Abstract
One characteristic of genomic plasticity is the presence of extrachromosomal circular DNA (eccDNA). High levels of eccDNA are associated with genomic instability, exposure to carcinogens and aging. We have recently reported developmentally regulated formation of eccDNA that occurs preferentially in pre-blastula Xenopus laevis embryos. Multimers of tandemly repeated sequences were over-represented in the circle population while dispersed sequences were not detected, indicating that circles were not formed at random from any chromosomal sequence. Here we present detailed mechanistic studies of eccDNA formation in a cell-free system derived from Xenopus egg extracts. We show that naked chromosomal DNA from sperm or somatic tissues serves as a substrate for direct tandem repeat circle formation. Moreover, a recombinant bacterial tandem repeat can generate eccDNA in the extract through a de novo mechanism which is independent of DNA replication. These data suggest that the presence of a high level of any direct tandem repeat can confer on DNA the ability to be converted into circular multimers in the early embryo irrespective of its sequence and that homologous recombination is involved in this process.
INTRODUCTION
Extrachromosomal circular DNA molecules (eccDNA, also known as small polydispersed circular DNA, spcDNA) consisting of chromosomal sequences have been observed in many eukaryotic tissues and cultured cells during the last three decades (1). The presence of these molecules reflects the plasticity of the eukaryotic genome and elevated levels of eccDNA are associated with aging, genomic instability and cancer (2). However, until recently not much was known about their biological role or the mechanism by which they were formed. This was mainly due to the lack of a suitable technique for detection of eccDNA from a small number of cells and the lack of a convenient physiological experimental system that produces eccDNA.
A neutral–neutral 2-dimensional (2D) gel electrophoresis technique facilitates the detection of eccDNA in relatively small amounts of genomic DNA (2,3). Using this method we have recently shown that such molecules are present and specific to the earliest steps of embryonic development of Xenopus laevis, the pre-blastula stage (4). The amount of eccDNA is dramatically reduced in later developmental stages and is not detected in adult tissues. The production of eccDNA does not proceed at random as multimers of the tandemly repeated sequence satellite 1 (5) are over-represented in the circle population while other sequences (such as rDNA and the dispersed repetitive JCC31 element) were not detected.
Analysis of eccDNA in different organisms showed that tandem repetitive sequences were over-represented. Indeed, circular multimers of tandem repeats have been observed in HeLa cells (6,7) and in Drosophila embryos (8–10).
In the yeast Saccharomyces cerevisiae extrachromosomal circles consisting of multimers of the rDNA gene cluster accumulate upon aging and in mutants of Sgs1, the homolog of the RecQ family helicases (11). In humans mutations in the members of this family cause genetic disorders, including genome instability and predisposition to cancer. Genetic studies show that homologous recombination through the Rad52 pathway is involved in formation of rDNA circles in yeast (12). However, in higher eukaryotes genetic and biochemical analyses of eccDNA formation remain to be performed.
We reported that the machinery involved in circle formation during X.laevis early development is stored in the egg (4). In the present work we have investigated the mechanism of circle formation using Xenopus egg extracts as a model system. This system is capable of generating eccDNA de novo upon incubation with demembranated sperm nuclei. We show that naked DNA, either from sperm or from somatic tissues, is an efficient substrate for circle formation in vitro. We also present evidence that sequences which are organized in tandem can be efficiently converted into circles by the egg extract. Incubation in egg extracts of a recombinant cloning vector consisting of direct tandem plasmid fragments results in the production of a series of circular multimers of the repeated unit. In addition, we show that DNA synthesis is involved in eccDNA formation in a process that does not depend on normal DNA replication.
MATERIALS AND METHODS
Detailed protocols available at http://www.igh.cnrs.fr/equip/mechali/. Egg extracts
Preparation of low and high speed egg extracts was performed as previously described (4,13).
Preparation of DNA
Total genomic DNA was prepared by rapidly homogenizing sperm cells, erythrocytes or spleen tissue in 30 mM EDTA, 1% SDS, 0.5% Triton X-100 and 0.3 M NaCl and then incubating at 37°C for 16 h with 0.1 mg/ml proteinase K. The DNA was extracted with phenol/chloroform, precipitated with ethanol and digested with 0.2 mg/ml RNase A for 2 h at 37°C. After ethanol precipitation and resuspension in 10 mM Tris, 1 mM EDTA (pH 8) DNA samples were ready for further manipulation.
When cell-free systems were used reactions were stopped by addition of 5 vol of 30 mM EDTA, 1% SDS, 0.5% Triton X-100, 0.3 M NaCl and total DNA was prepared as above.
Charomid 9-42 (14) was obtained from C.Jaulin (I.G.H.) and was further propagated in RecA– bacteria.
Neutral–neutral 2D electrophoresis
Separation of DNA in neutral–neutral 2D gels was performed according to Brewer and Fangman (15), with modifications as described (2,3). Briefly, the DNA was first separated on 0.4% agarose at low voltage in 1× TBE and then the gel was rinsed in TBE containing 0.3 µg/ml ethidium bromide (EtBr). The lane of choice was cut out and placed on a clean gel support at 90° to the direction of the first electrophoresis. The lane was cast with 1.1% agarose containing 0.3 µg/ml EtBr and was run in 1× TBE in the presence of 0.3 µg/ml EtBr. The first dimension was run overnight at 1 V/cm and the second at 4 V/cm for 5 h at room temperature.
Blotting and hybridization
Southern blot analyses were performed with Hybond N+ nylon membranes (Amersham, UK) using probes labeled with a Ready-To-Go kit (Pharmacia). Radiolabeled DNA was detected by autoradiography (Hyperfilm MP; Amersham) and with a PhosphorImager (Molecular Dynamics) for quantification with ImageQuant 1.1 software.
RESULTS
Naked sperm DNA is a substrate for eccDNA formation
In a previous work we presented a cell-free system in which eccDNA is formed de novo when incubating demembranated sperm nuclei in activated egg extracts (4). We used this system and the 2D gel detection assay (Fig. 1A) to characterize the mechanism by which the circles are formed. Satellite 1 DNA (5), an over-represented sequence in the population of circular DNA (4), was used as a probe for genomic circular DNA.
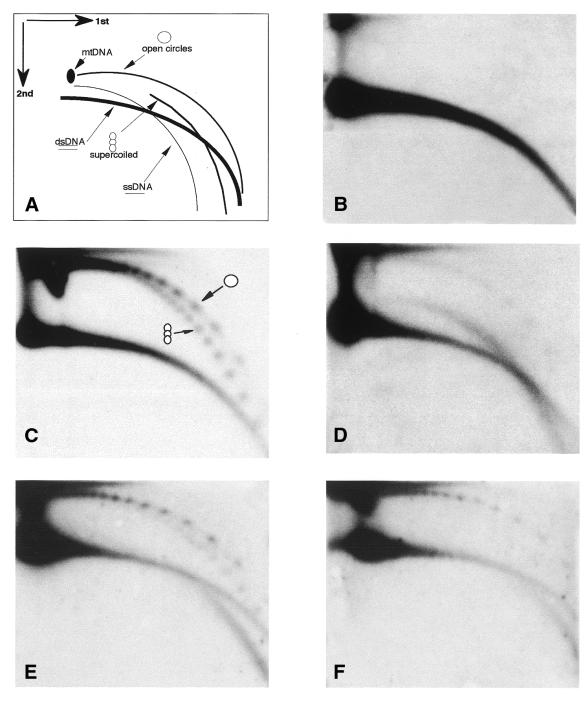
Figure 1.
Circular multimers of satellite 1 are formed in vitro from naked sperm DNA and DNA from adult tissues. (A) A scheme of 2D gel electrophoretic patterns of populations of linear and circular DNA molecules (2,3). Each arc consists of molecules that share the same structure but differ in their molecular mass. dsDNA, double-stranded DNA; ssDNA, single-stranded DNA; mtDNA, mitochondrial DNA; open circles, circular dsDNA; supercoiled, supercoiled dsDNA. (B–D) DNA was purified from sperm nuclei and treated with S1 nuclease to eliminate ssDNA. An aliquot of 300 ng was incubated for 2 h in 100 µl of low speed Xenopus egg extract. Following incubation the DNA was extracted, analyzed by 2D gel electrophoresis (C), along with non-incubated DNA (B), and hybridized with a satellite 1 probe. Sperm nuclei (270 ng DNA) were analyzed under the same conditions (D). Similar incubation conditions and 2D gel analysis were applied to 100 ng genomic DNA purified from Xenopus erythrocytes (E) and spleen (F).
We asked whether naked DNA could serve as a substrate for circle formation or whether some aspects of the structure of sperm nuclei is crucial for the generation of extrachromosomal circles. Total DNA purified from sperm nuclei and treated with S1 nuclease to remove any single-stranded (ss)DNA that might be present was incubated with egg extract and the reaction products were analyzed on a 2D gel. Hybridization with a satellite 1 DNA probe revealed massive formation of extrachromosomal circular multimers of satellite 1 which are not detected in the input DNA (Fig. 1B and C). Both circular and supercoiled forms were generated, as previously shown for total sperm nuclei (4). We conclude that the egg extract can form eccDNA from naked chromosomal DNA.
DNA incubated in egg extract assembles into chromatin and can form nuclei (16,17). However, production of eccDNA occurs rapidly in the extract and our data suggest that neither chromatin assembly nor formation of nuclei is a prerequisite for production of eccDNA (see below). The amount of satellite 1 circular multimers was higher when using naked DNA as substrate than with the demembranated sperm nuclei control in the same extract (compare Fig. 1C and D). Similarly, direct labeling experiments demonstrated higher amounts of labeled eccDNA when naked DNA was used as the template compared with a sperm nuclei template (data not shown), suggesting that naked DNA is more accessible than chromatin to the machinery that forms circular DNA.
Circular molecules can be generated in vitro from DNA of somatic tissues
eccDNA is specific to the early stages of embryogenesis and is not detected in somatic tissues (4). To test whether the DNA of differentiated somatic cells can serve as a template for circle formation we incubated DNA purified from adult erythrocytes (Fig. 1E) and spleen (Fig. 1F) or DNA from cultured Xenopus A6 cells (data not shown) in egg extract. In all cases circular multimers of satellite 1 appeared after incubation in egg extract. We conclude that DNA from adult tissues and sperm DNA are equally good substrates for circle formation activity in the egg. This finding rules out the possibility that sperm DNA is somehow marked during spermatogenesis for targeting by the circle forming machinery.
Recombinant bacterial tandem repeats are converted in vitro into circular multimers of the repeated unit
Tandem repeats are highly represented in populations of circular DNA in humans, yeast, Drosophila and Xenopus. We asked whether egg extracts can generate circular molecules using a defined template DNA containing tandem repeats. Charomid 9-42, a 42 kbp recombinant cloning vector that contains 18 tandem repeats of a 2 kb fragment of pBR322 and 5 kb of vector (14) was used as a model substrate (Fig. 2A). Charomid DNA was incubated in egg extracts in the presence of [α-32P]dCTP and the DNA products were analyzed by 2D gel electrophoresis. We detected clear spot ladders of supercoiled and relaxed circular molecules (Fig. 2B) that are multiples of 2 kb, as determined by known markers which were run in parallel (data not shown). In contrast, incubation of λ DNA in the extract does not generate small circles (Fig. 2C), nor does incubation of several other plasmids (data not shown). These results suggest that eccDNA consisting of multimers of the 2 kb tandem repeat are formed by the egg extract and that the presence of repeats is necessary for formation of circular molecules.
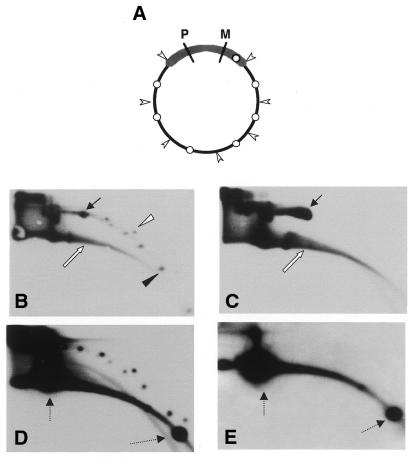
Figure 2.
Tandem repeats present in a recombinant vector are converted into circular multimers by low speed Xenopus egg extracts. Charomid 9-42, a cloning vector consisting of 18 tandem repeats of a 2 kb pBR322 fragment and a unique 5 kb vector sequence (14), was used. (A) Scheme showing the structure and restriction sites of the charomid. P, PvuI; M, MluI; arrowheads, AvaI sites separating the 2 kb repeats from each other and from the vector sequences; white circles, SalI sites in both the vector and each of the repeating units; thick line, vector sequence; thin line, multiple 2 kb repeats. For direct labeling (B and C), 300 ng of charomid (B) or λ DNA (C) were incubated for 90 min in 200 µl of low speed Xenopus egg extract supplemented with 50 µCi [α-32P]dCTP. After purification the DNA was separated on a 2D gel and exposed to film. Linear DNA (white arrow) and mitochondrial DNA (black arrow) are labeled in both cases (B and C). However, supercoiled (black arrowhead) and relaxed (white arrowhead) circular molecules are labeled only in the charomid reaction (B). For hybridization analysis (D and E), 300 ng of charomid DNA was incubated for 90 min in a low speed egg extract (in the absence of [α-32P]dCTP), purified and cleaved with MluI and PvuI, which cut in the vector sequence and leave the tandem repeats intact (D) (see also A). Charomid DNA that was not incubated in the extract was also cleaved (E). The DNA was separated on a 2D gel and hybridized with a charomid probe. Dashed arrows indicate the linear MluI–PvuI cleavage products.
To further confirm specific production of the 2 kb multimers we digested the reaction products with the restriction enzymes PvuI and MluI, which cleave the charomid only in the vector sequence, leaving the repeats intact (Fig. 2A). In this step the original cloning vector, as well as possible smaller variants that may have occurred in the bacteria, should be linearized but circular molecules consisting of the repeats should remain intact. The charomid DNA was incubated in egg extract and the digestion products analyzed by 2D gel electrophoresis followed by hybridization with a total charomid DNA probe. Figure 2D shows that the circular molecules were resistant to PvuI and MluI digestion. Such molecules were not present in the original charomid population (Fig. 2E), thus they were produced de novo in the extract. In addition, digestion with SalI, which cuts in the repeats, abrogated the circles (data not shown), confirming that they consisted of repeated charomid fragment. We conclude that the egg extract recognizes tandem repeats and can generate circular molecules from these repeated units.
Labeled circular molecules are formed in high speed egg extracts
Xenopus low speed extracts are efficient in DNA replication (18). eccDNA molecules produced in the extract contain newly synthesized DNA but their formation does not require DNA replication (4). This result suggests that eccDNA is excised from newly replicated chromosomal DNA in the Xenopus low speed egg extracts or, alternatively, that the newly synthesized DNA might be acquired by replication or repair after excision from the genome. To address this question we used high speed egg extracts in which membrane vesicle precursors are eliminated (18,19). These extracts cannot form nuclei upon addition of DNA and are unable to initiate double-stranded (ds)DNA replication (18,19). However, they still retain chromatin assembly activity (17,20) as well as the ability to convert ssDNA to dsDNA (19).
When demembranated sperm nuclei were incubated in high speed extracts, eccDNA molecules were produced in a similar manner to that observed in low speed extracts (Fig. 3A). Similar results were obtained with charomid DNA (Fig. 4) and with naked sperm DNA (data not shown). We conclude that the production of eccDNA does not require formation of nuclei nor the activity necessary for chromosomal DNA replication. However, when radiolabeled dCTP was added to the reaction the extrachromosomal circular molecules were labeled (Fig. 3B) and this reaction was sensitive to aphidicolin, an inhibitor of DNA polymerase α/δ (Fig. 3C). Note that phosphorimager quantification of the relative fraction of circular DNA from the total labeling (Figs 3, 4 and 6 and data not shown) indicates that a significant proportion, up to 33% of the labeling, is found in eccDNA. This suggests that eccDNA formation is a major DNA synthesis process in the absence of chromosomal replication.
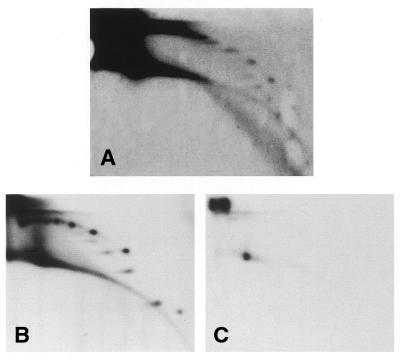
Figure 3.
High speed egg extracts can form circular DNA. (A) Sperm nuclei were incubated in a high speed egg extract for 90 min. DNA was purified and analyzed by 2D gel electrophoresis and hybridization with a satellite 1 probe. (B and C) Charomid DNA (100 ng) was incubated in 50 µl of a high speed egg extract supplemented with 25 µCi [α-32P]dCTP for 90 min, in the absence (B) or presence (C) of aphidicolin (50 µg/ml). DNA was purified and analyzed by 2D gel electrophoresis, blotted and directly exposed to film.
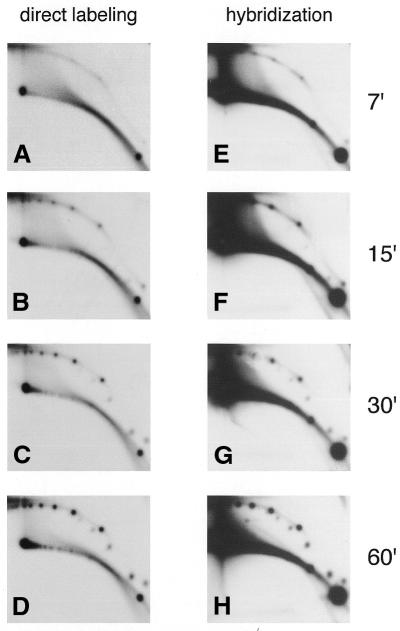
Figure 4.
Kinetics of circle formation in high speed extract. Charomid DNA (300 ng) was incubated in 100 µl of a high speed extract supplemented with 50 µCi [α-32P]dCTP. Aliquots were taken at 7, 15, 30 and 60 min incubation. DNA was purified, cleaved with MluI and PvuI and separated on a 2D gel, which was blotted and directly exposed to film to detect the newly labeled molecules (direct labeling) (A–D). The blot was then hybridized with a charomid probe to reveal the total charomid DNA in each sample (hybridization) (E–H).
Figure 6.
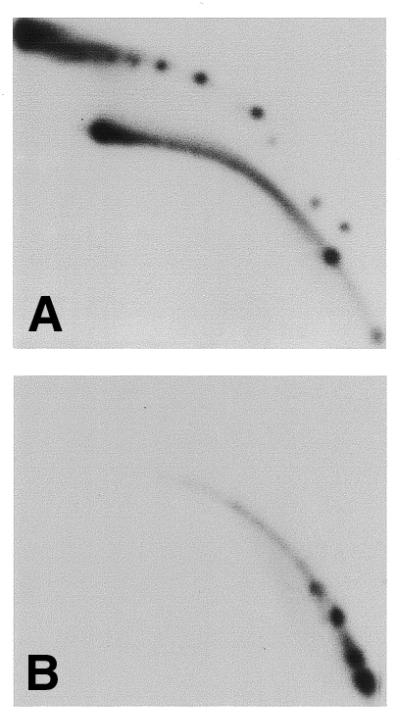
Labeled eccDNA is not a product of DNA replication. Charomid DNA (200 ng) was incubated in high speed extracts supplemented with 50 µCi [α-32P]dCTP for 90 min. The purified DNA was cleaved with MluI and PvuI and was then divided into two fractions. One fraction was digested with DpnI (B), whereas the second was not (A). The DNA was separated on a 2D gel, which was blotted and directly exposed to film.
Altogether, this series of experiments indicates that DNA synthesis is part of the process that leads to the formation of eccDNA. However, this activity is not related to the initiation of DNA replication on a dsDNA molecule, which does not occur in high speed extract.
Kinetics of eccDNA formation in high speed extracts
To further investigate the mechanism involved, the kinetics of eccDNA formation were analyzed by 2D gel electrophoresis by following either the total DNA population (by hybridization) or labeled molecules formed in the extract in the presence of [α-32P]dCTP. To avoid any background that could arise from DNA replication the reactions were performed in high speed extracts. Figure 4 further confirms that no signal for replicating intermediates, which migrate with characteristic features in these 2D gels, were detected (21). Labeled circular DNA molecules first appeared as open circles (Fig. 4A), whose amount increased with time (Fig. 4B), and, later on, supercoiled molecules were detected (Fig. 4C and D). Comparison of the data obtained by direct labeling (Fig. 4A–D) and hybridization (Fig. 4E–H) revealed identical results, with no additional non-labeled intermediates. These results show that DNA synthesis occurs concomitantly with production of eccDNA.
The population of spots that reflect multimers of discrete sizes appears relatively rapidly, with kinetics much faster than chromatin assembly, which takes 2–4 h under these conditions (20). Early time points also reveal a background of heterogeneous open circles centered around each spot (Fig. 4A and E) that remains at a constant level during the whole incubation, but represents a minor population at the end of the incubation (Fig. 4D and H). Pulse labeling experiments done at early or late incubation times have indicated that the heterogeneous open circles represent a sub-population of molecules formed during an early step of incubation. This population represents a repair mechanism involving end joining of a minor population of broken molecules present in the original charomid DNA which is not related to multimer formation (data not shown).
Circular DNA is formed in the absence of DNA replication
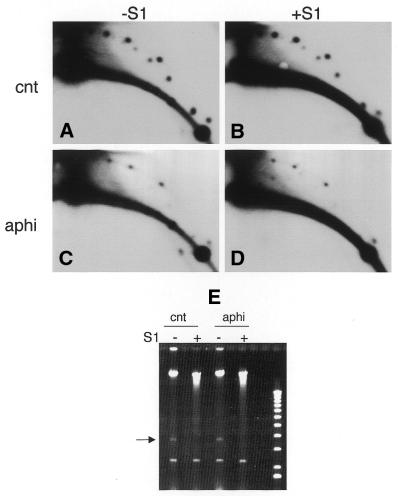
eccDNA can be formed in the presence of the DNA polymerase inhibitor aphidicolin (4), while Figure 3 shows that eccDNA contains newly synthesized DNA in the absence of chromosomal DNA replication. Therefore, although formation of extrachromosomal circles does not require DNA replication, some DNA synthesis may still be involved. Figure 5 confirms that in high speed extracts deficient in dsDNA replication extrachromosomal circles are still produced in the presence of aphidicolin. However, this population was much smaller (Fig. 5C) than the control (Fig. 5A).
Figure 5.
The level of eccDNA is strongly reduced in the presence of aphidicolin and is resistant to nuclease S1. Charomid DNA was incubated in high speed extract for 60 min in the absence (A and B) or presence of 50 µg/ml aphidicolin (C and D). DNA was purified and cleaved with MluI and PvuI. Single-stranded φX174 DNA (50 ng) was added to each sample and then they were divided into two aliquots. One aliquot of each sample was treated with nuclease S1 (+S1) (B and D) and the second was incubated with the nuclease reaction buffer (–S1) only (A and C). Reaction products were analyzed on 2D gels, blotted and hybridized with charomid probe. (E) Ethidium bromide staining of the first dimension showing the disappearance of φX174 ssDNA used as an internal control to monitor digestion by nuclease S1 (arrow).
To analyze whether the extrachromosomal circles contained single-stranded region we also digested the reaction products with S1 nuclease (Fig. 5B and D). The same patterns of charomid eccDNA were observed before and after digestion, indicating that the final eccDNA population did not contain single-stranded gaps, even in the presence of aphidicolin.
φX174 ssDNA was added to the sample as a digestion control (Fig. 5E). S1 nuclease activity was demonstrated by disappearance of the φX174 bands in the ethidium bromide stained gel (Fig. 5E), as well as following hybridization of the samples in Figure 5A–D with a φX174 probe (data not shown).
The charomid template grown in bacteria is dam methylated and is therefore sensitive to DpnI, which cleaves at multiple sites along the repeated sequence. DNA synthesis in eukaryotic systems is not accompanied by this methylation and thus one round of replication generates hemimethylated DNA, which is resistant to this enzyme. Therefore, if the labeled eccDNA molecules contain fully replicated DNA they should be resistant to DpnI. As shown in Figure 6, the labeled circles were sensitive to DpnI, indicating that they contained non-replicated parental dsDNA.
We conclude that in high speed extracts eccDNA is labeled by a process which does not involve DNA replication. A mechanism for eccDNA formation that involves exonuclease digestion followed by gap filling could account for the data observed. Exonuclease activity has been detected in the mature egg and in the early embryo (22) and this activity is followed by DNA gap filling in the extract. If such a mechanism is involved in eccDNA production filling in will be inhibited in the presence of aphidicolin, leading to a circle population prone to breakage in the exposed single-stranded region and linearization. This explanation is consistent with the decreased level of eccDNA in the presence of aphidicolin (Fig. 5C).
DISCUSSION
The production of eccDNA during early Xenopus development can be reproduced in vitro using Xenopus egg extracts and naked DNA or plasmids carrying tandem repeat sequences. eccDNA can also be generated with genomic DNA from either sperm nuclei or differentiated tissues. Therefore, specific imprinting of the sperm nuclei is not required for the reaction to occur during early embryogenesis. The efficiency of the reaction using a cosmid containing bacterial tandem repeats also argues in favor of a lack of sequence specificity for generation of the circles. Since recognition of substrates for eccDNA formation depends only on the organization and not the sequence of the DNA, an artificial molecule such as a charomid can serve as an efficient substrate for circle formation in vivo. Hence, this study offers a new system for studying eccDNA formation.
Our observations suggest that formation of eccDNA depends on a mechanism which involves intramolecular homologous recombination and does not require DNA replication. This is supported by a recent genetic analysis showing that formation of extrachromosomal circular rDNA in yeast occurs by homologous recombination, with formation of circular multimers of the ribosomal gene clusters through the RAD52 pathway (12).
Single-strand annealing (SSA) was reported as a recombination mechanism in the oocyte used to repair double-strand breaks (23). This mechanism might account for circular multimers of tandem repeats both in the presence and absence of DNA synthesis. However, such a mechanism requires two double-strand breaks to generate linear molecules that can be circularized and would require massive random chromosomal breakage in the early embryo. Chromosomal breakage has not been observed in the early embryo nor in the extracts. Moreover, if such natural random breakage occurred, other non-tandem sequences would generate circles of heterogeneous size by end joining, which is the major repair activity (22), and such circles were not detected in vivo (4) or in vitro following incubation of λ DNA in egg extracts (Fig. 2C). Alternatively, chromosomal breakage specific for tandem repeats could occur, generating substrates that could be repaired by SSA. This possibility is unlikely, but cannot be ruled out.
DNA synthesis clearly participates in the mechanism that leads to formation of eccDNA. The activity does not depend on dsDNA replication but is sensitive to aphidicolin. It does not require the presence of ssDNA in the original population, since S1 nuclease pretreatment of the DNA incubated in the extract did not quantitatively or qualitatively affect the reaction (Fig. 1). If ssDNA regions were formed as part of the process leading to eccDNA then complementary DNA strand synthesis activity present in the extract would effectively fill the gaps (19). In addition, resistance of the incubation products to nuclease S1 (Fig. 5) indicates the absence of accumulated single-stranded intermediates even in the presence of aphidicolin.
The partial inhibition of eccDNA formation by aphidicolin may be explained by an unbalanced equilibrium of exonuclease activity and gap filling activity at a step between excision of the repeat and its circularization or after circularization. In this case nicked molecules would be subjected to exonuclease activity without the possibility for filling in. This would then lead to their linearization in the extract and not to their accumulation. This idea is also consistent with the observation that supercoiled eccDNA is more severely affected by aphidicolin, since according to our kinetic experiment (Fig. 4) this population appears later in the reaction. If in the absence of DNA synthesis newly formed open circles are prone to linearization during their formation or shortly after, then less molecules would exist in the supercoiled form.
Our results show that the mechanism of eccDNA formation is highly active on tandem repeats of DNA. The use of a charomid DNA template, in combination with 2D gel analysis, permits rapid assay for the presence of eccDNA-forming activity during embryonic development. It may also provide a powerful system for analyzing mutants in which chromosomal stability is affected. The presence of this activity in the unfertilized egg as well as during early development demonstrates an unexpected plasticity, which may have repercussions at two levels. First, it may affect genome integrity in the population. Secondly, it may result in modifications affecting DNA repeats at the level of single individuals within embryos from the same parents. Tandem repeats comprise large regions of heterochromatin in eukaryotes. They are mostly found in the centromeric and telomeric regions, but also exist along the arms of the chromosomes. One of the characteristics of tandemly repeated sequences is their plasticity/instability and heterogeneity between individuals (24,25). Clusters of repetitive coding genes (such as rDNA, histones and tRNA) are also organized in tandem and exhibit certain levels of plasticity. This plasticity is traditionally explained by classical recombination mechanisms such as gene conversion and unequal crossing-over during meiosis (for a review see 25). Our data show a mitotic plasticity of tandem repeats and suggests a further unusual level of recombination that affects these sequences during early development.
Acknowledgments
ACKNOWLEDGEMENTS
We thank D.Fisher and C.Jaulin for critical reading of this manuscript. We also thank C.Jaulin for helpful discussions during the course of this work. This study has been supported by grants from the CNRS, Association pour la Recherche sur le Cancer, Ligue Nationale contre le Cancer and Fondation pour la Recherche Médicale (FRM). S.C. was supported by post-doctoral fellowships from EMBO and FRM.
References
- 1.Gaubatz J.W. (1990) Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat. Res., 237, 271–292. [DOI] [PubMed] [Google Scholar]
- 2.Cohen S., Regev,A. and Lavi,S. (1997) Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene, 14, 977–985. [DOI] [PubMed] [Google Scholar]
- 3.Cohen S. and Lavi,S. (1996) Induction of circles of heterogeneous sizes in carcinogen-treated cells: two-dimensional gel analysis of circular DNA molecules. Mol. Cell. Biol., 16, 2002–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S., Menut,S. and Méchali,M. (1999) Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol. Cell. Biol., 19, 6682–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam B.S. and Carroll,D. (1983) Tandemly repeated DNA sequences from Xenopus laevis. I. Studies on sequence organization and variation in satellite 1 DNA (741 base-pair repeat). J. Mol. Biol., 165, 567–585. [DOI] [PubMed] [Google Scholar]
- 6.Kiyama R., Matsui,H. and Oishi,M. (1986) A repetitive DNA family (Sau3A family) in human chromosomes: extrachromosomal DNA and DNA polymorphism. Proc. Natl Acad. Sci. USA, 83, 4665–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiyama R., Matsui,H., Okumura,K. and Oishi,M. (1987) A group of repetitive human DNA families that is characterized by extrachromosomal oligomers and restriction-fragment length polymorphism. J. Mol. Biol., 193, 591–597. [DOI] [PubMed] [Google Scholar]
- 8.Pont G., Degroote,F. and Picard,G. (1987) Some extrachromosomal circular DNAs from Drosophila embryos are homologous to tandemly repeated genes. J. Mol. Biol., 195, 447–451. [DOI] [PubMed] [Google Scholar]
- 9.Degroote F., Pont,G., Micard,D. and Picard,G. (1989) Extrachromosomal circular DNAs in Drosophila melanogaster: comparison between embryos and Kc0% cells. Chromosoma, 98, 201–206. [DOI] [PubMed] [Google Scholar]
- 10.Stanfield S. and Helinski,D.R. (1976) Small circular DNA in Drosophila melanogaster. Cell, 9, 333–345. [DOI] [PubMed] [Google Scholar]
- 11.Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- 12.Park P.U., Defossez,P.A. and Guarente,L. (1999) Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menut S., Lemaitre,J.M., Hair,A. and Méchali,M. (1999) DNA replication and chromatin assembly using Xenopus egg extracts. In Richter,J.D. (ed.) Advances in Molecular Biology: A Comparative Methods Approach to the Study of Oocytes and Embryos. Oxford University Press, pp. 198–226.
- 14.Saito I. and Stark,G.R. (1986) Charomids: cosmid vectors for efficient cloning and mapping of large or small restriction fragments. Proc. Natl Acad. Sci. USA, 83, 8664–8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- 16.Newport J. (1987) Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell, 48, 205–217. [DOI] [PubMed] [Google Scholar]
- 17.Laskey R.A., Mills,A.D. and Morris,N.R. (1977) Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell, 10, 237–243. [DOI] [PubMed] [Google Scholar]
- 18.Blow J.J. and Laskey,R.A. (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell, 47, 577–587. [DOI] [PubMed] [Google Scholar]
- 19.Méchali M. and Harland,R.M. (1982) DNA synthesis in a cell-free system from Xenopus eggs: priming and elongation on single-stranded DNA in vitro. Cell, 30, 93–101. [DOI] [PubMed] [Google Scholar]
- 20.Almouzni G. and Méchali,M. (1988) Assembly of spaced chromatin: involvement of ATP and DNA topoisomerase activity. EMBO J., 20, 4355–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyrien O. and Méchali,M. (1993) Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J., 12, 4511–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehman C.W., Clemens,M., Worthylake,D.K., Trautman,J.K. and Carroll,D. (1993) Homologous and illegitimate recombination in developing Xenopus oocytes and eggs. Mol. Cell. Biol., 13, 6897–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll D. (1996) Homologous genetic recombination in Xenopus: mechanism and implications for gene manipulation. Prog. Nucleic Acid Res. Mol. Biol., 54, 101–125. [DOI] [PubMed] [Google Scholar]
- 24.Vogt P. (1992) Code domains in tandem repetitive DNA sequence structures. Chromosoma, 101, 585–589. [DOI] [PubMed] [Google Scholar]
- 25.Vogt P. (1990) Potential genetic functions of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved “chromatin folding code”. Hum. Genet., 84, 301–336. [DOI] [PubMed] [Google Scholar]