Abstract
Mitochondrial oxidation of nutrients is tightly regulated in response to the cellular environment and changes in energy demands. In vitro studies evaluating the mitochondrial capacity of oxidizing different substrates are important for understanding metabolic shifts in physiological adaptations and pathological conditions, but may be influenced by the nutrients present in the culture medium or by the utilization of endogenous stores. One such influence is exemplified by the Crabtree effect (the glucose-mediated inhibition of mitochondrial respiration) as most in vitro experiments are performed in glucose-containing media. Here, using high-resolution respirometry, we evaluated the oxidation of endogenous or exogenous substrates by cell lines harboring different metabolic profiles. We found that a 1-h deprivation of the main energetic nutrients is an appropriate strategy to abolish interference of endogenous or undesirable exogenous substrates with the cellular capacity of oxidizing specific substrates, namely glutamine, pyruvate, glucose, or palmitate, in mitochondria. This approach primed mitochondria to immediately increase their oxygen consumption after the addition of the exogenous nutrients. All starved cells could oxidize exogenous glutamine, whereas the capacity for oxidizing palmitate was limited to human hepatocarcinoma Huh7 cells and to C2C12 mouse myoblasts that differentiated into myotubes. In the presence of exogenous glucose, starvation decreased the Crabtree effect in Huh7 and C2C12 cells and abrogated it in mouse neuroblastoma N2A cells. Interestingly, the fact that the Crabtree effect was observed only for mitochondrial basal respiration but not for the maximum respiratory capacity suggests it is not caused by a direct effect on the electron transport system.
Keywords: energy metabolism, fatty acid, glucose, glutamine, mitochondria, pyruvate, Crabtree effect, respirometry
Introduction
Nutrient availability elicits a series of adaptations that ensure the maintenance of cellular functions upon distinct environmental conditions. Energy and nutrient-sensing systems regulate these adaptations, controlling metabolic pathways, protein synthesis, autophagy, cell cycle progression, and mitochondrial biogenesis, morphology, and function (1).
When starved of nutrients, cells need, at a certain point, to mobilize endogenous substrates to maintain their energy charge at safe levels. Another strategy is the induction of the autophagy pathway, a mechanism to mobilize fatty acids, amino acids, and sugars from distinct organelles for their use as energy supplies (2, 3). One of the determinants of the mitochondrial substrate immediate preference is the differential assembly of mitochondrial supercomplexes (4), which are supramolecular structures formed by the association of the electron transport system (ETS)4 complexes. The formation of the supercomplexes increases the efficiency of ATP generation independently of mitochondrial biogenesis and de novo synthesis of the respiratory complexes (5). In a normal cellular context, all of these adaptations are tightly regulated in response to the cellular environment (growth factors, nutrient availability, etc.) and endogenous energy demand (1, 6). However, in some pathological conditions, such as cancer (7) and viral infection (8), these regulatory mechanisms are bypassed, and cells may change the preference for oxidizing specific energetic substrates despite the nutrient availability, deviating from normal metabolic routes. This ultimately may result in the impairment of cellular functions and/or damage to the organism. Until now, the understanding of the molecular mechanisms involved in reprogramming the ETS capacity for using determined metabolic fuels has been restricted due to methodological limitations caused by the influence of the nutrients already present in the culture medium. One example is the glucose-mediated inhibition of mitochondrial respiration, or the Crabtree effect, as most in vitro experiments are carried out in media containing glucose.
Regulatory phenomena of carbohydrate metabolism, such as the Pasteur, Warburg, and Crabtree effects, although mechanistically quite different from each other, share as a common outcome the reciprocal regulation of glycolytic and oxidative metabolisms. The Pasteur effect is well characterized in microorganisms such as yeasts and functions in the repression of fermentation in aerobic conditions (9). The Warburg effect is related to a long-term metabolic reprogramming that involves gene expression events. In this case, there is a switch to the glycolytic over the oxidative metabolism as is commonly observed in cancer cells (10). In comparison with the Warburg effect, the Crabtree effect is characterized as an immediate and reversible event, consisting in the suppression of respiration in the presence of glucose. Despite its description almost a century ago by Herbert Grace Crabtree (11), the mechanisms that mediate this phenomenon are still unknown (12).
Cellular respiration has been extensively studied in isolated mitochondria and permeabilized cells where the operation of mitochondrial complexes can be evaluated by addition of substrates that are directly metabolized in mitochondria, such as ADP, glutamate, malate, succinate, acylcarnitine, and others (13, 14). However, this approach bypasses the context of the entire cell and does not take into account overall cellular metabolism in terms of protein regulation/signaling. If it is of interest to study metabolism at a higher level, the recommended approach would involve the use of intact cells. In this regard, there are few studies that explore in detail the mitochondrial oxidation of individual substrates in intact cells. Consequently, the experimental design to address this issue still needs to be established depending on the scientific question being asked. In the present study, it was shown that short-term deprivation of the main energetic nutrients is a useful strategy to evaluate metabolic shifts in vitro using intact cells. We found that this procedure depletes most endogenous substrates in three different cell lines, allowing the evaluation of the effects of exogenous substrates on oxygen consumption rates (OCRs). We also explored the contribution of endogenous substrates to respiratory rates of non-starved and starved cells. Our findings suggest that the Crabtree effect is decreased in cells subjected to short-term starvation, which is possibly associated with the inhibition of the mitochondrial oxidation of glutamine. In summary, in addition to contributing to a better understanding of the Crabtree effect, our results presented here also pave the way for future investigations of the metabolic switches in pathological situations.
Results
1-h starvation makes the cells responsive to a specific oxidative substrate
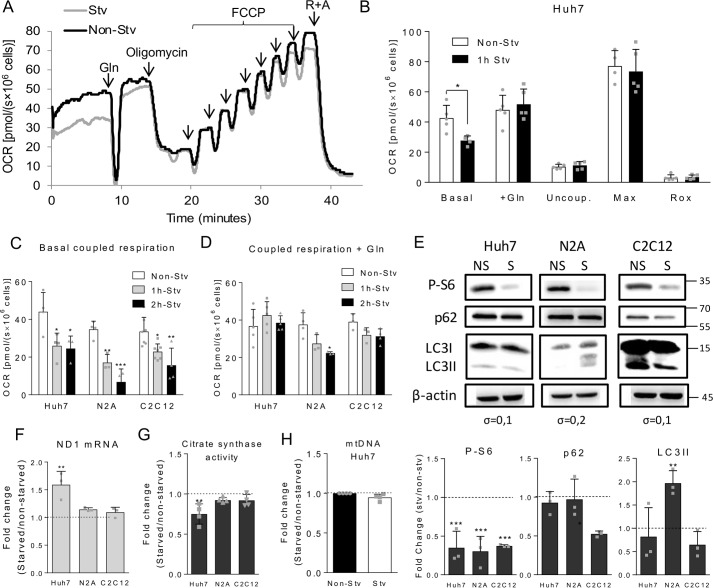
The first goal of this work was to evaluate the capacity of cells harboring distinct metabolic profiles, namely the N2A mouse neuroblastoma, C2C12 mouse myoblast, and Huh7 human hepatocarcinoma cell lines, to oxidize different substrates. For this, we analyzed the impact of adding an exogenous substrate to intact cells. Cells were resuspended in a culture medium in which the serum and main nutrients (glucose, pyruvate, and glutamine) were absent (DMEM-A). Fig. 1A shows representative OCR curves of Huh7 cells to which glutamine was reintroduced after the basal OCR was recorded in DMEM-A. Surprisingly, we did not observe a considerable increase in OCR after glutamine addition (see Fig. 1A, black line). This led us to speculate whether the cells maintained in complete medium would keep large amounts of endogenous substrates, which were sufficient to sustain the basal respiratory rates, thus hindering an increase in OCR related to the use of the added substrate. In order to circumvent this limitation, whether subjecting the cells to short-term deprivation of the main oxidative nutrients (1- or 2 h-incubation in DMEM-A) would make them responsive to the addition of a specific substrate was tested. We found that Huh7 cells starved for 1 h displayed lower basal respiratory rates when compared with non-starved cells (see Fig. 1A, gray line). After reintroduction of glutamine in the medium, in contrast to non-starved cells, the respiratory rates of starved cells significantly increased, restoring the OCR values observed for non-starved cells (Fig. 1B). These data suggest that short-term starvation would be a proper approach to evaluate the impact of a specific substrate in cellular oxidative metabolism.
Figure 1.
Effects of short-term nutrient deprivation on Huh7, N2A, and C2C12 cells. A, representative respirometry experiment using Huh7 cells subjected or not to 1-h starvation (Stv or Non-Stv, respectively) followed by reintroduction of 2.5 mm glutamine in the culture medium during OCR recording. After OCR stabilization, the following ETS modulators were added: 250 nm oligomycin to measure uncoupled respiration, sequential additions of FCCP to achieve maximum respiration (at 1 mm), and 0.5 mm rotenone and 3.65 μm antimycin (R+A) to determine Rox rates. B, OCR recordings of non-starved or starved Huh7 cells in DMEM-A in the following conditions: basal respiration, after addition of 2.5 mm glutamine (+Gln), after inhibition of ATP synthase with oligomycin (uncoupled respiration (Uncoup.), upon titration with FCCP (maximum respiration (Max)), and after inhibition of the respiratory complexes with antimycin and rotenone (Rox). C, coupled respiration (basal respiration minus uncoupled respiration values) of Huh7, C2C12, and N2A cells subjected to 1- and 2-h starvation. D, coupled respiration after glutamine reintroduction to non-starved, 1-, or 2-h-starved Huh7, N2A, or C2C12 cell suspensions. E, immunodetection by Western blotting of P-S6, p62, LC3I, LC3II (the cleaved form of LC3I), and β-actin in Huh7, N2A, and C2C12 cell lysates in non-starved (NS) and starved (S) conditions. The position of the molecular weight markers are indicated on the left, and the standard deviations of the differences between non-starved and starved conditions are shown below the β-actin blotting. The lower panels show the -fold change observed for the densitometric quantification of the bands between starved and non-starved conditions for p62, LC3II, and P-S6. F, relative mRNA expression of the mitochondrial gene ND1 between starved and non-starved Huh7, N2A, and C2C12 cells. G, relative activity of citrate synthase between starved and non-starved Huh7, N2A, and C2C12 cells. H, relative quantification of mitochondrial DNA between starved and non-starved Huh7 cells. For each graph, individual data points are represented with symbols. Error bars represent mean ± S.D., and asterisks indicate significant differences between non-starved and starved conditions for each cell line: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To further explore the respiratory profile in different conditions, we analyzed OCR after addition of typical modulators of the respiratory machinery. Oligomycin is an inhibitor of ATP synthase that allows determination of the oxygen consumption related to the proton leak through the mitochondrial inner membrane (uncoupled respiration). Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) disrupts the proton gradient, forcing the mitochondrial complexes to work in a maximal electron transport capacity (maximal respiration). Rotenone and antimycin are inhibitors of complexes I and III, respectively, which function in impairing electron transport through the respiratory chain, revealing the residual oxygen consumption (Rox). This represents the non-ETS contribution in OCR, including non-mitochondrial oxygen consumption and that performed by other mitochondrial processes (for instance by the activity of mitochondrial oxidases). The results showed that 1-h starvation affected neither the uncoupled, maximal mitochondrial respiration, nor Rox rates (Fig. 1B). Comparing the three different cell types, it was found that 1-h starvation was enough to significantly reduce the coupled respiratory rates (oxygen consumption rate associated to ATP synthesis, calculated by subtracting the uncoupled respiration from the basal OCR) in all cell lines tested (Fig. 1C). After glutamine addition, Huh7 and C2C12 cells were able to restore the coupled respiratory rates even when subjected to 2-h starvation (Fig. 1D). Conversely, N2A cells were much more sensitive to starvation, showing a reduced capacity to restore respiratory rates after 1-h starvation/restimulation and a significant impairment in OCR after 2 h of starvation. Based on these results, 1-h starvation was the condition chosen for subsequent assays. To mitigate the possibility that traces of amino acids present in DMEM-A could interfere in the analyses, we performed a similar experiment (starvation/restimulation with glutamine) using the balanced saline solution Krebs-Henseleit buffer instead of DMEM-A. Although the respiratory rates of N2A, C2C12, and Huh7 cells were slightly lower in Krebs-Henseleit buffer when compared with DMEM-A-starved cells, we observed the same response profile upon starvation and glutamine reintroduction in both conditions (not shown). This indicated that the other components of DMEM-A, including traces of amino acids, were not significantly used as oxidative substrates by the cells. So, starvation with DMEM-A allow cells to respond to substrate reintroduction similarly to cells grown in the complete absence of substrates (saline solution). We next explored whether 1-h starvation would induce other cellular adaptations in addition to the observed changes in mitochondrial respiration. Regarding the nutrient-sensing pathways, the results showed that mechanistic target of rapamycin (mTOR) pathway was down-regulated in Huh7, N2A, and C2C12 cells after 1-h starvation as shown by the decrease in phosphorylation of the ribosomal protein S6, a downstream effector of this pathway (Fig. 1E). Additionally, 1-h starvation did not induce autophagy in Huh7 and C2C12 cells but did in N2A cells as assessed by an increase in microtubule-associated protein 1A/1B light chain 3 II (LC3II) in this condition. This result is expected for neuronal cells as these cells typically do not store large amounts of substrates and as a consequence are more prone to suffer from lack of nutrients. Although a decrease in p62 levels has been observed in starved C2C12 cells, this event without the concomitant elevation of LC3II levels does not support the induction of autophagy in these cells. To evaluate whether starvation altered mitochondrial content in Huh7, N2A, and C2C12 cells, we measured the expression of the mitochondrial gene NADH:ubiquinone oxidoreductase core subunit 1 (ND1) and the activity of the mitochondrial enzyme citrate synthase. Neither N2A nor C2C12 cells displayed any change in either parameter. However, Huh7 cells showed increased expression of the ND1 gene, whereas citrate synthase activity decreased after 1-h starvation (Fig. 1, F and G). To further explore these contradictory results, we compared the relative amount of mitochondrial DNA between non-starved and starved conditions by quantitative PCR. The results showed that 1-h starvation did not induce mitochondrial biogenesis (Fig. 1H). Taken together, the results indicate that 1-h starvation elicits adaptations only in hepatic cells in terms of mitochondrial gene expression and enzymatic activity, suggesting that these cells are more prone to change mitochondrial metabolism in a condition of nutrient deprivation. In summary, the results show that, despite some differences among Huh7, N2A, and C2C12 cells, short-term (1-h) deprivation of the main oxidative nutrients makes cells prone to instantly respond to the reintroduction of a substrate, allowing OCR measurements related specifically to the added substrate.
1-h starvation depletes endogenous substrates in Huh7, N2A, and C2C12 cells without affecting cell viability
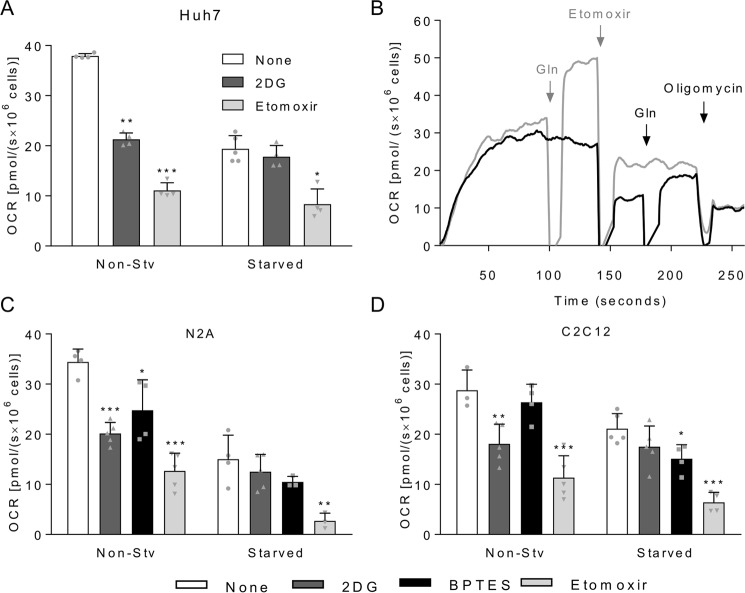
To assess the effect of 1-h starvation on the consumption of endogenous substrates, Huh7, N2A, and C2C12 cells were treated with 2-deoxy-d-glucose (2DG), which inhibits the glycolytic pathway by competing with glucose as the substrate of hexokinase; bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES), a selective inhibitor of isoform 1 of glutaminase (GLS1) (15), or etomoxir, an inhibitor of carnitine palmitoyltransferase I involved in the transport of fatty acyl-CoA into the mitochondrial matrix. Using these inhibitors, we evaluated the oxidation of endogenous glucose (or glycogen), glutamine, or palmitate (or triacylglycerols), respectively, in terms of OCR (Fig. 2). The respiratory rates of non-starved Huh7 cells decreased 44% in the presence of 2DG, whereas no significant changes in OCR were observed for starved cells, indicating the exhaustion of available glucose stocks after 1-h starvation (Fig. 2A). When Huh7 cells were incubated with etomoxir, OCR inhibition reached 70% for non-starved cells and, in a lesser extent, this was also observed after 1-h starvation, with 57% inhibition of OCR after etomoxir addition. This result is expected for hepatic cell lines, which usually accumulate large amounts of lipids in intracellular lipid droplets. However, to ensure the specificity of etomoxir in inhibiting only fatty acid oxidation, we tested whether etomoxir affected glutamine oxidation (Fig. 2B). Addition of glutamine to cells previously treated with etomoxir restores the OCR values to those observed for cells treated first with glutamine and then with etomoxir. This demonstrated that etomoxir does act as a specific inhibitor of fatty acid oxidation. Because of the lack of selective chemical inhibitors for GLS2, the isoform of glutaminase expressed in hepatocytes, it was not possible to determine the contribution of oxidation of endogenous glutamine in Huh7 cells. In the case of non-starved N2A cells, 2DG caused 41% OCR inhibition, whereas etomoxir and BPTES promoted 63 and 28% OCR inhibition, respectively (Fig. 2C). Starved N2A cells had no significant change in respiratory rates upon addition of 2DG and BPTES, indicating the exhaustion of the available glucose and glutamine stocks after 1-h starvation. Fatty acids were most likely still present and available in these cells as etomoxir promoted a significant decrease in respiratory rates, which almost reached the value of the uncoupled respiration. For non-starved C2C12 cells, BPTES had no significant effect on respiratory rates but promoted a 28% decrease in respiration of starved cells, indicating that endogenous glutamine is mobilized upon starvation but not when the cells are supplemented with this nutrient (Fig. 2D). Glycolysis inhibition led to a 37% decrease in OCR of non-starved C2C12 cells, whereas no changes were found for starved cells, suggesting that glucose/glycogen is the main endogenous substrate consumed during starvation in C2C12 myoblasts. For all cell lines tested, palmitate was shown to be the main substrate oxidized in mitochondria even in non-starved cells, responsible for 65% of coupled mitochondrial respiration. Conversely, in starved cells, the coupled OCR associated to fatty acid oxidation represented 57% for Huh7, 82% for N2A, and 70% for C2C12 cells. To eliminate the possibility that starvation causes cell death, cellular viability was evaluated, showing no significant changes between starved and non-starved cells for Huh7 (89.4 ± 4.4–89.7 ± 3.9; mean ± S.D.), N2A (89.0 ± 4.6–87.3 ± 4.2) and C2C12 cells (95.6 ± 2.9–93.2 ± 3.3). Taken together, these results showed that cells depend primarily on oxidation of endogenous fatty acids, whereas glucose stocks are nearly depleted after 1-h starvation. In addition, glutamine plays a minor role as an endogenous substrate in all three cell lines tested.
Figure 2.
Oxidation of endogenous substrates by non-starved or starved cells. A, coupled OCR for non-starved (Non-Stv) or 1-h-starved Huh7 cells. Cells were loaded in oxygraph chambers, and after basal respiration was recorded, cells were not treated (white bars) or treated with 5 mm 2DG (dark gray bars), 10 μm BPTES (black bars), or 200 μm etomoxir (light gray bars). After stabilization of the signal, OCR was recorded, and cells were treated with oligomycin for measurement of uncoupled OCR, which was used to calculate the coupled OCR. B, overlay of two representative respirometry experiments with starved Huh7 cells in DMEM-A. In the experiment shown with the black line, after basal respiration was recorded, etomoxir was added to the cells followed by the reintroduction of 2.5 mm glutamine and subsequent oligomycin addition. In the experiment shown with the gray line, after basal respiration was recorded, 2.5 mm glutamine was added to the cells followed by sequential addition of 200 μm etomoxir and oligomycin. C, same experiment as described in A except that the cells were N2A cells. D, same experiment as described in A except that the cells were C2C12 cells. For each graph, individual data points are represented with symbols. Error bars represent mean ± S.D. of three to five independent experiments, and asterisks indicate significant differences between non-treated coupled OCR and coupled OCR upon addition of the inhibitors: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
1-h starvation allows measuring the mitochondrial oxidation of specific substrates by Huh7, N2A, and C2C12 cells
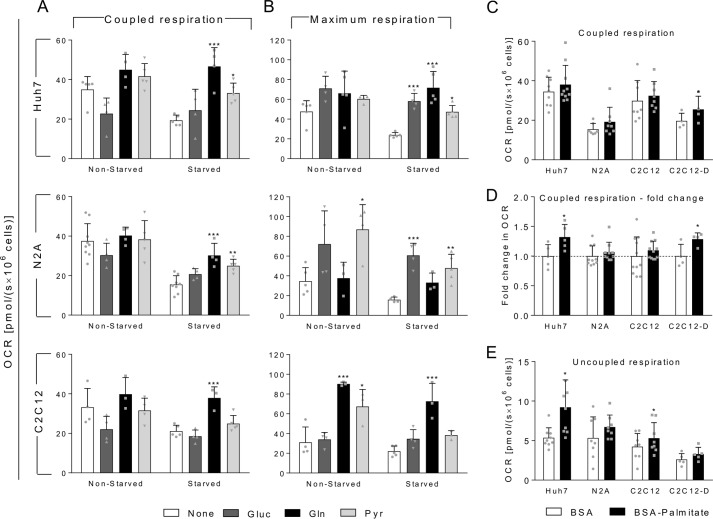
When non-starved Huh7, N2A, and C2C12 cells were incubated with exogenous glutamine or pyruvate, we could detect a slight but not significant increase in coupled OCR (Fig. 3A). Conversely, starved cells displayed a large increase in OCR upon exogenous addition of glutamine or pyruvate with the exception of starved-C2C12 cells, which did not show significant alteration in the OCR after addition of pyruvate but did with glutamine. Regarding the maximum respiration (Fig. 3B), Huh7 cells followed the same profile observed for basal coupled respiration when glucose, glutamine, or pyruvate was used as the exogenous substrate. This indicates that starvation does not affect the capacity of the cells to oxidize these substrates. In the case of N2A cells, the highest respiratory rates were achieved in the presence of glucose and pyruvate. In addition, the maximum OCR in the presence of glutamine was similar to that in the absence of substrates, suggesting that the mitochondrial oxidative machinery of N2A cells is not engaged to oxidize glutamine efficiently. Alternatively, starved and non-starved C2C12 cells are fully able to oxidize glutamine. However, both N2A and C2C12 cells showed a reduction in the maximum capacity of oxidizing pyruvate after starvation, but this limitation was not observed for coupled OCR, which represents the physiological condition. The oxidation of exogenous glucose by the cells was shown to be a more complex phenomenon, which will be explored in more detail in the next section.
Figure 3.
Oxidation of exogenous substrates by non-starved or starved cells. Non-starved or 1-h-starvedHuh7, N2A, or C2C12 cells were loaded in oxygraph chambers, and after basal respiration was recorded, cells were maintained without any exogenous substrate (None; white bars) or incubated with 5 mm glucose (Gluc; dark gray bars), 2.5 mm glutamine (Gln; black bars), or 10 mm pyruvate (Pyr; light gray bars). After stabilization of the signal, OCR was recorded, and cells were treated with oligomycin for measurement of uncoupled OCR and FCCP for measurement of the maximal respiration. A, coupled OCR for Huh7, N2A, or C2C12 cells as indicated in the figure. B, maximum OCR in the same conditions as in A. C, coupled OCR of Huh7 cells, N2A cells, C2C12 myoblasts, and C2C12 cells differentiated into myotubes (C2C12-D) in presence of BSA alone (white bars) or 200 μm BSA-palmitate (black bars). D, -fold change of coupled respiration calculated for each independent experiment shown in C. Error bars shown in BSA control represent the variations around the mean of the experiments calculated as -fold change. E, uncoupled OCR of Huh7 cells, N2A cells, C2C12 cells, and C2C12 cells differentiated into myotubes in the presence of BSA alone (white bars) or 200 μm BSA-palmitate (black bars). For each graph, individual data points are represented with symbols. Error bars represent mean ± S.D. of three to nine independent experiments, and asterisks indicate significant differences between basal coupled OCR and coupled OCR measured after introduction of the indicated substrates: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The oxidation of exogenous palmitate by Huh7, N2A, and C2C12 cells is shown separately due to the need of adapting the protocol for measuring OCR with this substrate. In this case, the cells were preincubated for 1 h in DMEM-A supplemented with either 0.5 mm carnitine, 200 μm palmitate conjugated with BSA, or BSA with 0.5 mm carnitine as control. This procedure was shown to increase the use of palmitate by Huh7 cells when compared with cells without the preincubation step (not shown). It is important to note here that during the preincubation period, cells are restricted from other substrates, such as glucose, glutamine, and pyruvate. Because undifferentiated C2C12 myoblasts are not good representatives of highly oxidative muscle cells, which are known to oxidize fatty acids efficiently, we decided to include differentiated C2C12 myotubes in this assay. At first glance, the results obtained for coupled respiration showed that only C2C12 myotubes were able to significantly oxidize exogenous palmitate (Fig. 3C). However, by analyzing the experiments separately, we observed that in each independent assay using Huh7 cells the presence of palmitate induced an increase in the basal respiration. To identify small variations in OCR that are observed in each experiment but are masked when the averages are calculated, we analyzed the results in terms of relative change in coupled OCR comparing cells incubated with BSA alone with those incubated with palmitate-BSA. This analysis revealed that Huh7 cells and differentiated C2C12 myotubes increased their coupled OCR by 32 and 28%, respectively, in presence of palmitate-BSA when compared with BSA alone. However, no changes were found for N2A and C2C12 myoblasts (Fig. 3D), indicating that only the Huh7 cells and C2C12 myotubes were able to oxidize exogenous palmitate when it is the only substrate provided. Another factor that contributes to the higher variation observed in these experiments is the uncoupling effect of palmitate, which is largely known to occur (16–18). Indeed, the palmitate uncoupling effect was statistically significant in Huh7 and C2C12 myoblasts (Fig. 3E). It is important to mention that we did not detect any difference in uncoupled OCR when other substrates were used (not shown). Additionally, the palmitate-induced increase in OCR cannot be compared with the results obtained for the other substrates because the molar palmitate concentrations used were much lower (200 μm compared with 10 mm pyruvate or 2.5 mm glutamine) as higher palmitate concentrations are known to cause lipotoxicity (19).
Thus, these results show that 1-h starvation caused the depletion of endogenous substrates, allowing the determination of OCR associated specifically with the use of an exogenous substrate. This occurred without changes in cellular viability and respiratory function as each cell type still retained the capacity of oxidizing the substrate provided as much as non-starved cells in almost all the cases studied.
Starvation reduces the Crabtree effect in Huh7 and C2C12 cells and abolishes it in N2A cells
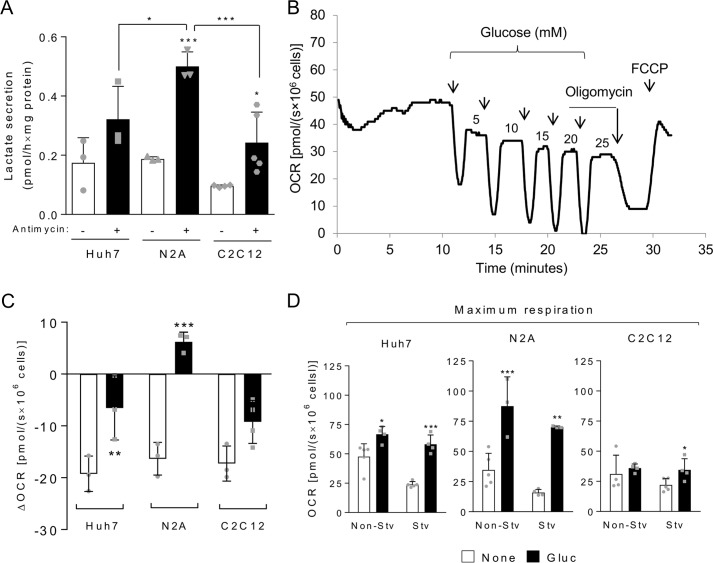
Glucose metabolism needed to be evaluated more carefully because of the Crabtree effect, the inhibition of mitochondrial respiration triggered by glucose, which is a complex process of which little is known. Thus, here we deeply explored the mitochondrial oxidation of glucose, taking into account the potential occurrence of the Crabtree effect. First, we analyzed the glycolytic activity of Huh7, N2A, and C2C12 cell lines by assessing lactate secretion in the absence or presence of antimycin, an inhibitor of complex III that blocks mitochondrial respiration, thus forcing the cells to their maximum glycolytic capacity. The three cell lines presented a moderate glycolytic activity that was enhanced by cell incubation with antimycin, and among them, N2A showed the highest glycolytic capacity (Fig. 4A), which is expected for neuroblasts. Interestingly, we could not detect lactate secretion during 1-h starvation for all cell lines tested (not shown). This result may be explained by a shift from lactate production to pyruvate consumption or by the depletion of glucose stocks during starvation.
Figure 4.
The Crabtree effect is reduced in starved cells and is not seen in maximal respiration conditions. A, secreted lactate measured in culture medium of cells incubated for 1 h in serum-free medium in the absence or presence of 3.65 μm antimycin as indicated. B, representative oxygraph experiment using Huh7 cells to which increasing amounts of glucose (arrows indicate glucose additions, and the numbers correspond to the final concentrations after each addition in millimolar) were added. C, differences in OCR before and after glucose addition (to a final concentration of 5 mm) for non-starved and starved Huh7, N2A, and C2C12 cells. D, maximum respiration measured for non-starved (Non-Stv) and starved (Stv) cells in the absence of exogenous substrates (None; white bars) or presence of 5 mm glucose (Gluc; black bars). For each graph, individual data points are represented with symbols. Error bars represent mean ± S.D. of three to five independent experiments, and asterisks indicate significant differences between non-starved and starved conditions or between coupled OCR without substrates and coupled OCR calculated after reintroduction of glucose: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The Crabtree effect could be clearly observed in Huh7, N2A, and C2C12 cells with a significant inhibition of respiration after glucose addition (Fig. 4, B and C). Remarkably, after 1-h starvation, the Crabtree effect was reduced in Huh7 and C2C12 cells and completely abolished in N2A. In this case, addition of 5 mm glucose to starved cells promoted an increase in mitochondrial OCR instead of an inhibition (Fig. 4C). Interestingly, both for non-starved and starved conditions, the Crabtree effect was not seen when maximum OCR was measured (Fig. 4D). This was evident in Huh7 and N2A cells but also occurred for C2C12 cells. In the presence of glucose, the maximum respiration in C2C12 cells was similar to that observed in its absence (Fig. 4D). To further investigate the Crabtree effect in our models, we evaluated whether glucose-induced inhibition of mitochondrial respiration would be reversed by subsequent addition of a substrate that each cell type oxidizes with the highest capacity, namely glutamine for Huh7 and pyruvate for N2A (Fig. 5). Interestingly, we found that, in the presence of glucose, the mitochondrial oxidation of glutamine, but not of pyruvate, is inhibited. Again, the Crabtree effect was not observed in conditions of maximum respiration because, even in the presence of glucose, Huh7 cells achieved an OCR similar to that obtained in the presence of glutamine alone (compare with Fig. 3B). A possible explanation for this result is that the Crabtree effect may specifically involve the glutamine utilization pathway in mitochondria.
Figure 5.
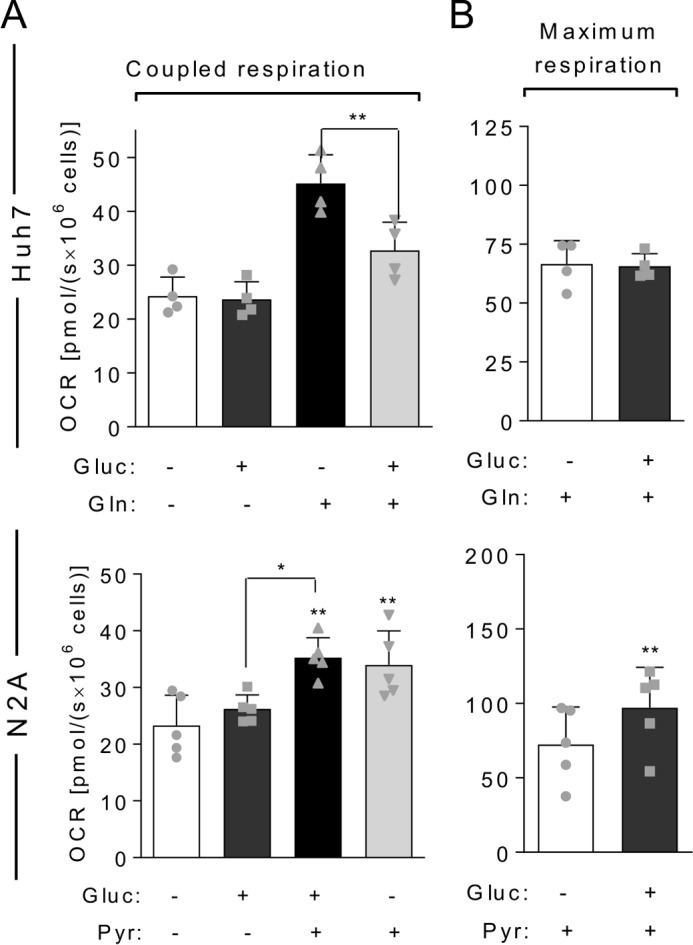
Glucose inhibits the oxidation of glutamine but not of pyruvate. Starved Huh7 or N2A cells were loaded in oxygraph chambers, and after basal respiration was recorded, cells were maintained without any exogenous substrate (−; white bars) or incubated with 5 mm glucose (Gluc) (dark gray bars); a substrate that cells have high capacity to use, 2.5 mm glutamine (Gln) for Huh7 or 10 mm pyruvate (Pyr) for N2A (black bars); or 5 mm glucose followed by the chosen substrate (light gray bars). After stabilization of the signal, OCR was recorded, and cells were treated with oligomycin for measurement of uncoupled OCR and FCCP for measurement of the maximal respiration. A, coupled OCR of Huh7 or N2A cells as indicated in the figure. B, maximum respiration achieved in the same experiments. For each graph, individual data points are represented with symbols. Error bars represent mean ± S.D. of four to five independent experiments, and asterisks show significant differences between coupled OCR without substrates and coupled OCR calculated after reintroduction of substrates: *, p < 0.05; **, p < 0.01.
Discussion
One of the main limitations of evaluating the capacity of a cell to oxidize a given substrate in vitro is the influence of the nutrients already present in the culture medium. This may limit the ability to ascertain the molecular mechanisms involved in the switch among metabolic fuels during adaptation to physiological situations as well as under pathological conditions. Here we developed a method to evaluate mitochondrial oxidation of endogenous and exogenous substrates by subjecting the cells to short-term complete deprivation of oxidative nutrients. We applied this procedure to cell lines harboring different metabolic profiles. The results showed that this can be a strategy to compare the cell's capacity of using specific substrates in different cellular contexts.
Traditional methods used to measure the oxidation of substrates in mitochondria are based on tracing isotopically labeled oxidative substrates by quantifying, for example, radiolabeled CO2 produced in the tricarboxylic acid cycle (20–22). This experimental approach, however, provides limited information about mitochondrial function itself. In this regard, respirometry is more informative. With the use of drugs that modulate ETS operation, in a typical respirometry experiment it is possible to discriminate oxygen consumption associated with ATP synthesis (coupled respiration) from that resulting from proton leak, reactive oxygen species production, or other cellular processes (for instance, activity of cellular oxidases). Also, the determination of maximum respiratory rates provides clues about the integrity of ETS and its maximum operating capacity. Furthermore, using the experimental procedure described here (depletion of endogenous substrates followed by the addition of a single substrate), we found that the maximum respiration can provide information about the capacity of ETS for oxidizing a specific substrate. Interestingly, the findings presented here show that this parameter depends on the substrate and varies among different cell types, suggesting that each cell is more capable of using some substrates more than others. Another advantage of respirometry over other methods can be exemplified by the detection of the uncoupling effect of palmitate (16–18), which was detected in our experiments but cannot be observed by using isotopically labeled tracers. It is also important to highlight that if one intends to quantify the amount of a given substrate being used in a determined condition, tracing isotopically labeled substrates in the presence of all other nutrients, under physiological concentrations, would be a more suitable approach. Here, however, determining the capacity of cells to oxidize a determined substrate without the interference of other substrates was successfully achieved using respirometric measurements after short-term starvation.
The effects of the 1-h starvation procedure described here can be summarized as (a) the reduction of basal coupled OCR without loss of cellular viability or the capacity to restore non-starved OCR levels upon reintroduction of substrates (especially glutamine but also pyruvate to a lesser extent) and (b) the decrease in oxidation of endogenous stocks of glucose with an increased percentage of OCR associated with oxidation of endogenous fatty acids. In addition, we demonstrated for the first time that the inhibition of OCR by glucose, the Crabtree effect, is reduced in starved cells. It is important to mention that these cellular changes seem to be made more likely by interference in the immediate mitochondrial behavior and cell signaling (such as the decrease in phosphorylation of the ribosomal protein S6 observed here), which can be easily reversed, and most likely does not impact the cellular phenotype.
One can starve cells in different ways: depriving serum, subtracting specific nutrients, and isolating primary cells from fasted animals. All forms of nutrient deprivation were shown to induce metabolic changes in the cells mostly related to increasing the capacity of mitochondrial oxidative metabolism. For instance, in the human glioblastoma cell line U251, 24–72-h serum deprivation induced the up-regulation of the synthesis of mitochondrial proteins and promoted cell metabolic reprogramming from glycolytic to oxidative metabolism (23). In an in vivo study, liver cells of 24-h-fasted mice showed an increased oxidative phosphorylation activity, an effect mediated by the NAD+-dependent deacetylase sirtuin 3, a mitochondrial sensor of nutrients (24). Another work demonstrated that 72-h deprivation of amino acids increased maximum OCR in HEK cells (25). Here we demonstrated that short-term starvation does not increase the basal or maximum respiratory capacity of cells, but it is important to note that in our experiments cells were starved for only 1 h, whereas in all other studies the effects of starvation were evaluated after much longer periods (from 24 to 72 h). Thus, our experimental conditions would be considered as acute starvation and probably do not promote significant alterations in protein expression and in other long-term processes. Another difference is that in the cited studies cells were subjected to mild starvation, whereas here we performed the experiments in harsher conditions in which practically all oxidative nutrients were removed.
The use of cell lines as model systems to recapitulate specific metabolic phenotypes can be limited, especially because they often carry genetic alterations that commonly induce shifts in their metabolic profiles. In this sense, our findings also provide some clues about the validity of the cell lines chosen here as models for metabolic studies. The N2A cell line was established in 1969 from a spontaneous brain tumor found in an albino mouse (26). These cells were shown to display many features found in the original tissue, such as the capacity to form synaptic contacts (27), presence of glutamate receptors and excitable membrane (28), expression of specific functional neuron enzymes (29), and ability to differentiate into neurons (30). Neurons characteristically are highly metabolically active cells that present poor capacity for nutrient storage (31). Here we observed that this is also true for N2A cells because they were much more sensitive to the effects of nutrient deprivation than Huh7 and C2C12 cells with autophagy being induced after 1-h starvation in N2A but not in the other cells. Additionally, we found that N2A cells showed a high capacity of oxidizing pyruvate in accordance with the known neuronal utilization of astrocyte-derived pyruvate, which rescues cells from harmful drops in energy charge (32, 33). Regarding glutamine metabolism, it is known that neurons can largely use this amino acid to synthesize the neurotransmitters glutamate and γ-aminobutyric acid (GABA) (34), explaining our observations that N2A cells have limited capacity of oxidizing glutamine in mitochondria compared with other substrates. In addition, among the cells lines tested, N2A displayed the highest glycolytic capacity, which is a typical feature of neuron cells. Therefore, N2A cells responded to nutrient deprivation and mitochondrial oxidation of substrates as expected for neuronal cells, validating this cell line as a good model for studying neuronal metabolic adaptations. C2C12 cells, which are committed to myogenesis to become multinucleated myotubes in culture under low-serum growth medium, are phenotypically assigned as proliferating myoblasts (35, 36). In contrast to N2A and Huh7 cells, C2C12 was the only cell line able to mobilize glutamine upon starvation, which is in accordance with the typical high protein content profile of muscle cells, a possible source of amino acids that can be used to supply the energetic demand in conditions of low nutrient availability. It is known that undifferentiated C2C12 cells are metabolically different from C2C12 myotubes, which are highly oxidative, reaching respiratory rates 4-fold higher than those of C2C12 myoblasts (37). In agreement with this, here we showed that C2C12 myoblasts are not able to oxidize exogenous palmitate, but after differentiation, the resulting myotubes acquire this ability as expected for muscle cells. Finally, the Huh7 hepatocarcinoma cell line is largely used as a model for studying drug toxicity (38–40) as these cells express and modulate functional cytochrome P450 and several other specific hepatic genes involved in drug metabolization. In addition, this cell line shows several other liver-related activities, such as the synthesis of lipoproteins, plasma proteins, and others (41–43). Huh7 cells are also a reliable model for studying hepatic viral infection, sustaining infection of hepatotropic viruses, such as hepatitis C virus (44) and dengue virus (45). Here we showed that Huh7 cells were able to oxidize exogenous palmitate, a characteristic feature of hepatocytes. Indeed, these cells have been used for studying lipid metabolism in different contexts (46–48) and seem to be a good model for this purpose. Thus, the observed differences in respirometric behavior among the cell lines tested here validate our method as a reliable approach for studying metabolic adaptations.
Our findings also have important implications for shedding light on the mechanisms involved in the Crabtree effect, which is still not well understood. Our results clearly demonstrated that glucose does not inhibit mitochondrial maximum OCR, suggesting that the Crabtree effect does not depend on the direct inhibition of complex I–IV of the mitochondrial ETS, but we do not discard the involvement of ATP synthase. Maximum respiration corresponds to the capacity of the mitochondrial respiratory complexes to carry electrons but bypasses the contribution of ATP synthase as the presence of FCCP allows protons to cross the inner mitochondrial membrane from the intermembrane space to the matrix independently of ATP synthase activity. In addition, in our experiments, ATP synthase was inhibited with oligomycin before determination of maximum respiration. In agreement with our results, many of the current hypotheses that try to explain the Crabtree effect suggest that it is directly or indirectly related to the coupled respiration. For instance, the lack of ADP availability or changes in phosphate potential in mitochondria are considered causative events of the Crabtree effect (12). Another hypothesis suggests that glucose induces an increase in the Ca2+ intake into mitochondria, which would in turn inhibit ATP synthase (12, 49). Conversely, the hypothesis that the glycolytic intermediate fructose 1,6-bisphosphate mediates the Crabtree effect by inhibiting mitochondrial complexes III and IV (50) is not supported by the findings presented here because it implies that the Crabtree effect would also be seen when the maximum respiration is measured. We do not exclude the possibility that fructose 1,6-bisphosphate may inhibit mitochondrial complexes III and IV in isolated mitochondria, but our data show that this is unlikely to happen in intact cells, at least for those we tested.
An intriguing result is that the glucose-induced inhibition of mitochondrial respiration occurs when glutamine is the only substrate provided, but not in the case of pyruvate. In accordance with our results, the reciprocal regulation of mitochondrial oxidation of glucose and glutamine has already been described using 14C-labeled substrates (22). One possibility is that glucose impairs the transport of glutamine into mitochondria, which does not occur during starvation due to reduced utilization of available glutamine (and other substrates) in this condition. In this regard, uncoupling protein 2, a member of the mitochondrial anion carrier protein family, harbors a mild uncoupling activity and other metabolic functions which harbors a mild uncoupling activity and other metabolic functions still not completely established, was recently shown to regulate the transport of substrates into mitochondria, enhancing glutaminolysis and limiting mitochondrial oxidation of acetyl-CoA-producing substrates, in a manner similar to glucose (51, 52). The role of uncoupling protein 2 in mediating the Crabtree effect is an interesting possibility to be explored further. Considering the obvious importance of better understanding the effects of glucose in cellular metabolism, more efforts should be made to unravel the molecular mechanisms of the Crabtree effect, an open question that continues to persist almost one century since its discovery.
Experimental procedures
Cell culture
Cells were cultivated in the following media: Huh7 cells in DMEM, N2A cells in α-minimum essential medium, and C2C12 cells in DMEM-high glucose, all supplemented with 10% fetal bovine serum (complete medium) in the presence of the antibiotics penicillin (100 units/ml) and streptomycin (60 μg/ml) in a humidified incubator at 37 °C and 5% CO2. For starvation, cells were washed in phosphate-buffered saline (PBS) and incubated in DMEM-A supplemented with 2.4 g/liter sodium bicarbonate and 2 g/liter HEPES in a humidified incubator at 37 °C and 5% CO2 for the periods indicated in each experiment.
Viability assays
Cell viability was determined using Muse® Count and Viability Assay kit (Millipore, catalog number MCH100102) in a Muse Cell Analyzer.
BSA conjugation with palmitate
Preparation of BSA-conjugated palmitate was performed as described by the protocol developed by Seahorse Bioscience Inc. (North Billerica, MA) (53).
Oxygraphy in intact cells
Approximately two million non-starved or starved cells in DMEM-A were charged in OROBOROS Oxygraph-2k chambers. After stabilization of the signal, basal OCR was recorded, and cells were incubated or not (endogenous OCR) with different substrates as indicated in each experiment. Then cells were treated with 250 nm oligomycin, and uncoupled OCR was recorded followed by titration with FCCP for determination of their maximum respiratory capacity. For determination of non-mitochondrial OCR, at the end of the experiment cells were treated with rotenone (0.5 mm) alone or rotenone plus antimycin (3.65 μm) to inhibit mitochondrial respiratory complexes I and III, respectively. Coupled OCR was calculated by subtracting uncoupled from basal OCR. Non-mitochondrial OCR was subtracted from all data before being used in the analyses. We did not detect significant differences in cell viability before and after the experiments.
Western blotting
Cells were washed with cold PBS and subjected to lysis in buffer A (20 mm HEPES, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA,1% Triton X-100, 10% glycerol, 1 mm PMSF, 2 μg/ml aprotinin, 2 μg/ml pepstatin A, and 2 μg/ml leupeptin, pH 7.3–7.4). Cell debris was removed by centrifugation, and proteins were subjected to SDS-PAGE/Western blotting with the following antibodies: phosphorylated p70 S6K (Thr-389) (catalog number 9206, Cell Signaling Technology), total p70 S6K (catalog number 2708, Cell Signaling Technology), phosphorylated S6 (P-S6) (Ser-240/244) (catalog number 2215, Cell Signaling Technology), total S6 (catalog number 2317, Cell Signaling Technology), LC3 (catalog number 3868, Cell Signaling Technology), p62 (catalog number ab56416, Abcam), and α-tubulin (catalog number sc-23498, Santa Cruz Biotechnology). Immunoreactive signals were visualized by enhanced chemiluminescence using SuperSignal chemiluminescent substrate (Life Technologies) with a UVITEC imaging system. The quantification of bands was done using Alliance software.
Quantitative PCR
Total RNA was extracted from the cells using an Illustra RNAspin Mini kit (GE Healthcare). Reverse transcription (RT) was carried out with 2 μg of total RNA using Superscript III reverse transcriptase (Invitrogen), and extraction of mitochondrial and genomic DNA was performed with DNAzol® reagent (Thermo Fisher). For quantitative PCR, expression levels were determined in triplicate in a StepOne Plus System using SYBR® Green PCR Master Mix (Applied Biosystems). mRNA data were normalized to ribosomal protein L19 expression, and mitochondrial DNA data were normalized to genomic DNA through amplification of a fragment of the triose-phosphate isomerase gene. For each reaction, primer efficiencies were calculated using LinRegPCR software (version 12.17), and relative quantitation was obtained according to Pfaffl (54). Reactions were performed with the following specific primers for human or mouse sequences: Mus musculus mitochondrial ND1, 5′-AACCCTAGCAGAAACAAACCG-3′ (forward) and 5′-CCGGCTGCGTATTCTACGTT-3′ (reverse); M. musculus RPL19, 5′-CTCGTGTCTGTACATAGCGG-3′ (forward) and 5′-CTAAACAAGAGGGCAACAGACAA-3′ (reverse); human mitochondrial ND1 TCAACCTACGCCCTGATCGG-3′ (forward) and 5′-GGAGAGGTTAAAGGAGCCACT-3′ (reverse); human RPL19, 5′-TGGGCTGATCATCCGCAAGCC-3′ (forward) and 5′-CCCATGTGCCTGCCCTTCCG-3′ (reverse); and human triose-phosphate isomerase, 5′-GAAGGTTGTTTTCGAGCAGACA-3′ (forward) and 5′-ATGGCCCACACAGGCTCATA-3′ (reverse).
Quantification of lactate in conditioned medium
Lactate measurement was performed by enzymatic assay in hydrazine/glycine buffer as described previously (55, 56). Data were normalized by protein content.
Citrate synthase activity
Citrate synthase activity was measured in reaction medium containing 5,5′-dithiobis(2-nitrobenzoic acid), acetyl coenzyme A, and cell lysate as described previously (57).
Statistical analyses
Bar graphs with two columns were analyzed with paired Student's t test, and bar graphs with three or more columns were analyzed by one-way analysis of variance followed by Bonferroni's multiple comparison post-test. All graphics and statistical analyses were performed using GraphPad Prism 5 software (version 5.01).
Author contributions
J. D. Z. and A. T. D. P. designed the experiments, interpreted the results, and wrote the manuscript. J. D. Z., L. O. F.-S., A. S. C., E. C.-L., and M. H. D. performed the experiments and analyzed the data. L. A. K. and A. G. contributed reagents, interpreted results, and assisted with the manuscript. All authors reviewed and approved the final version of the manuscript.
Acknowledgments
We thank Prof. Franklin David Rumjanek and Mariana Figueiredo Rodrigues for kindly providing the primers for human ND1 gene. We also thank Juan Pérez and Ana Sant'Anna for scientific discussions, Prof. Hugo Armelin for kindly allowing the use of his laboratory, and Glauce M. Barbosa for technical assistance.
This work was supported in part by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro Grant FAPERJ E-26/201.167/2014 and Conselho Nacional de Desenvolvimento Científico e Tecnológico Grant MCT/CNPq 306669/2013-7. The authors declare that they have no conflicts of interest with the contents of this article.
- ETS
- electron transport system
- OCR
- oxygen consumption rate
- FCCP
- carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
- p62
- protein 62
- LC3
- microtubule-associated protein 1A/1B light chain 3
- ND1
- NADH:ubiquinone oxidoreductase core subunit 1
- 2DG
- 2-deoxy-d-glucose
- BPTES
- bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide
- Rox
- residual oxygen consumption
- GLS
- glutaminase
- P-S6
- phosphorylated S6.
References
- 1. Yuan H. X., Xiong Y., and Guan K. L. (2013) Nutrient sensing, metabolism, and cell growth control. Mol. Cell 49, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rambold A. S., Cohen S., and Lippincott-Schwartz J. (2015) Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boya P., Reggiori F., and Codogno P. (2013) Emerging regulation and functions of autophagy. Nat. Cell Biol. 15, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lapuente-Brun E., Moreno-Loshuertos R., Acín-Pérez R., Latorre-Pellicer A., Colás C., Balsa E., Perales-Clemente E., Quirós P. M., Calvo E., Rodríguez-Hernández M. A., Navas P., Cruz R., Carracedo Á., López-Otín C., Pérez-Martos A., et al. (2013) Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 [DOI] [PubMed] [Google Scholar]
- 5. Schägger H., and Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang S. A., Pacold M. E., Cervantes C. L., Lim D., Lou H. J., Ottina K., Gray N. S., Turk B. E., Yaffe M. B., and Sabatini D. M. (2013) mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341, 1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weinberg S. E., and Chandel N. S. (2015) Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 11, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El-Bacha T., and Da Poian A. T. (2013) Virus-induced changes in mitochondrial bioenergetics as potential targets for therapy. Int. J. Biochem. Cell Biol. 45, 41–46 [DOI] [PubMed] [Google Scholar]
- 9. Racker E. (1974) History of the Pasteur effect and its pathobiology. Mol. Cell. Biochem. 5, 17–23 [DOI] [PubMed] [Google Scholar]
- 10. Diaz-Ruiz R., Rigoulet M., and Devin A. (2011) The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta 1807, 568–576 [DOI] [PubMed] [Google Scholar]
- 11. Crabtree H. G. (1929) Observations on the carbohydrate metabolism of tumours. Biochem. J. 23, 536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammad N., Rosas-Lemus M., Uribe-Carvajal S., Rigoulet M., and Devin A. (2016) The Crabtree and Warburg effects: do metabolite-induced regulations participate in their induction? Biochim. Biophys. Acta 1857, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 13. Pesta D., and Gnaiger E. (2012) High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol. Biol. 810, 25–58 [DOI] [PubMed] [Google Scholar]
- 14. Picard M., Taivassalo T., Ritchie D., Wright K. J., Thomas M. M., Romestaing C., and Hepple R. T. (2011) Mitochondrial structure and function are disrupted by standard isolation methods. PLoS One 6, e18317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robinson M. M., McBryant S. J., Tsukamoto T., Rojas C., Ferraris D. V., Hamilton S. K., Hansen J. C., and Curthoys N. P. (2007) Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). Biochem. J. 406, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carlsson C., Borg L. A., and Welsh N. (1999) Sodium palmitate induces partial mitochondrial uncoupling and reactive oxygen species in rat pancreatic islets in vitro. Endocrinology 140, 3422–3428 [DOI] [PubMed] [Google Scholar]
- 17. Andreyev A. Yu., Bondareva T. O., Dedukhova V. I., Mokhova E. N., Skulachev V. P., and Volkov N. I. (1988) Carboxyatractylate inhibits the uncoupling effect of free fatty acids. FEBS Lett. 226, 265–269 [DOI] [PubMed] [Google Scholar]
- 18. Lardy H. A., and Pressman B. C. (1956) Effect of surface active agents on the latent ATPase of mitochondria. Biochim. Biophys. Acta 21, 458–466 [DOI] [PubMed] [Google Scholar]
- 19. Egnatchik R. A., Leamy A. K., Noguchi Y., Shiota M., and Young J. D. (2014) Palmitate-induced activation of mitochondrial metabolism promotes oxidative stress and apoptosis in H4IIEC3 rat hepatocytes. Metabolism 63, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Moura M. B., Uppala R., Zhang Y., Van Houten B., and Goetzman E. S. (2014) Overexpression of mitochondrial sirtuins alters glycolysis and mitochondrial function in HEK293 cells. PLoS One 9, e106028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim-Muller J. Y., Kim Y. J., Fan J., Zhao S., Banks A. S., Prentki M., and Accili D. (2016) FoxO1 deacetylation decreases fatty acid oxidation in β-cells and sustains insulin secretion in diabetes. J. Biol. Chem. 291, 10162–10172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zielke H. R., Ozand P. T., Tildon J. T., Sevdalian D. A., and Cornblath M. (1978) Reciprocal regulation of glucose and glutamine utilization by cultured human diploid fibroblasts. J. Cell. Physiol. 95, 41–48 [DOI] [PubMed] [Google Scholar]
- 23. Liu Z., Sun Y., Tan S., Liu L., Hu S., Huo H., Li M., Cui Q., and Yu M. (2016) Nutrient deprivation-related OXPHOS/glycolysis interconversion via HIF-1α/C-MYC pathway in U251 cells. Tumour Biol. 37, 6661–6671 [DOI] [PubMed] [Google Scholar]
- 24. Liu L., Nam M., Fan W., Akie T. E., Hoaglin D. C., Gao G., Keaney J. F. Jr., and Cooper M. P. (2014) Nutrient sensing by the mitochondrial transcription machinery dictates oxidative phosphorylation. J. Clin. Investig. 124, 768–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson M. A., Vidoni S., Durigon R., Pearce S. F., Rorbach J., He J., Brea-Calvo G., Minczuk M., Reyes A., Holt I. J., and Spinazzola A. (2014) Amino acid starvation has opposite effects on mitochondrial and cytosolic protein synthesis. PLoS One 9, e93597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klebe R. J., and Ruddle F. H. (1969) Neuroblastoma: cell culture analysis of a differentiating stem cell system. J. Cell Biol. 43, 69A [Google Scholar]
- 27. Spoerri P. E., Glees P., and Dresp W. (1980) The time course of synapse formation of mouse neuroblastoma cells in monolayer cultures. Cell Tissue Res. 205, 411–421 [DOI] [PubMed] [Google Scholar]
- 28. Van der Valk J. B., and Vijverberg H. P. (1990) Glutamate-induced inward current in a clonal neuroblastoma cell line. Eur. J. Pharmacol. 185, 99–102 [DOI] [PubMed] [Google Scholar]
- 29. Thompson J. M., London E. D., and Johnson J. E. Jr. (1982) Ultrastructural, functional and biochemical characteristics of mouse and human neuroblastoma cell lines. Neuroscience 7, 1807–1815 [DOI] [PubMed] [Google Scholar]
- 30. Tremblay R. G., Sikorska M., Sandhu J. K., Lanthier P., Ribecco-Lutkiewicz M., and Bani-Yaghoub M. (2010) Differentiation of mouse Neuro 2A cells into dopamine neurons. J. Neurosci. Methods 186, 60–67 [DOI] [PubMed] [Google Scholar]
- 31. Spasić M. R., Callaerts P., and Norga K. K. (2009) AMP-activated protein kinase (AMPK) molecular crossroad for metabolic control and survival of neurons. Neuroscientist 15, 309–316 [DOI] [PubMed] [Google Scholar]
- 32. Matsumoto K., Yamada K., Kohmura E., Kinoshita A., and Hayakawa T. (1994) Role of pyruvate in ischaemia-like conditions on cultured neurons. Neurol. Res. 16, 460–464 [DOI] [PubMed] [Google Scholar]
- 33. Maus M., Marin P., Israël M., Glowinski J., and Prémont J. (1999) Pyruvate and lactate protect striatal neurons against N-methyl-D-aspartate-induced neurotoxicity. Eur. J. Neurosci. 11, 3215–3224 [DOI] [PubMed] [Google Scholar]
- 34. Sonnewald U., Westergaard N., Schousboe A., Svendsen J. S., Unsgård G., and Petersen S. B. (1993) Direct demonstration by [13C]NMR spectroscopy that glutamine from astrocytes is a precursor for GABA synthesis in neurons. Neurochem. Int. 22, 19–29 [DOI] [PubMed] [Google Scholar]
- 35. Blau H. M., Pavlath G. K., Hardeman E. C., Chiu C. P., Silberstein L., Webster S. G., Miller S. C., and Webster C. (1985) Plasticity of the differentiated state. Science 230, 758–766 [DOI] [PubMed] [Google Scholar]
- 36. Tannu N. S., Rao V. K., Chaudhary R. M., Giorgianni F., Saeed A. E., Gao Y., and Raghow R. (2004) Comparative proteomes of the proliferating C2C12 myoblasts and fully differentiated myotubes reveal the complexity of the skeletal muscle differentiation program. Mol. Cell. Proteomics 3, 1065–1082 [DOI] [PubMed] [Google Scholar]
- 37. Remels A. H., Langen R. C., Schrauwen P., Schaart G., Schols A. M., and Gosker H. R. (2010) Regulation of mitochondrial biogenesis during myogenesis. Mol. Cell. Endocrinol. 315, 113–120 [DOI] [PubMed] [Google Scholar]
- 38. Kogure T., Ueno Y., Iwasaki T., and Shimosegawa T. (2004) The efficacy of the combination therapy of 5-fluorouracil, cisplatin and leucovorin for hepatocellular carcinoma and its predictable factors. Cancer Chemother. Pharmacol. 53, 296–304 [DOI] [PubMed] [Google Scholar]
- 39. Sivertsson L., Ek M., Darnell M., Edebert I., Ingelman-Sundberg M., and Neve E. P. (2010) CYP3A4 catalytic activity is induced in confluent Huh7 hepatoma cells. Drug Metab. Dispos. 38, 995–1002 [DOI] [PubMed] [Google Scholar]
- 40. Choi S., Sainz B. Jr., Corcoran P., Uprichard S., and Jeong H. (2009) Characterization of increased drug metabolism activity in dimethyl sulfoxide (DMSO)-treated Huh7 hepatoma cells. Xenobiotica 39, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goldring C. E., Kitteringham N. R., Jenkins R., Lovatt C. A., Randle L. E., Abdullah A., Owen A., Liu X., Butler P. J., Williams D. P., Metcalfe P., Berens C., Hillen W., Foster B., Simpson A., et al. (2006) Development of a transactivator in hepatoma cells that allows expression of phase I, phase II, and chemical defense genes. Am. J. Physiol. Cell Physiol. 290, C104–C115 [DOI] [PubMed] [Google Scholar]
- 42. Nakabayashi H., Taketa K., Miyano K., Yamane T., and Sato J. (1982) Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42, 3858–3863 [PubMed] [Google Scholar]
- 43. Yao H., and Ye J. (2008) Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 cells. J. Biol. Chem. 283, 849–854 [DOI] [PubMed] [Google Scholar]
- 44. Sainz B. Jr., and Chisari F. V. (2006) Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma-derived cells. J. Virol. 80, 10253–10257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee Y. R., Lei H. Y., Liu M. T., Wang J. R., Chen S. H., Jiang-Shieh Y. F., Lin Y. S., Yeh T. M., Liu C. C., and Liu H. S. (2008) Autophagic machinery activated by dengue virus enhances virus replication. Virology 374, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heaton N. S., and Randall G. (2010) Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dávalos A., Goedeke L., Smibert P., Ramírez C. M., Warrier N. P., Andreo U., Cirera-Salinas D., Rayner K., Suresh U., Pastor-Pareja J. C., Esplugues E., Fisher E. A., Penalva L. O., Moore K. J., Suárez Y., et al. (2011) miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., and Takano T. (2004) Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta 1644, 47–59 [DOI] [PubMed] [Google Scholar]
- 49. Wojtczak L., Teplova V. V., Bogucka K., Czyz A., Makowska A., Wieckowski M. R., Duszyński J., and Evtodienko Y. V. (1999) Effect of glucose and deoxyglucose on the redistribution of calcium in Ehrlich ascites tumour and Zajdela hepatoma cells and its consequences for mitochondrial energetics. Further arguments for the role of Ca2+ in the mechanism of the Crabtree effect. Eur. J. Biochem. 263, 495–501 [DOI] [PubMed] [Google Scholar]
- 50. Díaz-Ruiz R., Avéret N., Araiza D., Pinson B., Uribe-Carvajal S., Devin A., and Rigoulet M. (2008) Mitochondrial oxidative phosphorylation is regulated by fructose 1,6-bisphosphate. A possible role in Crabtree effect induction? J. Biol. Chem. 283, 26948–26955 [DOI] [PubMed] [Google Scholar]
- 51. Pecqueur C., Bui T., Gelly C., Hauchard J., Barbot C., Bouillaud F., Ricquier D., Miroux B., and Thompson C. B. (2008) Uncoupling protein-2 controls proliferation by promoting fatty acid oxidation and limiting glycolysis-derived pyruvate utilization. FASEB J. 22, 9–18 [DOI] [PubMed] [Google Scholar]
- 52. Vozza A., Parisi G., De Leonardis F., Lasorsa F. M., Castegna A., Amorese D., Marmo R., Calcagnile V. M., Palmieri L., Ricquier D., Paradies E., Scarcia P., Palmieri F., Bouillaud F., and Fiermonte G. (2014) UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc. Natl. Acad. Sci. U.S.A. 111, 960–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seahorse Bioscience Inc. (2010) Preparation of bovine serum albumin (BSA)-conjugated palmitate. A substrate for measuring fatty acid oxidation [FAO] in the XF Analyzer, Seahorse Bioscience Inc., North Billerica, MA [Google Scholar]
- 54. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pereira da Silva A. P., El-Bacha T., Kyaw N., dos Santos R. S., da-Silva W. S., Almeida F. C., Da Poian A. T., and Galina A. (2009) Inhibition of energy-producing pathways of HepG2 cells by 3-bromopyruvate. Biochem. J. 417, 717–726 [DOI] [PubMed] [Google Scholar]
- 56. Hamilton S. D., and Pardue H. L. (1984) Quantitation of lactate by a kinetic method with an extended range of linearity and low dependence on experimental variables. Clin. Chem. 30, 226–229 [PubMed] [Google Scholar]
- 57. Ramos-Filho D., Chicaybam G., de-Souza-Ferreira E., Guerra Martinez C., Kurtenbach E., Casimiro-Lopes G., and Galina A. (2015) High intensity interval training (HIIT) induces specific changes in respiration and electron leakage in the mitochondria of different rat skeletal muscles. PLoS One 10, e0131766. [DOI] [PMC free article] [PubMed] [Google Scholar]