Abstract
Several mitochondrial tRNA mutations have been associated with hypertension, but their pathophysiology remains poorly understood. In this report, we identified a novel homoplasmic 3253T→C mutation in the mitochondrial tRNALeu(UUR) gene in a Han Chinese family with maternally inherited hypertension. The m.3253T→C mutation affected a highly conserved uridine at position 22 at the D-stem of tRNALeu(UUR), introducing a G-C base pairing (G13-C22) at the D-stem and a tertiary base pairing (C22-G46) between the D-stem and the variable loop. We therefore hypothesized that the m.3253T→C mutation altered both the structure and function of tRNALeu(UUR). Using cytoplasmic hybrid (cybrid) cell lines derived from this Chinese family, we demonstrated that the m.3253T→C mutation perturbed the conformation and stability of tRNALeu(UUR), as suggested by faster electrophoretic mobility of mutated tRNA relative to the wild-type molecule. Northern blot analysis revealed an ∼45% decrease in the steady-state level of tRNALeu(UUR) in the mutant cell lines carrying the m.3253T→C mutation, as compared with control cell lines. Moreover, an ∼35% reduction in aminoacylation efficiency of tRNALeu(UUR) was observed in the m.3253T→C mutant cells. These alterations in tRNALeu(UUR) metabolism impaired mitochondrial translation, especially for those polypeptides with a high proportion of Leu(UUR) codons, such as ND6. Furthermore, we demonstrated that the m.3253T→C mutation decreased the activities of mitochondrial complexes I and V, markedly diminished mitochondrial ATP levels and membrane potential, and increased the production of reactive oxygen species in the cells. In conclusion, our findings may provide new insights into the pathophysiology of maternally inherited hypertension.
Keywords: hypertension, mitochondrial disease, mitochondrial DNA (mtDNA), mitochondrial membrane potential, mitochondrial metabolism, mitochondrial respiratory chain complex, mutant, pathogenesis, RNA structure, transfer RNA (tRNA)
Introduction
Hypertension is a major public health problem, affecting ∼1 billion people worldwide and 265 million adults in China (1, 2). Hypertension is a major risk factor for coronary heart disease, stroke, congestive heart failure, and renal disease (3). The etiology of hypertension is not well understood because of multifactorial causes, including environment and inherited risk factors (4). Mitochondria can regulate various aspects of vascular function, thereby being critical for pathogenesis of hypertension (5–7). Maternal transmissions of hypertension have been implicated in some pedigrees, suggesting that the mutation(s) in mitochondrial DNA (mtDNA)3 is involved in the pathogenesis of hypertension (7–11). Human mtDNA encodes 13 subunits of the oxidative phosphorylation system, two rRNAs, and 22 tRNAs required for translation (12). Unlike canonical tRNAs, such as human cytosolic tRNAs, human mitochondrial tRNAs have specific features such as non-classical G-U pairs and mismatches (13, 14). There are three types of unusual secondary structures in the mitochondrial tRNAs (13–15). The tRNASer(UCN) has a noncanonical cloverleaf structure with only one base (A9) between the acceptor stem and D stem, a short D loop, and an extended anticodon stem with 6 bp. The tRNASer(AGY) lacks the entire D loop. The other tRNAs do not have the canonical D-loop/T-loop interaction (15, 16). Mitochondrial tRNAs are the hot spots for mutations associated with hypertension (17–22). These hypertension-associated tRNA mutations included the tRNAIle 4263A→G and 4295A→G mutations, tRNAMet 4435A→G mutation, tRNAAla 5655A→G mutation, and m.4401A→G mutation in the junction of tRNAMet and tRNAGln genes (22–27). These mutations have structural and functional consequences, including the processing of RNA precursors, nucleotide modification, and aminoacylation (24, 26–30). However, the pathophysiology underlying these tRNA mutations remains poorly understood. Thus, it is necessary to establish the link between hypertension and mitochondrial dysfunction caused by mitochondrial tRNA mutations and their cause/effect relation.
As part of a genetic screening program for hypertension in a cohort of 2,070 Han Chinese hypertensive subjects, we identified the T to C transition at position 3253 (m.3253T→C) of the tRNALeu(UUR) gene in one proband whose family exhibited a maternal transmission of hypertension (22). As shown in Fig. 1A, the m.3253T→C mutation was localized at high conserved uridine (U22) of the D stem of tRNALeu(UUR). It was anticipated that the m.3253T→C mutation introduced the G-C base pairing (G13-C22) at the D-stem and also formed a tertiary base pairing (C22-G46) with the G46 in the variable loop of tRNALeu(UUR) (30). Thus, the m.3253T→C mutation may alter the structure and function of tRNALeu(UUR), thereby causing mitochondrial dysfunction necessary for hypertension. In particular, the mutation may affect the stability and aminoacylation capacity of this tRNA. The functional significance of this tRNA mutation was investigated through cybrid cell lines generated by transferring mitochondria from lymphoblastoid cell lines derived from the Chinese family into mtDNA-less (ρ°) cells (31). These cell lines were assayed for the effect of the m.3253T→C mutation on the stability and aminoacylation capacity of tRNAs, mitochondrial translation, enzymatic activities of electron transport chain complexes, rate of O2 consumption, production of ATP, mitochondrial membrane potential, and reactive oxygen species (ROS).
Figure 1.
The analysis of the conformation and stability of tRNALeu(UUR). A, cloverleaf structure of human mitochondrial tRNALeu(UUR). Broken lines, anticipated tertiary base pairings, including G13-U22 and U22-G46. B, schematic model for the tertiary structure of tRNAGlu (Protein Data Bank code 2DER). Broken square, D-stem and variable loop region of tRNA. C, Northern blot analysis of tRNA under native conditions. Two μg of total mitochondrial RNA from various cell lines were electrophoresed through native polyacrylamide gel, electroblotted, and hybridized with DIG-labeled oligonucleotide probes specific for tRNALeu(UUR), tRNALys, tRNAMet, and tRNAIle, respectively. D, thermal stability of wild-type and mutant tRNALeu(UUR). ΔTm indicates the difference of Tm value between wild-type and mutant tRNALeu(UUR). The calculations were based on three independent experiments.
Results
Identification of the tRNALeu(UUR) 3253T→C mutation
The tRNALeu(UUR) 3253T→C mutation was identified in one proband (FY916 II-1) among 2,070 Chinese hypertensive but absent in 512 Chinese control subjects (22). As shown in Fig. 1A, the m.3253T→C mutation was localized at a highly conserved uridine (U22) of the D-stem of tRNALeu(UUR). As shown in Fig. 1B, it was anticipated that the m.3253T→C mutation produced the G-C base pairing (G13-C22) at the D-stem and formed a tertiary base pair (C22-G46) with the G46 in the variable loop of tRNALeu(UUR) (29, 30). Thus, it was anticipated that the U to C substitution at position 22 by the m.3253T→C mutation would alter the structure and function of tRNALeu(UUR). The Sanger sequence analysis of the entire mtDNA in the proband (II-1) and other four matrilineal relatives exhibited the identical m.3253T→C mutation and distinct sets of polymorphisms belonging to the Eastern Asian haplogroup D4 (supplemental Table S1). However, there were no other functionally significant variants in the mtDNAs. Further restriction fragment length polymorphism analysis showed that the m.3253T→C mutation was present in homoplasmy in all matrilineal relatives but absent in other members of the Chinese family (supplemental Fig. S1).
Clinical evaluation of the Chinese family
Members of the Chinese family carrying the m.3253T→C mutation underwent a physical examination, laboratory assessment of cardiovascular disease risk factors, and routine electrocardiography. Of five matrilineal relatives of pedigree FY916, as shown in supplemental Fig. S2, four (three males and one female) individuals suffered from hypertension, whereas none of the other nonmaternal relatives suffered from hypertension. None of the offspring of the three affected fathers exhibited hypertension. The clinical data are summarized in supplemental Table S2. The age at onset of hypertension ranged from 28 to 53 years, with an average of 40 years old. There was no evidence that any member of the family had any other cause to account for hypertension. However, none of the other clinical abnormalities were observed in the maternal kindred. These data indicated the maternal inheritance of hypertension in this family.
Altered conformation and stability of tRNALeu(UUR)
It was anticipated that the G13-C22 base pairing at the D-stem and the base pairing (C22-G46) by the m.3253T→C mutation led to structural alterations of tRNALeu(UUR). To test whether the m.3253T→C mutation affected the conformation of tRNALeu(UUR), total mitochondrial RNA were electrophoresed through a 10% polyacrylamide gel (native condition) in Tris borate-EDTA buffer and then electroblotted onto a positively charged nylon membrane for hybridization analysis with digoxigenin (DIG)-labeled oligodeoxynucleotide probes for tRNALeu(UUR), tRNALys, tRNAMet, and tRNAIle, respectively. As shown in Fig. 1C, electrophoretic patterns showed that the tRNALeu(UUR) in three mutant cybrid cell lines carrying the m.3253T→C mutation migrated much faster than those of three control cybrid cell lines lacking this mutation. These data indicated that the m.3253T→C mutation changed the conformation of tRNALeu(UUR).
The stability of the transcripts of wild-type and mutant tRNALeu(UUR) were examined by the measurement of the melting temperatures (Tm) by calculating the derivatives of the absorbance against a temperature curve. As shown in Fig. 1D, the Tm values for wild-type (U22) and mutant (C22) tRNALeu(UUR) transcripts were 40.07 and 41.02 °C, respectively. This suggested that the global folding of mutant tRNALeu(UUR) was more stable than that of wild-type tRNALeu(UUR). These data further suggested that the m.3253T→C mutation affected the stability of tRNALeu(UUR).
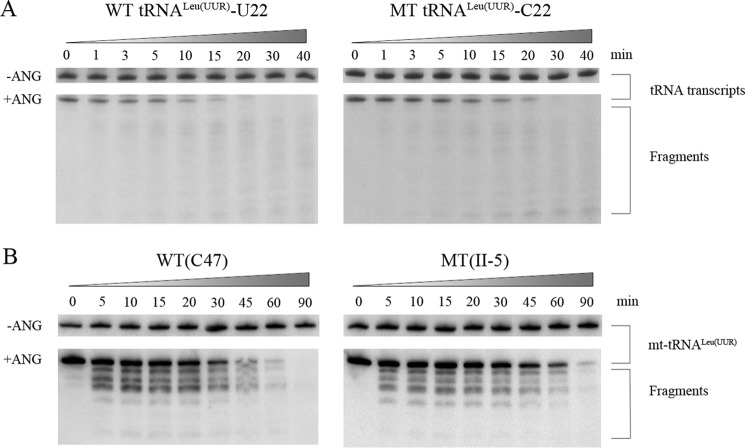
We further evaluated whether the m.3253T→C mutation perturbs the tertiary structure of tRNALeu(UUR) by analyzing the sensitivity of wild-type and mutant tRNALeu(UUR) to digestion with angiogenin (ANG) (32). ANG is a ribonuclease that cleaves tRNA in or near the anticodon loop under stress conditions (32, 33). The wild-type (U22) and mutant (C22) tRNALeu(UUR) obtained from in vitro transcription and from control and mutant cell lines were digested by angiogenin and followed by Northern blot analysis. As shown in Fig. 2, the wild-type (U22) tRNALeu(UUR) transcript and tRNALeu(UUR) obtained from the wild-type cell lines (C47) were more sensitive to angiogenin-mediated digestion than those in the mutant (C22) tRNALeu(UUR) transcript and mutant tRNALeu(UUR) obtained from the mutant cell lines (II-5), respectively.
Figure 2.
In vitro angiogenin cleavage assays. A, angiogenin digestion pattern of in vitro transcripts of WT (U22) and mutant (MT) (C22) tRNALeu(UUR). Two μg of purified tRNA transcripts were used for the ANG cleavage reaction over time (from 0 to 40 min). Cleavage products of tRNA transcripts were electrophoresed through a denaturing polyacrylamide gel and stained with methylene blue. B, angiogenin digestion pattern of tRNALeu(UUR) purified from control (C47) and mutant (II-5) lymphoblastoid cell lines. Two μg of RNAs were used for the ANG cleavage reaction at various lengths (from 0 to 90 min). Cleavage products of tRNAs were resolved in 15% denaturating polyacrylamide gels with 8 m urea, electroblotted, and hybridized with DIG-labeled oligonucleotide probe specific for the tRNALeu(UUR).
Marked decrease in the steady-state levels of tRNALeu(UUR)
To assess whether the m.3253T→C mutation affects the metabolism of tRNALeu(UUR), we subjected mitochondrial RNAs from mutant and control cybrid cell lines to Northern blots and hybridized them with DIG-labeled oligodeoxynucleotide probes for tRNALeu(UUR), tRNAGln, tRNAIle, tRNATrp, and tRNAVal, respectively. As shown in Fig. 3A, the steady-state level of tRNALeu(UUR) in the mutant cells was significantly decreased, as compared with those of control cell lines. For comparison, the average levels of tRNALeu(UUR) in the mutant cybrid cell lines were ∼58, 59, 49, and 57% of average values of three control cybrids after normalization to tRNAGln, tRNAIle, tRNATrp, and tRNAVal (p = 0.0007–0.0062), respectively.
Figure 3.
Northern blot analysis of tRNA under a denaturing condition. A, 2 μg of total mitochondrial RNA from various cell lines were electrophoresed through a denaturing polyacrylamide gel, electroblotted, and hybridized with DIG-labeled oligonucleotide probes specific for tRNALeu(UUR), tRNAGln, tRNAIle, tRNATrp, and tRNAVal, respectively. B, quantification of tRNA levels. Shown is the average relative tRNALeu(UUR) content per cell, normalized to the average content per cell of tRNAGln, tRNAIle, tRNATrp, and tRNAVal in three cybrid cell lines derived from one affected subject (II-5) and three cybrid cell lines derived from one Chinese control subject (C47). The values for the latter are expressed as percentages of the average values for the control cell lines. The calculations were based on three independent experiments. The error bars indicate two S.E. values. P, significance, according to the t test, of the differences between mutant and control cell lines.
Deficient aminoacylation of tRNALeu(UUR)
To evaluate whether the m.3253T→C mutation affects the aminoacylation of mitochondrial tRNAs, we examined the aminoacylation capacities of tRNALeu(UUR), tRNALeu(CUN), tRNAThr, tRNAIle, and tRNALys in control and mutant cell lines by the use of electrophoresis in an acidic polyacrylamide/urea gel system. As shown in Fig. 4A, the top band represented the charged tRNA, whereas the bottom band represented the uncharged tRNA. The electrophoretic mobility of either charged or uncharged tRNALeu(UUR) in cell lines carrying the m.3253T→C mutation migrated faster than those of control cell lines. To further distinguish nonaminoacylated tRNA from aminoacylated tRNA, samples of tRNAs were deacylated by being heated for 10 min at 60 °C at pH 8.3 and then run in parallel. As shown in Fig. 4B, only one band (uncharged tRNA) was present in both mutant and control cell lines after deacylation. However, there were no obvious differences in electrophoretic mobility of tRNALeu(CUN), tRNAThr, tRNAIle, and tRNALys between the control and mutant cell lines. Notably, the efficiency of aminoacylated tRNALeu(UUR) in the mutant cell lines was 68.7% relative to the average values of control cell lines (p < 0.001), whereas the levels of aminoacylation in tRNALeu(CUN), tRNAThr, tRNAIle, and tRNALys in mutant cell lines were comparable with those in the control cell lines (Fig. 4C).
Figure 4.
In vivo aminoacylation assays. A, 2 μg of total mitochondrial RNA purified from six cell lines under acid conditions were electrotrophoresed at 4 °C through an acid (pH 4.5) 10% polyacrylamide, 8 m urea gel, electroblotted, and hybridized with DIG-labeled oligonucleotide probes specific for tRNALeu(UUR), tRNAIle, tRNAThr, and tRNALeu(CUN), respectively. B, the samples from one control (C47–1) and one mutant (II-5–10) cell lines were deacylated (DA) by heating for 10 min at 60 °C at pH 8.3, electrophoresed, and hybridized with DIG-labeled oligonucleotide probes specific for the tRNALeu(UUR) and tRNALeu(CUN). C, quantification of aminoacylated proportions of tRNALeu(UUR) in the mutant and controls. The calculations were based on three independent experiments. Graph details and symbols are explained in the legend and key to Fig. 3.
Decreases in the levels of mitochondrial proteins
To examine whether the m.3253T→C mutation alters mitochondrial translation, we used Western blot analysis to examine the steady-state levels of eight respiratory complex subunits (encoded by mtDNA) in mutant and control cells with Tom20 (mitochondrial protein encoded by the nuclear gene) as a loading control. As shown in Fig. 5A, the levels of ND1, ND4, ND5, and ND6 (subunits 1, 4, 5, and 6 of NADH dehydrogenase) and ATP6 and ATP8 (subunits 6 and 8 of H+-ATPase) exhibited variable reductions in the mutant cell lines, whereas the levels of CYTB (apocytochrome b) and CO2 (subunit II of cytochrome c oxidase) in the mutant cell lines showed results similar to that for control cells. As shown in Fig. 5B, the overall levels of eight mitochondrial translation products in the mutant cell lines ranged from ∼78 to 87%, with an average of 83% (p = 0.001), relative to the mean value measured in the control cell lines. As shown in Fig. 5 and supplemental Table S3, mutant cell lines exhibited a marked reduction (50.3%) in the level of ND6 harboring with a higher proportion or number of leucine (UUR) codons and relatively mild reductions (6.8–44.5%) in the levels of ND1, ND4, ND5, ATP6, and ATP8, respectively. By contrast, there were similar levels of CYTB and CO2 between the mutant and control cell lines. However, the levels of polypeptide synthesis in mutant cells, relative to those in control cells, showed no significant correlation with either the number of codons or the proportion of leucine UUR (supplemental Table S3).
Figure 5.
Western blot analysis of mitochondrial proteins. A, 20 μg of total cellular proteins from various cell lines were electrophoresed through a denaturing polyacrylamide gel, electroblotted and hybridized with eight polypeptides (mtDNA-encoded subunits of respiratory complexes) and with Tom20 as a loading control. B, quantification of total mitochondrial protein levels. The levels of mitochondrial proteins in three mutant cell lines and three control cell lines were determined as described elsewhere (24, 40). C, quantification of eight polypeptides. The levels of ND1, ND4, ND5, ND6, CO2, CYTB, ATP6, and ATP8 in three mutant cell lines and three control cell lines were determined as described elsewhere (24, 40). Graph details and symbols are explained in the legend to Fig. 3.
Reduced activities of complex I and V
To investigate the effect of the m.3253T→C mutation on the oxidative phosphorylation, we measured the activities of respiratory complexes by isolating mitochondria from three mutant and three control cell lines. Complex I (NADH ubiquinone oxidoreductase) activity was determined by following the oxidation of NADH with ubiquinone as the electron acceptor (34–36). The activity of complex II (succinate ubiquinone oxidoreductase) exclusively encoded by the nuclear DNA was examined by the artificial electron acceptor DCPIP (37, 38). Complex III (ubiquinone cytochrome c oxidoreductase) activity was measured as the reduction of cytochrome c (III) using d-ubiquinol-2 as the electron donor. Complex IV (cytochrome c oxidase) activity was monitored by following the oxidation of cytochrome c (II). The activity of complex V (F1-ATP synthase) was monitored by following the NADH oxidation via conversion of phosphoenolpyruvate to pyruvate and then pyruvate to lactate by lactate dehydrogenase (38). As shown in Fig. 6, the activities of complexes I, II, III, IV, and V in the mutant cells carrying the m.3253T→C mutation were 45.2, 100.4, 103.0, 98.9, and 54.9% of the mean value measured in two control cell lines (p = 0.001, 0.853, 0.115, 0.183, and 0.001), respectively.
Figure 6.
Enzymatic activities of respiratory chain complexes. The activities of respiratory complexes were investigated by an enzymatic assay on complexes I, II, III, IV, and V in mitochondria isolated from various cell lines. The calculations were based on five independent experiments. Graph details and symbols are explained in the legend to Fig. 3.
Respiration deficiency
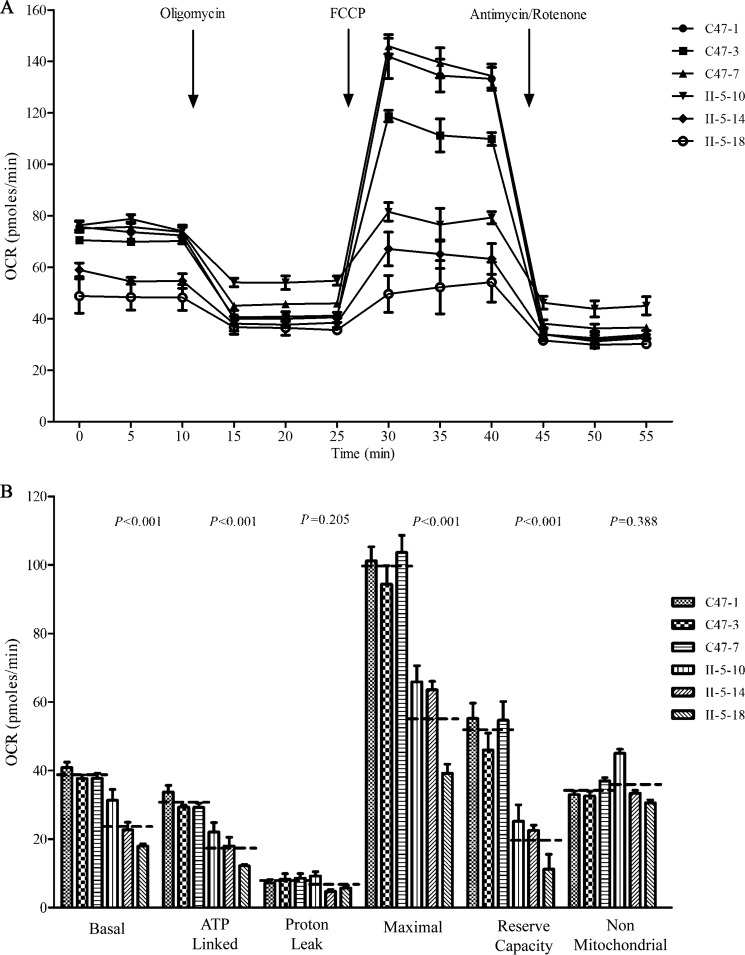
To evaluate whether the m.3253T→C mutation alters cellular bioenergetics, we examined the oxygen consumption rates (OCRs) of three mutant cell lines carrying the m.3253T→C mutation and three control cell lines. As shown in Fig. 7, the basal OCR in mutant cell lines was ∼61.8% (p < 0.001) relative to the mean value measured in the control cell lines. To investigate which of the enzyme complexes of the respiratory chain was affected in the mutant cell lines, oligomycin (to inhibit the ATP synthase), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (to uncouple the mitochondrial inner membrane and allow for maximum electron flux through the ETC), rotenone (to inhibit complex I), and antimycin A (to inhibit complex III) were added sequentially while measuring OCR. The difference between the basal OCR and the drug-insensitive OCR yields the amount of ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity, and non-mitochondrial OCR. As shown in Fig. 7, the ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity, and non-mitochondrial OCR in mutant cell lines were ∼56.6% (p < 0.001), 81.2% (p = 0.205), 56.3% (p < 0.001), 37.8% (p < 0.001), and 106.2% (p = 0.388), relative to the mean value measured in the control cell lines, respectively.
Figure 7.
Respiration assays. A, analysis of O2 consumption in the various cell lines using different inhibitors. The OCRs were first measured on 2 × 104 cells of each cell line under basal conditions and then with sequential additions of oligomycin (1.5 m), FCCP (0.5 m), rotenone (1.0 m), and antimycin A (1.0 m) at the indicated times to determine different parameters of mitochondrial functions. B, graphs present the ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity, and non-mitochondrial OCR in mutant and control cell lines. Non-mitochondrial OCR was determined as the OCR after rotenone/antimycin A treatment. Basal OCR was determined as OCR before oligomycin minus OCR after rotenone/antimycin A. ATP-lined OCR was determined as OCR before oligomycin minus OCR after oligomycin. Proton leak was determined as basal OCR minus ATP-linked OCR. Maximal was determined as the OCR after FCCP minus non-mitochondrial OCR. Reserve capacity was defined as the difference between maximal OCR after FCCP minus basal OCR. The average values of four independent experiments for each cell line are shown; the horizontal dashed lines represent the average value for each group. Graph details and symbols are explained in the legend to Fig. 3.
Reduced level in mitochondrial ATP production
The luciferin/luciferase assay was used to examine the capacity of oxidative phosphorylation in mutant and wild-type cell lines. Populations of cells were incubated in the media in the presence of glucose and in 2-deoxy-d-glucose with pyruvate (39, 40). In the presence of glucose, the levels of ATP production in mutant cells were comparable with those of control cells. In the presence of 2-deoxy-d-glucose with pyruvate, which could inhibit the glycolysis, the levels of ATP production in mutant cells ranged from 45 to 76%, with an average of 66% (p < 0.001) relative to the mean value measured in the control cells (Fig. 8).
Figure 8.

Measurement of cellular and mitochondrial ATP levels using bioluminescence assay. Cells were incubated with 10 mm glucose or 5 mm 2-deoxy-d-glucose plus 5 mm pyruvate to determine ATP generation under mitochondrial ATP synthesis. Average rates of ATP level per cell line are shown: ATP level in total cells (A) and ATP level in mitochondria (B). Four independent experiments were made for each cell line. Graph details and symbols are explained in the legend to Fig. 3.
Decrease in mitochondrial membrane potential
To examine whether the m.3253T→C mutation affects mitochondrial membrane potential (ΔΨm), a fluorescence probe JC-10 assay system was used to measure the ΔΨm in three mutant and three control cell lines. The ratio of fluorescence intensities excitation/emission = 490/590 and 490/530 nm (FL590/FL530) were recorded to delineate the ΔΨm of each sample. The relative ratios of the FL590/FL530 geometric mean between mutant and control cell lines were calculated to represent the level of ΔΨm, as described elsewhere (40). As shown in Fig. 9, the ΔΨm in the mutant cell lines carrying the m.3253T→C mutation ranged from 67 to 76%, with an average of 73% (p = 0.010) of the mean value measured in the control cell lines. In contrast, while in the presence of FCCP, the levels of ΔΨm in mutant cell lines were comparable with those of control cell lines (p = 0.886).
Figure 9.

Mitochondrial membrane potential analysis. ΔΨm was measured in two mutant and two control cell lines using a fluorescence probe JC-10 assay system. The ratio of fluorescence intensities excitation/emission = 490/590 and 490/530 nm (FL590/FL530) were recorded to delineate the ΔΨm level of each sample. The relative ratios of the FL590/FL530 geometric mean between mutant and control cell lines were calculated to reflect the level of ΔΨm. Shown are the relative ratio of JC-10 fluorescence intensities at excitation/emission = 490/530 and 490/590 nm in the absence (A) and presence (B) of 10 μm FCCP. The calculations were based on four independent experiments. Graph details and symbols are explained in the legend to Fig. 3.
Increase of ROS production
Flow cytometry was used to measure the levels of the ROS generation in the vital cells that were derived from three mutant cybrid cell lines carrying the m.3253T→C mutation and three control cybrid cell lines lacking the mutation. The assays were taken under normal conditions and then following H2O2 stimulation (40, 41). Geometric mean intensity was recorded to measure the production rate of ROS of each sample. And the ratio of geometric mean intensity between samples that were unstimulated or stimulated with H2O2 was calculated to delineate the reaction upon increasing levels of ROS under oxidative stress (40). As shown in Fig. 10, the levels of ROS generation in the mutant cell lines carrying the m.3253T→C mutation ranged from 119 to 146%, with an average of 13% (p < 0.001) of the mean value measured in the control cell lines.
Figure 10.

Measurement of ROS. The rates of production in ROS from two affected matrilineal relatives and two control individuals were analyzed by the BD-LSR II flow cytometer system with or without H2O2 stimulation. The relative ratio of intensity (stimulated versus unstimulated with H2O2) was calculated. The calculations were based on three independent experiments. Graph details and symbols are explained in the legend to Fig. 3.
Discussion
In the present study, we investigated the pathogenic mechanism of hypertension-associated m.3253T→C mutation in the tRNALeu(UUR) gene. The m.3253T→C mutation affected a highly conserved nucleotide (U22) of the D-stem in the tRNALeu(UUR), which is important for the tertiary structure and interaction with the components of translational machinery (15, 30). In fact, the base triplet of the compact core of tRNAs is normally made via the interaction R46·N22-N13 (where N represents any type of nucleotide, and R is a purine), whereas the R46·N22 contact is missing in class II tRNAs, such as tRNALeu, with a large variable region (30). The R46·N22-N13 interaction is critical for the intrinsic structure and the stability of the L-shaped tertiary structure of tRNAs as well as for the recognition by its cognate aminoacyl-tRNA synthetase (14, 30). However, human mitochondrial tRNALeu(UUR) exhibited an atypical structure where the nucleotide at position 22 (U) did not form the WC interaction with G13 in the D-stem and exhibited a relatively short variable region (13, 30). In the present study, the U to C substitution by the m.3253T→C mutation produced the G-C base pairing (G13-C22) at the D-stem. This mutation may also introduce the tertiary base pair (C22-G46) with the G46 in the variable loop of tRNALeu(UUR) and then cause the rearrangement of the tertiary interaction networks. As a result, the m.3253T→C mutation may correct the perturbed cloverleaf folds, restrict the accessible conformation space, and alter the function of tRNA. In particular, the altered tertiary structure caused by the m.3253T→C mutation affected the aminoacylation and stability of tRNALeu(UUR), as in the case of m.12311T→C mutation of tRNALeu(CUN) (42). Here, the m.3253T→C mutation changed the conformation of tRNALeu(UUR), as suggested by faster electrophoretic mobility of mutated tRNA with respect to the wild-type molecule, in contrast with the slower electrophoretic mobility of mutated tRNAs carrying tRNAThr 15927G→A and tRNAHis 12201T→C mutations (21, 40). In addition, the increased melting temperature was observed in mutant tRNALeu(UUR) (C22), as compared with its wild-type counterpart (U22). Furthermore, the fact that the mutant (C22) tRNALeu(UUR) was more resistant to angiogenin-mediated digestion than that in the wild type (U22) tRNALeu(UUR) further suggested that the m.3253T→C mutation altered the tertiary structure and stability of tRNALeu(UUR). In particular, the altered tertiary structure contributed to the decrease in the steady-state level of tRNALeu(UUR). Notably, the tRNALeu(UUR) carrying the m.3253T→C mutation was charged to a lesser extent by mitochondrial leucyl-tRNA synthetase, thereby altering aminoacylation. Here, 31% reduction in aminoacylated efficiency of tRNALeu(UUR) in mutant cells derived from this Chinese family was apparently responsible for a failure in tRNA metabolism, including the reduced level of tRNALeu(UUR). However, the reduced level of tRNALeu(UUR) in mutant cells carrying the m.3253T→C mutation was above the proposed threshold level (→70%) to produce a clinical phenotype associated with a mitochondrial tRNA mutation (24, 27, 43).
The failure in tRNA metabolism, including inefficient aminoacylation or shortage of tRNALeu(UUR), was responsible for the impairment of mitochondrial protein synthesis. Alternatively, the mutant tRNALeu(UUR) may faultily interact with the translational machinery, thereby altering mitochondrial protein synthesis (44, 45). In the present study, the reduced levels of mitochondrial proteins (an average decrease of ∼17%) were comparable with those in cell lines carrying the tRNAGlu 14692A→G and tRNAAsp 7551A→G mutations (46, 47). The variable decreases in the levels of ND1, ND4, ND5, ND6, ATP6, and ATP8 were observed in the mutant cell lines, whereas the levels of CYTB and CO2 in the mutant cell lines were comparable with those in control cell lines. Strikingly, mutant cell lines carrying the m.3253T→C mutation exhibited marked reductions (50.3 and 44.5%) in the levels of ND6 and ATP6, respectively. The impaired synthesis of ND1, ND4, ND5, and ND6 (subunits of complex I) and ATP6 and ATP8 (subunits of complex V) were specifically responsible for the reduced activities of complex I and complex V. These data were in contrast with the reduced activities of complexes I and IV in the mutant cell lines carrying the m.5655A→G and m.14692A→G mutations (24, 46). Furthermore, the impairment of mitochondrial translation led to the reduced rates in the basal OCR, ATP-linked OCR, maximal OCR, and reserve capacity in the mutant cell lines. The respiratory deficiency caused by the m.3253T→C mutation may increase the uncoupling of the oxidative pathway for ATP synthesis, oxidative stress, and subsequent failure of cellular energetic process (48). In the present study, a 34% drop in mitochondrial ATP production in mutant cybrids carrying the m.3253T→C mutation may be caused by the defective activities of complexes I and V. The reducing levels were comparable with those in cells carrying the deafness-associated m.12201T→C, m.14692A→G, and m.7551A→G mutations (40, 46, 47) but much lower than those in cell lines carrying the m.8344A→G and m.3243A→G mutations (49, 50). Furthermore, the deficient activities of respiratory chain complexes caused by tRNA mutations often alter mitochondrial membrane potentials (40, 51). In this investigation, the reductions in mitochondrial membrane potential in cybrid cell lines carrying the m.3253T→C mutation indicated the impaired pumping ability of hydrogen ions across the inner membrane and more electron leakage from the electron transport chain (24). The abnormal oxidative phosphorylation and mitochondrial membrane potential resulted in overproduction of reactive oxygen species and the subsequent failure of cellular energetic processes in cybrid cell lines carrying the m.3253T→C mutation. In turn, the increased production of ROS may produce damage of mitochondrial proteins, nucleic acids, and lipids, stimulating a forward-feeding loop of mitochondrial ROS generation and aggravated cell damages (52, 53). The skeletal and vascular smooth muscles may be preferentially involved because they were somehow exquisitely sensitive to inefficient metabolism, subtle imbalance in cellular redox state, and increased level of free radicals (54). The mild mitochondrial dysfunction observed in the cell lines carrying the m.3253T→C mutation suggested that this mutation is an inherited risk factor necessary for the development of hypertension but may by itself be insufficient to produce a clinical phenotype. The nuclear genetic or epigenetic factors may contribute to the development of clinical phenotype in subjects carrying the m.3253T→C mutation (55, 56). In particular, the tissue-specific effect of this tRNA mutation may be attributed to the tissue-specific RNA metabolism or the involvement of nuclear modifier genes (57, 58).
In summary, our findings suggest the pathogenic mechanism leading to an impaired oxidative phosphorylation in cybrid cell lines carrying the hypertension-associated tRNALeu(UUR) 3253T→C mutation. The m.3253T→C mutation altered the tertiary structure and function of this tRNA. A failure in tRNA metabolism impaired mitochondrial translation and respiration. As a result, this respiratory deficiency reduced mitochondrial ATP production and the increasing production of oxidative reactive species. An inefficient metabolism caused by the mitochondrial dysfunction in skeletal and vascular smooth muscles may lead to the elevation of systolic blood pressure. Thus, our findings may provide new insights into the pathophysiology of maternally inherited hypertension.
Experimental procedures
Subjects
A Chinese family with hypertension was recruited from the Hypertension Clinic, Wenzhou Medical University, China, as detailed previously (22). This study was in compliance with the Declaration of Helsinki. Informed consent, blood samples, and clinical evaluations were obtained from all participating family members under protocols approved by the ethics committees of Zhejiang University and the Wenzhou Medical University. Diagnosis of hypertension was based on the criteria of the American Heart Association (59). All available members of this family were evaluated at length to identify both personal and family medical histories of hypertension and other clinical abnormalities, as detailed elsewhere (22, 27). The 512 Chinese control subjects were obtained from a panel of unaffected subjects of Han Chinese ancestry from the same region.
Mutational analysis of mitochondrial DNAs
Genomic DNA was isolated from whole blood of participants using a QIAamp DNA Blood Mini Kit (Qiagen, catalog no. 51104). The subject's DNA fragments spanning the mitochondrial tRNALeu(UUR) gene were PCR-amplified by the use of oligodeoxynucleotides corresponding to mtDNA at positions 3150–3980 (12, 60). Each fragment was purified and subsequently analyzed by direct sequencing. These sequence results were compared with the updated consensus Cambridge sequence (GenBankTM accession number NC_012920) (12). The entire mtDNAs of five matrilineal relatives (II-1, II-4, II-5, III-3, and IV-2) of this Chinese family and a Chinese control subject (C47) were PCR-amplified in 24 overlapping fragments using sets of oligonucleotide primers, as described previously (61). These sequence results were compared with the updated consensus Cambridge sequence, as described above.
To quantify the m.3253T→C mutation, the PCR segment (795 bp) was amplified using genomic DNA as the template and oligodeoxynucleotides corresponding to mtDNA at positions 2718–3513 and subsequently digested with restriction enzyme BsteEII as the m.3253T→C mutation created the site for this enzyme. Equal amounts of various digested samples were then analyzed by electrophoresis through 3% agarose gel. The proportions of digested and undigested PCR product were determined by the ImageQuant program after ethidium bromide staining to determine whether the m.3253T→C mutation is in homoplasmy in these subjects.
Cell lines and culture conditions
Lymphoblastoid cell lines derived from one affected subject (II-5) carrying the m.3253T→C mutation and one genetically unrelated Chinese control subject (C47) lacking the mutation but belonging to same mtDNA haplogroup were immortalized by transformation with the Epstein–Barr virus, as described elsewhere (62). Lymphoblastoid cells were grown in RPMI 1640 medium with 10% FBS. The BrdU-resistant 143B.TK− cell line was grown in DMEM with 100 μg of BrdU/ml and 5% FBS. The mtDNA-less ρ°206 cell line was derived from 143B.TK− (31, 63) and was maintained in the same conditions as the parental line with the addition of 50 μg of uridine/ml. Lymphoblastoid cell lines derived from one affected subject (II-5) and the control subject (C47) were enucleated and subsequently fused to a large excess of mtDNA-less human ρ°206 cells, derived from the 143B.TK− cell line (31, 63). The cybrid clones were isolated by growing the fusion mixtures in selective DMEM containing BrdU and lacking uridine (31). Presumptive cybrids derived from each donor cell line were isolated and subsequently analyzed for the presence and level of the m.3254T→C mutation, number of mtDNA, and karyotype (24, 64). Three mutant cybrids (II-5-10, II-5-14, and II-5-18) carrying the homoplasmic m.3254T→C mutation and three control cybrids (C47-1, C47-3, and C47-7) lacking the mutation with similar mtDNA copy numbers and the same karyotype as 143B cell lines were used for the biochemical characterization.
UV melting assays
Unmodified tRNAs were made by in vitro transcription by T7 RNA polymerase according to previous protocols (65). Transcribed tRNA were diluted in 50 mm sodium phosphate buffer (pH 7.0) including 50 mm NaCl and 0.1 mm EDTA. Melting temperature curves were measured at 260 nm with a heating rate of 0.5 °C/min from 25 to 95 °C via an Agilent Cary 100 UV spectrophotometer.
In vitro angiogenin cleavage assay
In vitro transcriptions of mitochondrial tRNALeu(UUR) (wild type and mutant) were performed as described previously (65). Two μg of purified tRNA transcripts or mitochondrial RNAs were used for the cleavage reaction with 2.5 μg/ml recombinant ANG in buffer containing 30 mm HEPES, pH 7.4, 30 mm NaCl, 5 mm MgCl2, and 0.01% bovine serum albumin. Mixtures were incubated at 37 °C for the indicated times and quenched by adding 5 μl of loading buffer. Cleavage products of tRNA transcripts were electrophoresed through a denaturing polyacrylamide gel and stained with methylene blue. Cleavage products of human mitochondrial RNAs were respectively resolved in 15% denaturating polyacrylamide gels with 8 m urea and electroblotted, and the gels were treated according to the Northern blot analysis procedure described above.
Mitochondrial tRNA analysis
Total mitochondrial RNAs were obtained from mitochondria isolated from mutant and control cell lines (∼2.0 × 108 cells) using the TOTALLY RNATM kit (Ambion), as described previously (66). For the tRNA Northern blot analysis, 2 μg of total mitochondrial RNA were electrophoresed through a urea-denaturing 10% PAGE with 8 m urea in Tris borate-EDTA buffer. The gels were electroblotted onto a positively charged nylon membrane (Roche Applied Science) for hybridization analysis with DIG-labeled oligodeoxynucleotide probes for tRNALeu(UUR), tRNAGln, tRNAIle, tRNATrp, and tRNAVal, as detailed previously (24, 34, 60, 67). DIG-labeled oligodeoxynucleotides were generated by using a DIG oligonucleotide tailing kit (Roche Applied Science). The hybridization and quantification of density in each band were performed as detailed previously (24, 34). For the aminoacylation assays, total mitochondrial RNAs were isolated under acid conditions, and 2 μg of total mitochondrial RNAs was electrophoresed at 4 °C through an acid (pH 5.0) 10% polyacrylamide, 8 m urea gel to separate the charged and uncharged tRNA, as detailed elsewhere (45, 67). To further distinguish nonaminoacylated tRNA from aminoacylated tRNA, samples of tRNAs were deacylated by being heated for 10 min at 60 °C at pH 8.3 and then run in parallel (24, 67). The gels were then electroblotted onto a positively charged nylon membrane (Roche Applied Science) for hybridization analysis with oligodeoxynucleotide probes as described above. Quantification of density in each band was performed as detailed previously (67).
For the tRNA mobility shift assay, 2 μg of total mitochondrial RNA were electrophoresed through a 10% polyacrylamide native gel at 4 °C with 50 mm Tris-glycine buffer. After electrophoresis, the gels were treated according to the procedure for Northern blot analysis described above.
Western blot analysis
Western blot analysis was performed as detailed previously (24, 40). Five μg of proteins obtained from mitochondria that were isolated from cybrid cell lines were denatured and loaded onto SDS-polyacrylamide gels. Afterward, the gels were electroblotted onto a PVDF membrane for hybridization. Antibodies were obtained from different companies, including Abcam (ND1 (ab74257), ND5 (ab92624), CO2 (ab110258), and Tom20 (ab56783)), Santa Cruz Biotechnology, Inc. (ND4 (sc-20499-R) and ND6 (sc-20667)), and Proteintech (CYTB (55090-1-AP), ATP6 (55313-1-AP), and ATP8 (26723-1-AP)). Peroxidase Affini Pure goat anti-mouse IgG and goat anti-rabbit IgG (Jackson) were used as a secondary antibody, and protein signals were detected using the ECL system (CWBIO). Quantification of density in each band was performed as detailed previously (24, 40).
Assays of activities of respiratory complexes
The enzymatic activities of complex I, II, III, IV, and V were assayed as detailed before (34–38).
Measurements of oxygen consumption
The rates OCRs in lymphoblastoid cell lines were measured with a Seahorse Bioscience XF-96 extracellular flux analyzer (Seahorse Bioscience), as detailed previously (24, 40, 68).
ATP measurements
The CellTiter-Glo® luminescent cell viability assay kit (Promega) was used for the measurement of mitochondrial ATP levels, as detailed previously (24, 40).
Assessment of mitochondrial membrane potential
The JC-10 assay kit-microplate (Abcam) was used for detecting the membrane potential of mitochondrial, according to a modification of the manufacturer's instructions (40, 69).
Measurement of ROS production
ROS measurements were performed following the procedures detailed previously (24, 30, 41).
Author contributions
M. X. G. designed the experiments. M. Z., M. W., Q. H., and X. J. performed the experiments and contributed to data analysis in Figs. 1, 2, and 4–7. L. X., Z. L., W. S., and Y. C. performed the experiments and contributed to data analysis in Figs. 3, 4, and 8–10. H. L. performed the clinical evaluation. P. J. performed data analysis. M. Z. prepared the initial draft of the manuscript. M.-X. G. wrote the final version of the manuscript. All authors reviewed the manuscript.
Supplementary Material
This work was supported by National Basic Research Priorities Program of China Grant 2014CB541704 (to M. X. G.); National Natural Science Foundation of China Grants 81600326 (to M. Z.), 31371270 (to P. J.), 31401070 (to L. X.), and 81500611 (to M. W.); Postdoctoral Science Foundation of China Grant 2016M591987 (to M. Z.); and Fundamental Research Funds for the Central Universities Grant 2017QNA7026 (to M. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1–S3 and Figs. S1 and S2.
- mtDNA
- mitochondrial DNA
- ROS
- reactive oxygen species
- DIG
- digoxigenin
- ANG
- angiogenin
- OCR
- oxygen consumption rate
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- ΔΨm
- mitochondrial membrane potential.
References
- 1. Moser M. (1999) World Health Organization-International Society of Hypertension guidelines for the management of hypertension: do these differ from the U.S. recommendations? Which guidelines should the practicing physician follow? J. Clin. Hypertens. 1, 48–54 [PubMed] [Google Scholar]
- 2. Wu Y., Huxley R., Li L., Anna V., Xie G., Yao C., Woodward M., Li X., Chalmers J., Gao R., Kong L., Yang X., China NNHS Steering Committee, and China NNHS Working Group (2008) Prevalence, awareness, treatment, and control of hypertension in China: data from the China National Nutrition and Health Survey 2002. Circulation 118, 2679–2686 [DOI] [PubMed] [Google Scholar]
- 3. Chobanian A. V., Bakris G. L., Black H. R., Cushman W. C., Green L. A., Izzo J. L. Jr., Jones D. W., Materson B. J., Oparil S., Wright J. T. Jr., Roccella E. J., Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. National Heart, Lung, and Blood Institute, and National High Blood Pressure Education Program Coordinating Committee (2003) Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension 42, 1206–1252 [DOI] [PubMed] [Google Scholar]
- 4. Lifton R. P., Gharavi A. G., and Geller D. S. (2001) Molecular mechanisms of human hypertension. Cell 104, 545–556 [DOI] [PubMed] [Google Scholar]
- 5. Dromparis P., and Michelakis E. D. (2013) Mitochondria in vascular health and disease. Annu. Rev. Physiol. 75, 95–126 [DOI] [PubMed] [Google Scholar]
- 6. Eirin A., Lerman A., and Lerman L. O. (2015) Mitochondria: a pathogenic paradigm in hypertensive renal disease. Hypertension 65, 264–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marian A. J. (2011) Mitochondrial genetics and human systemic hypertension. Circ. Res. 108, 784–786 [DOI] [PubMed] [Google Scholar]
- 8. Wilson F. H., Hariri A., Farhi A., Zhao H., Petersen K. F., Toka H. R., Nelson-Williams C., Raja K. M., Kashgarian M., Shulman G. I., Scheinman S. J., and Lifton R. P. (2004) A cluster of metabolic defects caused by mutation in a mitochondrial tRNA. Science 306, 1190–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watson B. Jr., Khan M. A., Desmond R. A., and Bergman S. (2001) Mitochondrial DNA mutations in black Americans with hypertension-associated end-stage renal disease. Am. J. Kidney Dis. 38, 529–536 [DOI] [PubMed] [Google Scholar]
- 10. Zinner S. H., Levy P. S., and Kass E. H. (1971) Familial aggregation of blood pressure in childhood. N. Engl. J. Med. 284, 401–404 [DOI] [PubMed] [Google Scholar]
- 11. Havlik R. J., and Feinleib M. (1982) Epidemiology and genetics of hypertension. Hypertension 4, III121–III127 [DOI] [PubMed] [Google Scholar]
- 12. Andrews R. M., Kubacka I., Chinnery P. F., Lightowlers R. N., Turnbull D. M., and Howell N. (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147. [DOI] [PubMed] [Google Scholar]
- 13. Florentz C., Sohm B., Tryoen-Tóth P., Pütz J., and Sissler M. (2003) Human mitochondrial tRNAs in health and disease. Cell Mol. Life Sci. 60, 1356–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Helm M., Brulé H., Friede D., Giegé R., Pütz D., and Florentz C. (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6, 1356–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suzuki T., Nagao A., and Suzuki T. (2011) Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 45, 299–329 [DOI] [PubMed] [Google Scholar]
- 16. Zheng J., Ji Y., and Guan M. X. (2012) Mitochondrial tRNA mutations associated with deafness. Mitochondrion 12, 406–413 [DOI] [PubMed] [Google Scholar]
- 17. Ruiz-Pesini E., Lott M. T., Procaccio V., Poole J. C., Brandon M. C., Mishmar D., Yi C., Kreuziger J., Baldi P., and Wallace D. C. (2007) An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 35, D823–D828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abbott J. A., Francklyn C. S., and Robey-Bond S. M. (2014) Transfer RNA and human disease. Front. Genet. 5, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu C., Yang Q., Hwang S. J., Sun F., Johnson A. D., Shirihai O. S., Vasan R. S., Levy D., and Schwartz F. (2012) Association of genetic variation in the mitochondrial genome with blood pressure and metabolic traits. Hypertension 60, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y., Li Y., Wang X., Ma Q., Zhu C., Li Z., Yin T., Yang J., Chen Y., and Guan M. (2016) Mitochondrial tRNA mutations in Chinese hypertensive individuals. Mitochondrion 28, 1–7 [DOI] [PubMed] [Google Scholar]
- 21. Jia Z., Wang X., Qin Y., Xue L., Jiang P., Meng Y., Shi S., Wang Y., Qin Mo J., and Guan M. X. (2013) Coronary heart disease is associated with a mutation in mitochondrial tRNA. Hum. Mol. Genet. 22, 4064–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xue L., Wang M., Li H., Wang H., Jiang F., Hou L., Geng J., Lin Z., Peng Y., Zhou H., Yu H., Jiang P., Mo J. Q., and Guan M. X. (2016) Mitochondrial tRNA mutations in 2070 Chinese Han subjects with hypertension. Mitochondrion 30, 208–221 [DOI] [PubMed] [Google Scholar]
- 23. Liu Y., Li R., Li Z., Wang X. J., Yang L., Wang S., and Guan M. X. (2009) Mitochondrial transfer RNAMet 4435A>G mutation is associated with maternally inherited hypertension in a Chinese pedigree. Hypertension 53, 1083–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang P., Wang M., Xue L., Xiao Y., Yu J., Wang H., Yao J., Liu H., Peng Y., Liu H., Li H., Chen Y., and Guan M. X. (2016) A hypertension-associated tRNAAla mutation alters tRNA metabolism and mitochondrial function. Mol. Cell Biol. 36, 1920–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z., Liu Y., Yang L., Wang S., and Guan M. X. (2008) Maternally inherited hypertension is associated with the mitochondrial tRNAIle A4295G mutation in a Chinese family. Biochem. Biophys. Res. Commun. 367, 906–911 [DOI] [PubMed] [Google Scholar]
- 26. Li R., Liu Y., Li Z., Yang L., Wang S., and Guan M. X. (2009) Failures in mitochondrial tRNAMet and tRNAGln metabolism caused by the novel 4401A>G mutation are involved in essential hypertension in a Han Chinese family. Hypertension 54, 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang S., Li R., Fettermann A., Li Z., Qian Y., Liu Y., Wang X., Zhou A., Mo J. Q., Yang L., Jiang P., Taschner A., Rossmanith W., and Guan M. X. (2011) Maternally inherited essential hypertension is associated with the novel 4263A>G mutation in the mitochondrial tRNAIle gene in a large Han Chinese family. Circ. Res. 108, 862–870 [DOI] [PubMed] [Google Scholar]
- 28. Ruiz-Pesini E., and Wallace D. C. (2006) Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum. Mutat. 27, 1072–1081 [DOI] [PubMed] [Google Scholar]
- 29. Suzuki T., and Suzuki T. (2014) A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 42, 7346–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Giegé R., Juhling F., Pütz J., Stadler P., Sauter C., and Florentz C. (2012) Structure of transfer RNAs: similarity and variability. RNA 3, 37–61 [DOI] [PubMed] [Google Scholar]
- 31. King M. P., and Attadi G. (1996) Mitochondria-mediated transformation of human rho0 cells. Methods Enzymol. 264, 313–334 [DOI] [PubMed] [Google Scholar]
- 32. Yamasaki S., Ivanov P., Hu G. F., and Anderson P. (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saxena S. K., Rybak S. M., Davey R. T. Jr, Youle R. J., and Ackerman E. J. (1992) Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily. J. Biol. Chem. 267, 21982–21986 [PubMed] [Google Scholar]
- 34. Zhang J., Jiang P., Jin X., Liu X., Zhang M., Xie S., Gao M., Zhang S., Sun Y. H., Zhu J., Ji Y., Wei Q. P., Tong Y., and Guan M. X. (2014) Leber's hereditary optic neuropathy caused by the homoplasmic ND1 m.3635G>A mutation in nine Han Chinese families. Mitochondrion 18, 18–26 [DOI] [PubMed] [Google Scholar]
- 35. Jiang P., Jin X., Peng Y., Wang M., Liu H., Liu X., Zhang Z., Ji Y., Zhang J., Liang M., Zhao F., Sun Y. H., Zhang M., Zhou X., Chen Y., et al. (2016) The exome sequencing identified the mutation in YARS2 encoding the mitochondrial tyrosyl-tRNA synthetase as a nuclear modifier for the phenotypic manifestation of Leber's hereditary optic neuropathy-associated mitochondrial DNA mutation. Hum. Mol. Genet. 25, 584–596 [DOI] [PubMed] [Google Scholar]
- 36. Li Y., D'Aurelio M., Deng J. H., Park J. S., Manfredi G., Hu P., Lu J., and Bai Y. (2007) An assembled complex IV maintains the stability and activity of complex I in mammalian mitochondria. J. Biol. Chem. 282, 17557–17562 [DOI] [PubMed] [Google Scholar]
- 37. Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Péquignot E., Munnich A., and Rötig A. (1995) Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 11, 144–149 [DOI] [PubMed] [Google Scholar]
- 38. Thorburn D. R., Chow C. W., and Kirby D. M. (2004) Respiratory chain enzyme analysis in muscle and liver. Mitochondrion 4, 363–375 [DOI] [PubMed] [Google Scholar]
- 39. De Meirleir L., Seneca S., Lissens W., De Clercq I., Eyskens F., Gerlo E., Smet J., and Van Coster R. (2004) Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J. Med. Genet. 41, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gong S., Peng Y., Jiang P., Wang M., Fan M., Wang X., Zhou H., Li H., Yan Q., Huang T., and Guan M. X. (2014) A deafness-associated tRNAHis mutation alters the mitochondrial function, ROS production and membrane potential. Nucleic Acids Res. 42, 8039–8048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mahfouz R., Sharma R., Lackner J., Aziz N., and Agarwal A. (2009) Evaluation of chemiluminescence and flow cytometry as tools in assessing production of hydrogen peroxide and superoxide anion in human spermatozoa. Fertil. Steril. 92, 819–827 [DOI] [PubMed] [Google Scholar]
- 42. Hao R., Zhao M. W., Hao Z. X., Yao Y. N., and Wang E. D. (2005) A T-stem slip in human mitochondrial tRNALeu(CUN) governs its charging capacity. Nucleic Acids Res. 33, 3606–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guan M. X., Enriquez J. A., Fischel-Ghodsian N., Puranam R. S., Lin C. P., Maw M. A., and Attardi G. (1998) The deafness-associated mtDNA 7445 mutation, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase ND6 subunit gene expression. Mol. Cell Biol. 18, 5868–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chomyn A., Enriquez J. A., Micol V., Fernandez-Silva P., and Attardi G. (2000) The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol. Chem. 275, 19198–19209 [DOI] [PubMed] [Google Scholar]
- 45. Enriquez J. A., Chomyn A., and Attardi G. (1995) MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNALys and premature translation termination. Nat. Genet. 10, 47–55 [DOI] [PubMed] [Google Scholar]
- 46. Wang M., Liu H., Zheng J., Chen B., Zhou M., Fan W., Wang H., Liang X., Zhou X., Eriani G., Jiang P., and Guan M. X. (2016) A deafness- and diabetes-associated tRNA mutation causes deficient pseudouridinylation at position 55 in tRNAGlu and mitochondrial dysfunction. J. Biol. Chem. 291, 21029–21041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang M., Peng Y., Zheng J., Zheng B., Jin X., Liu H., Wang Y., Tang X., Huang T., Jiang P., and Guan M. X. (2016) A deafness-associated tRNAAsp mutation alters the m1G37 modification, aminoacylation and stability of tRNAAsp and mitochondrial function. Nucleic Acids Res. 44, 10974–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wallace D. C. (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. James A. M., Sheard P. W., Wei Y. H., and Murphy M. P. (1999) Decreased ATP synthesis is phenotypically expressed during increased energy demand in fibroblasts containing mitochondrial tRNA mutations. Eur. J. Biochem. 259, 462–469 [DOI] [PubMed] [Google Scholar]
- 50. Pallotti F., Baracca A., Hernandez-Rosa E., Walker W. F., Solaini G., Lenaz G., Melzi D'Eril G. V., Dimauro S., Schon E. A., and Davidson M. M. (2004) Biochemical analysis of respiratory function in cybrid cell lines harbouring mitochondrial DNA mutations. Biochem. J. 384, 287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szczepanowska J., Malinska D., Wieckowski M. R., and Duszynski J. (2012) Effect of mtDNA point mutations on cellular bioenergetics. Biochim. Biophys. Acta 1817, 1740–1746 [DOI] [PubMed] [Google Scholar]
- 52. Hayashi G., and Cortopassi G. (2015) Oxidative stress in inherited mitochondrial diseases. Free Radic. Biol. Med. 88, 10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sena L. A., and Chandel N. S. (2012) Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Addabbo F., Montagnani M., and Goligorsky M. S. (2009) Mitochondria and reactive oxygen species. Hypertension 53, 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vasan R. S., Beiser A., Seshadri S., Larson M. G., Kannel W. B., D'Agostino R. B., and Levy D. (2002) Residual lifetime risk for developing hypertension in middle-aged women and men: the Framingham Heart Study. JAMA 287, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 56. Chen C., Chen Y., and Guan M. X. (2015) A peep into mitochondrial disorder: multifaceted from mitochondrial DNA mutations to nuclear gene modulation. Protein Cell 6, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dittmar K. A., Goodenbour J. M., and Pan T. (2006) Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen D., Li F., Yang Q., Tian M., Zhang Z., Zhang Q., Chen Y., Guan M. X. (2016) The defective expression of gtpbp3 related to tRNA modification alters the mitochondrial function and development of zebrafish. Int. J. Biochem. Cell Biol. 77, 1–9 [DOI] [PubMed] [Google Scholar]
- 59. Joint National Committee on Prevention Detection Evaluation and Treatment of High Blood Pressure (1997) The sixth report of the Joint National Committee on Prevention, Detection, Evaluation and Treatment of High Blood Pressure. Arch. Intern. Med. 157, 2413–2446 [DOI] [PubMed] [Google Scholar]
- 60. Li R., and Guan M. X. (2010) Human mitochondrial leucyl-tRNA synthetase corrects mitochondrial dysfunctions due to the tRNALeu(UUR) A3243G mutation, associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like symptoms and diabetes. Mol. Cell Biol. 30, 2147–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rieder M. J., Taylor S. L., Tobe V. O., and Nickerson D. A. (1998) Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 26, 967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miller G., and Lipman M. (1973) Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. U.S.A. 70, 190–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. King M. P., and Attardi G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246, 500–503 [DOI] [PubMed] [Google Scholar]
- 64. Guan M. X., Fischel-Ghodsian N., and Attardi G. (2001) Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum. Mol. Genet. 10, 573–580 [DOI] [PubMed] [Google Scholar]
- 65. Li Y., Chen J., Wang E., and Wang Y. (1999) T7 RNA polymerase transcription of Escherichia coli isoacceptors tRNALeu. Sci. China C Life Sci. 42, 185–190 [DOI] [PubMed] [Google Scholar]
- 66. King M. P., and Attardi G. (1993) Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J. Biol. Chem. 268, 10228–10237 [PubMed] [Google Scholar]
- 67. Enríquez J. A., and Attardi G. (1996) Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 264, 183–196 [DOI] [PubMed] [Google Scholar]
- 68. Dranka B. P., Benavides G. A., Diers A. R., Giordano S., Zelickson B. R., Reily C., Zou L., Chatham J. C., Hill B. G., Zhang J., Landar A., and Darley-Usmar V. M. (2011) Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med. 51, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen Y. B., Aon M. A., Hsu Y. T., Soane L., Teng X., McCaffery J. M., Cheng W. C., Qi B., Li H., Alavian K. N., Dayhoff-Brannigan M., Zou S., Pineda F. J., O'Rourke B., Ko Y. H., et al. (2011) Bcl-xL regulates mitochondrial energetics by stabilizing the inner membrane potential. J. Cell Biol. 195, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.