Abstract
Cancer cells with specific genetic alterations may be highly dependent on certain nutrients for survival, which can inform therapeutic strategies to target these cancer-specific metabolic vulnerabilities. The glutamate/cystine antiporter solute carrier family 7 member 11 (SLC7A11, also called xCT) is overexpressed in several cancers. Contrasting the established pro-survival roles of SLC7A11 under other stress conditions, here we report the unexpected finding that SLC7A11 overexpression enhances cancer cell dependence on glucose and renders cancer cells more sensitive to glucose starvation-induced cell death and, conversely, that SLC7A11 deficiency by either knockdown or pharmacological inhibition promotes cancer cell survival upon glucose starvation. We further show that glucose starvation induces SLC7A11 expression through ATF4 and NRF2 transcription factors and, correspondingly, that ATF4 or NRF2 deficiency also renders cancer cells more resistant to glucose starvation. Finally, we show that SLC7A11 overexpression decreases whereas SLC7A11 deficiency increases intracellular glutamate levels because of SLC7A11-mediated glutamate export and that supplementation of α-ketoglutarate, a key downstream metabolite of glutamate, fully restores survival in SLC7A11-overexpressing cells under glucose starvation. Together, our results support the notion that both glucose and glutamate have important roles in maintaining cancer cell survival and uncover a previously unappreciated role of SLC7A11 to promote cancer cell dependence on glucose. Our study therefore informs therapeutic strategies to target the metabolic vulnerability in tumors with high SLC7A11 expression.
Keywords: cell death, glucose, glutamate, reactive oxygen species (ROS), tumor metabolism, SLC7A11, xCT
Introduction
To survive, cancer cells require an adequate supply of nutrients to meet their biosynthetic and bioenergetics needs and to maintain appropriate redox balance (1–3). Two of the most abundant and, arguably, the most important nutrients supporting cancer cell survival are glucose and glutamine (4). Glucose provides the major carbon source to generate energy (in the form of ATP) as well as precursors for the synthesis of macromolecules, including fatty acids, nucleotides, and amino acids. When transported into cells, glucose is metabolized to acetyl-CoA through a series of metabolic reactions, including glycolysis (1). Acetyl-CoA then enters into the tricarboxylic acid (TCA)4 cycle (also known as the citric acid cycle or the Krebs cycle), in which the carbon derived from glucose is oxidized to CO2, coupled with the generation of the high-energy electrons NADH and FADH2, which are subsequently utilized to generate ATP through electron transport and oxidative phosphorylation in mitochondria. In addition, the TCA cycle provides important precursors for many biosynthetic pathways. For example, α-ketoglutarate (αKG), a key intermediate in the TCA cycle, can serve as a precursor for the amino acid glutamate. Glutamine, on the other hand, is the most abundant amino acid in cell culture medium. Intracellular glutamine is first converted to glutamate, which then provides an important source for both carbon and nitrogen in various biosynthetic reactions (5). For example, glutamate can be converted to αKG through either a transamination or deamination reaction, which serves as an important anaplerotic reaction to replenish αKG and to maintain the TCA cycle.
Although nutrient supplies often are in excess under in vitro culturing conditions, nutrient availability could be limiting within a tumor mass under in vivo conditions because of the poor tumor vasculature in the tumor microenvironment and the increase of nutrient uptake in cancer cells, which eventually induces metabolic stress in cancer cells (6). Cancer cells often engage strategies of metabolic adaptation or metabolic flexibility to survive and grow under metabolic stress. One common strategy of metabolic adaptation employed by cancer cells is a shift to utilize other types of nutrient when one nutrient is limited. For example, when glucose is limited, tumor cells often enhance glutamine metabolism and promote glutamine-dependent αKG formation to maintain the TCA cycle and to support bioenergetics and biosynthesis in cancer cells (7, 8). Similarly, it has been shown that defective mitochondrial pyruvate transport induces metabolic flexibility and promotes glutamine metabolism to maintain the TCA cycle and cell survival (9, 10).
Specific genetic alterations in cancer cells may reprogram their metabolism networks and render such cancer cells highly dependent on one particular nutrient for survival, thus limiting their metabolic flexibility (2). For example, AKT hyperactivation promotes glucose uptake and glycolysis and renders cancer cells highly dependent on glucose for survival (11). Correspondingly, cancer cells with AKT hyperactivation are sensitive to glucose starvation (12). On the other hand, the Myc transcription factor promotes glutamine metabolism through transcriptional regulation of various enzymes involved in glutaminolysis, leading to glutamine dependence, and cancer cells with high Myc levels or activity are exquisitely sensitive to glutamine withdrawal (13, 14). These findings may have important therapeutic implications, as they suggest that tumors with specific genetic alterations may be particularly sensitive to drugs blocking the supply or metabolism of the nutrient on which cancer cells depend. For example, treatment of inhibitors of glutaminase, the key enzyme to convert glutamine to glutamate, in a Myc-induced liver cancer mouse model significantly decreased tumor development and prolonged animal survival (15), suggesting the utility of glutaminase inhibitors in the treatment of Myc-driven cancers. Thus, a more in-depth understanding of the interplays among different metabolism pathways and the underlying mechanism of cancer cell dependence on nutrients may provide novel insights into cancer therapies.
The amino acid transport system xc− is an antiporter that imports extracellular cystine coupled to the efflux of intracellular glutamate (16). The transport system xc− exists as a heterodimer, consisting of solute carrier family 7 member 11 (SLC7A11, also known as xCT), the catalytic subunit of the transport system xc− that mediates the transport function, and solute carrier family 3 member 2 (SLC3A2, also known as 4F2hc or CD98hc), the chaperone that recruits SLC7A11 to the plasma membrane (17). Because of the reduced microenvironment inside cells, intracellular cystine is quickly converted to cysteine, which then serves as a rate-limiting precursor for synthesis of the antioxidant tripeptide glutathione. SLC7A11 plays important roles in maintaining intracellular glutathione levels and protecting cells from oxidative stress-induced cell death (17). Consistent with this, many studies have shown that SLC7A11 is up-regulated in human cancers, and its inhibitors, such as sulfasalazine, have an anti-tumor effect (18, 19). However, the potential role of SLC7A11-mediated glutamate export function in cancer biology remains much less understood. In this study, we revealed that SLC7A11-mediated efflux of intracellular glutamate reduces metabolic flexibility in cancer cells and makes cancer cells more dependent on glucose for survival. Our study may provide a conceptual framework to target tumors with high SLC7A11 expression by drugs that block glucose uptake or metabolism.
Results
Glucose starvation induces SLC7A11 through ATF4 and NRF2
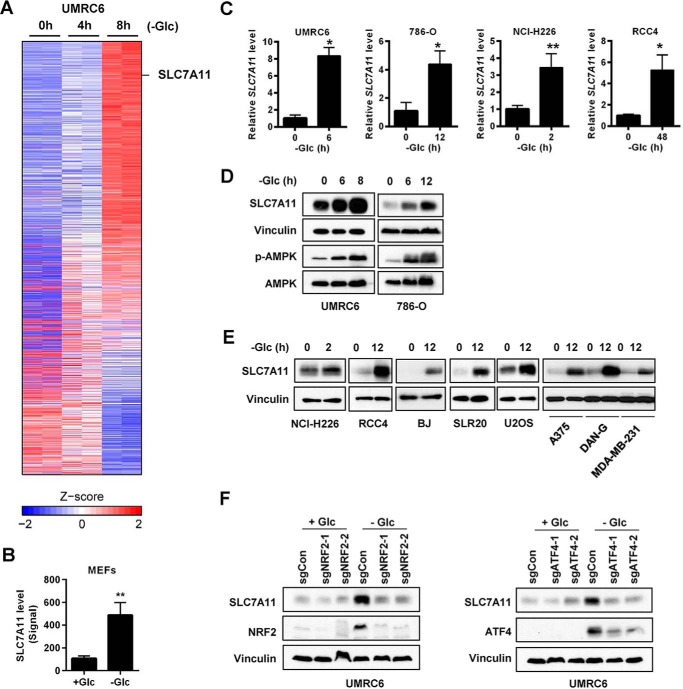
Analysis of microarray or RNA sequencing data generated from our previous studies (20, 21) revealed that glucose starvation induced SLC7A11 expression (Fig. 1, A and B). Real-time PCR analysis confirmed glucose starvation-induced SLC7A11 expression in several cancer cell lines, including UMRC6, 786-O, and RCC4 (all renal cancer cell lines) and NCI-H226 (a mesothelioma cell line) (Fig. 1C). Western blotting confirmed that glucose starvation increased AMPK phosphorylation as expected and also induced SLC7A11 protein levels in UMRC6 and 786-O cells (Fig. 1D). We further confirmed glucose starvation-induced SLC7A11 expression in several other cancer cell lines from different tumor types (Fig. 1E), suggesting that glucose starvation induces SLC7A11 expression in a diverse array of cancer cells irrespective of their tumor origins.
Figure 1.
Glucose starvation induces SLC7A11 through ATF4 and NRF2. A, heatmap representation of genes regulated upon glucose (Glc) starvation in UMRC6 cells. B, normalized SLC7A11 level from a mouse microarray using mouse embryonic fibroblasts (MEFs) treated with and without glucose for 24 h. C–E, glucose starvation–induced SLC7A11 expression in different cells was analyzed by RT-PCR (C) and Western blot analysis (D and E). F, the effects of NRF2 and ATF4 deletion on SLC7A11 induction upon glucose starvation in UMRC6 cells were examined by Western blotting. *, p < 0.05; **, p < 0.01.
Next we sought to identify the transcription factors that regulate glucose starvation-induced SLC7A11 expression. Previous studies showed that two stress-inducible transcription factors, ATF4 and NRF2, bind on the cis-regulatory elements of the SLC7A11 gene and cooperatively regulate SLC7A11 transcription (22). To examine whether ATF4 and/or NRF2 are involved in the regulation of glucose starvation-induced SLC7A11 expression, we utilized CRISPR technology and generated ATF4- and NRF2-deficient cells in UMRC6 cells by two independent single guide RNAs (sgRNAs). Western blotting revealed that the protein levels of ATF4 or NRF2 were extremely low under basal conditions (with 25 mm glucose), and glucose starvation significantly increased the protein levels of both ATF4 and NRF2 (Fig. 1F), which is consistent with the stress-inducible nature of these two transcription factors (23, 24). We found that the protein levels of ATF4 and NRF2 were dramatically decreased in the corresponding sgRNA-infected cells under glucose starvation, and, notably, glucose starvation-induced SLC7A11 expression was almost abolished in either ATF4 or NRF2 sgRNA-infected cells (Fig. 1F). Taken together, our data suggest that glucose starvation induces SLC7A11 expression through ATF4 and NRF2 transcription factors.
SLC7A11 sensitizes cancer cells to glucose starvation–induced cell death
The aforementioned data prompted further studies on the potential role of SLC7A11 in the regulation of cancer cell dependence on glucose. In some cellular contexts, cancer cells are dependent on glucose for survival, as glucose provides redox balance; correspondingly, it has been shown that long-term glucose starvation results in cell death associated with increased reactive oxygen species (ROS) levels, and treatment with the antioxidant N-acetylcysteine (NAC) abolishes glucose starvation–induced cell death in these cells (20, 25). Because SLC7A11-mediated cystine uptake promotes the biosynthesis of glutathione, an important antioxidant, we hypothesized that SLC7A11-high cancer cells should be less sensitive to glucose starvation (thus less dependent on glucose for survival), as these cells have better capabilities to detoxify glucose withdrawal–induced ROS.
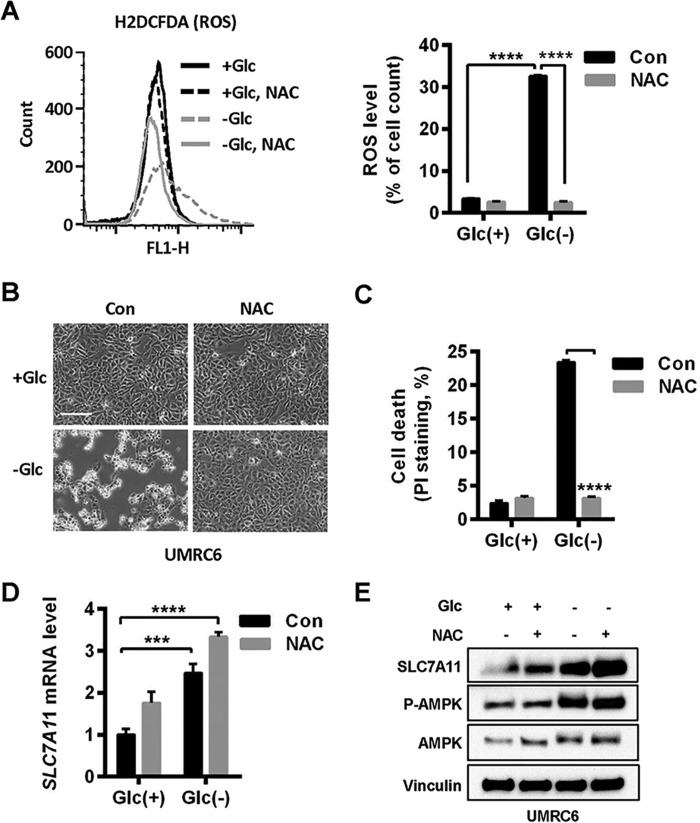
To test this hypothesis, we first examined whether glucose starvation–induced SLC7A11 expression depends on ROS. We first confirmed that NAC treatment normalized glucose starvation-induced ROS levels (Fig. 2A) and significantly attenuated glucose starvation–induced cell death in UMRC6 cells (Fig. 2, B and C). Surprisingly, we found that, in the same cell line, NAC treatment did not attenuate even slightly increased glucose starvation–induced SLC7A11 expression (Fig. 2, D and E). Our data showed that glucose starvation induces SLC7A11 expression independent of ROS, at least in the cell line used in our study, and further suggested that there is a dissociation between SLC7A11 and its antioxidant function under glucose starvation.
Figure 2.
Glucose starvation induces SLC7A11 independent of ROS. A, cellular ROS levels in UMRC6 cells after glucose (Glc) withdrawal for 6 h were determined by H2DCFDA flow cytometer analysis. To prevent ROS generation, 2 mm NAC was added to the glucose-free medium. Right panel, data represent mean ± S.D. (n = 3). Con, control. B and C, UMRC6 cells cultured in glucose-free medium for 6 h were monitored under inverted microscopy (B), and cell death was analyzed by PI staining (C). Scale bar = 100 μm. D and E, SLC7A11 mRNA and protein levels in UMRC6 cells upon glucose starvation and NAC treatment were analyzed by RT-PCR (D) and Western blotting (E). ***, p < 0.001; ****, p < 0.0001.
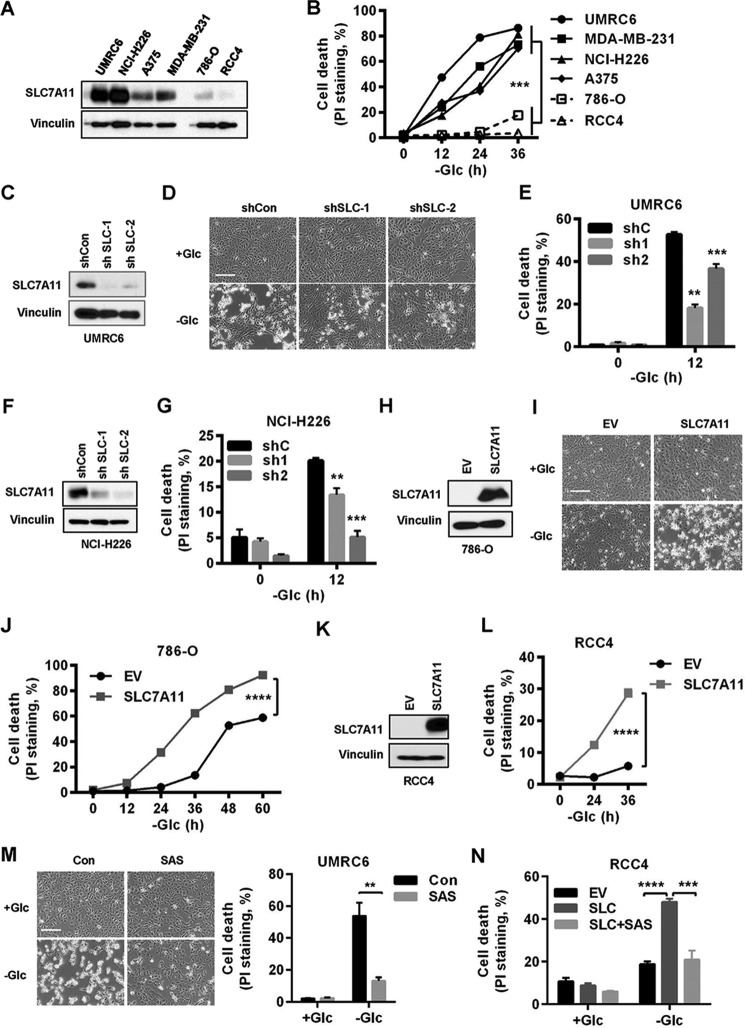
In addition, when we correlated SLC7A11 protein levels across a panel of cancer cell lines with sensitivities of these cell lines to glucose starvation-induced cell death, we observed that cancer cells with high SLC7A11 levels were more sensitive, rather than more resistant, as we hypothesized, to glucose starvation–induced cell death than cells with low SLC7A11 levels (Fig. 3, A and B). To study whether SLC7A11 plays any causal role in the regulation of glucose starvation–induced cell death, we generated SLC7A11 knockdown cells in UMRC6 cells (Fig. 3C), which harbor high SLC7A11 expression and are sensitive to glucose starvation–induced cell death (Fig. 3, A and B). Notably, SLC7A11 knockdown in UMRC6 cells significantly attenuated glucose starvation–induced cell death (Fig. 3, D and E). We confirmed this observation with SLC7A11 knockdown in another SCL7A11-high/glucose starvation–sensitive cell line, NCI-H226 (Fig. 3, F and G). Importantly, SLC7A11 knockdown efficiency correlated with the corresponding sensitivities to glucose starvation in these cells. Conversely, overexpression of SLC7A11 in RCC4 and 786-O cells (which exhibit low SLC7A11 expression and are resistant to glucose starvation; Fig. 3, A and B) markedly potentiated glucose starvation–induced cell death (Fig. 3, H–J for 786-O cells and K and L for RCC4 cells). Consistent with the data from genetic approaches described above, we further showed that pharmacologic inhibition of SLC7A11 transporter activity by sulfasalazine significantly attenuated glucose starvation–induced cell death in either UMRC6 cells (Fig. 3M) or SLC7A11-overexpressing RCC4 cells (Fig. 3N). Taken together, our results revealed that SLC7A11 renders cancer cells more dependent on glucose and, correspondingly, sensitizes cancer cells to glucose starvation–induced cell death.
Figure 3.
SLC7A11 sensitizes cancer cells to glucose starvation–induced cell death. A, SLC7A11 protein levels in different cancer cell lines were determined by Western blotting. B, cell death upon glucose (Glc) starvation for 0–36 h in different cancer cell lines was analyzed by PI staining. C, SLC7A11 protein levels in SLC7A11 knockdown UMRC6 cells. D and E, cell death in SLC7A11 knockdown UMRC6 cells upon glucose withdrawal was monitored (D) and analyzed by PI staining (E). Scale bar = 100 μm. F, SLC7A11 protein levels in SLC7A11 knockdown NCI-H226 cells. G, cell death upon glucose withdrawal was analyzed by PI staining for SLC7A11 knockdown NCI-H226 cells. H, SLC7A11 protein levels in SLC7A11-overexpressing 786-O cells. EV, empty vector. I and J, cell death in SLC7A11-overexpressing 786-O cells upon glucose withdrawal was monitored (I) and analyzed by PI staining (J). Scale bar = 100 μm. K, SLC7A11 protein levels in SLC7A11-overexpressing RCC4 cells. L, cell death in SLC7A11-overexpressing RCC4 cells upon glucose withdrawal was analyzed by PI staining. M and N, cell death in UMRC6 cells (M) or SLC7A11-overexpressing RCC4 cells (N) upon glucose withdrawal and treatment with 1 mm sulfasalazine (SAS) was monitored and analyzed by PI staining. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
ATF4 and NRF2 promote glucose starvation-induced cell death at least partly through SLC7A11
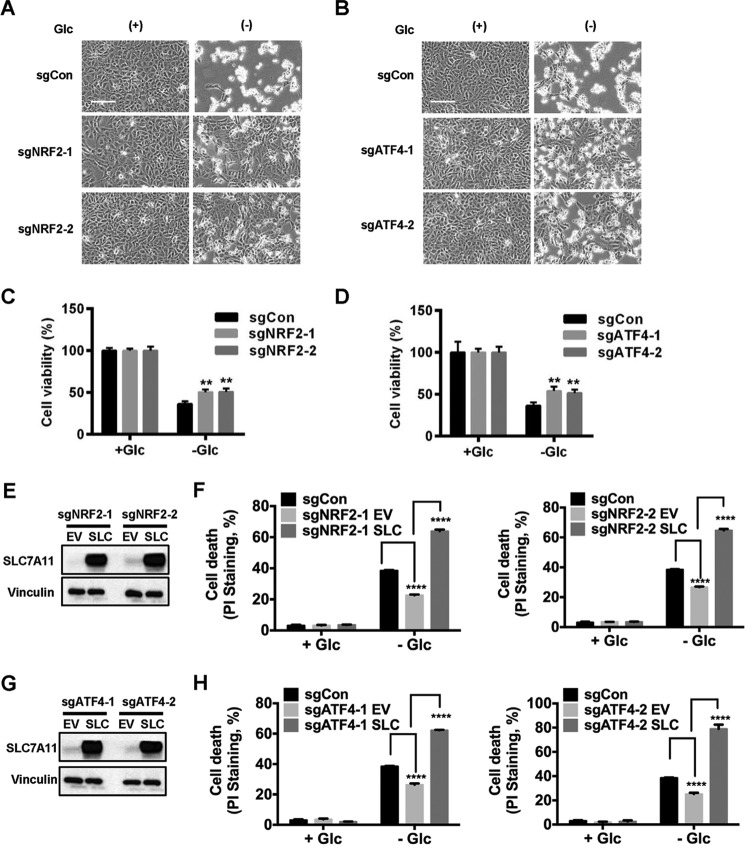
Because our data showed that glucose starvation induces SLC7A11 expression through ATF4 and NRF2, we next sought to determine the roles of ATF4 and NRF2 in glucose starvation-induced cell death. To test this, we cultured UMRC6 cells infected with control, ATF4, or NRF2 sgRNA (Fig. 1, E and F) in glucose-containing or glucose-free medium. Cell morphological analysis revealed that, although ATF4 or NRF2 deficiency did not affect cell viability under 25 mm glucose conditions, there were significantly fewer dead cells in ATF4- or NRF2-deficient cells than in control cells under glucose starvation conditions (Fig. 4, A and B). Cell viability assay confirmed that ATF4 or NRF2 deficiency partially attenuated glucose starvation-induced cell death in UMRC6 cells (Fig. 4, C and D). Next we studied whether SLC7A11 plays any causal role in the cell death resistance phenotype in ATF4- or NRF2-deficient cells under glucose starvation. We showed that overexpression of SLC7A11 in ATF4- or NRF2-deficient cells significantly resensitized cells to glucose starvation–induced cell death (Fig. 4, E–H). Together, our data reveal that ATF4 and NRF2 promote glucose starvation–induced cell death at least partly through SLC7A11.
Figure 4.
ATF4 and NRF2 promote glucose starvation–induced cell death. A–D, NRF2- and ATF4-deficient cells were cultured in glucose (Glc)-free medium for 6 h. Shown are representative images (A and B) and quantification of cell viability (C and D) of the indicated cell lines that were cultured in glucose-free medium for 6 h. Scale bar = 100 μm. E and G, SLC7A11 protein levels in SLC7A11-overexpressing NRF2-deficient (E) or ATF4-deficient cells (G). EV, empty vector. F and H, cell death in UMRC6 cells with the indicated genotypes after 5-h glucose starvation was analyzed by PI staining. **, p < 0.01; ****, p < 0.0001.
SLC7A11 regulation of glutamate efflux underlies SLC7A11-mediated increased sensitivity to glucose starvation
The aforementioned data prompted us to further study the underlying mechanisms by which SLC7A11 promotes glucose starvation–induced cell death. Under glucose depletion conditions, other nutrients, most notably glutamine, can partly compensate for glucose to maintain the TCA cycle and to enable glucose-independent cell survival (5, 7, 8). Glutamine is first metabolized to glutamate, and glutamate then serves as a precursor for several important metabolism processes, including the TCA cycle, nucleotide biosynthesis, and glutathione biosynthesis (5). Because SLC7A11 functions as an antiporter by exporting intracellular glutamate in exchange with extracellular cystine (17), we reasoned that SLC7A11 overexpression may partially deplete intracellular glutamate levels through exporting glutamate, thus rendering cells more dependent on glucose for survival. Correspondingly, such cells would be more vulnerable to glucose starvation, as they lack glutamine/glutamate-derived metabolic adaptation.
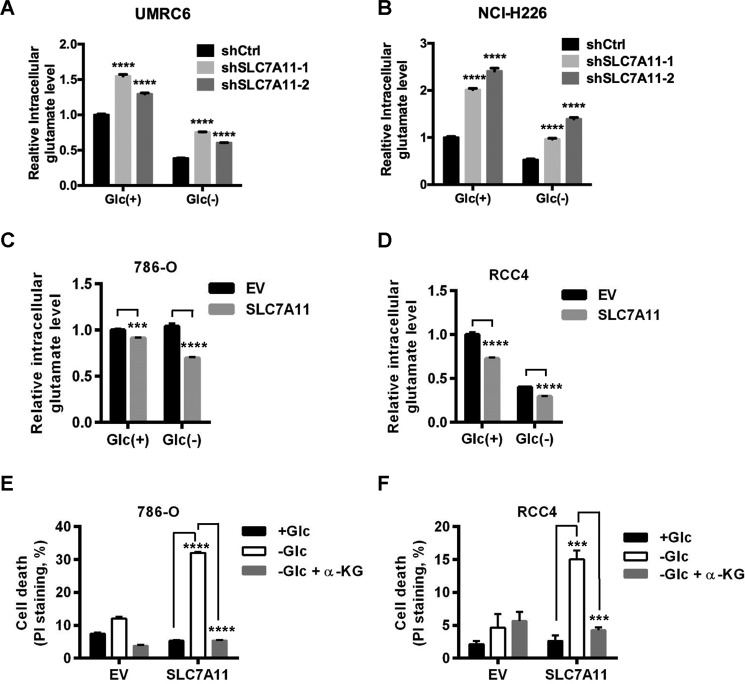
To test this hypothesis, we first examined the effects of SLC7A11 deficiency on intracellular glutamate levels. Consistent with the known role of SLC7A11 in exporting glutamate (16), our results showed that SLC7A11 knockdown increased intracellular glutamate levels (Fig. 5, A and B. Note that SCL7A11 knockdown efficiency correlated with corresponding intracellular glutamate levels in these cells. Also see Fig. 3, C and F). Conversely, we showed that SLC7A11 overexpression decreased intracellular glutamate levels (Fig. 5, C and D). One of the most important metabolites downstream of glutamate is αKG, which serves as a key intermediate in the TCA cycle. To further study whether the decreased levels of glutamate, and likely αKG, in SLC7A11-overexpressing cells underlie the increased sensitivities of these cells to glucose starvation, we examined whether supplementation of dimethyl αKG, a cell-permeable form of αKG, in SLC7A11-overexpressing cells would dampen SLC7A11 overexpression-induced cell death under glucose starvation. Indeed, our results revealed that dimethyl αKG treatment in SLC7A11-overexpressing 786-O or RCC4 cells fully restored cell survival to a level similar as that in corresponding empty vector cells under glucose starvation conditions (Fig. 5, E and F). Collectively, our results strongly suggested that SLC7A11 regulation of glutamate efflux and the corresponding decrease in intracellular glutamate/αKG levels are at least partially responsible for the increased sensitivity to glucose starvation in SLC7A11-overexpressing cells.
Figure 5.
SLC7A11 regulation of glutamate efflux underlies SLC7A11-mediated increased sensitivity to glucose starvation. A and B, intracellular glutamate levels were determined in SLC7A11 knockdown UMRC6 cells after 3 h of glucose (Glc) withdrawal (A) or NCI-H226 cells after 1 h of glucose withdrawal (B). All values were normalized to control shRNA (shCon) cells in complete medium with 25 mm glucose. C and D, intracellular glutamate levels were determined in SLC7A11-overexpressing 786-O cells after 8 h of glucose withdrawal (C) or RCC4 cells after 14 h of glucose withdrawal (D). All values were normalized to empty vector (EV) cells in complete medium with 25 mm glucose. E and F, SLC7A11-overexpressing 786-O cells (E) or RCC4 cells (F) were cultured in complete medium or glucose-free medium with and without 5 mm dimethyl-αKG for 24 h. Cell death was measured by PI staining. ***, p < 0.001; ****, p < 0.0001.
Discussion
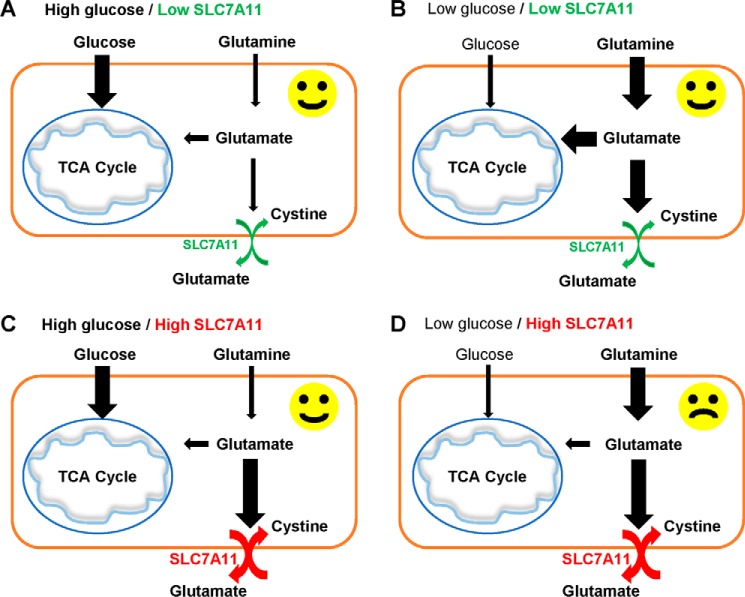
Previous studies have shown that decreased flux from glucose into the TCA cycle, such as under conditions of glucose deprivation or defective mitochondrial pyruvate transport, induces metabolic flexibility and promotes compensatory glutamine metabolism to maintain the TCA cycle and cell survival in various cancer cells (7–10) (Fig. 6, A and B). Our study reinforces the important roles of metabolic flexibility by both glucose and glutamine in the maintenance of cancer cell survival and suggests a model in which SLC7A11 can limit such metabolic flexibility in cancer cells by decreasing intracellular glutamate, a key metabolite downstream of glutamine, as SLC7A11 mediates the efflux of intracellular glutamate, resulting in partial depletion of intracellular glutamate in cancer cells. We propose that, because of decreased intracellular glutamate levels, cancer cells with high SLC7A11 have more limited metabolic flexibility and more reliance on glucose for survival than those with low SLC7A11 (Fig. 6C). Correspondingly, SLC7A11-high cancer cells are much more vulnerable than their SLC7A11-low counterparts when glucose is limited (Fig. 6D). A recent study made similar observations as those shown in our study (26). It will be important to further test this model by metabolite profiling and flux analyses in future studies.
Figure 6.
Model for the role of SLC7A11 in glucose dependence. See “Discussion” for a detailed description.
Our study provides several important implications regarding both the biological roles of SLC7A11-mediated amino acid transport and therapies to target metabolic vulnerabilities in cancer treatment. First, most current studies of SLC7A11 have focused on its role in regulating cystine import, glutathione biosynthesis, and redox balance. Our study shows that SLC7A11-mediated glutamate efflux also plays important roles in tumor biology, but such roles are only revealed under glucose-limiting conditions. It will be interesting to further explore the biological impact of SLC7A11-mediated glutamate efflux under other stress conditions. Second, previous studies revealed that, in different cancer cell lines, SLC7A11 overexpression generally confers resistance to cell death induced by various stress conditions, such as oxidative stress, genotoxic stress, and proteasome inhibition, whereas SLC7A11 deficiency increases sensitivity to stress-induced cell death (22, 27, 28). It is generally accepted that SLC7A11-medated cystine uptake and glutathione biosynthesis underlie its pro-survival function under these stress conditions. Thus, the pro-cell death function of SLC7A11 in the context of glucose starvation revealed in this study is rather unexpected and unique. How SLC7A11 exerts such context-dependent roles in the regulation of cell death and survival remains to be further explored. Finally, because SLC7A11 is highly expressed in several types of human cancer (18, 19), our study further suggests that cancer cells or tumors with high SLC7A11 expression may be sensitive to drugs that block glucose metabolism, such as glycolysis inhibitors.
Experimental procedures
Cell culture studies
The HEK293T cells and other cancer cell lines used in this study were obtained from the ATCC. Plasmid transfections were performed using Lipofectamine 3000 (Life Technologies). Lentiviruses were produced in HEK293T cells with packing mixture (ViraPower lentiviral expression system, Invitrogen) and used to infect target cells according to the instructions of the manufacturer. For glucose starvation experiments, cells were cultured in DMEM with different concentrations of glucose (0 or 25 mm) plus 10% (v/v) dialyzed FBS, as described in our previous publication (29).
Cell death and viability measurement
To measure cell death, the cells were stained with propidium iodide (PI) followed by FACS analysis as described previously (30, 31). Briefly, cells were washed with cold PBS twice, and the cell number was adjusted to 1 × 106. The cells were resuspended in 100 μl of PBS containing 2 μg/ml of PI. The cells were incubated for 30 min at room temperature and analyzed with an LSRII flow cytometer (BD Biosciences). The CCK8 kit (Sigma, 96992) was used to measure the viability of cells in a 96-wll plate. 7500 cells/well were seeded and incubated overnight. Then cells were washed once with PBS, followed by glucose-free medium treatment for 6 h. All medium was then removed, and fresh medium containing CCK8 reagent was replaced. The plate was incubated for 1 h, and A450 was measured by microplate reader (FLUOstar Omega, BMG Labtech).
ROS measurement
Cells were incubated in a 60-mm dish containing 4 μm CM-H2DCFDA (Thermo Fisher, C6827). After incubation for 30 min at 37 °C, cells were washed with PBS and trypsinized, followed by PI stating in PBS for 5 min. Then cells were subjected to flow cytometry analysis using a cytometer (Accuri C6, BD Biosciences).
Constructs and reagents
shRNAs targeting human SLC7A11 (V2LHS_204910, V2LHS_251161) and the SLC7A11 cDNA-containing pDONR223 plasmid were obtained from the MD Anderson Cancer Center shRNA and ORFeome Core Facility. SLC7A11 was cloned into the plenti6-V5 destination vector using gateway LR clonase II enzyme mixture (Thermo Fisher Scientific, 11791-020). sgRNA targeting NRF2 and ATF4 was cloned into the pLenti-CRISPR-V2 plasmid. Dimethyl α-ketoglutarate and N-acetyl-l-cysteine were purchased from Sigma (349631 and A7250, respectively). Sulfasalazine was obtained from Santa Cruz Biotechnology (sc-204312).
Real-time PCR
Real-time PCR was performed as described previously (32, 33). Briefly, total RNA was extracted from cells using RNeasy (Qiagen), and first-strand cDNA was prepared with a high-capacity cDNA reverse transcription kit (Applied Biosystems, ABI). Real-time PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen) or TaqMan Universal PCR Master Mix (ABI) and was run on Stratagene MX3000P. For quantification of gene expression, the 2−ΔΔCt method was used. β-Actin expression was used for normalization.
Western blot analysis
Cultured cells were lysed with Nonidet P-40 buffer (150 mm sodium chloride, 1.0% Nonidet P-40, and 50 mm Tris (pH 8.0)) containing complete mini protease inhibitors (Roche) and phosphatase inhibitor mixture (Calbiochem). Western blots were obtained utilizing 20–40 μg of lysate protein. The following antibodies were used in this study: monoclonal anti-vinculin antibody (Sigma-Aldrich, V4505, 1:10,000 dilution), SLC7A11/xCT (D2M7A) rabbit mAb (Cell Signaling Technology, 12619, 1:5000 dilution), phospho-AMPKα (Thr-172, 40H9) rabbit mAb (Cell Signaling Technology, 2535S, 1:1000 dilution), AMPKα (D63G4) rabbit mAb (Cell Signaling Technology, 5832S, 1:1000 dilution), ATF-4 (D4B8) rabbit mAb (Cell Signaling Technology, 11815, 1:1000 dilution), NRF2 (D1Z9C) XP® rabbit mAb (Cell Signaling Technology, 12721, 1:1000 dilution), and α-tubulin antibody (Cell Signaling Technology, 2144, 1:5000 dilution).
Intracellular glutamate detection in cell lysates
The Glutamate-GloTM assay kit (Promega, J7021) was used to measure intracellular glutamate levels. Briefly, the cells were seeded in a 96-well plate, followed by glucose starvation as described above. Cells were then washed with ice-cold PBS and lysed with 0.6 n HCl. The lysing reagent was inactivated using 1 m Tris base. Glutamate dehydrogenase uses the glutamate present in the lysate and NAD+ to produce αKG and NADH. In the presence of NADH, a pro-luciferin reductase substrate is converted by reductase to luciferin, which is then used by Ultra-GloTM recombinant luciferase to produce light. The luminescent signal was subsequently measured by a Gen5 microplate reader (Biotek). All values were normalized to control (empty vector or control shRNA) cells in complete medium.
Statistical analysis
The results of the cell culture experiments were collected from three independent cultures for each sample. Data are presented as means ± S.D. Statistical significance (p value) of different columns was calculated using unpaired Student's t test. Statistical significance (p value) of different groups was calculated using two-way analysis of variance (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
Oligonucleotide and primer sequences
Oligonucleotide and primer sequences were as follows: SLC7A11 forward RT-PCR, 5′-ATGCAGTGGCAGTGACCTTT-3′; SLC7A11 reverse RT-PCR, 5′-GGCAACAAAGATCGGAACTG-3′; β-actin forward RT-PCR, 5′-CGGAACCGCTCATTGCC-3′; β-actin reverse RT-PCR, 5′-ACCCACACTGTGCCCATCTA-3′; ATF4 sgRNA1 forward, 5′-CACCGAGGTCTCTTAGATGATTACC-3′; ATF4 sgRNA1 reverse, 5′-AAACGGTAATCATTCAAGAGACCTC-3′; ATF4 sgRNA2 forward, 5′-CACCGAGTCCCTCCAACAACAGCAA-3′; ATF4 sgRNA2 reverse, 5′-AAACTTGCTGTTGTTGGAGGGACTC-3′; NRF2 sgRNA2 forward, 5′-CACCGTTACAACTAGATGAAGAGAC-3′; NRF2 sgRNA2 reverse, 5′-AAACGTCTCTTCATCTAGTTGTAAC-3′; NRF2 sgRNA3 forward, 5′-CACCGCAGATCCACTGGTTTCTGAC-3′; NRF2 sgRNA3 reverse, 5′-AAACGTCAGAAACCAGTGGATCTGC-3′; control sgRNA forward, 5′-CACCGGCACTACCAGAGCTAACTCA-3′; and control sgRNA reverse, 5′-AAACTGAGTTAGCTCTGGTAGTGCC-3′.
Author contributions
P. K. initiated the project and generated most of the data in Figs. 3; 4, E–H; and 5. Y. Z. generated most of the data in Figs. 1, 2, and 4, A–D. J. S. and W. L. conducted the computational analysis in Fig. 1A. B. G. and Y. Z. supervised the study. P. K., Y. Z., and B. G. designed the experiments and wrote the manuscript. All authors commented on the manuscript.
Acknowledgments
We thank all members of the B. G. laboratory for advice and technical assistance. We also thank Deepak Nagrath for helpful discussions.
This work was supported by the Andrew Sabin Family Fellow Award, an institutional research grant from the University of Texas MD Anderson Cancer Center, National Institutes of Health Grant CA181196, the Anna Fuller Fund, and Ellison Medical Foundation Grant AG-NS-0973-13) (to B. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TCA
- tricarboxylic acid
- αKG
- α-ketoglutarate
- sgRNA
- single guide RNA
- ROS
- reactive oxygen species
- NAC
- N-acetylcysteine
- PI
- propidium iodide
- RT-PCR
- real-time PCR
- CM-H2DCFDA
- chloromethyl 2′,7′-dichlorodihydrofluorescein diacetate.
References
- 1. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vander Heiden M. G., and DeBerardinis R. J. (2017) Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DeBerardinis R. J., and Chandel N. S. (2016) Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ward P. S., and Thompson C. B. (2012) Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hensley C. T., Wasti A. T., and DeBerardinis R. J. (2013) Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest. 123, 3678–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones R. G., and Thompson C. B. (2009) Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 23, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vincent E. E., Sergushichev A., Griss T., Gingras M. C., Samborska B., Ntimbane T., Coelho P. P., Blagih J., Raissi T. C., Choinière L., Bridon G., Loginicheva E., Flynn B. R., Thomas E. C., Tavare J. M., et al. (2015) Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol. Cell 60, 195–207 [DOI] [PubMed] [Google Scholar]
- 8. Yang C., Sudderth J., Dang T., Bachoo R. M., Bachoo R. G., McDonald J. G., and DeBerardinis R. J. (2009) Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 69, 7986–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang C., Ko B., Hensley C. T., Jiang L., Wasti A. T., Kim J., Sudderth J., Calvaruso M. A., Lumata L., Mitsche M., Rutter J., Merritt M. E., and DeBerardinis R. J. (2014) Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 56, 414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vacanti N. M., Divakaruni A. S., Green C. R., Parker S. J., Henry R. R., Ciaraldi T. P., Murphy A. N., and Metallo C. M. (2014) Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol. Cell 56, 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elstrom R. L., Bauer D. E., Buzzai M., Karnauskas R., Harris M. H., Plas D. R., Zhuang H., Cinalli R. M., Alavi A., Rudin C. M., and Thompson C. B. (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 [DOI] [PubMed] [Google Scholar]
- 12. Buzzai M., Bauer D. E., Jones R. G., Deberardinis R. J., Hatzivassiliou G., Elstrom R. L., and Thompson C. B. (2005) The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid β-oxidation. Oncogene 24, 4165–4173 [DOI] [PubMed] [Google Scholar]
- 13. Stine Z. E., Walton Z. E., Altman B. J., Hsieh A. L., and Dang C. V. (2015) MYC, metabolism, and cancer. Cancer Discov. 5, 1024–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., and Thompson C. B. (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiang Y., Stine Z. E., Xia J., Lu Y., O'Connor R. S., Altman B. J., Hsieh A. L., Gouw A. M., Thomas A. G., Gao P., Sun L., Song L., Yan B., Slusher B. S., Zhuo J., et al. (2015) Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Invest. 125, 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato H., Tamba M., Ishii T., and Bannai S. (1999) Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 274, 11455–11458 [DOI] [PubMed] [Google Scholar]
- 17. Lim J. C., and Donaldson P. J. (2011) Focus on molecules: the cystine/glutamate exchanger (system xc−). Exp. Eye Res. 92, 162–163 [DOI] [PubMed] [Google Scholar]
- 18. Lewerenz J., Hewett S. J., Huang Y., Lambros M., Gout P. W., Kalivas P. W., Massie A., Smolders I., Methner A., Pergande M., Smith S. B., Ganapathy V., and Maher P. (2013) The cystine/glutamate antiporter system xc− in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 18, 522–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bhutia Y. D., Babu E., Ramachandran S., and Ganapathy V. (2015) Amino acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res. 75, 1782–1788 [DOI] [PubMed] [Google Scholar]
- 20. Dai F., Lee H., Zhang Y., Zhuang L., Yao H., Xi Y., Xiao Z. D., You M. J., Li W., Su X., and Gan B. (2017) BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc. Natl. Acad. Sci. U.S.A. 114, 3192–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin A., Yao J., Zhuang L., Wang D., Han J., Lam E. W., TCGA Research Network, and Gan B. (2014) The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene 33, 3183–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye P., Mimura J., Okada T., Sato H., Liu T., Maruyama A., Ohyama C., and Itoh K. (2014) Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition. Mol. Cell. Biol. 34, 3421–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., and Gorman A. M. (2016) The integrated stress response. EMBO Rep. 17, 1374–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sykiotis G. P., and Bohmann D. (2010) Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 3, re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Z., Rahme G. J., Chatterjee P. D., Havrda M. C., and Israel M. A. (2017) ID2 promotes survival of glioblastoma cells during metabolic stress by regulating mitochondrial function. Cell Death Dis. 8, e2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shin C. S., Mishra P., Watrous J. D., Carelli V., D'Aurelio M., Jain M., and Chan D. C. (2017) The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat. Commun. 8, 15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Polewski M. D., Reveron-Thornton R. F., Cherryholmes G. A., Marinov G. K., Cassady K., and Aboody K. S. (2016) Increased expression of system xc− in glioblastoma confers an altered metabolic state and temozolomide resistance. Mol. Cancer Res. 14, 1229–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Banjac A., Perisic T., Sato H., Seiler A., Bannai S., Weiss N., Kölle P., Tschoep K., Issels R. D., Daniel P. T., Conrad M., and Bornkamm G. W. (2008) The cystine/cysteine cycle: a redox cycle regulating susceptibility versus resistance to cell death. Oncogene 27, 1618–1628 [DOI] [PubMed] [Google Scholar]
- 29. Liu X., Xiao Z. D., Han L., Zhang J., Lee S. W., Wang W., Lee H., Zhuang L., Chen J., Lin H. K., Wang J., Liang H., and Gan B. (2016) LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat. Cell Biol. 18, 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu X., and Gan B. (2016) lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle 15, 3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin A., Piao H. L., Zhuang L., dos Sarbassov D., Ma L., and Gan B. (2014) FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacological inhibition of the PI3K-AKT pathway. Cancer Res. 74, 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee H., Dai F., Zhuang L., Xiao Z. D., Kim J., Zhang Y., Ma L., You M. J., Wang Z., and Gan B. (2016) BAF180 regulates cellular senescence and hematopoietic stem cell homeostasis through p21. Oncotarget 7, 19134–19146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gan B., Lim C., Chu G., Hua S., Ding Z., Collins M., Hu J., Jiang S., Fletcher-Sananikone E., Zhuang L., Chang M., Zheng H., Wang Y. A., Kwiatkowski D. J., Kaelin W. G. Jr., et al. (2010) FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18, 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]