is critical for its function (Fig. 3C). Equivalent to depletion of IBTKα, overexpression of IBTKα also suppressed the toxicity of palmitate (Fig. 3D).
Figure 3.
IBTKα is required for the induction of autophagy. A, shCTRL, shIBTKα, shCHOP, and shATG5 HepG2 cells were treated with either vehicle (−) or palmitate (+) for 12 h, and the indicated proteins were measured by immunoblot analyses. B, shCTRL, shIBTKα, shCHOP, and shATG5 HepG2 cells were treated with palmitate for 24 h, and cell viability was measured by LDH release. C, control (shCTRL) and shIBTKα HepG2 cells, or those overexpressing (OE) a FLAG-tagged IBTKα, were treated with vehicle (V), palmitate (PA), or chloroquine (CQ) for 24 h, and the indicated proteins were measured by immunoblot analyses. D, shCTRL, shIBTKα, and OE-IBTKα HepG2 cells were treated with palmitate for 24 h, and cell viability was determined by LDH release. E, shCTRL and shIBTKα HepG2 cells were treated with palmitate or vehicle for 12 h, and the indicated gene transcripts were measured by qPCR. F, HepG2 cells stably transduced with GFP-LC3b were treated with vehicle, chloroquine, or palmitate for 12 h, and co-localization with LAMP2 was visualized using immunofluorescence microscopy and quantified at the pixel level.
Earlier, we reported that knockdown of IBTKα in mouse embryo fibroblasts in the absence of environmental stress and to a lesser extent in HepG2 cells can lead to increased apoptosis (9). In support of this idea, we determined that the small amounts of caspase 3/7 activity measured in HepG2 cells void of stress were modestly elevated by knockdown of IBTKα (supplemental Fig. 1). A similar result was determined for HepG2 cells treated with bafilomycin A1. By contrast, the modest amounts of caspase 3/7 activity measured in HepG2 cells exposed to palmitate (Fig. 2A) were significantly lowered by depletion of IBTKα (supplemental Fig. 1). These results combined with our lipotoxicity results indicate that IBTKα can have differential effects on cell viability depending on the nature and extent of the environmental stress and the affected cell types.
IBTKα mRNA levels and translation were increased by the PERK/CHOP pathway in response to palmitate (Fig. 1). Corresponding expression measurements of the IBTKα-depleted cells indicated that although the levels of IBTKα mRNA and protein were lowered as expected, CHOP and its known transcriptional target genes MAP1LC3B and SQSTM1 were fully induced after palmitate treatment, indicating that IBTKα functions as a unique downstream effector in the UPR (Fig. 3, A and E). Of note, mTORC1 remained repressed in all knockdowns following palmitate treatment, as measured by phosphorylation of S6 kinase, yet autophagy was not induced (Fig. 3A). These findings indicate that IBTKα is required for the induction of autophagy during activation of the UPR by lipotoxicity, and depletion of IBTKα or its upstream regulator CHOP rescues hepatocytes from exposure to saturated FFAs.
Both shIBTKα and control (shCTRL) cells were next assayed for accumulation of LC3b in the presence or absence of bafilomycin A1 or chloroquine (Fig. 3B). Depletion of IBTKα decreased conversion of LC3b-I to LC3b-II, as well as lowered accumulation of P62 upon treatment with chloroquine. Similar results were also observed upon overexpression of IBTKα (Fig. 3C). These results suggest that IBTKα is not only a downstream UPR target but is also an essential effector in a pathway leading to the induction of autophagy prior to LC3b lipidation.
To further test the proposed role of IBTKα in phagophore initiation, we expressed GFP-LC3b in HepG2 cells treated with chloroquine or vehicle and measured the accumulation of LC3b as judged by immunofluorescence microscopy (Fig. 3F). In the control HepG2 (shCTRL) cells, there was a distinct punctate pattern upon chloroquine treatment. However, knockdown of either ATG5 or IBTKα sharply lowered the detected punctate GFP-LCb3. These results support the idea that IBTKα is critical for phagophore initiation.
We next investigated the ultrastructural changes by electron microscopy that occurred in WT, CHOP-KO, and IBTKα-KO HepG2 cells treated with bafilomycin A1, palmitate, or vehicle (supplemental Figs. S2–S4). Deletion of IBTKα in cells in the absence of stress resulted in loss of ER organization and an accumulation of damaged mitochondria (supplemental Figs. S2C and S3). Bafilomycin A1 or palmitate treatment of IBTKα-KO cells, but not control cells, led to dilation of the ER, suggesting that IBTKα is important for ER organization and general trafficking in the adaptive response to ER stress. WT HepG2 cells treated with bafilomycin A1 showed accumulation of large autophagolysosomes (supplemental Fig. 2A); however, no autophagosomes were present in either CHOP-KO or IBTKα-KO HepG2 cells treated with bafilomycin A1 (supplemental Figs. 2, B and C, S3 and S4). Exposure of WT HepG2 cells to palmitate also resulted in the appearance of lipid vacuoles throughout the cytoplasm, but a lack of autophagolysosomes, consistent with the observation that there was aberrant LC3b trafficking to the lysosome (supplemental Fig. 2A). By comparison, treatment of CHOP-KO and IBTKα-KO cells with palmitate resulted in decreased amounts of neutral lipid accumulation, still without the appearance of autophagosomes (supplemental Figs. 2, B and C, S3 and S4). These results support the idea that the UPR plays a direct role in phagophore formation through a signaling pathway involving CHOP and IBTKα.
IBTKα induces autophagy by binding to a multisubunit protein complex, including LC3b at the ERES
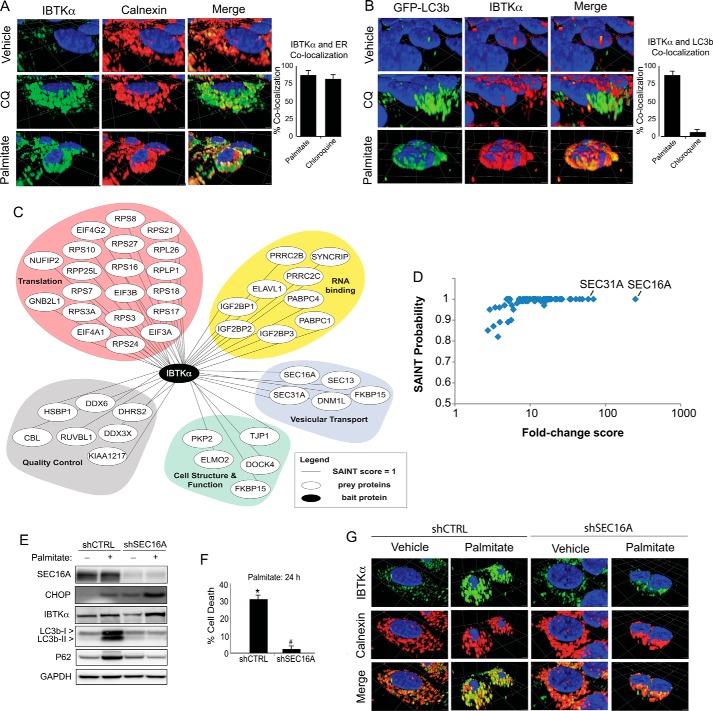
Phagophores have been suggested to form at the ER (28–32). Because both initiation of autophagy and ER morphology are dependent upon IBTKα (Fig. 3), we wanted to address the processes by IBTKα that can contribute to phagophore formation. We first utilized immunocytochemistry to determine the cellular location of IBTKα in WT HepG2 cells treated with chloroquine, palmitate, or vehicle by staining for endogenous IBTKα (Fig. 4A). IBTKα was primarily co-localized with the ER marker calnexin independent of stress. To assess whether IBTKα is also located at the site of autophagosome formation, we used GFP-LC3b HepG2 cells and followed a similar experimental design. IBTKα co-localized with only 5% of LC3b in both the vehicle and chloroquine-treated cells (Fig. 4B). However, upon treatment with palmitate, which would block autophagic flux, IBTKα co-localized with 90% of LC3b. These findings indicate that IBTKα co-localizes with phagophores initiating the ER, but not with mature autophagosomes at the lysosome.
Figure 4.
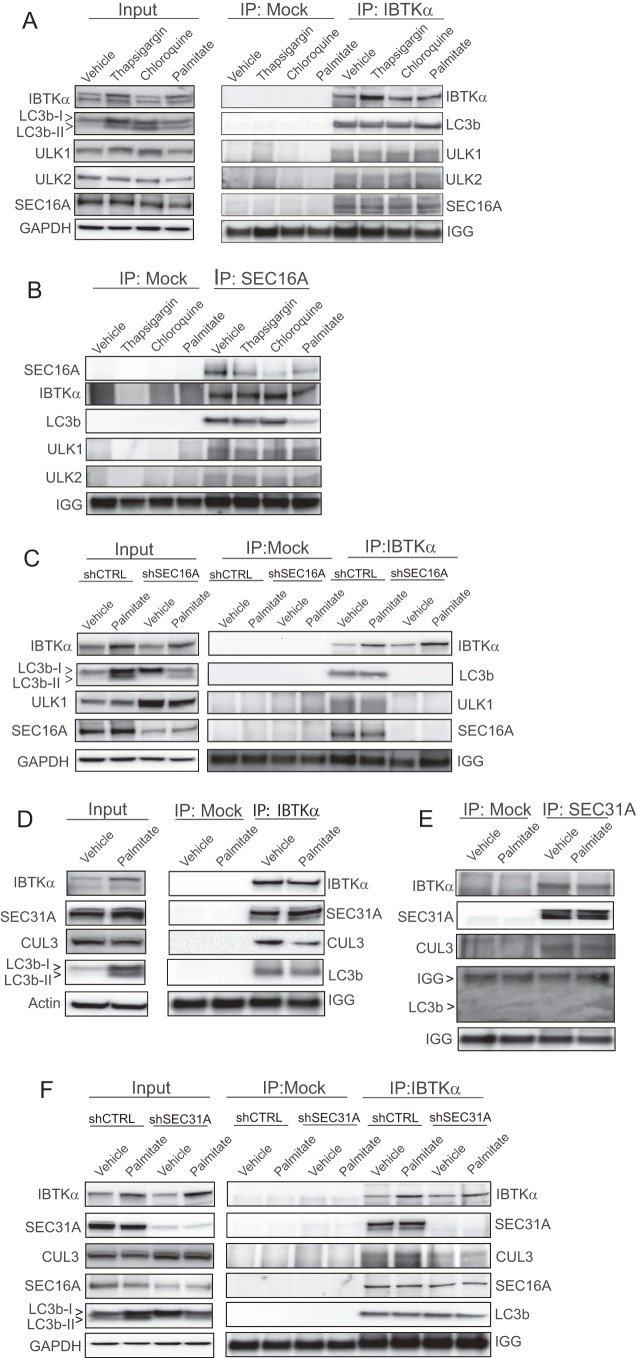
IBTKα associates with protein complex at the ERES and induces formation of phagophores. A, HepG2 cells were treated with vehicle, chloroquine (CQ), or palmitate, and IBTKα and the ER marker calnexin were visualized using immunofluorescence microscopy. Co-localization of IBTKα and ER was quantified at the pixel level. B, HepG2 cells stably expressing GFP-LC3b were treated with vehicle, chloroquine, or palmitate, and co-localization with IBTKα was visualized using immunofluorescence microscopy and quantified. C, HepG2 cells were treated with vehicle, chloroquine, thapsigargin, or palmitate. Cell lysates were used in immunoprecipitation experiments with endogenous IBTKα as bait. The network represents proteins, and their functional classes, pulled down following LC/MS analysis of eluents. D, diagram showing SAINT score against fold change spectral abundance for proteins identified in LC/MS analysis of IBTKα immunoprecipitation experiment. E, shCTRL and shSEC16A HepG2 cells were treated with either vehicle (−) or palmitate (+) for 12 h and the indicated proteins were measured by immunoblot analyses. F, control (shCTRL) and shSEC16A HepG2 cells were treated for 24 h and viability was measured by LDH release. G, shCTRL and shSEC16A HepG2 cells were treated with palmitate or vehicle for 12h and co-localization of IBTKα and canexin was visualized using immunofluorescence microscopy.
To better understand the mechanisms by which IBTKα is involved in phagophore initiation at the ER membrane, we treated WT HepG2 cells with palmitate, thapsigargin, chloroquine, or vehicle and carried out immunoprecipitations using antibody specific for endogenous IBTKα. Isolated proteins were then digested with trypsin, followed by mass spectrometry using multidimensional protein identification technology (MudPIT) (Fig. 4, C and D, and supplemental Table 1). RAW data files from each MudPIT step were used for FASTA database searching using SEQUEST HT, and the resulting dataset was filtered to allow for a false discovery rate of ≤1%. SAINT analysis was performed on the resulting peptide-spectrum matching counts from the database searches to identify proteins that displayed significant association with IBTKα prepared from WT cells compared with affinity carried out using IgG control pulldowns. A total of 73 proteins were identified that met the SAINT probability score of ≥0.8. These IBTKα-associated proteins include those involved in protein synthesis, quality control, cell structure and function, RNA binding, and vesicular transport (Fig. 4D). Among the proteins pulled down with IBTKα with the greatest degree of confidence were SEC16A and SEC31A, two essential proteins situated in the ERES, with functions in COPII-vesicle trafficking and suggested roles in phagophore initiation (32–34). The pulldown results were consistent with the notion that IBTKα is part of a larger complex that plays a role in phagophore formation at the ERES.
We next addressed whether the IBTKα-binding partner SEC16A also plays a direct role in phagophore formation and lipotoxicity. We treated HepG2 cells depleted for SEC16A expression with either palmitate or vehicle and measured changes in conversion of LC3b-I to LC3b-II as well as cell death (Fig. 4, E and F). Depletion of SEC16A decreased LC3b lipidation and P62 accumulation, coincident with enhanced resistance to lipotoxicity. Additionally, immunocytochemical analyses of the SEC16A-depleted cells HepG2 cells revealed an altered ER morphology and consequent changes in the pattern of IBTKα localization (Fig. 4G). These results indicate that SEC16A is important for ER organization, IBTKα localization, and induction of autophagy.
To further test whether IBTKα associates with SEC16A and LC3b, we treated WT HepG2 cells with thapsigargin, chloroquine, palmitate, or vehicle and then immunoprecipitated IBTKα, followed by immunoblot analyses of associated proteins (Fig. 5A). We found that independent of stress, IBTKα was complexed with LC3b, along with SEC16A, and ULK1/2, which have been suggested to interact directly with LC3b at the ERES and be essential for phagophore formation (32–36). These same protein interactions were observed using reciprocal SEC16A pulldowns, and loss of SEC16A expression resulted in disruption of the IBTKα complex (Fig. 5, B and C). These findings suggest that IBTKα associates with a complex, including SEC16A, and that this complex is essential for recruitment of a key phagophore-inducing complex involving LC3b and ULK1/2. Although IBTKα can be bound to this multisubunit complex independent of stress as judged by pulldown experiments, exposure to saturated FFAs enhances expression of IBTKα through the UPR and as a consequence its localization to this complex in the ERES.
Figure 5.
IBTKα associates with SEC proteins. A and B, HepG2 cells were treated for 12 h with the indicated stress compounds, and cell lysates were used for immunoprecipitation with IgG control (mock), IBTKα, or SEC16A followed by immunoblot analysis. On the left of A, input indicates the immunoblot analyses of total lysates. C, shCTRL and shSEC16A HepG2 cells were treated for 12 h with vehicle or palmitate, and cell lysates were used for immunoprecipitation with IgG control (mock) or IBTKα followed by immunoblot analysis. On the left of C, input indicates the immunoblot analyses of total lysates. D and E, HepG2 cells were treated with palmitate or vehicle for 12 h, and cell lysates were used for immunoprecipitation with IgG control (mock), IBTKα, or SEC16A, followed by immunoblot analysis to measure the indicated proteins. On the left of D, input indicates immunoblot analyses carried out using total lysates. F, shCTRL and shSEC31A HepG2 cells were treated with vehicle or palmitate for 12 h, and cell lysates were used for immunoprecipitation with IgG control (mock) or IBTKα, followed by immunoblot analyses to measure the indicated proteins.
We also tested the association between IBTKα and SEC31A and potentially additional members of the complex by immunoprecipitating IBTKα from HepG2 cells treated with palmitate or vehicle, followed by immunoblot analyses of the bound proteins. IBTKα was associated with SEC31A and CUL3, along with LC3b independent of treatment with saturated FFAs (Fig. 5, D and F). However, reciprocal pulldown of SEC31A indicated that LC3b is not part of this complex (Fig. 5E). Although knockdown of SEC31A lowered binding of IBTKα with CUL3, it did not disrupt association of IBTKα with SEC16A or LC3b (Fig. 5E). Knockdown of SEC31A also lowered LC3bI to LC3bII conversion and reduced P62 levels in HepG2 cells treated with palmitate (Figs. 5E and 6A). These results indicate that IBTKα can associate with distinct complexes at the ERES that participate in phagophore initiation.
Figure 6.
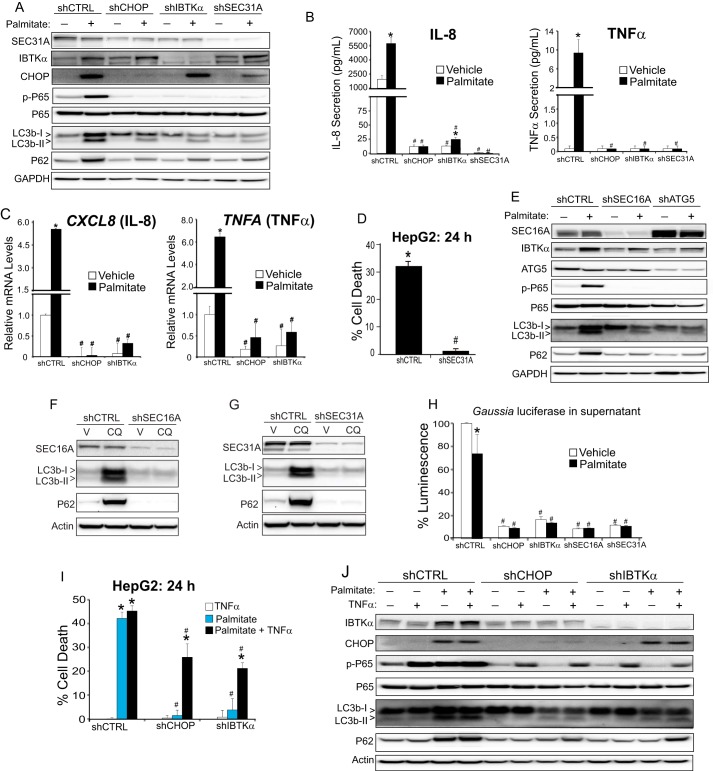
IBTKα and the UPR activate NF-κB and secretion of cytokines triggering lipotoxicity. A, control (shCTRL), shCHOP, shIBTKα, and shSEC31A HepG2 cells were treated with vehicle (−) or palmitate (+) for 12 h, and cell lysates were used for immunoblot analyses to determine the levels of the indicated proteins. B, shCTRL, shCHOP, shIBTKα, and shSEC31A HepG2 cells were treated with vehicle or palmitate for 12 h, and IL-8 and TNFα release was measured by sandwich ELISA. C, shCTRL, shCHOP, and shIBTKα HepG2 cells were treated with vehicle or palmitate for 12 h, and the levels of CXCL8 (IL-8) and TNFA (TNFα) mRNAs were measure by qPCR. D, shCTRL and shSEC31A HepG2 cells were treated with palmitate for 24 h, and cell viability was measured by LDH release. E, shCTRL, shSEC16A, and shATG5 HepG2 cells were treated with either vehicle (−) or palmitate (+) for 12 h, and cell lysates were used for immunoblot analyses to determine the levels of the indicated proteins. F, shCTRL and shSEC16A HepG2 cells were treated with chloroquine (CQ) or vehicle (V) for 24 h, and the indicated proteins were measured by immunoblot. G, HepG2 cells depleted for SEC31A (shSEC31A) or control (shCTRL) were treated with chloroquine or vehicle for 24 h, and the indicated proteins were measured by immunoblot. H, shCTRL, shCHOP, shIBTKα, shSEC16A, and shSEC31A HepG2 cells were transfected with a Gaussia luciferase reporter and treated with vehicle or palmitate for 12 h. Luciferase activity was measured in the supernatant. I, cultured shCTRL, shCHOP, and shIBTKα HepG2 cells were treated for 24 h with recombinant TNFα and palmitate, alone or in combination, as indicated, and cell viability was measured by LDH release. J, shCTRL, shCHOP, and shIBTKα HepG2 cells were treated for 24 h with recombinant TNFα and/or palmitate, as indicated, and the indicated proteins were measured by immunoblot analyses.
IBTKα and the UPR activate NF-κB and secretion of cytokines triggering lipotoxicity
Previously, we determined that treatment of hepatocytes with saturated FFAs induces the PERK/CHOP pathway in the UPR, leading to activation of NF-κB by a process involving phosphorylation of the subunit P65/RELA at serine 536 (10). Induced NF-κB contributes to increased expression and subsequent secretion of TNFα and IL-8; TNFα secreted from hepatocytes functions in a paracrine fashion, facilitating hepatocyte cell death and also further amplification of NF-κB activation, whereas both TNFα and IL-8 contribute to inflammation (10). The precise mechanisms by which CHOP contributes to activation of NF-κB in hepatocytes treated with saturated FFAs is uncertain. It is noteworthy that expression of the inhibitory IκBα was not significantly affected by loss of CHOP (10).
We reasoned that CHOP function in phagophore initiation and accumulation of suggested toxic autophagosome intermediates may be contributors to CHOP activation of NF-κB and its downstream target genes. To test this idea, we determined whether depletion of IBTKα, or one of its interacting partners at the ERES-SEC31A, also prevented induction of NF-κB-directed gene expression. Similar to that observed for knockdown of CHOP, depletion of either IBTKα or SEC31A blocked P65 phosphorylation at serine 536, as well as induced secretion of TNFα and IL-8 and mRNA expression of these genes (Fig. 6, A–C). Finally, knockdown of SEC31A also thwarted death of HepG2 cells upon prolonged exposure to palmitate as observed for depletion of CHOP and IBTKα (Fig. 6D).
We next addressed whether SEC31A, along with SEC16A, has an important role in phagophore formation in HepG2 cells. As noted earlier, knockdown of SEC16A lowered the levels of LC3B-II and P62 in HepG2 cells treated with palmitate (Figs. 4E and 6E). Furthermore, depletion of SEC16A sharply lowered LC3b-II upon a block in autophagic flux during treatment with chloroquine (Fig. 6F). Similar results were observed upon depletion of SEC31A (Fig. 6G). These results support the idea that both SEC16A and SEC31A participate in initiation of autophagy.
SEC16A and SEC31A have well documented functions in the formation of COPII vesicles at the ERES (37), along with their suggested role in phagophores during assembly (32–34). Therefore, the role of these SEC proteins, and by inference CHOP and IBTKα, for activation of NF-κB and hepatotoxicity by saturated FFAs could result from the functions of these genes in facilitating secretion of TNFα and/or phagophore formation. ATG5 is central for phagophore formation, as visualized by lowered LC3b lipidation and P62 accumulation in ATG5-depleted HepG2 cells treated with palmitate (Figs. 3A and 6E). Of importance, knockdown of ATG5 blocked P65 phosphorylation upon treatment with the saturated FFA, strongly supporting the model that NF-κB activation during lipotoxicity is dependent upon the induction of autophagy (Fig. 6E). These findings indicate that lowered secretion of TNFα and IL-8 in the IBTKα-depleted cells is not merely a consequence of loss of COPII-directed secretion, which occurs with disruption of the protein complexes featuring SEC16A and SEC31A at the ERES (37).
To address whether general secretion also involves IBTKα and associated proteins at the ERES that facilitate initiation of autophagy, we transfected control and HepG2 cells depleted for IBTKα, CHOP, SEC16A, or SEC31A with a Gaussia luciferase reporter construct and measured secreted luciferase activity in the cell culture media following treatment with either vehicle of palmitate (Fig. 6H). Depletion of each of the four gene targets resulted in a reduction of Gaussia luciferase secretion. These findings suggest that the assembled multisubunit protein complexes containing IBTKα at the ERES are integral to secretion processes emanating from the ER, as well as the formation of autophagosomes.
We previously showed that secreted TNFα is central for hepatocellular death upon exposure to saturated FFAs (10). We addressed this idea by adding recombinant TNFα to WT, shCHOP, and shIBTKα HepG2 cells in the presence or absence of palmitate treatment. There was significant death of shCHOP and shIBTKα cells only after treatment with TNFα in combination with palmitate (Fig. 6I), which was accompanied by increased phosphorylation of P65 (Fig. 6J). These results indicate that IBTKα is critical for cytokine expression and secretion and consequent inhibition of autophagic flux during metabolic stress.
UPR and NF-κB are induced in human liver biopsies of NASH patients
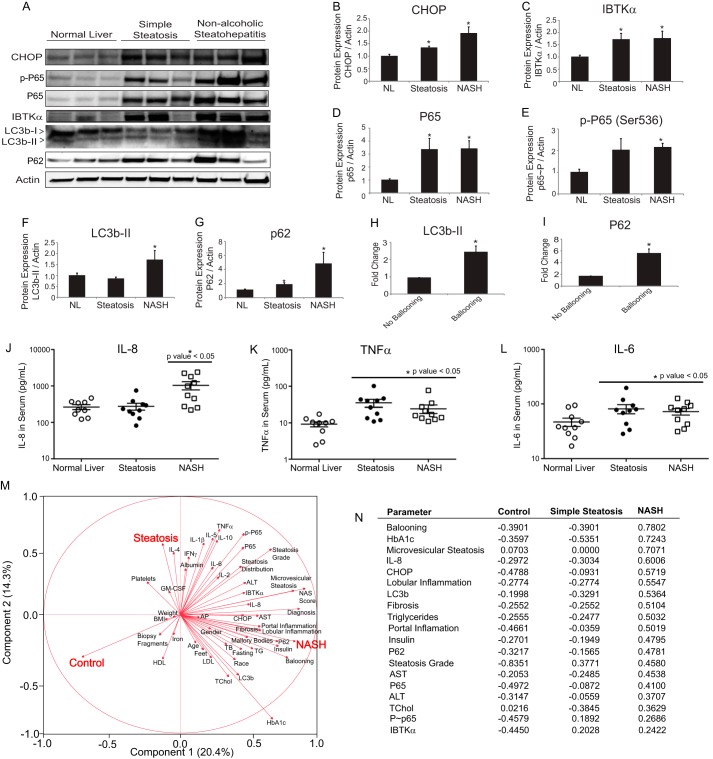
To address whether our proposed UPR signaling model derived from the HepG2 and primary human hepatocyte studies is truly relevant to disease progression in human liver, we investigated induction of the UPR and autophagy in a set of liver biopsy samples from patients undergoing bariatric surgery. These patients were matched for body mass index (BMI), gender, and age (supplemental Table 2). Patients were categorized after histological evaluation and standard scoring for NAFLD, including steatosis grade, degree of fibrosis, and presence of hepatocellular ballooning and lobular inflammation according to the NASH clinical research network proposed system (38). Levels of CHOP, P65, phosphorylated P65, and IBTKα were increased in the livers of patients with NAFLD (simple steatosis and NASH) relative to normal control (Fig. 7, A–E). Additionally, inhibition of autophagic flux was observed in NASH patients as measured by LC3b-II conversion and P62 levels (Fig. 7, F and G), consistent with severe hepatocellular injury markers such as ballooning (Fig. 7, H and I). These findings indicate that the UPR, including CHOP and IBTKα, and NF-κB are activated in patients with NAFLD, and as the disease progresses from simple steatosis to NASH, there is a strong correlation with inhibition of autophagic flux.
Figure 7.
UPR and NF-κB are up-regulated in NAFLD and NASH. A, representative immunoblot measurements for the indicated proteins in lysates prepared from human liver biopsy samples of patients with normal liver, simple steatosis, or NASH. B–G, protein quantification of immunoblots from lysates of human liver biopsy samples (n = 10 per group). Data are shown as means ± S.E. of the mean (S.E.). H and I, protein quantification of immunoblots for LC3b-II and P62 from lysates prepared from human liver biopsy samples of patients grouped by histological presence of ballooning. Data are shown as mean ± S.E. J–L, circulating levels of IL-8, TNFα, and IL-6 from serum of patients to matched liver biopsy samples. M, principal component analysis that combined all cytokine data, histological and clinical chemistry findings, as well as protein quantification from liver biopsy samples. N, abridged correlation table from principal component analysis. See supplemental Tables 2 and 3 for additional information.
To address whether UPR and NF-κB activation are associated with increased cytokine secretion, we performed a cytokine panel on serum from patients (Fig. 7, J–L, and supplemental Table 3). Levels of IL-6, IL-8, and TNFα were elevated in NASH patients, whereas only IL-6 and TNFα were elevated in those with simple steatosis. These results, along with all other metadata collected on the human samples, were utilized to carry out a principle component analysis (Fig. 7, M and N). From this analysis, we identified a mixture of cell-based and serum biomarkers that are closely related to NASH, steatosis, or both NAFLD states. For example, CHOP, IL-8, AST, LC3b, and P62 correlate strongly with NASH, whereas IBTKα, phosphorylated P65, and TNFα correlate with both NAFLD states (simple steatosis and NASH).
Discussion
Accumulation of saturated FFAs and activation of the UPR activation are common features associated with NAFLD; however, how the UPR contributes to the progression from simple steatosis to NASH is not clear (7). This report indicates that activation of the UPR and IBTKα, in combination with inhibition of autophagic flux during prolonged exposure to saturated FFAs, is as a key driver of hepatocellular death and the pathophysiology of NASH (Fig. 7A). IBTKα expression was induced as part of the UPR by both translational and transcriptional control mechanisms (Fig. 1), and loss of IBTKα in human hepatocytes thwarted induction of autophagy and enhanced survival of hepatocytes exposed to saturated FFAs (Fig. 2).
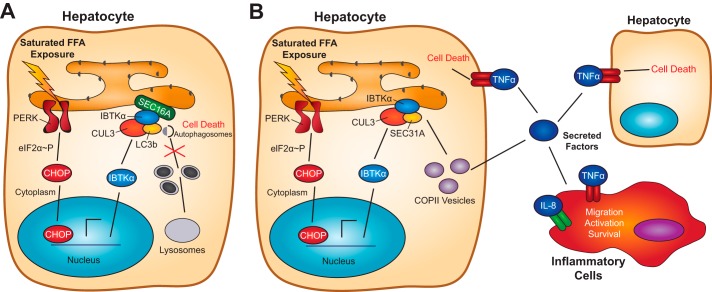
Translational control during ER stress allows for rapid enhanced expression of IBTKα and consequent assembly of a multisubunit complex at the ERES that included key factors, such as LC3b, ULK1/2, SEC16A, and SEC31A that served to promote phagophore initiation (Fig. 4). In response to saturated FFAs, the UPR and CHOP trigger a signaling pathway involving NF-κB, which induces transcriptional expression of key cytokines that contribute to inflammation and cell death (Figs. 6 and 8B). Among these cytokines, TNFα is suggested to function as an autocrine and paracrine factor that contributes to cell death and amplifies NF-κB-directed gene expression (Fig. 8B).
Figure 8.
Model for the role of IBTKα in the regulation of autophagy and pathogenesis of NASH. A, model for the UPR and IBTKα regulation of autophagy induction at the ERES during metabolic stress. Saturated FFAs induce the UPR, featuring induced transcriptional expression of IBTKα by CHOP and preferential translation by eIF2α-P. IBTKα assembles in a multisubunit complex with SEC16A, ULK1/2, and LC3b at the ERES, culminating in induction of phagophores. Saturated FFAs also block the autophagic flux, contributing to hepatocyte death. B, model for the UPR and IBTKα regulation of secretion through COPII vesicles during metabolic stress. Induced secretion of key cytokines, including TNFα and IL-8, play a key role in hepatocyte cell death and inflammation during lipotoxicity.
Enhanced levels of IBTKα are also suggested to promote formation of vesicles from the ER that facilitate secretion of these cytokines (Fig. 6). Lowered levels of secreted reporter Gaussian luciferase upon loss of IBTKα, as well as SEC16A and SEC31A, indicate that IBTKα facilitates at least a portion of the secretome. IBTKα may function as a multidomain adapter protein in the assembly of the protein complex at the ERES and/or help direct E3 ubiquitin ligase CUL3 ubiquitylation of proteins required for assembly or function of the complex. Importantly, loss of SEC31A did not disrupt IBTKα association with SEC16A or LC3b, suggesting that there are distinct complexes, including IBTKα, that are situated at the ERES (Fig. 8). Autophagy initiation and COPII-directed vesicle transport are suggested to be linked, as depletion of IBTKα prevents both the induction of autophagy and secretion of the secreted luciferase reporter and cytokines. The full complement of proteins functioning in conjunction with IBTKα in the ERES complexes is currently unknown. Furthermore, it is not known whether these distinct complexes are restricted to either formation of phagophores or to protein secretion or, alternatively, there are some common protein assembly steps at the ERES that contribute to both processes.
Autophagy is often thought of as a cell survival pathway that directs damaged organelles and misfolded proteins for degradation in lysosomes (39). However, autophagy can play a direct role in cell death, and recent studies have suggested that there are two main processes: 1) type II autophagic cell death, which consists of cell death accompanied by accumulation of large secondary vacuoles, and 2) autosis, which is characterized by a disappearance of the ER and general self-eating (40). Our results suggest that saturated FFAs induce neither type II cell death nor autosis, as autophagy is a key driver in hepatocellular death without the appearance of secondary lysosomes.
A key feature of our study is the localization of IBTKα to ERES through a complex that includes LC3b, SEC16A, and SEC31A (Fig. 4). When the hepatocytes were exposed to palmitate, LC3b-II was retained at the endoplasmic reticulum with IBTKα, suggesting either a defect in phagophore elongation or trafficking from the ERES to the lysosome. Further support that a block in autophagy flux rather than reduced autophagy per se is critical for the lipotoxicity of hepatocytes is that loss of ATG5, which facilitated phagophore formation, and CHOP that can direct expression of genes critical to formation of autophagosomes provides for resistance to saturated FFAs. These findings suggest that saturated FFAs induce a novel form of autophagic cell death, perhaps a subset of autosis, which allows for the lipidation of LC3b but interferes with LC3b trafficking from the ER to lysosomes. Importantly, this block in autophagic flux is coincident with increased inflammation and TNFα signaling thereby linking key features of NASH pathogenesis in a linked signaling cascade (Fig. 8B). Although it is possible that this form of cell death is unique to metabolic stress, it is also likely to be a driver in other diseases states or even drug-induced liver injury in which hepatocellular death and inflammation are prominent pathologies.
Induction of the UPR and a block in autophagic flux were also supported by our analysis of human liver biopsy samples, which indicated that levels of IBTKα, CHOP, as well as total and phosphorylated P65 were increased in both simple steatosis and in NASH (Fig. 7). Of importance, levels of LC3b-II and P62 were significantly elevated in NASH, which were strongly associated with hepatocellular injury markers, such as ballooning. Therefore, we have identified biomarkers suggested to be specific for NASH, as well as others associated with general NAFLD. Furthermore, analyses of the liver biopsy samples indicate that our in vitro model correlates well with the events occurring in NAFLD. Our principal component analysis combining protein quantification, histology, and serum analysis indicates that key factors and pathways described herein will provide new insight into the progression of simple steatosis to NASH.
In summary, we identified IBTKα as a novel preferentially translated member of the UPR during metabolic stress, which plays a role in both phagophore initiation and formation of transport vesicles at the ERES, linking autophagy and secretion to the pathogenesis of NASH. Furthermore, our study indicates that aberrant autophagy and the consequent inhibition of autophagic flux are key drivers in the pathogenesis of NASH at both the cellular level and as correlative biomarkers in patient samples. At present, no approved therapies exist to treat NASH, and there is a need a for better mechanistic understanding of cellular targets that repress the associated pathologies and progression of NASH. The ability to target both hepatocellular death and inflammation through IBTKα in the treatment of simple steatosis and/or NASH could have therapeutic benefits, and further work in the validation of this target is warranted.
Experimental procedures
Stable gene knockdowns and knock-outs
Stable knockdown and control cells were generated by transducing HepG2 cells with lentivirus carrying shRNA from Sigma against shCHOP (TRCN0000364393 and TRCN0000007263), shIBTKα (TRCN0000082575 and TRCN0000082577), shATG5 (TRCN0000151963 and TRCN0000151474), shSEC16A (TRCN0000246017), and shSEC31A (TRCN0000436177). Stable knockouts were constructed by using a plasmid from Sigma expressing the guide RNA, CAS9, long terminal repeats, and puromycin for CHOP (HS0000185403, GGAAATCGAGCGCCTGACCAGG), PERK (Hs0000302986, AATTATCAGCACTTTAGATGGG), or IBTKα (HS0000392536, GCTTTGGATCTTGTAATGAAGG). Stable N-terminal GFP-LC3b cells were constructed using a plasmid (EX-T0824-Lv103) from GeneCopoeia (Rockville, MD). Following transduction, cells taking up virus were selected using 10 μg/ml puromycin, and knock-out cells were sorted for single cells using flow cytometry. For the Gaussia luciferase assay, PSV40-gluc control plasmid from New England Biolabs (Ipswich, MA) was transfected to cells, and supernatant was used to measure secreted luciferase.
Cell culture and measurements of cell viability
Human hepatoma HepG2 cells were purchased from ATCC and were both cultured and treated with 600 μm FFAs as described previously (10). Furthermore, to invoke ER stress or to alter autophagy, cells were treated with 1 μm thapsigargin, 2 μm tunicamycin, 50 μm chloroquine, or 0.1 μm bafilomycin A1, as indicated. Cell viability was measured using lactate dehydrogenase (LDH) (41). Data were normalized to total LDH release by 10% Triton X-100. Recombinant TNFα (PHC03015L; Life Technologies, Inc.) was formulated in phosphate-buffered saline (PBS) with BSA and applied to cells as indicated in the figure legends. Caspase 3/7 activity was measured using the Apo-ONE homogeneous caspase 3/7 assay from Promega (Madison, WI).
Polysome profiling
HepG2 control and PERK-KO knock-out cells were cultured in the presence of vehicle, 600 μm palmitate, or 1 μm thapsigargin for 6 h. 50 μg/ml cycloheximide was added to each culture dish for 10 min prior to collection. Cell lysates were collected and subjected to centrifugation in a 10–50% sucrose gradient in a Beckman SW41Ti rotor for 2 h at 4 °C at 40,000 rpm. To measure RNA abundance, an absorbance of 254 nm was monitored as described (42). ATF4, CHOP, and IBTKα mRNA levels were measured by qPCR in each of the seven collected fractions as described (20). Firefly luciferase mRNA was added to each fraction to facilitate normalization in the cDNA and qPCR analysis for the transcript measurements as described. Data are represented as the percentage of transcript found in each fraction relative to the total for each mRNA, and a percentage change in large polysomes (fractions 5–7) with treatment relative to vehicle was measured.
Measurements of mRNA by qPCR
Following the indicated compound treatments, RNA was isolated using TRIzol reagent (Life Technologies, Inc.), and cDNA synthesis was performed using the TaqMan RT kit (Life Technologies, Inc.). Primers utilized for measuring mRNA levels are in supplemental Table 4, and transcripts were normalized to GAPDH.
Immunoblot analysis and ELISAs
Protein lysates were collected and quantified using Pierce BCA Protein Assay kit (Thermo Fisher Scientific). Proteins were separated by electrophoresis using 4–12% BisTris gels via SDS-PAGE, transferred to nitrocellulose filters, and blocked for 1 h at room temperature. Membranes were incubated overnight with the following primary antibodies: from Cell Signaling Technology (Beverly, MA), ATF4 (11815S), GAPDH (2118S), p65 (8242), p-P65 (3033), CUL3 (2759S), ULK1 (8054S), PERK (5683), ATG5 (2630S), S6K (9202S), P-S6K (9208S), calnexin (26795), and cleaved caspase 3 (9664L); from Santa Cruz Biotechnology (Dallas, TX), CHOP (sc-7351) and LAMP2 (sc-18822); from Sigma, β-actin (A5441); from Novus, LC3b (NB1002220), SEC16A (NB183016), IBTKα (NBP15033 and NBP188512), p62/SQSTM1 (H00008878-M01), and ULK2 (NBP133136); from Thermo Fisher Scientific, calnexin (MA3-027); and from Abcam (Cambridge, MA), SEC31A (AB8660). For human serum samples, the cytokine human magnetic 10-plex panel from Thermo Fisher Scientific was utilized on the Lumiex MAGPIX from EMD Millipore (Billerica, MA). Human IL-8 and TNFα in the conditioned media of HepG2 cells were measured using R&D Systems (Minneapolis, MN) Quantikine ELISA kits D8000C and DTA00C, respectively.
Immunoprecipitation
For immunoprecipitation analyses, cells were trypsinized, washed once with PBS, and flash-frozen following 5 min of incubation on ice in a hypotonic lysis buffer containing 20 mm Hepes, 2 mm MgCl2, and 10% glycerol. Following a rapid thaw in a pre-warmed water bath, NaCl (150 mm final concentration) was added to cell suspensions on ice for 5 min, followed by clarification by centrifugation using a microcentrifuge at 10,000 × g for 15 min. Supernatant was transferred to protein G Dynabeads coated with the following primary antibodies: from Abcam, SEC16A (ab70722) and SEC31A (AB8660); from Novus, IBTKα (NBP188512) and IgG control (3900S). After an overnight incubation in a cold room with continuous rotation to ensure mixing, beads were washed with a buffer containing 20 mm Hepes, 2 mm MgCl2, 10% glycerol, and 150 mm NaCl. Supernatant was removed, and proteins associated with beads were eluted with either urea for LC/MS analysis or with 1× SDS-PAGE buffer for immunoblot analysis.
Mass spectrometry and quantitative proteomics analysis
Mass spectrometry analysis of isolated protein complexes pulled down with antibodies against native IBTKα and peptide sequence matching via SEQUEST HT was performed using Proteome DiscovererTM 1.4 (Thermo Fisher Scientific), as described previously (43). Three biological replicates were carried out for the IBTKα pulldown as compared with IgG control. For each biological replicate, four technical replicates were performed. Protein sequence matching was performed against a human FASTA database from Uniprot (March 16, 2014) containing additional laboratory contaminant proteins, such as proteolytic enzymes. Nonspecific interactions were eliminated using the statistical model Significance Analysis of Interactome (SAINT) and a stringent SAINT threshold of p > 0.8 (44–46). This statistical tool assigns the number of peptide identifications for each target protein to a probability distribution, which is then used to measure the likelihood of biologically accurate protein interactions.
Cell imaging
For all cellular imaging, cultured HepG2 cells were treated with the indicated compounds, then fixed with 1× Prefer from Anatech (San Diego), and permeabilized with 0.1% Triton X-100 for 10 min. This was followed by overnight incubations with primary antibody in PBS containing 0.1% BSA, and then a 2-h incubation with the corresponding secondary Alexa Fluor conjugates and 10 μg/ml Hoechst (33342) from Life Technologies, Inc. Images were acquired by spinning disk confocal microscopy on the Opera (PerkinElmer Life Sciences) using a ×60 water objective, which were transferred to Volocity (PerkinElmer Life Sciences) for three-dimensional rendering of the 0.5 μm z-stack sections and data quantification. Samples collected for electron microscopy were processed and imaged by DSimaging, LLC (West Lafayette, IN).
Human NAFLD samples
Samples were collected under a study protocol that was reviewed and approved by the Medical College of Wisconsin's Institutional Review Board (previous institution for S. G.). Each participating subject gave a written Informed Consent for participation. Subjects were morbidly obese (BMI ≥40 kg/m2 or >35 kg/m2 with significant co-morbidities) with prior unsuccessful attempts to lose and maintain weight, and who underwent bariatric surgery. A protocol intraoperative liver biopsy was performed on all patients for histological phenotyping. Patients with alcohol intake >20 g/day and those with other liver diseases based on positive disease-specific serological tests and suggestive liver histology were excluded. Patients using drugs associated with NAFLD prior to liver biopsy were excluded. An experienced pathologist read the liver biopsy according to the NASH Clinical Research Network working group system (National Institutes of Health) (38).
Statistical analysis
Data are depicted as ± S.D. unless otherwise noted. Differences between multiple groups were analyzed using analysis of variance and a post hoc Tukey's honest significant difference. For all figures, p < 0.05 was considered statistically significant and is depicted using an asterisk, whereas treatment groups in knockdown/knock-out experiments considered statistically significant relative to the respective shCTRL treatment are depicted by a number sign (#).
Author contributions
J. A. W. conceived the study, designed, performed, and analyzed experiments and wrote the manuscript. S. K. Y. designed and performed translational control experiments in Fig. 1. A. L. M. performed and analyzed proteomics data. S. G. provided human liver biopsy and serum samples and consulted with the principal component analysis. J. L. S. and H. C. M. designed and analyzed experiments and contributed to the preparation of the manuscript. R. C. W. conceived and coordinated the study, designed and analyzed experiments, and wrote the manuscript. All authors approved the final manuscript version.
Supplementary Material
Acknowledgments
We thank Naga Chalasani, Richard Day, and members of the Wek laboratory for helpful discussions. We also thank Thomas Baker, Armando Irizarry, Frank Dorsey, and Brianna Paisley at Eli Lilly for their support.
This work was supported in part by National Institutes of Health Grant GM049164 (to R. C. W.) and the Ralph W. and Grace M. Showalter Research Trust Fund. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S4 and Figs. S1–S4.
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- UPR
- unfolded protein response
- IBTKα
- inhibitor of Bruton's tyrosine kinase
- ER
- endoplasmic reticulum
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone
- BisTris
- 2-[bis(2-hydroxyethyl) amino]-2-(hydroxymethyl)propane-1,3-diol
- ERES
- endoplasmic reticulum exit site
- LDH
- lactate dehydrogenase
- qPCR
- quantitative PCR.
References
- 1. Masuoka H. C., and Chalasani N. (2013) Nonalcoholic fatty liver disease: an emerging threat to obese and diabetic individuals. Ann. N.Y. Acad. Sci. 1281, 106–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Satapathy S. K., and Sanyal A. J. (2015) Epidemiology and natural history of nonalcoholic fatty liver disease. Semin. Liver Dis. 35, 221–235 [DOI] [PubMed] [Google Scholar]
- 3. Yeh M. M., and Brunt E. M. (2014) Pathological features of fatty liver disease. Gastroenterology 147, 754–764 [DOI] [PubMed] [Google Scholar]
- 4. Ricchi M., Odoardi M. R., Carulli L., Anzivino C., Ballestri S., Pinetti A., Fantoni L. I., Marra F., Bertolotti M., Banni S., Lonardo A., Carulli N., and Loria P. (2009) Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J. Gastroenterol. Hepatol. 24, 830–840 [DOI] [PubMed] [Google Scholar]
- 5. Berlanga A., Guiu-Jurado E., Porras J. A., and Auguet T. (2014) Molecular pathways in non-alcoholic fatty liver disease. Clin. Exp. Gastroenterol. 7, 221–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malhi H., and Kaufman R. J. (2011) Endoplasmic reticulum stress in liver disease. J. Hepatol. 54, 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fu S., Watkins S. M., and Hotamisligil G. S. (2012) The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 15, 623–634 [DOI] [PubMed] [Google Scholar]
- 8. Walter P., and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 9. Baird T. D., and Wek R. C. (2012) Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 3, 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Willy J. A., Young S. K., Stevens J. L., Masuoka H. C., and Wek R. C. (2015) CHOP links endoplasmic reticulum stress to NF-κB activation in the pathogenesis of nonalcoholic steatohepatitis. Mol. Biol. Cell 26, 2190–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. González-Rodríguez A., Mayoral R., Agra N., Valdecantos M. P., Pardo V., Miquilena-Colina M. E., Vargas-Castrillón J., Lo Iacono O., Corazzari M., Fimia G. M., Piacentini M., Muntané J., Boscá L., García-Monzón C., Martín-Sanz P., and Valverde Á. M. (2014) Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 5, e1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Musso G., Cassader M., and Gambino R. (2016) Non-alcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat. Rev. Drug Discov. 15, 249–274 [DOI] [PubMed] [Google Scholar]
- 13. Gual P., Gilgenkrantz H., and Lotersztajn S. (2017) Autophagy in chronic liver diseases: the two faces of Janus. Am. J. Physiol. Cell Physiol. 312, C263–C273 [DOI] [PubMed] [Google Scholar]
- 14. Lin C. W., Zhang H., Li M., Xiong X., Chen X., Chen X., Dong X. C., and Yin X. M. (2013) Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J. Hepatol. 58, 993–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levine B., and Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Høyer-Hansen M., and Jäättelä M. (2007) Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 14, 1576–1582 [DOI] [PubMed] [Google Scholar]
- 17. B'chir W., Maurin A. C., Carraro V., Averous J., Jousse C., Muranishi Y., Parry L., Stepien G., Fafournoux P., and Bruhat A. (2013) The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 41, 7683–7699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rzymski T., Milani M., Pike L., Buffa F., Mellor H. R., Winchester L., Pires I., Hammond E., Ragoussis I., and Harris A. L. (2010) Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29, 4424–4435 [DOI] [PubMed] [Google Scholar]
- 19. Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., Ogawa S., Kaufman R. J., Kominami E., and Momoi T. (2007) ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14, 230–239 [DOI] [PubMed] [Google Scholar]
- 20. Baird T. D., Palam L. R., Fusakio M. E., Willy J. A., Davis C. M., McClintick J. N., Anthony T. G., and Wek R. C. (2014) Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol. Biol. Cell 25, 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin L., Pahuja K. B., Wickliffe K. E., Gorur A., Baumgärtel C., Schekman R., and Rape M. (2012) Ubiquitin-dependent regulation of COPII coat size and function. Nature 482, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pisano A., Ceglia S., Palmieri C., Vecchio E., Fiume G., de Laurentiis A., Mimmi S., Falcone C., Iaccino E., Scialdone A., Pontoriero M., Masci F. F., Valea R., Krishnan S., Gaspari M., et al. (2015) CRL3IBTK regulates the tumor suppressor Pdcd4 through ubiquitylation coupled to proteasomal degradation. J. Biol. Chem. 290, 13958–13971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han J., Back S. H., Hur J., Lin Y. H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., and Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alkhouri N., Carter-Kent C., and Feldstein A. E. (2011) Apoptosis in nonalcoholic fatty liver disease: diagnostic and therapeutic implications. Expert Rev. Gastroenterol. Hepatol. 5, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo M., Lu Z., Sun H., Yuan K., Zhang Q., Meng S., Wang F., Guo H., Ju X., Liu Y., Ye T., Lu Z., and Zhai Z. (2010) Nuclear entry of active caspase-3 is facilitated by its p3-recognition-based specific cleavage activity. Cell Res. 20, 211–222 [DOI] [PubMed] [Google Scholar]
- 26. Amir M., and Czaja M. J. (2011) Autophagy in nonalcoholic steatohepatitis. Expert Rev. Gastroenterol. Hepatol. 5, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Noda N. N., and Inagaki F. (2015) Mechanisms of autophagy. Annu. Rev. Biophys. 44, 101–122 [DOI] [PubMed] [Google Scholar]
- 28. Tooze S. A., and Yoshimori T. (2010) The origin of the autophagosomal membrane. Nat. Cell Biol. 12, 831–835 [DOI] [PubMed] [Google Scholar]
- 29. Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., and Yamamoto A. (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 [DOI] [PubMed] [Google Scholar]
- 30. Ylä-Anttila P., Vihinen H., Jokitalo E., and Eskelinen E. L. (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180–1185 [DOI] [PubMed] [Google Scholar]
- 31. Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., Griffiths G., and Ktistakis N. T. (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanchez-Wandelmer J., Ktistakis N. T., and Reggiori F. (2015) ERES: sites for autophagosome biogenesis and maturation? J. Cell Sci. 128, 185–192 [DOI] [PubMed] [Google Scholar]
- 33. Graef M., Friedman J. R., Graham C., Babu M., and Nunnari J. (2013) ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 24, 2918–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan D., Cai Y., Wang J., Zhang J., Menon S., Chou H. T., Ferro-Novick S., Reinisch K. M., and Walz T. (2013) The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. U.S.A. 110, 19432–19437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alemu E. A., Lamark T., Torgersen K. M., Birgisdottir A. B., Larsen K. B., Jain A., Olsvik H., Øvervatn A., Kirkin V., and Johansen T. (2012) ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 287, 39275–39290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., and Ohsumi Y. (2001) Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 12, 3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zanetti G., Pahuja K. B., Studer S., Shim S., and Schekman R. (2011) COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14, 20–28 [DOI] [PubMed] [Google Scholar]
- 38. Kleiner D. E., Brunt E. M., Van Natta M., Behling C., Contos M. J., Cummings O. W., Ferrell L. D., Liu Y. C., Torbenson M. S., Unalp-Arida A., Yeh M., McCullough A. J., Sanyal A. J., and Nonalcoholic Steatohepatitis Clinical Research Network. (2005) Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 [DOI] [PubMed] [Google Scholar]
- 39. Green D. R., and Levine B. (2014) To be or not to be? How selective autophagy and cell death govern cell fate. Cell 157, 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y., and Levine B. (2015) Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 22, 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Q., Jones T. W., Brown P. C., and Stevens J. L. (1990) The mechanism of cysteine conjugate cytotoxicity in renal epithelial cells. Covalent binding leads to thiol depletion and lipid peroxidation. J. Biol. Chem. 265, 21603–21611 [PubMed] [Google Scholar]
- 42. Teske B. F., Baird T. D., and Wek R. C. (2011) Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol. 490, 333–356 [DOI] [PubMed] [Google Scholar]
- 43. Mosley A. L., Hunter G. O., Sardiu M. E., Smolle M., Workman J. L., Florens L., and Washburn M. P. (2013) Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcriptional elongation. Mol. Cell. Proteomics 12, 1530–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., et al. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Choi H., Liu G., Mellacheruvu D., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2012) Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr. Protoc. Bioinformatics Chapter 8, Unit 8.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.