Abstract
Interferon γ (IFNγ) is a pleiotropic protein secreted by immune cells. IFNγ signals through the IFNγ receptor, a protein complex that mediates downstream signaling events. Studies into IFNγ signaling have provided insight into the general concepts of receptor signaling, receptor internalization, regulation of distinct signaling pathways, and transcriptional regulation. Although IFNγ is the central mediator of the adaptive immune response to pathogens, it has been shown to be involved in several non-infectious physiological processes. This review will provide an introduction into IFNγ signaling biology and the functional roles of IFNγ in the autoimmune response.
Keywords: autoimmune disease, autoimmunity, interferon, signal transducers and activators of transcription 1 (STAT1), signaling
Introduction
According to the National Institutes of Health, autoimmune diseases (ADs)3 are one of the top 10 leading causes of death in female children and women in all age groups up to 64 years of age (1). Excess secretion of IFNs has been associated with development of human ADs (2). Interferons (IFNs) comprise a family of proteins classified as type I (IFN-α, -β, -ϵ, -κ, and -ω), type II (IFN-γ, herein IFNγ), and type III (IFN-λ1–4) that have pleiotropic roles in immunity, cancer biology, and autoimmunity (2). Here, we aim to provide a basic understanding of the functions of the type II IFNγ and the IFNγ-signaling pathway (hereafter referred to as IFN signaling) in a contextual and timing perspective related to the autoimmune environment and the development of ADs.
Biological role of IFNγ on inflammation
Since Wheelock (3) reported that IFNγ inhibited viral replication, IFNγ has become an essential regulator of several immune processes, including vaccine-mediated responses and pro-inflammatory CD4+ T helper 1 (Th1) cell responses (4). As an effector cytokine of Th1 immunity, IFNγ is the key regulator of macrophage activation via the Janus kinase (JAK)/signal transducer and activators of transcription (STAT) pathway (Fig. 1A) (5). Normally, in the early phases of the host immune response, production of IFNγ by natural killer cells, CD4+T helper 1 (Th1) cells, and CD8+ T cells aims to improve antigen recognition in antigen-presenting cells (APCs) such as macrophages and dendritic cells. IFNγ activates macrophages toward the “M1” phenotype, which is characterized by the expression of high levels of pro-inflammatory cytokines such as Il-1β, IL-12, IL-23, and TNF-α; high production of reactive nitrogen and oxygen intermediates; promotion of Th1 T cell response; and strong inflammatory activity (6). APCs express major histocompatibility complex (MHC) class I and II proteins and activate cross-presentation antigenic pathways. In parallel, IFNγ signaling generates other cytokines and inflammatory factors to sustain inflammation, maintain Th1 responses, and inhibit differentiation of regulatory T cells, CD4+ T helper 2 cells (Th2), and Th17 cells (7). Despite these amplification steps, IFNγ signaling is generally short-lived to elicit functional recovery of homeostasis, including tissue repair and reestablishment of tissue physiology.
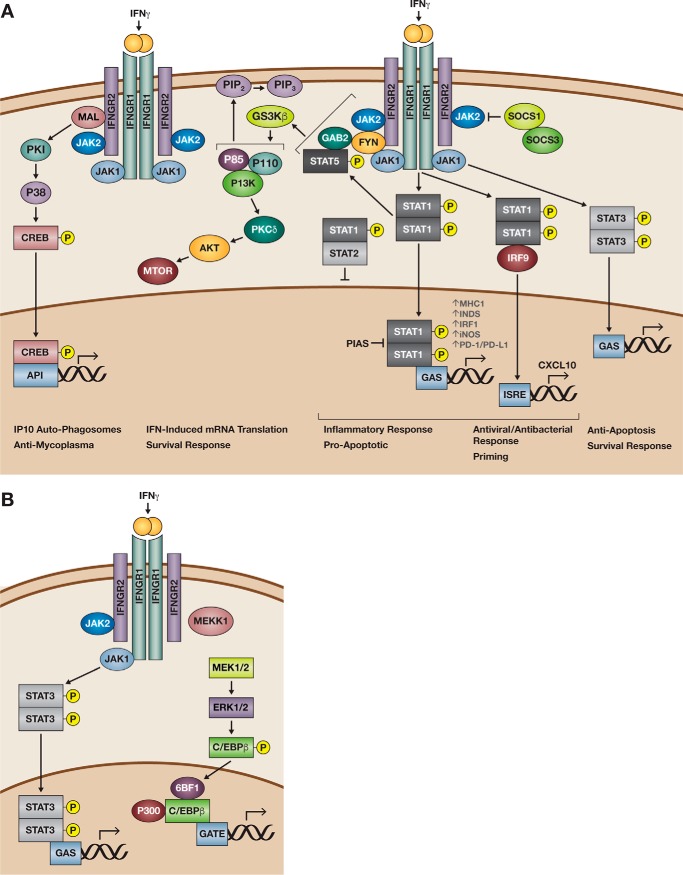
Figure 1.
Overview of the STAT-1 canonical and non-canonical signaling pathways elicited by IFNγ. As a dimer, IFNγ (orange) binds the IFNGR, which is composed of the IFNGR1 and IFNGR2 subunits, the kinases Jak1/Jak2. A, in canonical IFNγ signaling, phosphorylation of Jak1 and JAK2 results in the phosphorylation of STAT1 (center). A STAT1 homodimer translocates to the nucleus and binds to GAS found in the promoters of IFNγ-regulated genes such as HLA-A, NOS-2, IRF1, PDCD1, and CD274. Recruitment of adaptor proteins associated with IFNGR2 such as MAL and Fyn results in non-canonical STAT1 signaling. MAL-dependent IFN-γ receptor (IFNGR) signaling elicits signaling via MAPK p38 phosphorylation to up-regulate expression of the chemokine IP-10, antimycoplasma proteins, and formation of autophagosomes (left). GSK3β activation and Fyn elicit pSTAT5 recruitment to activate PI3K to regulate cell membrane permeabilization. Alternatively, IFNγ activation of GSK3β via PKCδ activates AKT/mTOR regulation of survival responses. Nevertheless, upon STAT activation, control mechanisms aiming to regulate signaling target either the JAK catalytic sites with SOCS proteins (upper right corner) or blockade of the STAT dimers binding to GAS sites with PIAS or through binding with un-phosphorylated STAT2. Alternatively, IFN priming increases IRF9, which is recruitment to STAT1 dimers for binding to ISRE sites. Antiviral and antibacterial responses benefit from this mechanism. B, in cells not expressing STAT1, STAT3 can be phosphorylated by Jak1/Jak2 resulting in translocation of the STAT3 dimer to GAS sites. Moreover, IFNγ activation of ERK results in C/EBPβ activation and binding to a novel IFN-response element (GATE). Figure has been adapted and modified from Refs. 17 and 29.
IFNγ signaling: Canonical and non-canonical pathways
In the context of inflammation, IFNγ induces a rapid response via the JAK/STAT or canonical pathway. However, in the context of ADs, maintenance of chronically high levels of IFNs leads to activation of both canonical and non-canonical pathways, albeit in a cell- and context-specific manner.
Canonical IFNγ-signaling pathways
In the canonical pathway of IFN signaling, IFNγ dimerizes and binds to the two IFNGR1 receptors. The IFNGR is composed of two distinct chains, the high affinity IFNGR1 (α) and a low affinity IFNGR2 (β) (8). The identification of a glycosylation-deficient mutant residue in the IFNGR1 detailed two key steps preceding initiation of IFNγ signaling (9). In the first step, IFNγ binding induces the receptors to undergo a conformational change in lipid nano-domains, whereby the box 1 domains on the IFNGRs are brought into proximity of each other allowing recruitment of two JAKs, JAK1 and JAK2, to the IFNGR1 and IFNGR2 chains, respectively. This recruitment step occurs independently of their enzymatic activities. In the second step, JAK1/2 activation induces a second conformational change that allows STAT1 to associate with the IFNγ–IFNGR complex. In turn, JAK1 and JAK2 phosphorylate the transcription factor STAT1 (pSTAT1) forming a homodimer that translocates to the nucleus (9). At this moment, the released IFNGRs associated with the cortical-actin network and prepared for alternative regulation via receptor trafficking and endocytosis. Importantly, these events demonstrate that receptor internalization may not be required for IFNγ signaling to take place.
In the nucleus, pSTAT1 binds with high affinity to DNA sequences termed the γ-interferon-activated site (GAS) to initiate transcription of interferon-stimulated genes (ISGs) (Fig. 1A) (11). At the same time, acetylation of pSTAT1 sets the timer for STAT1 inactivation via complex formation between acetylated STAT1 and the protein-tyrosine phosphatase T cell protein-tyrosine phosphatase (TCP45) (12). In turn, histone deacetylase 3 (HDAC3) deacetylates STAT1, thus permitting a new cycle of phosphorylation and re-stimulation (13). Hence, the rapid increase of pSTAT1 after IFNγ stimulation elicits a reversible and dynamic response to rapidly restore homeostasis.
Non-canonical IFNγ signaling pathways
The observation that IFNγ is capable of inducing gene expression in STAT1−/− bone marrow-derived macrophages suggested that IFNγ can act independently of STAT-1 or in an alternative non-canonical fashion (14). Generally, the activation of non-canonical pathways appears to be later rather than earlier after STAT1 activation. Nevertheless, there is evidence suggesting that non-canonical pathways could be activated in the absence or presence of Stat1 in a context-dependent manner. In the absence of STAT1 (Fig. 1B), IFNγ can activate STAT3, in a JAK-STAT-dependent process that results in activation of GAS-regulated genes (15, 16). Moreover, the absence of Stat1 in primary fibroblasts or neurons led to enhanced ERK activation following IFNγ addition, implying that the cell-specific availability of signal transducers can diversify the cellular response following IFN engagement (17). STAT-independent IFNγ signaling can occur via activation of other MAPKs, such as PyK2, ERK1/2, and JNK (18, 19); the adaptor proteins CrkL and small G protein Rap1 (20); and the Src homology 2 domain-containing protein-tyrosine phosphatases SHP-1 and SHP-2 (18, 21). Note, IFNγ activation of different kinases such as ERK1/ERK2 (MAPKs) (22) or glycogen synthase kinase 3 (GSK3β) signaling (23) results in activation of different transcription factors. For example, IFNγ activation of ERK regulates binding of CCAAT/enhancer-binding protein-β (C/EBPβ) to a novel IFNγ response element called GATE (22). GATE has little homology with GAS and binds to different transacting factors such as C/EBPβ. Recent evidence found that phosphorylation of C/EBPβ involves IFN-stimulated proteolytic processing of ATF6, and ERK1 and ERK2 are necessary to control autophagy of several infectious agents (24). Conversely, IFNγ activation of GSK3β via the phosphoinositide 3-kinase-AKT pathway regulates CREB/AP1-dependent DNA binding to suppress IL-10 production (23).
Conversely, in the presence of Stat1, activation of canonical and non-canonical pathways could happen simultaneously (Fig. 1A). However, the outcomes of such activation are cell- and context-specific. For example, in mice infected with systemic Dengue, Stat1-dependent pathways were required for early viral control, but Stat1-independent pathways were later required for viral clearance (25). Furthermore, in a mouse model of encephalitis, the activation of Stat1-dependent and Stat1-independent pathways advanced IFNγ-induced reduction of myelin sheath thickness in the CNS despite Stat1 knockdown (26). Apparently, initiation of these alternative signaling pathways starts at the JAK activation sites in IFNGR1 with the recruitment of adaptor molecules such as MyD88 adaptor-like (Mal) (27) or the Fyn kinase (28). MAL is encoded by the gene Toll-interleukin 1 receptor domain containing adaptor protein (TIRAP). Noticeably, Mal-dependent IFNGR signaling required phosphorylation of the mitogen-activated protein kinases (MAPK) p38, not Stat1, for phagosome maturation and killing of intracellular infectious organisms such as Mycobacterium tuberculosis. Moreover, defects in the Mal-dependent IFNγ-signaling pathway due to non-synonymous single nucleotide polymorphism at S180L in the human TIRAP gene explains best the increased susceptibility of SLE patients for mycobacterial and pneumococcal infections (27). Similarly, recruitment of the Src kinase Fyn results in the formation of a complex that allows IFNγ to activate Stat5b via PI3K signaling (29). The ability to activate Stat5 while preserving IFNγ activation of STAT1-dependent immune events represents an advantageous adaptation mechanism to regulate macromolecular permeability in enteric epithelium with low STAT1 levels. Moreover, IFNγ activation of AKT and mTOR via PI3K improved mRNA translation of IFNγ-regulated genes complementing STAT1-dependent mechanisms (30). Thus, IFNγ can regulate complex processes beyond their known short-term effects. Therefore, the overall biological effect of IFNγ signaling likely results from a well-adjusted combination of Stat1-dependent and Stat1-independent mechanisms activated sequentially during progression of the inflammatory disease.
Alternative mechanisms regulating the IFNγ signaling via endocytosis
As diverse as the IFNγ signaling downstream pathways could be, all actions start generally at the IFNGR. As human IFNγ does not signal in mouse or rat cells, evidence that microinjection of human IFNγ elicited antiviral activity in murine cells suggested that the IFNGR provides species specific responses to IFNγ (31). Further evidence showing that retention of IFNγ within cells also resulted in an IFNγ-dependent signaling leaded researchers to further claim that the IFNGR drives species specificity responses (32, 33). Thus, the IFNGR holds the key to control the activity of IFNγ among species (34).
The regulation of IFNγ signaling involves essential management mechanisms that regulate the differential expression and cell-surface localization of the IFNGR chains (Fig. 2). IFNGR1 is usually expressed in excess, whereas IFNGR2 is more tightly regulated in most but not all cell types (35, 36). In fact, the absence of IFNGR2 on the surface of Th1 cells supports the model that regulation of its surface expression regulates responsiveness to IFNγ (37). Moreover, several viruses such as the vaccinia virus and myxoma virus encode and secrete IFNγ receptor mimetics, which are peptides with significant sequence similarity to the human and mouse receptors for IFNγ, to neutralize IFNγ activity (38). These data suggest that cell regulation of IFNGR accounts for cell-specific differences in response to IFNγ and how a target cell becomes unresponsive to further IFNγ stimulation.
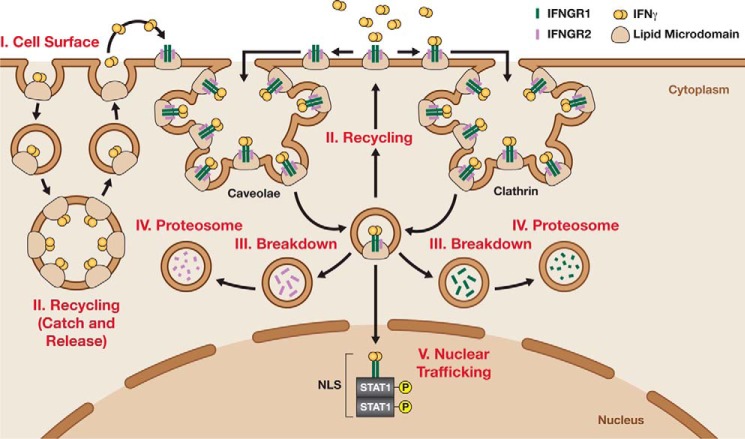
Figure 2.
Endocytic mechanisms regulating IFNγ signaling via differential expression of the IFNGR. IFNGR1 (green) and IFNGR2 (purple) are associated in lipid micro-domains on the cell surface. In the presence of IFNγ dimers (orange), the cytokine receptor complex is redirected to clathrin or caveolae sites at the cell surface. Once there, IFNγ either binds the receptor or is recruited to PS-rich domains. Then, the IFNG complexes are endocytosed either through clathrin-dependent events or clathrin-independent events that involve caveolae-dependent mechanisms. IFNγ–PS complexes are slowly recycled back to the cell surface, and IFNγ is released in an autocrine-like manner (left). In contrast, IFNγ–IFNGR complexes are endocytosed and broke down before being recycled or degraded. Noticeably, IFNGR2 is enzymatically cleaved and separated for degradation via proteasomes. Meanwhile, IFNGR1 can be recycled to the cell surface or targeted for degradation. Alternatively, the IFNG–IFNGR–p-STAT1 complex could remain stable and continue translocating to the nucleus using NLS.
Recent data showing that the IFNGR1 and IFNGR2 are loosely associated in sphingolipid-rich areas on the cell surface, called lipid micro-domains, provided mechanistic details over their complex formation (39, 40). As shown in Fig. 2, upon IFNγ dimer binding to IFNGR1 and IFNGR2, a ternary complex is formed within the micro-domains in preparation to deliver downstream signaling from the cell surface to the nucleus (41). Note, IFNγ–IFNGR complex readiness is independent of receptor internalization (42). Upon Stat1 activation and internalization, the two IFNGRs are differentially processed within cells using clathrin-dependent or clathrin-independent mechanisms (43). Clathrin-dependent endocytosis utilizes the protein clathrin to mediate endocytosis and is the primary mechanism by which the IFNGR1 is recycled to the cell surface (42). Infectious organisms such as herpesvirus K3-5 or Trypanosoma cruzi down-regulate the surface expression of host IFNGR by increasing their endocytosis rates, which leads to suppression of cell-mediated immunity (44, 45). Conversely, clathrin-independent endocytosis uses sphingolipid-rich caveolae, also known as lipid rafts, to regulate IFNGR2 at the cell surface (46). Human T cells use this pathway to limit sensitivity to IFNγ as a strategy for dampening the host immune response (47).
Recently, a novel mechanism explains how membrane cell dynamics modulates cell-to-cell communication via cytokines (Fig. 2). Surprisingly, positively charged regions of IFNγ, IL-12, and IL-23 were proved to interact directly with negatively charged cell-surface phosphatidylserines (PS) on tumor cells (48). The cytokine–PS complexes are endocytosed, possibly via caveolae-dependent mechanisms, to be slowly recycled back to the cell surface in an autocrine-like manner. Once released, cytokines bind their respective receptors. This mechanism explains how tumor cells manage to extend a response after a short-lived cytokine exposure.
Another less understood and controversial regulatory mechanism proposes that the whole IFNγ/IFNGR structure translocates to the nuclei and defines species selectiveness (32). Independent reports showed that such an event is possible due to a putative nuclear localization sequence (NLS) contained both in IFNγ and the IFNGR structures (49). It has been suggested that those NLS allows IFNγ to regulate STAT1 trafficking within cells (50–52). As the concept of a nuclear localization step awaits further verification, its relevance on IFNγ signaling remains incomplete. Together, controlling the IFNGR expression at the cell surface is a straightforward control mechanism available to all cells to regulate responses to IFNγ.
Chronic exposure to IFNγ leads to ADs
In ADs, immune cells are exposed, often simultaneously, to more than one IFN causing integration of IFN signaling (53). Recently, it was determined that such exposure could go back up to 3.0 to 4.5 years before individuals are even diagnosed with ADs such as SLE (54) or rheumatoid arthritis (RA) (55), respectively. In this type of relapsing-remitting ADs, immune cells are constitutively and constantly exposed to waves of significantly “high” (relapse or flare state) or “low” (remission state) levels of type I and type II IFNs during distinct periods of time (56, 57). Pre-exposure to low sub-activating concentrations of IFNs sensitizes cells to produce enhanced responses to extracellular inflammatory stimuli that include IFNs themselves, as well as other cytokines such as tumor necrosis factor α (TNFα) or toll-like receptor activators (58). This process, known as priming, is characterized by the intracellular accumulation of STATs. Contrary to acute inflammation, primed immune cells such as M1 macrophages are the predominant phenotype at sites of inflammation for ADs such as RA, multiple sclerosis, and lupus nephritis (59, 60).
IFN priming elicits post-transcriptional and/or epigenetic changes that promote synergism for gene induction and regulation after subsequent exposure to type I and type II IFNs (Fig. 1). Noticeably, IFN priming involves increased association of similar or different types of IFN receptors (53). Under this mechanism, type I IFNs augment IFNγ signaling via association of type I and II IFN receptor subunits. Conversely, IFNγ priming for IFNα signaling occurs but involves only STAT1 and not STAT2 or STAT3 (61). Priming for production of large amounts of type I IFNs is mediated by an autoamplification loop in which IFNα induces expression of the transcription factor IFN regulator factor 7 (Irf7) that activates IFNα gene promoters (61). This step creates a robust priming effect, where IFNγ enhances positive signaling by recruiting other Irfs. For example, the formation of the heterotrimeric transcription factor complex known as ISGF3 between Stat1 and IFN-regulatory factor 9 (Irf9) required type I IFN priming and prolonged IFNγ activation (Fig. 1A) (62, 63). Those complexes elicited Stat1 regulation over gene promoters with GAS and/or interferon-stimulated response elements (ISRE) (64) allowing regulation of genes with either one or both elements such as the IFNγ-regulated cytokine CXCL10/Cxcl10 (IP-10) (65). Consequently, IFN priming elicits regulation of complex biological process via epigenetic remodeling and enrichment of STAT1-binding motifs in mice (66). For example, microglial reactive oxygen species production in response to IFNs required simultaneous modification of three mechanisms, including up-regulation of the NADPH oxidase subunit NOX2, up-regulation of NO production, and the reduction of intracellular GSH levels (67).
Recently, it was reported that un-phosphorylated STATs play roles modulating the IFN signaling response. For example, un-phosphorylated STAT1 (U-STAT1) is capable of regulating a set of ISGs that offer some protection against various viruses but rendered cells resistant to chemotherapy and radiation (68, 69). Studies in mouse models of arthritis and experimental autoimmune encephalomyelitis (EAE) determined that U-Stat1 regulated ISGs that are regularly induced later rather than earlier after stimulation with either IFNγ or IFNβ (68). Moreover, evidence suggests that IFNγ priming involves regulation of microRNAs, as suppression of miR-3473b limited activation of primed macrophages (70).
To counterbalance the effect of IFN priming and receptor activation, cells also activated precise complementary inhibitory mechanisms at different levels (Fig. 1). At the plasma membrane, suppressor of cytokine signaling proteins block receptor activation by binding to the activated JAK catalytic sites, thus turning off downstream signals. Moreover, at the cytoplasm, an increase in un-phosphorylated STAT2 binds pStat1 to diminish its nuclear translocation during continuous IFNγ stimulation possibly eliciting adaptation to long-term IFNγ stimulation (71). In the nuclei, protein inhibitors of activated STAT (PIAS) associate with activated STAT dimers via their zinc-binding ring finger domain in the center of the molecule, preventing them from binding to the DNA (72). Thus, a primed innate immune system modulates the functions of IFNs and defines the host response to underlying triggers of autoimmune disorders.
Understanding the role of IFNγ in ADs with mouse models
Several mouse models of ADs, such as SLE, RA, collagen-induced arthritis (CIA), and EAE have proven the essential role of IFNγ both in promoting and suppressing different stages during AD progression. For example, it is known that both ifng−/− and stat1−/− mice are highly susceptible to EAE (73, 74). In contrast, it has been shown that MHC-II induced by IFNγ-hyperproducing T cells is important on the disease course of CIA and RA (75). These data suggest that IFNγ via the JAK/Stat1 pathway modulates AD progression. However, targeting the function of IFNγ at different disease stages is essential to understand its biological role. Indeed, administration of mouse IFNγ applied at very early stages of EAE aggravates the disease because of highly producing IFNγ CD4+ T cells (76). However, at a later stage, administration of IFNγ reduces the severity of EAE when it is mediated by CD4+ Th17 cells (76). Similarly, IFNγ is required, even in the presence of complete Freund's adjuvant, at the onset and development of severe arthritis following immunization with glucose-6-phosphate isomerase (77). The described evidence indicates that CIA and EAE are more Th17 cell-mediated disease models instead of pure Th1 cell-mediated diseases, where IFNγ exerts a dynamic and context-dependent response. Although IFNγ may be a reasonable target for clinical trials, the lack of biomarkers defining when to start or stop intervention limits the therapeutic value of IFNγ. Specifically, there is a need for markers that determine the availability of IFNγ-producing cells at different time points during the immune stimulus. These limitations probably account for the current narrow therapeutic index and inadequate clinical utility of IFNγ (78). As a result, most of the ongoing preclinical development is concentrated on its inducers, in particular IL-12 or IL-18 (79).
Despite the benefits from mouse models, human ADs are chronic inflammation processes instead of short-term and self-limited diseases induced in preclinical models. To address this issue, our laboratory designed a mouse model carrying deletion of the ifng 3′-untranslated region adenylate uridylate-rich element (ARE). The persistent serum levels of IFNγ, due to increased stability of mRNA, result in gradual establishment of SLE-like autoimmunity (80). More importantly, we have provided evidence that heterozygous ARE-del mice share similar levels of autoantibodies and demonstrate a histological score of tissue damage in target organs like that seen in homozygous ARE-del mice that express twice the systemic IFNγ levels (80, 81). This evidence suggests the existence of a threshold level of IFNγ that, when crossed, results in a more severe pathology. Similarly, in another model of lupus, mutation of ring finger and CCCH-type domains 1 (rc3h1/roquin) reduced the rate of decay of IFNγ mRNA and increased IFNγ signaling. As a result, germinal center B cells increased their numbers and production of autoantibodies (82). Despite the significant overlap between genes induced by either type I and type 2 IFNs, these models strongly suggest that chronic exposure to IFNγ causes or contributes to SLE-like disease. Hence, development of models for long-term inflammation could be essential for understanding the differences between disease stages.
IFN-mediated chronic inflammation, checkpoint inhibitors, and immunotherapy in ADs
Traditionally, “high” serum levels of IFNγ (referred herein as IFNγ levels) are associated with pro-inflammatory active disease, whereas “low” IFNγ levels are associated with anti-inflammatory inactive autoimmune disease. As levels of IFNγ are compared with healthy individuals, there is no consensus of what “high” and “low” IFNγ levels mean. Data from pre-clinical and clinical studies in mice and humans showed administration of type 1 IFNs (IFNα and -β) generally exert a linear dose response, whereas exogenous IFNγ exhibits a “bell-shaped” dose-response curve (83). A “bell-shaped” dose response is characterized by induction of stimulatory effects at low doses until it reaches a summit point where additional dosing cause inhibitory activity and deleterious effects (84). These data suggest the function(s) of endogenous and exogenous IFNs are probably defined by the dynamics between systemic and local inflammatory environments. In the local environment, stimulatory and inhibitory pathways are activated to limit inflammation and destruction of self-tissues (85). Recent evidence showed that IFNγ is the main regulator of programmed cell death protein 1 (PD-1) and its two ligands, PD-L1 (B7-H1 or CD274) and PD-L2 (B7-DC or CD273) (86). This fact suggests that chronic inflammation influences the local co-inhibitory pathways in autoimmunity. In ADs, PD-1 (CD279) is the most studied co-inhibitory receptor, although the role of other checkpoint inhibitors such as cytotoxic T lymphocyte associated protein 4 (CTLA4, CD152) is more controversial (87). PD-1 and CTLA4 have critical, multifaceted roles in regulating the balance among T cell activation, tolerance, and immuno-mediated tissue damage. Evidence showed that polymorphisms at the mouse Pdcd1 and Ctla4 promoters have been associated with susceptibility to develop AD such as in SLE and AR (56). These facts reminded us that insufficient co-inhibition can also promote the development and progression of AD.
Immunotherapy targeting checkpoint inhibitors, such as PD-L1 and CTLA4, is one of the most rapidly growing fields in cancer therapy. Nevertheless, the issue with checkpoint blockade is the occurrence of associated toxicities termed immune-related adverse events (irAEs). Generally, AD patients are thought to be at higher risk of developing hematological malignancies such as Hodgkin's lymphoma or cervical cancer due to the underlying dysregulation of their immune system and anti-inflammatory treatment regime (88). Because of this, therapeutic use of check point inhibitors in patients with ADs developing tumors had mixed results so far. In a small cohort of AD patients that develop melanoma, CTLA4 blockade induces tumor growth inhibition, but 25–50% developed mild to moderate exacerbations of their AD or experienced conventional CTLA4-induced irAEs, respectively (10). Nevertheless, the high risk of IrAEs makes the use of checkpoint inhibitors controversial even in patients with certain autoimmune diseases (pernicious anemia, Crohn's disease, ulcerative colitis, SLE, and psoriasis) that develop tumors. Therefore, understanding the regulation, function, and importance of the IFN signaling detailed in this review could help to unravel new therapeutic options for ADs and other chronic diseases, including cancer.
Summary
Overall, there is a consensus that the effect of IFNγ signaling is largely controlled in a multidimensional fashion where timing, exposure levels, target organ, and cellular environment define the outcome of the immune response to IFNγ expression. As such, interventions altering the IFNγ activity will be approached with great attention and thoughtfulness as to both local and systemic effects in a wide variety of disease states. This cautious approach arises because IFNγ impacts the balance between protection and development of autoimmune responses. Hence, regardless of the activated pathways, IFNγ will directly or indirectly determine the dynamics of inflammation in subjects with underlying autoimmunity. Although this review focused on immune cells and autoimmunity, these same principles are starting to be extended to define the role of IFNγ in other chronic conditions such as cancer and immunotherapeutic approaches to treat cancer. Specifically, the functions of IFNγ are being reexamined to better understand how it is impacting defined pathogenic or controlled outcomes. Considering IFNγ priming and cross-regulation as an existing pre-condition during the different stages of autoimmune diseases or in certain types of cancers will help to develop a better understanding of the role of this important immunoregulatory molecule in disease initiation, progression, and treatment.
This work was supported in part by the Intramural Research Program of the NCI, Cancer and Inflammation Program, Center for Cancer Research, National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- AD
- autoimmune disease
- C/EBPβ
- CCAAT/enhancer-binding protein-β
- JAK
- Janus kinase
- APC
- antigen-presenting cell
- GAS
- γ-interferon-activated site
- ISG
- interferon-stimulated gene
- RA
- rheumatoid arthritis
- SLE
- systemic lupus erythematosus
- EAE
- experimental autoimmune encephalomyelitis
- ARE
- adenylate uridylate-rich element
- irAE
- immune-related adverse event
- mTOR
- mechanistic target of rapamycin
- IFNGR
- IFN-γ receptor
- PS
- phosphatidylserine
- ISRE
- interferon-stimulated response element
- PIAS
- protein inhibitors of activated STAT
- NLS
- nuclear localization sequence
- STAT
- signal transducers and activators of transcription
- CIA
- collagen-induced arthritis.
References
- 1. NIAID, National Institutes of Health (2002) Autoimmune Diseases Coordinating Committee–Autoimmune Diseases Research. United States Department of Health and Human Services, Bethesda, MD [Google Scholar]
- 2. Pollard K. M., Cauvi D. M., Toomey C. B., Morris K. V., and Kono D. H. (2013) Interferon-γ and systemic autoimmunity. Discov. Med. 16, 123–131 [PMC free article] [PubMed] [Google Scholar]
- 3. Wheelock E. F. (1965) Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science 149, 310–311 [PubMed] [Google Scholar]
- 4. Billiau A., and Matthys P. (2009) Interferon-γ: a historical perspective. Cytokine Growth Factor Rev. 20, 97–113 [DOI] [PubMed] [Google Scholar]
- 5. Cherwinski H. M., Schumacher J. H., Brown K. D., and Mosmann T. R. (1987) Two types of mouse helper T cell clone. III. Further differences in lymphokine synthesis between Th1 and Th2 clones revealed by RNA hybridization, functionally monospecific bioassays, and monoclonal antibodies. J. Exp. Med. 166, 1229–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sica A., and Mantovani A. (2012) Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shachar I., and Karin N. (2013) The dual roles of inflammatory cytokines and chemokines in the regulation of autoimmune diseases and their clinical implications. J. Leukocyte Biol. 93, 51–61 [DOI] [PubMed] [Google Scholar]
- 8. Randal M., and Kossiakoff A. A. (2001) The structure and activity of a monomeric interferon-γ:α-chain receptor signaling complex. Structure 9, 155–163 [DOI] [PubMed] [Google Scholar]
- 9. Blouin C. M., Hamon Y., Gonnord P., Boularan C., Kagan J., Viaris de Lesegno C., Ruez R., Mailfert S., Bertaux N., Loew D., Wunder C., Johannes L., Vogt G., Contreras F. X., Marguet D., et al. (2016) Glycosylation-dependent IFN-γR partitioning in lipid and actin nanodomains is critical for JAK activation. Cell 166, 920–934 [DOI] [PubMed] [Google Scholar]
- 10. Johnson D. B., Sullivan R. J., Ott P. A., Carlino M. S., Khushalani N. I., Ye F., Guminski A., Puzanov I., Lawrence D. P., Buchbinder E. I., Mudigonda T., Spencer K., Bender C., Lee J., Kaufman H. L., et al. (2016) Ipilimumab therapy in patients with advanced melanoma and preexisting autoimmune disorders. JAMA Oncol. 2, 234–240 [DOI] [PubMed] [Google Scholar]
- 11. Decker T., Kovarik P., and Meinke A. (1997) GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 17, 121–134 [DOI] [PubMed] [Google Scholar]
- 12. Krämer O. H., Knauer S. K., Greiner G., Jandt E., Reichardt S., Gührs K. H., Stauber R. H., Böhmer F. D., and Heinzel T. (2009) A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 23, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen X., Barozzi I., Termanini A., Prosperini E., Recchiuti A., Dalli J., Mietton F., Matteoli G., Hiebert S., and Natoli G. (2012) Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl. Acad. Sci. U.S.A. 109, E2865–E2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gil M. P., Bohn E., O'Guin A. K., Ramana C. V., Levine B., Stark G. R., Virgin H. W., and Schreiber R. D. (2001) Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. U.S.A. 98, 6680–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qing Y., and Stark G. R. (2004) Alternative activation of STAT1 and STAT3 in response to interferon-γ. J. Biol. Chem. 279, 41679–41685 [DOI] [PubMed] [Google Scholar]
- 16. van Boxel-Dezaire A. H., and Stark G. R. (2007) Cell type-specific signaling in response to interferon-γ. Curr. Top. Microbiol. Immunol. 316, 119–154 [DOI] [PubMed] [Google Scholar]
- 17. O'Donnell L. A., Henkins K. M., Kulkarni A., Matullo C. M., Balachandran S., Pattisapu A. K., and Rall G. F. (2015) Interferon γ induces protective non-canonical signaling pathways in primary neurons. J. Neurochem. 135, 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramana C. V., Gil M. P., Han Y., Ransohoff R. M., Schreiber R. D., and Stark G. R. (2001) Stat1-independent regulation of gene expression in response to IFN-γ. Proc. Natl. Acad. Sci. U.S.A. 98, 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsuzawa T., Fujiwara E., and Washi Y. (2014) Autophagy activation by interferon-γ via the p38 mitogen-activated protein kinase signalling pathway is involved in macrophage bactericidal activity. Immunology 141, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alsayed Y., Uddin S., Ahmad S., Majchrzak B., Druker B. J., Fish E. N., and Platanias L. C. (2000) IFN-γ activates the C3G/Rap1 signaling pathway. J. Immunol. 164, 1800–1806 [DOI] [PubMed] [Google Scholar]
- 21. You M., and Zhao Z. (1997) Positive effects of SH2 domain-containing tyrosine phosphatase SHP-1 on epidermal growth factor- and interferon-γ-stimulated activation of STAT transcription factors in HeLa cells. J. Biol. Chem. 272, 23376–23381 [DOI] [PubMed] [Google Scholar]
- 22. Hu J., Roy S. K., Shapiro P. S., Rodig S. R., Reddy S. P., Platanias L. C., Schreiber R. D., and Kalvakolanu D. V. (2001) ERK1 and ERK2 activate CCAAAT/enhancer-binding protein-β-dependent gene transcription in response to interferon-γ. J. Biol. Chem. 276, 287–297 [DOI] [PubMed] [Google Scholar]
- 23. Grimes C. A., and Jope R. S. (2001) CREB DNA binding activity is inhibited by glycogen synthase kinase-3β and facilitated by lithium. J. Neurochem. 78, 1219–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gade P., Ramachandran G., Maachani U. B., Rizzo M. A., Okada T., Prywes R., Cross A. S., Mori K., and Kalvakolanu D. V. (2012) An IFN-γ-stimulated ATF6–C/EBP-β–signaling pathway critical for the expression of Death Associated Protein Kinase 1 and induction of autophagy. Proc. Natl. Acad. Sci. U.S.A. 109, 10316–10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shresta S., Sharar K. L., Prigozhin D. M., Snider H. M., Beatty P. R., and Harris E. (2005) Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J. Immunol. 175, 3946–3954 [DOI] [PubMed] [Google Scholar]
- 26. Lin W., and Lin Y. (2010) IFN-γ inhibits central nervous system myelination through both STAT1-dependent and STAT1-independent pathways. J. Neurosci. Res. 88, 2569–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ní Cheallaigh C., Sheedy F. J., Harris J., Muñoz-Wolf N., Lee J., West K., McDermott E. P., Smyth A., Gleeson L. E., Coleman M., Martinez N., Hearnden C. H., Tynan G. A., Carroll E. C., Jones S. A., et al. (2016) A common variant in the adaptor mal regulates interferon γ signaling. Immunity 44, 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uddin S., Sher D. A., Alsayed Y., Pons S., Colamonici O. R., Fish E. N., White M. F., and Platanias L. C. (1997) Interaction of p59fyn with interferon-activated Jak kinases. Biochem. Biophys. Res. Commun. 235, 83–88 [DOI] [PubMed] [Google Scholar]
- 29. Smyth D., Phan V., Wang A., and McKay D. M. (2011) Interferon-γ-induced increases in intestinal epithelial macromolecular permeability requires the Src kinase Fyn. Lab. Invest. 91, 764–777 [DOI] [PubMed] [Google Scholar]
- 30. Kaur S., Sassano A., Dolniak B., Joshi S., Majchrzak-Kita B., Baker D. P., Hay N., Fish E. N., and Platanias L. C. (2008) Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc. Natl. Acad. Sci. U.S.A. 105, 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trilling M., Le V. T., Zimmermann A., Ludwig H., Pfeffer K., Sutter G., Smith G. L., and Hengel H. (2009) γ interferon-induced interferon regulatory factor 1-dependent antiviral response inhibits vaccinia virus replication in mouse but not human fibroblasts. J. Virol. 83, 3684–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahmed C. M., Burkhart M. A., Mujtaba M. G., Subramaniam P. S., and Johnson H. M. (2003) The role of IFNγ nuclear localization sequence in intracellular function. J. Cell Sci. 116, 3089–3098 [DOI] [PubMed] [Google Scholar]
- 33. Sancéau J., Sondermeyer P., Béranger F., Falcoff R., and Vaquero C. (1987) Intracellular human γ-interferon triggers an antiviral state in transformed murine L cells. Proc. Natl. Acad. Sci. U.S.A. 84, 2906–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith M. R., Muegge K., Keller J. R., Kung H. F., Young H. A., and Durum S. K. (1990) Direct evidence for an intracellular role for IFN-γ. Microinjection of human IFN-γ induces Ia expression on murine macrophages. J. Immunol. 144, 1777–1782 [PubMed] [Google Scholar]
- 35. Bernabei P., Coccia E. M., Rigamonti L., Bosticardo M., Forni G., Pestka S., Krause C. D., Battistini A., and Novelli F. (2001) Interferon-γ receptor 2 expression as the deciding factor in human T, B, and myeloid cell proliferation or death. J. Leukocyte Biol. 70, 950–960 [PubMed] [Google Scholar]
- 36. Regis G., Conti L., Boselli D., and Novelli F. (2006) IFNγR2 trafficking tunes IFNγ-STAT1 signaling in T lymphocytes. Trends Immunol. 27, 96–101 [DOI] [PubMed] [Google Scholar]
- 37. Bach E. A., Szabo S. J., Dighe A. S., Ashkenazi A., Aguet M., Murphy K. M., and Schreiber R. D. (1995) Ligand-induced autoregulation of IFN-γ receptor β chain expression in T helper cell subsets. Science 270, 1215–1218 [DOI] [PubMed] [Google Scholar]
- 38. Upton C., Mossman K., and McFadden G. (1992) Encoding of a homolog of the IFN-γ receptor by myxoma virus. Science 258, 1369–1372 [DOI] [PubMed] [Google Scholar]
- 39. Marchetti M., Monier M. N., Fradagrada A., Mitchell K., Baychelier F., Eid P., Johannes L., and Lamaze C. (2006) Stat-mediated signaling induced by type I and type II interferons (IFNs) is differentially controlled through lipid microdomain association and clathrin-dependent endocytosis of IFN receptors. Mol. Biol. Cell 17, 2896–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Subramaniam P. S., and Johnson H. M. (2002) Lipid microdomains are required sites for the selective endocytosis and nuclear translocation of IFN-γ, its receptor chain IFN-γ receptor-1, and the phosphorylation and nuclear translocation of STAT1α. J. Immunol. 169, 1959–1969 [DOI] [PubMed] [Google Scholar]
- 41. Krause C. D., Lavnikova N., Xie J., Mei E., Mirochnitchenko O. V., Jia Y., Hochstrasser R. M., and Pestka S. (2006) Preassembly and ligand-induced restructuring of the chains of the IFN-γ receptor complex: the roles of Jak kinases, Stat1 and the receptor chains. Cell Res. 16, 55–69 [DOI] [PubMed] [Google Scholar]
- 42. Farrar M. A., Campbell J. D., and Schreiber R. D. (1992) Identification of a functionally important sequence in the C terminus of the interferon-γ receptor. Proc. Natl. Acad. Sci. U.S.A. 89, 11706–11710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sadir R., Lambert A., Lortat-Jacob H., and Morel G. (2001) Caveolae and clathrin-coated vesicles: two possible internalization pathways for IFN-γ and IFN-γ receptor. Cytokine 14, 19–26 [DOI] [PubMed] [Google Scholar]
- 44. Kierszenbaum F., Mejia Lopez H., Tanner M. K., and Sztein M. B. (1995) Trypanosoma cruzi-induced decrease in the level of interferon-γ receptor expression by resting and activated human blood lymphocytes. Parasite Immunol. 17, 207–214 [DOI] [PubMed] [Google Scholar]
- 45. Li Q., Means R., Lang S., and Jung J. U. (2007) Downregulation of γ interferon receptor 1 by Kaposi's sarcoma-associated herpesvirus K3 and K5. J. Virol. 81, 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deckert M., Ticchioni M., and Bernard A. (1996) Endocytosis of GPI-anchored proteins in human lymphocytes: role of glycolipid-based domains, actin cytoskeleton, and protein kinases. J. Cell Biol. 133, 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rigamonti L., Ariotti S., Losana G., Gradini R., Russo M. A., Jouanguy E., Casanova J. L., Forni G., and Novelli F. (2000) Surface expression of the IFN-γ R2 chain is regulated by intracellular trafficking in human T lymphocytes. J. Immunol. 164, 201–207 [DOI] [PubMed] [Google Scholar]
- 48. Oyler-Yaniv J., Oyler-Yaniv A., Shakiba M., Min N. K., Chen Y. H., Cheng S. Y., Krichevsky O., Altan-Bonnet N., and Altan-Bonnet G. (2017) Catch and release of cytokines mediated by tumor phosphatidylserine converts transient exposure into long-lived inflammation. Mol. Cell 66, 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bader T., and Weitzerbin J. (1994) Nuclear accumulation of interferon γ. Proc. Natl. Acad. Sci. U.S.A. 91, 11831–11835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Larkin J. 3rd., Subramaniam P. S., Torres B. A., and Johnson H. M. (2001) Differential properties of two putative nuclear localization sequences found in the carboxyl terminus of human IFN-γ. J. Interferon Cytokine Res. 21, 341–348 [DOI] [PubMed] [Google Scholar]
- 51. Subramaniam P. S., Green M. M., Larkin J. 3rd, Torres B. A., and Johnson H. M. (2001) Nuclear translocation of IFN-γ is an intrinsic requirement for its biologic activity and can be driven by a heterologous nuclear localization sequence. J. Interferon Cytokine Res. 21, 951–959 [DOI] [PubMed] [Google Scholar]
- 52. Subramaniam P. S., Larkin J. 3rd, Mujtaba M. G., Walter M. R., and Johnson H. M. (2000) The COOH-terminal nuclear localization sequence of interferon γ regulates STAT1α nuclear translocation at an intracellular site. J. Cell Sci. 113, 2771–2781 [DOI] [PubMed] [Google Scholar]
- 53. Hu X., and Ivashkiv L. B. (2009) Cross-regulation of signaling pathways by interferon-γ: implications for immune responses and autoimmune diseases. Immunity 31, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lu R., Munroe M. E., Guthridge J. M., Bean K. M., Fife D. A., Chen H., Slight-Webb S. R., Keith M. P., Harley J. B., and James J. A. (2016) Dysregulation of innate and adaptive serum mediators precedes systemic lupus erythematosus classification and improves prognostic accuracy of autoantibodies. J. Autoimmun. 74, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nielen M. M., van Schaardenburg D., Reesink H. W., van de Stadt R. J., van der Horst-Bruinsma I. E., de Koning M. H., Habibuw M. R., Vandenbroucke J. P., and Dijkmans B. A. (2004) Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 50, 380–386 [DOI] [PubMed] [Google Scholar]
- 56. Zhang Q., and Vignali D. A. (2016) Co-stimulatory and co-inhibitory pathways in autoimmunity. Immunity 44, 1034–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Banchereau J., and Pascual V. (2006) Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25, 383–392 [DOI] [PubMed] [Google Scholar]
- 58. Borges da Silva H., Fonseca R., Alvarez J. M., and D'Império Lima M. R. (2015) IFN-γ priming effects on the maintenance of effector memory CD4(+) T cells and on phagocyte function: evidences from infectious diseases. J. Immunol. Res. 2015, 202816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brühl H., Cihak J., Plachý J., Kunz-Schughart L., Niedermeier M., Denzel A., Rodriguez Gomez M., Talke Y., Luckow B., Stangassinger M., and Mack M. (2007) Targeting of Gr-1+,CCR2+ monocytes in collagen-induced arthritis. Arthritis Rheum. 56, 2975–2985 [DOI] [PubMed] [Google Scholar]
- 60. Viallard J. F., Pellegrin J. L., Ranchin V., Schaeverbeke T., Dehais J., Longy-Boursier M., Ragnaud J. M., Leng B., and Moreau J. F. (1999) Th1 (IL-2, interferon-γ (IFN-γ)) and Th2 (IL-10, IL-4) cytokine production by peripheral blood mononuclear cells (PBMC) from patients with systemic lupus erythematosus (SLE). Clin. Exp. Immunol. 115, 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tassiulas I., Hu X., Ho H., Kashyap Y., Paik P., Hu Y., Lowell C. A., and Ivashkiv L. B. (2004) Amplification of IFN-α-induced STAT1 activation and inflammatory function by Syk and ITAM-containing adaptors. Nat. Immunol. 5, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 62. Lees J. R. (2015) Interferon γ in autoimmunity: a complicated player on a complex stage. Cytokine 74, 18–26 [DOI] [PubMed] [Google Scholar]
- 63. Rauch I., Rosebrock F., Hainzl E., Heider S., Majoros A., Wienerroither S., Strobl B., Stockinger S., Kenner L., Müller M., and Decker T. (2015) Noncanonical effects of IRF9 in intestinal inflammation: more than type I and type III interferons. Mol. Cell. Biol. 35, 2332–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Begitt A., Droescher M., Meyer T., Schmid C. D., Baker M., Antunes F., Knobeloch K.-P., Owen M. R., Naumann R., Decker T., and Vinkemeier U. (2014) STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat. Immunol. 15, 168–176 [DOI] [PubMed] [Google Scholar]
- 65. Brownell J., Bruckner J., Wagoner J., Thomas E., Loo Y.-M., Gale M. Jr., Liang T. J., and Polyak S. J. (2014) Direct, interferon-independent activation of the CXCL10 promoter by NF-κB and interferon regulatory factor 3 during hepatitis C virus infection. J. Virol. 88, 1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hoeksema M. A., Scicluna B. P., Boshuizen M. C., van der Velden S., Neele A. E., Van den Bossche J., Matlung H. L., van den Berg T. K., Goossens P., and de Winther M. P. (2015) IFN-γ priming of macrophages represses a part of the inflammatory program and attenuates neutrophil recruitment. J. Immunol. 194, 3909–3916 [DOI] [PubMed] [Google Scholar]
- 67. Spencer N. G., Schilling T., Miralles F., and Eder C. (2016) Mechanisms underlying interferon-γ-induced priming of microglial reactive oxygen species production. PLoS ONE 11, e0162497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cheon H., and Stark G. R. (2009) Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. U.S.A. 106, 9373–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Minn A. J. (2015) Interferons and the immunogenic effects of cancer therapy. Trends Immunol. 36, 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu C., Xue Y., Wang P., Lin L., Liu Q., Li N., Xu J., and Cao X. (2014) IFN-γ primes macrophage activation by increasing phosphatase and tensin homolog via downregulation of miR-3473b. J. Immunol. 193, 3036–3044 [DOI] [PubMed] [Google Scholar]
- 71. Ho J., Pelzel C., Begitt A., Mee M., Elsheikha H. M., Scott D. J., and Vinkemeier U. (2016) STAT2 is a pervasive cytokine regulator due to its inhibition of STAT1 in multiple signaling pathways. PLoS Biol. 14, e2000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goropevšek A., Holcar M., and Avčin T. (2017) The role of STAT signaling pathways in the pathogenesis of systemic lupus erythematosus. Clin. Rev. Allergy Immunol. 52, 164–181 [DOI] [PubMed] [Google Scholar]
- 73. Bettelli E., Sullivan B., Szabo S. J., Sobel R. A., Glimcher L. H., and Kuchroo V. K. (2004) Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J. Exp. Med. 200, 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Willenborg D. O., Fordham S. A., Staykova M. A., Ramshaw I. A., and Cowden W. B. (1999) IFN-γ is critical to the control of murine autoimmune encephalomyelitis and regulates both in the periphery and in the target tissue: a possible role for nitric oxide. J. Immunol. 163, 5278–5286 [PubMed] [Google Scholar]
- 75. Bäcklund J., Li C., Jansson E., Carlsen S., Merky P., Nandakumar K.-S., Haag S., Ytterberg J., Zubarev R. A., and Holmdahl R. (2013) C57BL/6 mice need MHC class II Aq to develop collagen-induced arthritis dependent on autoreactive T cells. Ann. Rheum. Dis. 72, 1225–1232 [DOI] [PubMed] [Google Scholar]
- 76. Wildbaum G., Zohar Y., and Karin N. (2010) Antigen-specific CD25-Foxp3-IFN-γhighCD4+ T cells restrain the development of experimental allergic encephalomyelitis by suppressing Th17. Am. J. Pathol. 176, 2764–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schubert D., Maier B., Morawietz L., Krenn V., and Kamradt T. (2004) Immunization with glucose-6-phosphate isomerase induces T cell-dependent peripheral polyarthritis in genetically unaltered mice. J. Immunol. 172, 4503–4509 [DOI] [PubMed] [Google Scholar]
- 78. Lee S., and Margolin K. (2011) Cytokines in cancer immunotherapy. Cancers 3, 3856–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Adorini L. (2003) Cytokine-based immunointervention in the treatment of autoimmune diseases. Clin. Exp. Immunol. 132, 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hodge D. L., Berthet C., Coppola V., Kastenmüller W., Buschman M. D., Schaughency P. M., Shirota H., Scarzello A. J., Subleski J. J., Anver M. R., Ortaldo J. R., Lin F., Reynolds D. A., Sanford M. E., Kaldis P., et al. (2014) IFN-γ AU-rich element removal promotes chronic IFN-γ expression and autoimmunity in mice. J. Autoimmun. 53, 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bae H. R., Leung P. S., Tsuneyama K., Valencia J. C., Hodge D. L., Kim S., Back T., Karwan M., Merchant A. S., Baba N., Feng D., Park O., Gao B., Yang G. X., Gershwin M. E., and Young H. A. (2016) Chronic expression of interferon γ leads to murine autoimmune cholangitis with a female predominance. Hepatology 64, 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lee S. K., Silva D. G., Martin J. L., Pratama A., Hu X., Chang P.-P., Walters G., and Vinuesa C. G. (2012) Interferon-γ excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity 37, 880–892 [DOI] [PubMed] [Google Scholar]
- 83. Talmadge J. E., Tribble H. R., Pennington R. W., Phillips H., and Wiltrout R. H. (1987) Immunomodulatory and immunotherapeutic properties of recombinant γ-interferon and recombinant tumor necrosis factor in mice. Cancer Res. 47, 2563–2570 [PubMed] [Google Scholar]
- 84. Reynolds A. R. (2010) Potential relevance of bell-shaped and u-shaped dose-responses for the therapeutic targeting of angiogenesis in cancer. Dose Response 8, 253–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ravetch J. V., and Lanier L. L. (2000) Immune inhibitory receptors. Science 290, 84–89 [DOI] [PubMed] [Google Scholar]
- 86. Nishimura H., and Honjo T. (2001) PD-1: an inhibitory immunoreceptor involved in peripheral tolerance. Trends Immunol. 22, 265–268 [DOI] [PubMed] [Google Scholar]
- 87. Romo-Tena J., Gómez-Martín D., and Alcocer-Varela J. (2013) CTLA-4 and autoimmunity: new insights into the dual regulator of tolerance. Autoimmun. Rev. 12, 1171–1176 [DOI] [PubMed] [Google Scholar]
- 88. Liu J., Blake S. J., Smyth M. J., and Teng M. W. (2014) Improved mouse models to assess tumour immunity and irAEs after combination cancer immunotherapies. Clin. Transl. Immunol. 3, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]