Abstract
Matrix metalloproteinases (MMPs) are considered important in articular cartilage breakdown during osteoarthritis (OA). Similarly, the endocannabinoid system (ECS) is implicated in joint function and modulation of nociceptive processing. Functional interplay between ECS and MMPs has been recently indicated. Here, we tested if changes in the expression of selected MMPs and major ECS elements temporally correlate with the intensity of OA-related pain. Knee OA was induced in male Wistar rats by intra-articular sodium monoiodoacetate injection. OA-like pain behavior was tested using the dynamic weight bearing. Joint tissue samples at different time points after OA induction were subjected to gene (quantitative polymerase chain reaction) and protein (Western blot) expression analyses. Monoiodoacetate-induced nocifensive responses in rats showed a biphasic progression pattern. The alterations in expression of selected MMPs elegantly corresponded to the two-stage development of OA pain. The most substantial changes in the expression of the ECS system were revealed at a later stage of OA progression. Alterations within ECS are involved in the process of adaptation to persistent painful stimuli. The accumulation of MMPs in osteoarthritic cartilage may have a role in the biphasic progression of OA-related pain. Temporal association of changes in ECS and MMPs expression shows a potential therapeutic approach that utilizes the concept of combining indirect ECS-mediated MMP inhibition and ECS modulation of pain transduction.
Keywords: osteoarthritis, monoiodoacetate, pain, matrix metalloproteinases, endocannabinoid system
Introduction
Osteoarthritis (OA) is a chronic degenerative condition of articular cartilage, which is composed of chondrocytes surrounded by ample extracellular matrix (ECM).1 The physiological function of healthy cartilage relies on the structural integrity and sustainable remodeling of the ECM. In OA, the ECM stability of articular cartilage is compromised by extensive proteolytic breakdown associated with alterations in protein synthesis leading to progressive cartilage loss and joint dysfunction.1,2
The matrix metalloproteinase (MMP) family of proteases is considered to play a key role in cartilage degradation.2 The expression of multiple genes encoding MMPs was found to be altered in osteoarthritic cartilage.3 Importantly, enhanced protein expression of MMPs and their increased proteolytic activity in osteoarthritic cartilage has also been shown.4–6 The imbalanced ratio of MMPs and their endogenous regulators, namely tissue inhibitors of metalloproteinases (TIMPs), may drive ECM catabolism.7 The MMP-neutralizing capacity of endogenous TIMPs appears to be insufficient in OA cartilage.8 Moreover, despite remarkable results obtained from OA animal models, synthetic MMP inhibitors have shown little or no clinical efficacy.8,9
The endocannabinoid system (ECS) plays a unique neuromodulatory role in pain processing.10 The ECS consists of the cannabinoid receptors CB1 and CB2; their endogenous ligands (endocannabinoids; ECs), including the main and most widely studied to date, anandamide (AEA); and the enzymes involved in EC synthesis and degradation.10 ECs, especially AEA, have shown analgesic properties in different animal pain models.10 Thus, a huge scientific effort has been invested to pharmacologically increase the available reservoir of AEA, mainly by inhibiting its degradation.11,12 The constitutive expression of both types of cannabinoid receptors on chondrocytes has been found.13 Moreover, the activity of ECS in knee joints contributes to the regulation of synovial blood flow and joint pain.14,15 A growing body of evidence suggests that ECS may serve as an emerging therapeutic target for OA pain.16,17 Indeed, both the systemic and local administration of ECS modulators in OA animal models have shown anti-nociceptive and anti-arthritic effects in the affected knee.15,16,18
Functional interplay between ECS and MMPs has been recently proposed.19–21 Several scientific reports indicate that the modulation of CB1 and CB2 receptors may result in the inhibition of MMP activity.19–21 Interactions between ECM-degrading enzymes and the ECS in articular cartilage homeostasis has been less intensively studied, nonetheless ECS-mediated MMPs inhibition has been also suggested.22 Of importance, both the systems were implicated to play a role in chronic pain pathophysiology.23,24
The correlation between the expression of MMPs and the ECS during the development of OA pain and its implications remain unclear. Taking into account the above premises, we hypothesized that alterations in MMPs and ECS expression during OA progression may be intertwined and reflect the severity of pain. Therefore, we investigated OA-related pain behavior in osteoarthritic rats and confronted it with the expression of ECM-degrading enzymes and key ECS components in disease-affected joints. Understanding the changes within these two regulatory systems in the context of chronic OA pain could lay the foundation for a new therapeutic approach targeting both MMPs inhibition and ECS-mediated analgesia.
Methods
Animals
Male 60-day old Wistar rats (Charles River, Hamburg Germany) initially weighing 225–250 g were housed 5 per cage under a standard 12/12 h light/dark cycle and had free access to food and water. We ran our behavioral analyses in the morning (between 8:00 to 11:00 a.m.) in the animal house (conventional maintained in open conditions and specified pathogen free [maintained in barrier conditions] standards). We ensured all animals had the opportunity to exhibit their natural behaviors, we looked after their psychological and physical well-being to minimize any discomfort. No important adverse events were observed. The procedures on animals were performed following the recommendations of the International Association of Studies on Pain and 3R policy.25 The study was approved by the Local Ethics Committee of the Institute of Pharmacology PAS (approval number 938/2012). Across the experiments, we compared non-treated animals (further referred to as intact animals) and osteoarthritic animals at different time points after treatment. Our previous results showed no significant changes in joint hypersensitivity and dynamic weight bearing (DWB) tests in saline-injected rats.16 Therefore, a vehicle-treated group was not included in the experiment. For biochemical assays, rats were randomly allocated to the groups. For behavioral study, the same set of rats were assessed before and after intervention and served as their own controls.
OA rat model
The rats were deeply anesthetized with 5% isoflurane (Forane®, Baxter Healthcare Corporation, Deerfield, IL, USA) in 100% O2 (3 L/min) until the flexor withdrawal reflex was abolished. The skin covering the right knee joint was shaved and swabbed with 100% ethanol. A 27-gauge needle was introduced into the joint cavity through the patellar ligament, and 50 μl containing 3 mg of sodium monoiodoacetate (MIA; Sigma-Aldrich, Saint Louis, MO, USA) in 0.9% saline was injected intra-articular to induce OA-like lesions. The MIA model was chosen due to its particular usefulness for studying joint pain and multiple similarities in the mechanisms of cartilage degeneration to those observed in human OA cartilage.26,27
Behavioral testing of hind limb incapacitance
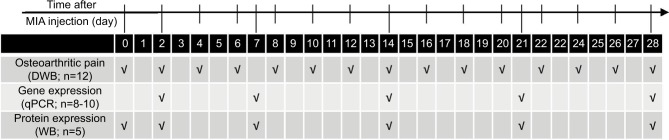
Rats were monitored for OA-related pain symptoms before MIA injection and on every second day after the procedure during the 28 days of disease development (the experimental design is presented in Figure 1). Hind limb incapacitance was measured using the DWB test (Bioseb, Vitrolles, France) as previously described.16,28 Due to the observer-independent character of the measurement, DWB was not performed in a blinded fashion. The rat was allowed to move freely within the apparatus for 5 min while the pressure data from transducers-equipped floor and live recordings from combined video camera were transmitted and collected. Data validation was performed using the DWB software v1.3 (Bioseb). Mean values for the weight borne by each of the hind limbs (in grams) were calculated, normalized to body mass, presented as a percentage difference between these limbs and utilized as an index of knee joint pain.
Figure 1.
Outline of the experiment design.
Notes: Rats were monitored for OA-related pain symptoms by DWB before MIA injection and every second day after the procedure during the 28-day period of the experiment. Gene expression analysis by qPCR was conducted on joint tissue samples collected on days 2, 7, 14, 21 and 28 post-MIA injection. For proteomic assessment, the joint tissue samples were collected before MIA injection and 2, 7, 14, 21 and 28 days after OA induction and processed for WB.
Abbreviations: DWB, dynamic weight bearing; MIA, monoiodoacetate; OA, osteoarthritis; qPCR, quantitative polymerase chain reaction; WB, Western blotting.
RNA isolation and quantitative polymerase chain reaction (qPCR)
Joint tissue samples consisted of meniscus and articular cartilage of the tibia and femur (synovium and bone were discarded; isolation of all specimens was performed by one researcher only). The specimens were isolated on days 2, 7, 14, 21 and 28 post-MIA injection (Figure 1). The samples were placed in individual tubes, immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation. The samples were thawed on ice and homogenized in 1 mL of TRIzol reagent (Invitrogen, Waltham, MA, USA). RNA isolation was performed according to the manufacturer’s protocol. The total RNA concentration was assessed using a Nanodrop ND-1000 spectrophotometer (Labtech International, Uckfield, UK). RNA quality was determined by chip-based capillary electrophoresis using the RNA 6000 Nano LabChip Kit and Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Total RNA concentrations in each sample were equalized to 1 μg/μL. Reverse transcription (RT) to cDNA was performed using iScript RT Supermix (Bio-Rad, Hercules, CA, USA). qPCR reactions were performed using iTaq Universal Probes Supermix (Bio-Rad), primer sets (TaqMan Gene Expression Assays, Life Technologies, Carlsbad, CA, USA; details in Table 1) and run on Real-Time PCR CFX96 Touch System (Bio-Rad). The cycle threshold (CT) values were computed automatically using the CFX Manager software (Bio-Rad) with the default parameters. Mean CT values were investigated for all genes of interest and are listed in Table S1. To examine the variation in cDNA amounts across the groups, the expression levels were normalized to the CT value of the Hprt1 reference gene. RNA abundance was calculated as 2−(normalized ΔCt). We considered that due to changes in cellular composition during MIA-induced OA development, the RNAs populations extracted from the intact cartilage and MIA-treated cartilage might be significantly different.29–31 Therefore, we did not use intact animals as a control group in the qPCR analysis. A group of samples collected at day 2 post-MIA injection was used as a control instead. We believe that the profile of changes in mRNA abundance levels from day 2 to day 28 is highly informative and reflects transcription dynamics over the course of OA. To facilitate a comparison between the gene and protein levels, statistical significance against day 2 was also presented for the Western blotting (WB) results.
Table 1.
List of TaqMan assays used for quantitative polymerase chain reaction gene expression analysis
| Gene name | Gene symbol | Gene expression Assay ID | Assay location (RefSeq ID) | Amplicon length |
|---|---|---|---|---|
| Hypoxanthine phosphoribosyltransferase 1 | Hprt1 | Rn01527840_m1 | 673 (NM_012583.2) | 64 |
| SRY (sex determining region Y)-box 9 | Sox9 | Rn01751070_m1 | 1177 (XM_003750950.3) | 102 |
| Cartilage oligomeric matrix protein | Comp | Rn00563255_m1 | 1132 (NM_012834.1) | 87 |
| Matrix metallopeptidase 3 | Mmp3 | Rn00591740_m1 | 1379 (X02601.1) | 67 |
| Matrix metallopeptidase 9 | Mmp9 | Rn00579162_m1 | 2016 (NM_031055) | 72 |
| Matrix metallopeptidase 13 | Mmp13 | Rn01448194_m1 | 652 (NM_133530.1) | 65 |
| TIMP metallopeptidase inhibitor 1 | Timp1 | Rn01430873_g1 | 240 (NM_053819.1) | 118 |
| TIMP metallopeptidase inhibitor 2 | Timp2 | Rn00573232_m1 | 466 (NM_021989.2) | 70 |
| Cannabinoid receptor 1 | Cb1 | Rn02758689_s1 | 1235 (NM_012784.4) | 92 |
| Cannabinoid receptor 2 | Cb2 | Rn04342831_s1 | 847 (NM_020543.4) | 99 |
| N-acyl phosphatidylethanolamine phospholipase D | Napepld | Rn01786262_m1 | 1054 (NM_199381.1) | 71 |
| Phospholipase A2 | Pla2g2a | Rn00668379_g1 | 104 (NM_031598.3) | 79 |
| Phospholipase C, beta 1 | Plcb1 | Rn01514511_m1 | 729 (NM_001077641.1) | 58 |
| Glycerophosphodiester phosphodiesterase 1 | Gde1 | Rn00583529_m1 | 945 (NM_032615.2) | 71 |
| Fatty acid amide hydrolase | Faah | Rn00577086_m1 | 376 (NM_024132.3) | 63 |
| Prostaglandin-endoperoxide synthase 2 | Ptgs2 | Rn01483828_m1 | 312 (NM_017232.3) | 112 |
| Arachidonate 12-lipoxygenase | Alox12 | Rn01461082_m1 | 1210 (NM_001105798.1) | 79 |
| Arachidonate 15-lipoxygenase | Alox15 | Rn00696151_m1 | 144 (NM_031010.2) | 82 |
Protein isolation and WB analysis
For proteomic assessment, joint tissue samples (as described above) of ipsilateral knees were collected before MIA injection and on days 2, 7, 14, 21 and 28 after OA induction (Figure 1). Cartilage samples were frozen in liquid nitrogen and stored at −80°C until protein isolation. Using the Rotor Stator Homogenizer (IKA®-Werke, Staufen, Germany), the samples were homogenized in RIPA buffer (Sigma-Aldrich) with cocktails of proteases and phosphatases inhibitors (1:200; Sigma-Aldrich), and cleared by centrifugation (14,000 × g for 30 min). The protein concentration in the supernatant was determined using the BCA Protein Assay Kit (Sigma-Aldrich). Samples containing 20 μg of protein were heated for 8 min at 80°C in Laemmli 4× loading buffer (Bio-Rad) and resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis SDS-PAGE (Criterion™ TGX™ Precast Gels, Bio-Rad). After gel electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes (Trans-Blot System, Bio-Rad). The blots were blocked using 2.5% albumin and 2.5% non-fat dry milk in TBS with 0.1% Tween-20 (Bio-Rad,) for 2 h at 37°C. The blots were incubated with antibody of interest overnight at 4°C and then incubated with peroxidase-conjugated secondary antibody for 2 h in RT (all antibodies are detailed in Table 2). Immuno-complexes were detected using an ECL substrate (Clarity™, Bio-Rad) visualized using the LAS-1000 system (Fujifilm, Tokyo, Japan). A relative level of immunoreactivity was quantified using the Image Gauge software (Fujifilm). The WB experiment results were normalized and compared with those from the corresponding intact samples (non-treated animals). Due to changing joint architecture and dynamic tissue recomposition during MIA-induced OA development, we were not able to obtain a stable protein reference. Consequently, we did not employ a loading control in our analysis. The high quality of the WB analyses is confirmed by the immunoblots presented in Figure S1.
Table 2.
List of antibodies used for Western blotting experiments
| Antibody description | Supplier | Species and type | Dilution | Cat no. |
|---|---|---|---|---|
| Primary antibodies | ||||
| Anti-COMP | Abcam | Rb mAb | 1:1000 | ab128893 |
| Anti-SOX9 | Millipore | Rb pAb | 1:1000 | AB5535 |
| Anti-MMP2 (recognizes both pro- and active form) | Abcam | Ms mAb | 1:100 | ab2462 |
| Anti-MMP3 (recognizes both pro- and active form) | Aviva System Biology | Rb pAb | 1:500 | OAAF01874 |
| Anti-MMP9 (recognizes both pro- and active form) | Millipore | Rb pAb | 1:500 | AB19016 |
| Anti-MMP13 (recognizes both pro- and active form) | Abcam | Rb pAb | 1:1000 | ab39012 |
| Anti-TIMP1 (recognizes both pro- and active form) | Millipore | Rb pAb | 1:200 | AB770 |
| Anti-CB1 | Cayman Chemicals | Rb pAb | 1:300 | 101500 |
| Anti-CB2 | Cayman Chemicals | Rb pAb | 1:500 | 101550 |
| Anti-FAAH | Cayman Chemicals | Rb pAb | 1:500 | 101600 |
| Anti-NAPE-PLD | Cayman Chemicals | Rb pAb | 1:500 | 10305 |
| Anti-15LOX2 | Abcam | Rb pAb | 1:200 | ab23691 |
| Anti-COX2 | Novus Biologicals | Rb pAb | 1:2000 | B110–1948 |
| Secondary antibodies | ||||
| Horse anti-Ms IgG conjugated with HRP | Vector Laboratories | Hr pAb | 1:2000 | PI-2000 |
| Goat anti-Rb IgG conjugated with HRP | Vector Laboratories | Gt pAb | 1:5000 | PI-1000 |
Abbreviations: COMP, cartilage oligomeric matrix protein; Gt, goat; Hr, horse; HRP, horseradish peroxidase; mAb, monoclonal antibody; MMP, matrix metalloproteinase; Ms, mouse; pAb, polyclonal antibody; Rb, rabbit; TIMP, tissue inhibitors of metalloproteinase; FAAH, fatty acid amide hydrolase; NAPE-PLD, N-acyl phosphatidylethanolamine phospholipase D.
Statistics
All statistical analyses were performed using Prism V.5 (GraphPad Software, La Jolla, CA, USA). The results are plotted as the mean ± SEM (standard error of the mean). Differences in the weight distribution (DWB test, n=12), transcriptomic (qPCR assay, n=8–10) and proteomic (WB analysis, n=5) results between the groups were assessed by one-way analysis of variance (ANOVA) followed by Bonferroni post hoc test. The number of animals was minimized to comply with 3R policy, but with respect to preserving statistical significance. All samples were included for analysis, and no outliers were detected. The Pearson correlation was computed with the weight distribution data and expression levels of the measured transcripts and proteins. The interpretation of the results should accommodate a limited number of data points in the analysis (5 time points). For graphical representation of qPCR data, trend lines (locally weighted scatterplot smoothing; LOWESS) were added. The threshold of significance was set at p<0.05. (*) denotes significant differences vs. intact animals (before OA induction), (#) vs. day 2 post-MIA injection, ($) vs. indicated bar.
Results
Development of pain-related behavior and changes in cartilage turnover during the progression of MIA-induced OA
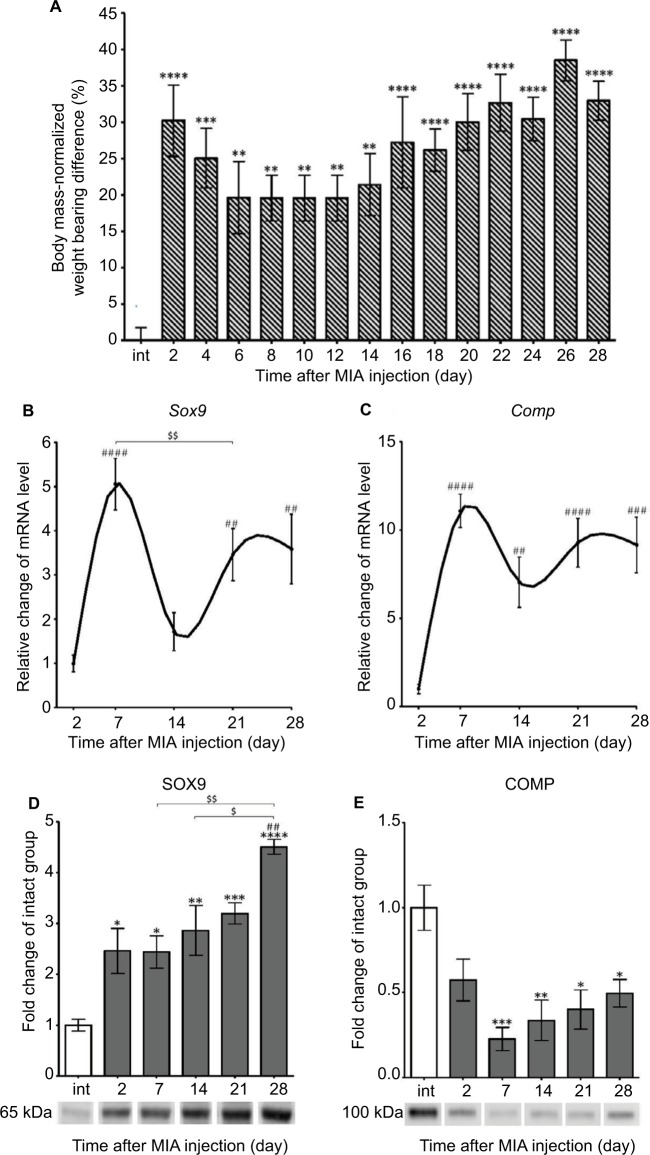
We used the DWB test to assess pain-like behavior in osteoarthritic rats. Our results revealed significant alterations in paw loading during OA progression (Figure 2A). MIA administration to the rat knee resulted in a persistent pain state characterized by a biphasic profile of pain-like behavior (Figure 2A). After an initial phase of an enormously increased weight-bearing deficit at days 2 and 4, a slight alleviation of symptoms was noticed between days 6 and 12 (Figure 2A). From days 14 to 28, we observed the second phase of chronic, progressive pain (Figure 2A).
Figure 2.
Progression of MIA-induced OA on the behavioral and molecular levels.
Notes: (A) Disturbed weight bearing for the hind limbs of rats injected with 3 mgs MIA. Loading incapacitance was measured via the DWB test before MIA injection and every second day during 28 days of OA development. The bars represent body mass-normalized weight bearing difference between MIA-treated ipsilateral and non-injected contralateral hind limbs (n=12). (B, C) Alterations in gene expression of cartilage turnover markers in osteoarthritic rats during disease progression. Transcripts levels for Sox9 and Comp in joint tissue samples were measured by quantitative polymerase chain reaction. mRNA samples were collected 2, 7, 14, 21 and 28 days after OA induction. The results are normalized to Hprt1 and presented as a relative change of mRNA levels in comparison to day 2 (n=8–10). (D, E) Changes in SOX9 and COMP protein expression measured by Western blotting assay. Samples were collected before MIA injection and 2, 7, 14, 21 and 28 days after OA induction. The results are presented as fold change (mean ± SEM) in comparison to the intact group (n=5). Representative bands from each group are presented below the charts. The data were analyzed with one-way analysis of variance followed by Bonferroni’s multiple comparison post hoc test; values with p<0.05 were considered significant. *Denotes significant differences vs intact animals (before OA induction) at p<0.05, **denotes significant differences vs intact animals (before OA induction) at p<0.01, ***denotes significant differences vs intact animals (before OA induction) at p<0.005, ****denotes significant differences vs intact animals (before OA induction) at p<0.001. ##denotes significant differences vs day 2 post-MIA injection at p<0.01, ###denotes significant differences vs day 2 post-MIA injection at p<0.005, ####denotes significant differences vs day 2 post-MIA injection at p<0.001, $denotes significant differences vs indicated bar at p<0.05, $$denotes significant differences vs indicated bar at p<0.01.
Abbreviations: COMP, cartilage oligomeric matrix protein; DWB, dynamic weight bearing; MIA, monoiodoacetate; OA, osteoarthritis.
Next, we determined the cartilage integrity and its metabolic state in OA joint tissue samples. Therefore, the expression of disease progression markers such as the transcriptional factor SOX9 (an important regulator of chondrogenesis) and cartilage oligomeric matrix protein (COMP; a protein crucial for cartilage structure) was measured by qPCR.32,33 Our results showed that Sox9 as well as Comp genes had two phases of up-regulation, peaking at day 7 and between days 21 and 28 compared with day 2 (Figure 2B, C; Table 3). Interestingly, we noticed a robust and gradual increase in SOX9 protein from day 2, with the maximal expression level observed on day 28 (Figure 2D). The expression of the COMP protein showed U-shaped regulation, with its minimum at day 7, and never returned to the control level of intact animals (Figure 2E).
Table 3.
Summary of gene expression alterations revealed in rat knee model during progression of OA
| Gene description | Time after MIA injection
|
|||
|---|---|---|---|---|
| Day 7 | Day 14 | Day 21 | Day 28 | |
| Progression markers | ||||
| Sox9 | ↑↑↑↑ | ↑↑ | ↑↑ | |
| Comp | ↑↑↑↑ | ↑↑ | ↑↑↑↑ | ↑↑↑ |
| MMPs/TIMPs | ||||
| Mmp3 | – | ↑↑↑↑ | ↑↑↑↑ | – |
| Mmp9 | – | ↑↑ | – | – |
| Mmp13 | – | – | ↑ | ↑↑ |
| Timp1 | ↑ | – | ↑ | ↑ |
| Timp2 | ↑↑↑↑ | ↑ | ↑↑ | ↑↑↑↑ |
| ECS receptors | ||||
| Cb1 | – | – | – | – |
| Cb2 | – | – | – | – |
| EC synthesis | ||||
| Napepld | – | – | – | – |
| Pla2g2a | – | – | – | – |
| Plcb1 | – | – | – | ↑ |
| Gde1 | – | – | – | ↑↑↑↑ |
| EC degradation | ||||
| Faah | – | – | – | |
| Ptgs2 | – | – | – | ↑↑ |
| Alox 12 | – | – | – | |
| Alox 15 | ↓ | ↓ | – | ↓ |
Notes: Transcripts abundance levels of selected genes in joint tissue samples were assessed by quantitative polymerase chain reaction. Total RNA samples were collected 2, 7, 14, 21 and 28 days after OA induction. The results are presented as relative increase (↑) or decrease (↓) of mRNA levels in comparison to day 2 (n=8–10). Data were analyzed with one-way analysis of variance followed by Bonferroni’s multiple comparison test. (↑/↓) denotes significant differences vs. day 2 post MIA injection, (↑) for p<0.05, (↑↑) for p<0.01, (↑↑↑) for p<0.005, (↑↑↑↑) for p<0.001.
Abbreviations: EC, endocannabinoid; ECS, endocannabinoid system; MIA, monoiodoacetate; MMP, matrix metalloproteinase; OA, osteoarthritis; TIMP, tissue inhibitors of metalloproteinase.
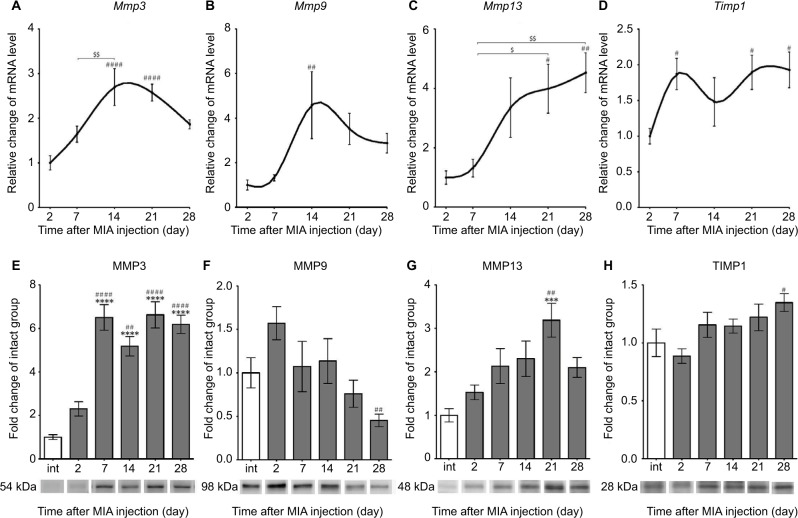
Alterations in gene and protein expression of selected MMPs and TIMPs in OA-affected cartilage
The expression of selected MMPs and TIMPs that are particularly important for OA pathogenesis was evaluated in the cartilage of MIA-injected rat knees. We found a significant up-regulation of Mmp3 and Mmp9 transcripts at day 14, followed by a gradual decrease until day 28 (Figure 3A, B; Table 3). The Mmp13 gene also presented pronounced elevation at day 14, and its mRNA level remained increased until the end of the testing period (Figure 3C; Table 3). Interestingly, the expression profiles of Timp1 and Timp2 resemble the profiles of Sox9 and Comp, with two maxima at day 7 and between days 21 and 28 (Figure 3D; Table 3).
Figure 3.
Changes in transcript and protein expression levels of MMPs in OA-affected rat cartilage during disease progression.
Notes: (A–D) Transcripts abundance levels for Mmp3, Mmp9, Mmp13 and Timp1 genes in joint tissue samples were measured by quantitative polymerase chain reaction. Total RNA samples were collected 2, 7, 14, 21 and 28 days after OA induction. The results are presented as relative changes in the mRNA levels compared with day 2 (n=8–10). (E–H) Protein expression of the selected MMPs in osteoarthritic knee joints during 28 days of disease progression. The expression of MMP3, MMP9, MMP13 and TIMP1 was determined by immunoblotting. Samples were collected before MIA injection and 2, 7, 14, 21 and 28 days after OA induction. The results are presented as fold of change (mean ± SEM) in comparison to the intact group (n=5). Representative bands from each group are presented below the charts. Statistical analysis was performed using one-way analysis of variance followed by Bonferroni post hoc test; values with p<0.05 were considered significant. ***Denotes significant differences vs int (before OA induction) at p<0.005, ****denotes significant differences vs int (before OA induction) at p<0.001, #denotes significant differences vs day 2 post-MIA injection at p<0.05, ##denotes significant differences vs day 2 post-MIA injection at p<0.01, ###denotes significant differences vs day 2 post-MIA injection at p<0.001, $denotes significant differences vs indicated bar at p<0.05, $$denotes significant differences vs indicated bar at p<0.01.
Abbreviations: int, intact animals; MIA, monoiodoacetate; MMP, matrix metalloproteinase; OA, osteoarthritis; SEM, standard error of the mean; TIMP, tissue inhibitors of metalloproteinases.
Consequently, proteomic analysis of selected MMPs and TIMPs was performed. Our data revealed that MMP3 protein expression (54 kDa pro-form) was remarkably elevated from day 7 and was maintained at a high level until day 28 in the intact control (Figure 3E; active 45 and 28 kDa forms of MMP3 were not investigated in this study).34 In contrast, MMP9 (98 kDa pro-form) presented a tendency to increase in level at day 2 and then significantly decrease at day 28 compared with day 2 (Figure 3F; active 82 kDa form of MMP9 was not observed).35 The expression of MMP13 (48 kDa active form) showed a gradual increase until day 21 (Figure 3G; 60 kDa pro-enzyme was far less abundant and not taken into consideration).36 Furthermore, the protein expression of TIMP1 (28 kDa active form) remained unchanged throughout the experiment, except a minor up-regulation at day 28 compared with day 2 (Figure 3H). Notably, we noticed a remarkable increase in protein level of MMP2 (63 kDa active form) at days 2, 21 and 28, corresponding well to the biphasic pain profile observed in the DWB test (Figure S2; 72 kDa pro-form was not observed).35 The alterations in MMP2 levels and changes in the DWB showed significant positive correlation (r=0.92, p<0.05; Table S2).
Alterations in gene and protein expression of ECS components in OA-affected cartilage
The main components of the ECS were subjected to an examination of the gene and protein expression in cartilage during OA progression. We assessed the transcript and protein abundance levels for cannabinoid receptors, enzymes engaged in AEA synthesis and degradation (listed in Table 3).37 We found that the mRNA levels for the majority of ECS genes (except Alox15) remained relatively stable throughout OA development (Table 3). The expression level of Alox15 mRNA was significantly diminished at days 7, 14 and 28 (Table 3). The ECS genes Plcb1, Gde1 and Ptgs2 showed increased mRNA abundance levels at day 28 compared with day 2 (Table 3).
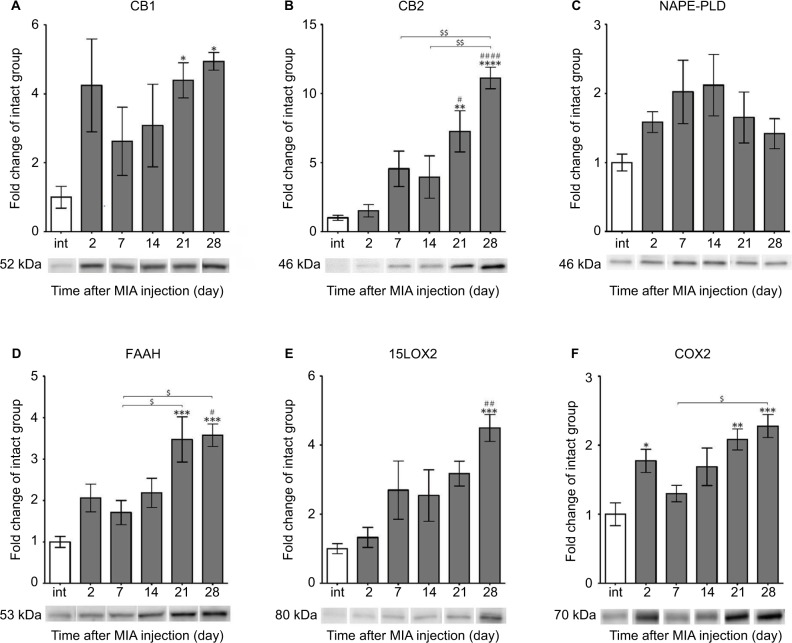
Next, we performed WB analyses of the selected ECS proteins. We showed that the level of the CB1 receptor (52 kDa) was highly augmented at days 21 and 28 (Figure 4A). Similarly, CB2 protein showed substantial up-regulation at days 21 and 28 (Figure 4B). The changes were statistically significant in reference to both the control and early time points of the experiment (Figure 4B). The NAPE-PLD protein levels revealed no significant differences between the experimental groups (Figure 4C). We also found that the profile of FAAH enzyme expression was similar to CB2, with accumulating abundance levels at day 21 and day 28 (Figure 4D). The 15LOX2 enzyme profile showed tendency to increase, with a strong up-regulation at day 28 (Figure 4E). The COX2 protein exhibited biphasic accumulation with peaks at days 2 and 28 (Figure 4F). The expression levels of certain ECS proteins, CB1 and NAPE-PLD, significantly correlated with the DWB data (r=0.88, p<0.05 and r=−0.88, p<0.05 respectively; Table S2), It is noteworthy that the other two proteins exhibited high correlation coefficients with p-value close to significant (FAAH: r=0.80, p=0.05 and COX2: r=0.79, p=0.06; Table S2).
Figure 4.
Alterations in protein expression of ECS components in knee joints associated with the progression of MIA-induced OA rat model.
Notes: Protein expression of CB1 and CB2 receptors (A, B), NAPE-PLD, FAAH enzymes (C, D), 15LOX2, COX2 enzymes (E, F) levels were determined by immunoblotting. Samples were collected before OA induction and 2, 7, 14, 21 and 28 days after MIA injection. The results are presented as fold of change (mean ± SEM) in comparison to the intact group (n=5). Representative bands from each group are presented below the charts. Statistical analysis was performed using one-way analysis of variance followed by Bonferroni post hoc test; values with p<0.05 were considered significant. *Denotes significant differences vs int (before OA induction) at p<0.05, **denotes significant differences vs int (before OA induction) at p<0.01, ***denotes significant differences vs int (before OA induction) at p<0.005, ****denotes significant differences vs int (before OA induction) at p<0.001, #denotes significant differences vs day 2 post-MIA injection at p<0.05, ##denotes significant differences vs day 2 post-MIA injection at p<0.01, ###denotes significant differences vs day 2 post-MIA injection at p<0.001, $denotes significant differences vs indicated bar at p<0.05, $$denotes significant differences vs indicated bar at p<0.01.
Abbreviations: ECS, endocannabinoid system; MIA, monoiodoacetate; OA, osteoarthritis; FAAH, fatty acid amide hydrolase; NAPE-PLD, N-acyl phosphatidylethanolamine phospholipase D; SEM, standard error of the mean; int, intact animals.
Discussion
In this study, we have investigated the regulation of ECs and MMPs expression during OA development. Both ECs and MMPs are considered promising targets for the pharmacological treatment of OA symptoms, including OA-related pain. The intra-articular injection of MIA produces weight-bearing asymmetry, which is considered as an indication of nociceptive pain similar to this observed in OA patients. We reported that the progress of weight-bearing asymmetry is biphasic, possibly reflecting the initial, inflammation-driven phase of pain followed by the second, chronic phase that is connected to pathomorphological alterations within the joint. The observed two-stage progression of the disease is consistent with our previous results and with other reports showing biphasic pain profile following injection of 3 mg of MIA in rats.16,38 Importantly, the altered weight distribution is sustained throughout the 6-week period of testing indicating chronic pain development.38
The MIA injection into the rat knee substantially affects cartilage homeostasis. Our findings confirm this, and show that both tested molecular markers of disease progression (Sox9 and Comp) exhibited two phases of increased gene expression corresponding to biphasic pain development. Interestingly, the first peak in transcriptional activation of these markers was observed slightly later than indicated by the DWB test. It was previously suggested that the matrix synthesis suppression manifested as decreased Sox9 levels, contributes to the progression of cartilage destruction in the MIA OA model.39 In contrast, our Sox9 expression results imply the enhanced regulation of chondrogenesis and promotion of regeneration on the affected side during the transient intensifications of pain episodes in this OA model. Our data also indicate a significant up-regulation of the SOX9 protein in OA-affected cartilage in a later phase of OA progression. However, both the SOX9 gene and its protein were shown to be decreased in the terminally damaged cartilage from OA patients.40 It can therefore be speculated that in patients who are at advanced stages of the disease, the SOX9-dependent recovery mechanisms are greatly diminished.41
A pronounced reduction in COMP protein expression indicates the irreversible breakdown of cartilage structure, which may contribute to the development of chronic OA-related pain. The average pain index in MIA-induced OA is closely correlated to the grade of histological abnormalities.27 Importantly, degenerative menisci from OA patients displays a reduced protein expression for COMP.42 Several clinical studies have reported a strong association between pain degree and degenerative OA features.43,44 These observations denote that despite etiological differences, the MIA-induced rat model of OA is characterized by similar pathomorphology and pain-related behavioral features as OA in humans.
Furthermore, our results indicate robust regulation in the expression of various MMPs during OA development. In general, we observed an enhancement in MMPs genes transcription with the highest expression in mRNA abundance levels at day 14, which reflects the initiation of the chronic phase of OA-related pain. Our results are in accordance with a study on rat MIA-injected cartilage showing elevated levels of Mmp2, Mmp3, Mmp13, Timp1 and Timp2 transcripts.45 Interestingly, a study of terminally damaged human cartilage identified increased MMP2, MMP9, MMP13 mRNA levels but decreased MMP3 level.3 Here, we report stage-specific patterns of changes in the protein expression of the selected MMPs. Globally; the accumulation of MMP2, MMP3 and MMP13 enzymes is detected at the early and late phases of the disease, which corresponds well with the periods of maximum pain. Of importance, MMP2 and MMP13 have been found in active forms. The results suggest that the intensified degradation of cartilage is directly translated to a more severe perception of pain. In line with above, it was proposed that the serum MMPs levels are related to particular OA stages.46 However, the phase-specific association of cartilaginous MMPs protein levels has not been reported. Moreover, our data indicate disrupted equilibrium between MMPs and TIMPs that may lead to progressive cartilage deterioration.7 In accordance, in synovial fluids of OA patients, the increase in the MMP3 level is markedly higher than the TIMP1 level.47 The results account for the chondroprotective potential of MMP inhibitors in MIA model of OA.48,49 Nonetheless, no MMP inhibitor is available for clinical use, predominantly because of inadequate efficacy.8,9
The major components of ECS showed only moderate gene expression changes in the osteoarthritic cartilage. However, we demonstrated that MIA injection into the rat knee leads to a tremendous increase in protein levels of cannabinoid receptors (CB1 and CB2) and AEA-degrading enzymes (FAAH, 15LOX2, and COX2). The induction was the most pronounced in the late phase of disease progression. In accordance, the expression levels of CB1, CB2 and FAAH in the synovial tissue biopsies from OA patients were similarly altered.50 Our result suggests that alterations in ECS in the state of chronic OA pain are twofold. First, up-regulation of cannabinoid receptors suggests that ECS in the OA-affected knee joint is highly activated and strive to counteract episodes of persistent pain. From the other site, increased expression of FAAH, 15LOX2, and COX2 may translate into decreased level of endogenous AEA and consequently, to enhanced severity of chronic OA pain. Interestingly, we observed that a biphasic profile of changes in COX2 expression also corresponds to a two-stage pattern of pain. Besides AEA inactivation, COX2 is also involved in the production of prostaglandins. Therefore, the increased COX2 abundance at day 2 confirms that the initial phase of OA pain is driven by inflammation. Moreover, the importance of COX2 up-regulation in human OA-affected cartilage has been already suggested.51 Furthermore, AEA, which is in nature an analgesic compound, may undergo oxidative metabolism by COX2 and 15LOX2, resulting in the production of pronociceptive bioactive metabolites.52,53 This way disrupted ECS-dependent pain processing may further manifest in increased pain sensation in later stage of OA.
Functional linkage between MMPs and ECS in cartilage homeostasis has been recently suggested. Several lines of evidence indicate that the modulation of CB1 and CB2 receptors may lead to the indirect inhibition of MMP activity. For instance, in vitro experiments have shown that synthetic cannabinoid agonist WIN 55,212-2 time- and dose-dependently decreased the gene expression of MMP3, MMP13, TIMP1 and TIMP2 in human chondrocytes.22 Another experiment indicated that ajulemic acid, a synthetic, non-psychoactive cannabinoid, suppressed the production of MMP1, MMP3, and MMP9 in human fibroblast-like synovial cells that may have accounted for its anti-arthritic effect.54
Accumulating data across different research fields also supports this notion. Research on arteriosclerosis indicated that the expression of CB2 was inversely correlated with MMP9 content in human atherosclerotic plaques.21 Similarly, a preclinical study revealed the reduction of MMP9 content in atherosclerotic mouse plaques after peritoneal injection of JWH-133 (the selective CB2 agonist).21 Similarly, preincubation with JWH-133 reduced release of MMP9 by intra-plaque neutrophils cultured in vitro.21 Furthermore, cannabinoids inhibited glioma metastasis and cell invasion by down-regulation of MMP2.5 Finally, the treatment with CB1 agonist (arachidonyl-2′-chloroethylamide) improved the outcomes in spinal cord injury, and its effect was mediated by attenuating MMP9 proteolytic activity.20
The direct inhibition of MMPs using small molecules proved to be insufficient in clinical conditions.8,9 Therefore, the idea of using cannabinoid receptors as mediators of MMPs activity appears to be very tempting. In our study, OA-related alterations in MMP expression were observed from the very beginning. Thus, early interventions targeting the local reservoir of CB1 and CB2 receptors may not only exert an analgesic effect during the inflammatory phase of the disease but may also prevent cartilage breakdown. Consequently, the development of chronic pain associated with the later stages of OA would be attenuated. Notably, transcriptional and protein alterations can be considered as two different levels of molecular regulation. The protein synthesis and degradation might be compensated; therefore, the transcriptional effects do not necessarily have a direct influence on the measured protein levels. The observed changes in gene expression may have functional implications at the later time-points. The second phase of the disease progression is characterized by a robust up-regulation of FAAH, COX2 and 15LOX2, with a concomitant increase in CB1 and CB2 expression. Aiming at the attenuation of AEA-degrading enzymes to increase the pool of endogenous AEA may provide an additional therapeutic solution. Importantly, pharmacological blockade of FAAH, COX2 or 15LOX2 proved to be effective and exert analgesic effect in animal models of OA.55,56 Moreover, the elevated levels of CB1 and CB2 could be utilized as mediators for MMPs inhibition in order to slow down cartilage deterioration. Importantly, a dual-acting compound that concomitantly targets the CB2 receptor and FAAH enzyme is already available, and its therapeutic potential needs to be explored.57 Indeed, a multi-target approach poses a rational strategy in drug research and development.58
In summary, this work is the first report describing the relationship between the development of OA-related pain and expression of functional markers of MMPs and ECS activity in the osteoarthritic knee. Our results identified molecular factors associated with biphasic progression of OA-related pain. We also speculate about potential sources of enhanced nociceptive signaling during development of OA pain. Our correlation data, together with literature premises about ECS-mediated MMPs neutralization activity opens a new way to search for a disease-modifying osteoarthritis drug. We assume that indirect ECS-driven MMP inhibition with simultaneous local activation of ECS would be useful for managing both enhanced pain signaling and cartilage deterioration. This study supports the utility of alternative biological mechanisms favoring the use of endocannabinoid compounds in the treatment of OA-related chronic pain. However, further thorough evaluation is needed to confirm this new mechanism, molecular pathways engaged and its therapeutic potential.
Supplementary materials
Raw images of all the WB immunoblots.
Notes: A set of 30 samples was loaded on two gels as indicated in the scheme (A). Gels and respective membrane were processed simultaneously and in parallel during the WB procedure. (B–N) Images of all performed immunoblots. Molecular weights of the investigated proteins and standard ladder are indicated on the left and right, respectively. For some immunoblots with >1 visible band, bands taken into consideration have been indicated with white arrows.
Abbreviations: COMP, cartilage oligomeric matrix protein; MMP, matrix metalloproteinase; TIMP, tissue inhibitors of metalloproteinase; WB, Western blotting; FAAH, fatty acid amide hydrolase; NAPE-PLD, N-acyl phosphatidylethanolamine phospholipase D.
MMP2 protein expression measurement in OA-affected rat cartilage during disease progression.
Notes: Altered protein expression of MMP2 in OA rats during 28 days of disease progression determined by immunoblotting. Samples were collected before MIA injection and 2, 7, 14, 21 and 28 days after OA induction. The results are presented as fold of change (mean ± SEM) in comparison to the intact group (n=5). Representative bands from each group are presented below the charts. Statistical analysis was performed using one-way analysis of variance followed by Bonferroni post hoc test; values with p<0.05 were considered significant. *Denotes significant differences vs. int (before OA induction).
Abbreviation: MIA, monoiodoacetate; MMP, matrix metalloproteinase; OA, osteoarthritis; int, intact animals; SEM, standard error of the mean.
Table S1.
Mean CT values for all genes under investigation
| Gene description | Gene symbol | Time after MIA injection
|
||||
|---|---|---|---|---|---|---|
| Day2 | Day 7 | Day 14 | Day 21 | Day 28 | ||
| Reference gene | Hprt1 | 25.13±0,12 | 25.38±0.17 | 25.78±0.21 | 25.78±0.17 | 26.04±0.14 |
| Progression markers | Sox9 | 32.56±0.37 | 29.86±0.23 | 32.53±0.55 | 31.41±0.42 | 31.16±0.27 |
| Comp | 25.65±0.41 | 21.67±0.26 | 23.16±0.41 | 22.95±0.44 | 22,72±0.23 | |
| MMPs/TIMPs | Mmp3 | 24.27±0.30 | 23.57±0.27 | 23.47±0.38 | 23.37±0.27 | 24.03±0.15 |
| Mmp9 | 27.27±0,55 | 25.74±0.25 | 25.64±0.58 | 25.50±0.38 | 25.57±0.19 | |
| Mmp13 | 26.36±0.46 | 25.95±0.37 | 24.60±0.54 | 24.28±0.36 | 24.46±0.18 | |
| Timp1 | 24.23±0.28 | 23.81±0.57 | 24.57±0.50 | 24.04±0.35 | 24.16±0.16 | |
| Timp2 | 24.94±0.18 | 23.29±0.23 | 24.34±0.34 | 24.14±0.29 | 24.09±0.17 | |
| ECS receptors | Cb1 | 32.56±0.18 | 32.46±0.28 | 33.11±0.33 | 32.90±0.17 | 33.35±0.20 |
| Cb2 | 32.03±0.30 | 32.54±0.43 | 33.66±0.54 | 33.28±0.31 | 33.82±0.20 | |
| EC synthesis | Napepld | 30.55±0.20 | 30.62±0.40 | 31.23±0.36 | 31.19±0.27 | 31.21±0.13 |
| Pla2g2a | 27.66±0.27 | 28.71±0.39 | 28.14±0.45 | 28.61±0.32 | 29.02±0.19 | |
| Plcb1 | 28.53±0.25 | 28.26±0.47 | 28.51±0.37 | 28.54±0.43 | 28.40±0.24 | |
| Gde1 | 25.64±0.19 | 25.87±0.50 | 26.06±0.38 | 25.71±0.26 | 25.50±0.17 | |
| EC degradation | Faah | 33.62±0.32 | 33.20±0.31 | 34.19±0.41 | 34.01±0.32 | 34.25±0.25 |
| Ptgs2 | 30.02±0.32 | 28.93±0.53 | 30.21±0.49 | 29.77±0.36 | 29.11±0.22 | |
| Alox 12 | 31.23±0.59 | 31.48±0.52 | 31.70±0.76 | 31.20±0.46 | 32.21±0.21 | |
| Alox 15 | 28.61±0.62 | 30.13±0.58 | 30.38±0.74 | 30.50±0.56 | 31.10±0.28 | |
Notes: Transcripts abundance levels of selected genes in joint tissue samples were assessed by quantitative polymerase chain reaction. Total RNA samples were collected 2, 7, 14, 21 and 28 days after osteoarthritis induction. The CT values are presented as a mean and SEM (n = 8–10).
Abbreviations: CT, cycle treshold; EC, endocannabinoid; ECS, endocannabinoid system; MMP, matrix metalloproteinase; SEM, standard error of the mean; TIMP, tissue inhibitors of metalloproteinase.
Table S2.
Correlation matrix for DWB data and expression levels
| Δ weight (%m) vs. Sox9 | Δ weight (%m) vs. Comp | Δ weight (%m) vs. Mmp3 | Δ weight (%m) vs. Mmp9 | Δ weight (%m) vs. Mmp13 | Δ weight (%m) vs. Timp1 | ||
|---|---|---|---|---|---|---|---|
 weight (%m) vs gene expression weight (%m) vs gene expression |
r | 0.05944 | −0.04753 | −0.09938 | −0.09954 | 0.4122 | 0.2161 |
| 95% confidence interval | −0.8685 to 0.8949 | −0.8924 to 0.8714 | −0.9026 to 0.8582 | −0.9026 to 0.8582 | −0.7389 to 0.9493 | −0.8232 to 0.9225 | |
| R square | 0.003533 | 0.002259 | 0.009877 | 0.009908 | 0.1699 | 0.04668 | |
| P (one-tailed) | 0.4622 | 0.4698 | 0.4368 | 0.4367 | 0.2452 | 0.3635 | |
| p-value summary | ns | ns | ns | ns | ns | ns | |
| Significant? (α = 0.05) | No | No | No | No | No | No | |
| Number of XY pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
|
| |||||||
| Δ weight (%m) vs. Cnr1 | Δ weight (%m) vs. Cnr2 | Δ weight (%m) vs. Faah | Δ weight (%m) vs. Napepld | Δ weight (%m) vs. Alox15 | Δ weight (%m) vs. Ptsg2 | ||
|
| |||||||
| r | −0.5224 | −0.03812 | −0.1887 | 0.005075 | 0.3837 | 0.3332 | |
| 95% confidence interval | −0.9615 to 0.6677 | −0.8905 to 0.8736 | −0.9182 to 0.8322 | −0.8812 to 0.8835 | −0.7539 to 0.9458 | −0.7778 to 0.9394 | |
| R square | 0.2729 | 0.001453 | 0.03561 | 2.58E-05 | 0.1472 | 0.111 | |
| P (one-tailed) | 0.1832 | 0.4757 | 0.3806 | 0.4968 | 0.2619 | 0.2919 | |
| p-value summary | ns | ns | ns | ns | ns | ns | |
| Significant? (α = 0.05) | No | No | No | No | No | No | |
| Number of XY pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
|
| |||||||
 weight (%m) vs gene expression weight (%m) vs gene expression |
Δ weight (%m) vs. Timp2 | Δ weight (%m) vs. Gde1 | Δ weight (%m) vs. Plcb1 | Δ weight (%m) vs. Alox12 | Δ weight (%m) vs. Pla2g2a | Δ weight (%m) vs. TIMP1 | |
|
| |||||||
| r | 0.04022 | 0.5007 | 0.3764 | 0.399 | 0.2813 | 0.2321 | |
| 95% confidence interval | −0.8731 to 0.8909 | −0.6837 to 0.9593 | −0.7575 to 0.9449 | −0.7460 to 0.9477 | −0.7995 to 0.9322 | −0.8177 to 0.9250 | |
| R square | 0.001618 | 0.2507 | 0.1417 | 0.1592 | 0.07911 | 0.05387 | |
| P (one-tailed) | 0.4744 | 0.1951 | 0.2662 | 0.2529 | 0.3233 | 0.3536 | |
| p-value summary | ns | ns | ns | ns | ns | ns | |
| Significant? (α = 0.05) | No | No | No | No | No | No | |
| Number of XY pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
|
| |||||||
| Δ weight (%m) vs. SOX9 | Δ weight (%m) vs. COMP | Δ weight (%m) vs. MMP3 | Δ weight (%m) vs. MMP2 | Δ weight (%m) vs. MMP9 | Δ weight (%m) vs. MMP13 | ||
|
| |||||||
| r | 0.5237 | 0.6089 | 0.07722 | 0.9253 | −0.4425 | 0.3316 | |
| 95% confidence interval | −0.6667 to 0.9617 | −0.5909 to 0.9701 | −0.8640 to 0.8984 | 0.2340 to 0.9952 | −0.9528 to 0.7216 | −0.7785 to 0.9392 | |
| R square | 0.2743 | 0.3707 | 0.005963 | 0.8562 | 0.1958 | 0.11 | |
| P (one-tailed) | 0.1825 | 0.1379 | 0.4509 | 0.0121 | 0.2278 | 0.2928 | |
| p-value summary | ns | ns | ns | * | ns | ns | |
| Significant? (α = 0.05) | No | No | No | Yes | No | No | |
| Number of XY pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
|
| |||||||
| Δ weight (%m) vs. CB1 | Δ weight (%m) vs. CB2 | Δ weight (%m) vs. FAAH | Δ weight (%m) vs. NAPE- PLD | Δ weight (%m) vs. 15LOX2 | Δ weight (%m) vs. COX2 | ||
|
| |||||||
| r | 0.8844 | 0.5222 | 0.8038 | −0.8895 | 0.364 | 0.7891 | |
| 95% confidence interval | 0.009190 to 0.9924 | −0.6679 to 0.9615 | −0.2701 to 0.9865 | −0.9927 to -0.03346 | −0.7636 to 0.9434 | −0.3071 to 0.9854 | |
| R square | 0.7821 | 0.2726 | 0.6461 | 7.91E-01 | 0.1325 | 0.6226 | |
| P (one-tailed) | 0.0232 | 0.1834 | 0.0506 | 0.0217 | 0.2735 | 0.0563 | |
| p-value summary | * | ns | ns | * | ns | ns | |
| Significant? (alpha = 0.05) | Yes | No | No | Yes | No | No | |
| Number of XY Pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
Notes: Correlation matrix for DWB data and expression levels. Pearson correlation analysis has been performed: 1) between behavioral effects and alterations in gene expression levels, and 2) between behavioral effects and alterations in protein expression levels (grey shading); p<0.05 has been considered significant.
Abbreviations: COMP, cartilage oligomeric matrix protein; MMP, matrix metalloproteinase; TIMP, tissue inhibitors of metalloproteinase; FAAH, fatty acid amide hydrolase; NAPE-PLD, N-acyl phosphatidylethanolamine phospholipase D.
Acknowledgments
This work was supported by the National Science Centre, Poland, Grant SONATA BIS NCN/2012/07/E/NZ7/01269, ETIUDA NCN/2015/16/T/NZ7/00052 and statutory funds from the Institute of Pharmacology PAS, Department of Pain Pharmacology. Natalia Malek is recipient of a scholarship from KNOW sponsored by the Ministry of Science and Higher Education, Republic of Poland.
Footnotes
Authors’ contributions
AP performed the WB assays and DWB tests, analyzed the WB and DWB results, collected and assembled all of the data, conducted the statistical analyses, interpreted the data and wrote the paper; MKos performed the qPCR experiments and documented and analyzed the qPCR data; NM induced the MIA model, isolated knee joint samples, participated in study design; MKor interpreted the data, critically evaluated the ideas, drafted the manuscript, and provided substantive support; KS obtained funding, conceived and designed the study, was involved in interpretation of the data, drafting the manuscript and revisions for intellectual content. All authors discussed the results and its implications at all stages and have approved the final version of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Buckwalter J, Mankin HJ, Grodzinsky AJ. Articular cartilage and osteoarthritis. Instructional course lectures. 2005;54:465–480. [PubMed] [Google Scholar]
- 2.Ishiguro N, Kojima T, Poole AR. Mechanism of cartilage destruction in osteoarthritis. Nagoya J Med Sci. 2002;65(3–4):73–84. [PubMed] [Google Scholar]
- 3.Kevorkian L, Young DA, Darrah C, et al. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004;50(1):131–141. doi: 10.1002/art.11433. [DOI] [PubMed] [Google Scholar]
- 4.Billinghurst RC, Dahlberg L, Ionescu M, et al. Enhanced cleavage of type II collagen by collagenases in osteoarthritic articular cartilage. J Clin Invest. 1997;99(7):1534–1545. doi: 10.1172/JCI119316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roach HI, Yamada N, Cheung KS, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52(10):3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 6.Wu W, Billinghurst RC, Pidoux I, et al. Sites of collagenase cleavage and denaturation of type II collagen in aging and osteoarthritic articular cartilage and their relationship to the distribution of matrix metalloproteinase 1 and matrix metalloproteinase 13. Arthritis Rheum. 2002;46(8):2087–2094. doi: 10.1002/art.10428. [DOI] [PubMed] [Google Scholar]
- 7.Dean DD, Martel-Pelletier J, Pelletier JP, Howell DS, Woessner JF. Evidence for metalloproteinase inhibitor imbalance in human osteoarthritic cartilage. J Clin Invest. 1989;84(2):678–685. doi: 10.1172/JCI114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meszaros E, Malemud CJ. Prospects for treating osteoarthritis: enzyme-protein interactions regulating matrix metalloproteinase activity. Ther Adv Chronic Dis. 2012;3(5):219–229. doi: 10.1177/2040622312454157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Close DR. Matrix metalloproteinase inhibitors in rheumatic diseases. Ann Rheum Dis. 2001;60(Suppl 3):iii62–iii7. doi: 10.1136/ard.60.90003.iii62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guindon J, Hohmann AG. The endocannabinoid system and pain. CNS Neurol Disord Drug Targets. 2009;8(6):403–421. doi: 10.2174/187152709789824660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piomelli D, Hohmann AG, Seybold V, Hammock BD. A lipid gate for the peripheral control of pain. J Neurosci. 2014;34(46):15184–15191. doi: 10.1523/JNEUROSCI.3475-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodhams SG, Sagar DR, Burston JJ, Chapman V. The role of the endocannabinoid system in pain. Pain control. 2015;227:119–143. doi: 10.1007/978-3-662-46450-2_7. [DOI] [PubMed] [Google Scholar]
- 13.Mbvundula EC, Bunning RA, Rainsford KD. Arthritis and cannabinoids: HU-210 and Win-55,212–2 prevent IL-1alpha-induced matrix degradation in bovine articular chondrocytes in-vitro. J Pharm Pharmacol. 2006;58(3):351–358. doi: 10.1211/jpp.58.3.0009. [DOI] [PubMed] [Google Scholar]
- 14.McDougall JJ, Yu V, Thomson J. In vivo effects of CB2 receptor-selective cannabinoids on the vasculature of normal and arthritic rat knee joints. Br J Pharmacol. 2008;153(2):358–366. doi: 10.1038/sj.bjp.0707565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuelert N, Johnson MP, Oskins JL, Jassal K, Chambers MG, McDougall JJ. Local application of the endocannabinoid hydrolysis inhibitor URB597 reduces nociception in spontaneous and chemically induced models of osteoarthritis. Pain. 2011;152(5):975–981. doi: 10.1016/j.pain.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Malek N, Mrugala M, Makuch W, et al. A multi-target approach for pain treatment – dual inhibition of fatty acid amide hydrolase and TRPV1 in a rat model of osteoarthritis. Pain. 2015;156(5):890–903. doi: 10.1097/j.pain.0000000000000132. [DOI] [PubMed] [Google Scholar]
- 17.La Porta C, Bura S, Negrete R, Maldonado R. Involvement of the endocannabinoid system in osteoarthritis pain. Eur J Neurosci. 2014;39(3):485–500. doi: 10.1111/ejn.12468. [DOI] [PubMed] [Google Scholar]
- 18.Yao BB, Hsieh GC, Frost JM, et al. In vitro and in vivo characterization of A-796260: a selective cannabinoid CB2 receptor agonist exhibiting analgesic activity in rodent pain models. Br J Pharmacol. 2008;153(2):390–401. doi: 10.1038/sj.bjp.0707568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blázquez C, Salazar M, Carracedo A, et al. Cannabinoids inhibit glioma cell invasion by down-regulating matrix metalloproteinase-2 expression. Cancer Res. 2008;68(6):1945–1952. doi: 10.1158/0008-5472.CAN-07-5176. [DOI] [PubMed] [Google Scholar]
- 20.Hong J, Nandiwada V, Jones V, et al. CB1 cannabinoid receptor agonist inhibits matrix metalloproteinase activity in spinal cord injury: A possible mechanism of improved recovery. Neurosci Lett. 2015;597:19–24. doi: 10.1016/j.neulet.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Montecucco F, Di Marzo V, da Silva RF, et al. The activation of the cannabinoid receptor type 2 reduces neutrophilic protease-mediated vulnerability in atherosclerotic plaques. Eur Heart J. 2012;33(7):846–856. doi: 10.1093/eurheartj/ehr449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn S, Wilkinson J, Crawford A, Le Maitre C, Bunning R. Cannabinoid WIN-55,212–2 mesylate inhibits interleukin-1β induced matrix metalloproteinase and tissue inhibitor of matrix metalloproteinase expression in human chondrocytes. Osteoarthr Cartil. 2014;22(1):133–144. doi: 10.1016/j.joca.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 23.Lakhan SE, Avramut M. Matrix metalloproteinases in neuropathic pain and migraine: Friends, enemies, and therapeutic targets. Pain Res Treat. 2012;2012:952906. doi: 10.1155/2012/952906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manzanares J, Julian M, Carrascosa A. Role of the cannabinoid system in pain control and therapeutic implications for the management of acute and chronic pain episodes. Curr Neuropharmacol. 2006;4(3):239–257. doi: 10.2174/157015906778019527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann M. Ethical guidelines for investigations of experimental pain in conscious animals. Pain. 1983;16(2):109–110. doi: 10.1016/0304-3959(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 26.Janusz MJ, Little CB, King LE, et al. Detection of aggrecanase- and MMP-generated catabolic neoepitopes in the rat iodoacetate model of cartilage degeneration. Osteoarthritis Cartilage. 2004;12(9):720–728. doi: 10.1016/j.joca.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi K, Imaizumi R, Sumichika H, et al. Sodium iodoacetate-induced experimental osteoarthritis and associated pain model in rats. J Vet Med Sci. 2003;65(11):1195–1199. doi: 10.1292/jvms.65.1195. [DOI] [PubMed] [Google Scholar]
- 28.Tétreault P, Dansereau MA, Doré-Savard L, Beaudet N, Sarret P. Weight bearing evaluation in inflammatory, neuropathic and cancer chronic pain in freely moving rats. Physiol Behav. 2011;104(3):495–502. doi: 10.1016/j.physbeh.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Goldring MB, Marcu KB. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res Ther. 2009;11(3):224. doi: 10.1186/ar2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelttari K, Barbero A, Martin I. A potential role of homeobox transcription factors in osteoarthritis. Ann Transl Med. 2015;3(17):254. doi: 10.3978/j.issn.2305-5839.2015.09.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandell LJ, Aigner T. Articular cartilage and changes in arthritis. An introduction: cell biology of osteoarthritis. Arthritis Res. 2001;3(2):107–113. doi: 10.1186/ar148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 33.Rousseau JC, Garnero P. Biological markers in osteoarthritis. Bone. 2012;51(2):265–277. doi: 10.1016/j.bone.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Sun S, Bay-Jensen AC, Karsdal MA, et al. The active form of MMP-3 is a marker of synovial inflammation and cartilage turnover in inflammatory joint diseases. BMC Musculoskelet Disord. 2014;15:93. doi: 10.1186/1471-2474-15-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gepstein A, Shapiro S, Arbel G, Lahat N, Livne E. Expression of matrix metalloproteinases in articular cartilage of temporomandibular and knee joints of mice during growth, maturation, and aging. Arthritis Rheum. 2002;46(12):3240–3250. doi: 10.1002/art.10690. [DOI] [PubMed] [Google Scholar]
- 36.Cowell S, Knäuper V, Stewart ML, et al. Induction of matrix metalloproteinase activation cascades based on membrane-type 1 matrix metalloproteinase: associated activation of gelatinase A, gelatinase B and collagenase 3. Biochem J. 1998;331(Pt 2):453–458. doi: 10.1042/bj3310453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malek N, Kucharczyk M, Staro;wicz K. Alterations in the anandamide metabolism in the development of neuropathic pain. Biomed Res Int. 2014;2014:686908. doi: 10.1155/2014/686908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bove SE, Calcaterra SL, Brooker RM, et al. Weight bearing as a measure of disease progression and efficacy of anti-inflammatory compounds in a model of monosodium iodoacetate-induced osteoarthritis. Osteoarthr Cartilage. 2003;11(11):821–830. doi: 10.1016/s1063-4584(03)00163-8. [DOI] [PubMed] [Google Scholar]
- 39.Nam J, Perera P, Liu J, et al. Sequential alterations in catabolic and anabolic gene expression parallel pathological changes during progression of monoiodoacetate-induced arthritis. PLoS One. 2011;6(9):e24320. doi: 10.1371/journal.pone.0024320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haag J, Gebhard PM, Aigner T. SOX9 gene expression in human osteoarthritic cartilage. Pathobiology. 2008;75(3):195–199. doi: 10.1159/000124980. [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, Im GI. SOX trio decrease in the articular cartilage with the advancement of osteoarthritis. Connect Tissue Res. 2011;52(6):496–502. doi: 10.3109/03008207.2011.585409. [DOI] [PubMed] [Google Scholar]
- 42.López-Franco M, López-Franco O, Murciano-Antón MA, et al. Meniscal degeneration in human knee osteoarthritis: in situ hybridization and immunohistochemistry study. Arch Orthop Trauma Surg. 2016;136(2):175–183. doi: 10.1007/s00402-015-2378-4. [DOI] [PubMed] [Google Scholar]
- 43.Duncan R, Peat G, Thomas E, Hay E, McCall I, Croft P. Symptoms and radiographic osteoarthritis: not as discordant as they are made out to be? Ann Rheum Dis. 2006;66(1):86–91. doi: 10.1136/ard.2006.052548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neogi T, Felson D, Niu J, et al. Association between radiographic features of knee osteoarthritis and pain: results from two cohort studies. BMJ. 2009;339:b2844. doi: 10.1136/bmj.b2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong YJ, Kim I, Cho JH, et al. Anti-osteoarthritic effects of the litsea japonica fruit in a rat model of osteoarthritis induced by monosodium iodoacetate. PLoS One. 2015;10(8):e0134856. doi: 10.1371/journal.pone.0134856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burrage PS, Brinckerhoff CE. Molecular targets in osteoarthritis: metalloproteinases and their inhibitors. Curr Drug Targets. 2007;8(2):293–303. doi: 10.2174/138945007779940098. [DOI] [PubMed] [Google Scholar]
- 47.Lohmander LS, Hoerrner LA, Dahlberg L, Roos H, Bjornsson S, Lark MW. Stromelysin, tissue inhibitor of metalloproteinases and proteoglycan fragments in human knee joint fluid after injury. J Rheumatol. 1993;20(8):1362–1368. [PubMed] [Google Scholar]
- 48.Baragi VM, Becher G, Bendele AM, et al. A new class of potent matrix metalloproteinase 13 inhibitors for potential treatment of osteoarthritis: evidence of histologic and clinical efficacy without musculoskeletal toxicity in rat models. Arthritis Rheum. 2009;60(7):2008–2018. doi: 10.1002/art.24629. [DOI] [PubMed] [Google Scholar]
- 49.Janusz MJ, Hookfin EB, Heitmeyer SA, et al. Moderation of iodoacetate-induced experimental osteoarthritis in rats by matrix metalloproteinase inhibitors. Osteoarthr Cartil. 2001;9(8):751–760. doi: 10.1053/joca.2001.0472. [DOI] [PubMed] [Google Scholar]
- 50.Richardson D, Pearson RG, Kurian N, et al. Characterisation of the cannabinoid receptor system in synovial tissue and fluid in patients with osteoarthritis and rheumatoid arthritis. Arthritis Res Ther. 2008;10(2):R43. doi: 10.1186/ar2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amin AR, Attur M, Patel RN, et al. Superinduction of cyclooxygenase-2 activity in human osteoarthritis- affected cartilage. Influence of nitric oxide. J Clin Invest. 1997;99(6):1231–1237. doi: 10.1172/JCI119280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forsell PK, Brunnström Å, Johannesson M, Claesson HE. Metabolism of anandamide into eoxamides by 15-lipoxygenase-1 and glutathione transferases. Lipids. 2012;47(8):781–791. doi: 10.1007/s11745-012-3684-z. [DOI] [PubMed] [Google Scholar]
- 53.Fowler CJ. The contribution of cyclooxygenase-2 to endocannabinoid metabolism and action. Br J Pharmacol. 2007;152(5):594–601. doi: 10.1038/sj.bjp.0707379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson DR, Stebulis JA, Rossetti RG, Burstein SH, Zurier RB. Suppression of fibroblast metalloproteinases by ajulemic acid, a nonpsychoactive cannabinoid acid. J Cell Biochem. 2007;100(1):184–190. doi: 10.1002/jcb.21046. [DOI] [PubMed] [Google Scholar]
- 55.Fowler CJ, Naidu PS, Lichtman A, Onnis V. The case for the development of novel analgesic agents targeting both fatty acid amide hydrolase and either cyclooxygenase or TRPV1. Br J Pharmacol. 2009;156(3):412–419. doi: 10.1111/j.1476-5381.2008.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grim TW, Ghosh S, Hsu K, Cravatt BF, Kinsey SG, Lichtman AH. Combined inhibition of FAAH and COX produces enhanced anti-allodynic effects in mouse neuropathic and inflammatory pain models. Pharmacol Biochem Behav. 2014;124:405–411. doi: 10.1016/j.pbb.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chicca A, Caprioglio D, Minassi A, et al. Functionalization of β-caryophyllene generates novel polypharmacology in the endocannabinoid system. ACS Chem Biol. 2014;9(7):1499–1507. doi: 10.1021/cb500177c. [DOI] [PubMed] [Google Scholar]
- 58.Lu JJ, Pan W, Hu YJ, Wang YT. Multi-target drugs: The trend of drug research and development. PLoS One. 2012;7(6):e40262. doi: 10.1371/journal.pone.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw images of all the WB immunoblots.
Notes: A set of 30 samples was loaded on two gels as indicated in the scheme (A). Gels and respective membrane were processed simultaneously and in parallel during the WB procedure. (B–N) Images of all performed immunoblots. Molecular weights of the investigated proteins and standard ladder are indicated on the left and right, respectively. For some immunoblots with >1 visible band, bands taken into consideration have been indicated with white arrows.
Abbreviations: COMP, cartilage oligomeric matrix protein; MMP, matrix metalloproteinase; TIMP, tissue inhibitors of metalloproteinase; WB, Western blotting; FAAH, fatty acid amide hydrolase; NAPE-PLD, N-acyl phosphatidylethanolamine phospholipase D.
MMP2 protein expression measurement in OA-affected rat cartilage during disease progression.
Notes: Altered protein expression of MMP2 in OA rats during 28 days of disease progression determined by immunoblotting. Samples were collected before MIA injection and 2, 7, 14, 21 and 28 days after OA induction. The results are presented as fold of change (mean ± SEM) in comparison to the intact group (n=5). Representative bands from each group are presented below the charts. Statistical analysis was performed using one-way analysis of variance followed by Bonferroni post hoc test; values with p<0.05 were considered significant. *Denotes significant differences vs. int (before OA induction).
Abbreviation: MIA, monoiodoacetate; MMP, matrix metalloproteinase; OA, osteoarthritis; int, intact animals; SEM, standard error of the mean.
Table S1.
Mean CT values for all genes under investigation
| Gene description | Gene symbol | Time after MIA injection
|
||||
|---|---|---|---|---|---|---|
| Day2 | Day 7 | Day 14 | Day 21 | Day 28 | ||
| Reference gene | Hprt1 | 25.13±0,12 | 25.38±0.17 | 25.78±0.21 | 25.78±0.17 | 26.04±0.14 |
| Progression markers | Sox9 | 32.56±0.37 | 29.86±0.23 | 32.53±0.55 | 31.41±0.42 | 31.16±0.27 |
| Comp | 25.65±0.41 | 21.67±0.26 | 23.16±0.41 | 22.95±0.44 | 22,72±0.23 | |
| MMPs/TIMPs | Mmp3 | 24.27±0.30 | 23.57±0.27 | 23.47±0.38 | 23.37±0.27 | 24.03±0.15 |
| Mmp9 | 27.27±0,55 | 25.74±0.25 | 25.64±0.58 | 25.50±0.38 | 25.57±0.19 | |
| Mmp13 | 26.36±0.46 | 25.95±0.37 | 24.60±0.54 | 24.28±0.36 | 24.46±0.18 | |
| Timp1 | 24.23±0.28 | 23.81±0.57 | 24.57±0.50 | 24.04±0.35 | 24.16±0.16 | |
| Timp2 | 24.94±0.18 | 23.29±0.23 | 24.34±0.34 | 24.14±0.29 | 24.09±0.17 | |
| ECS receptors | Cb1 | 32.56±0.18 | 32.46±0.28 | 33.11±0.33 | 32.90±0.17 | 33.35±0.20 |
| Cb2 | 32.03±0.30 | 32.54±0.43 | 33.66±0.54 | 33.28±0.31 | 33.82±0.20 | |
| EC synthesis | Napepld | 30.55±0.20 | 30.62±0.40 | 31.23±0.36 | 31.19±0.27 | 31.21±0.13 |
| Pla2g2a | 27.66±0.27 | 28.71±0.39 | 28.14±0.45 | 28.61±0.32 | 29.02±0.19 | |
| Plcb1 | 28.53±0.25 | 28.26±0.47 | 28.51±0.37 | 28.54±0.43 | 28.40±0.24 | |
| Gde1 | 25.64±0.19 | 25.87±0.50 | 26.06±0.38 | 25.71±0.26 | 25.50±0.17 | |
| EC degradation | Faah | 33.62±0.32 | 33.20±0.31 | 34.19±0.41 | 34.01±0.32 | 34.25±0.25 |
| Ptgs2 | 30.02±0.32 | 28.93±0.53 | 30.21±0.49 | 29.77±0.36 | 29.11±0.22 | |
| Alox 12 | 31.23±0.59 | 31.48±0.52 | 31.70±0.76 | 31.20±0.46 | 32.21±0.21 | |
| Alox 15 | 28.61±0.62 | 30.13±0.58 | 30.38±0.74 | 30.50±0.56 | 31.10±0.28 | |
Notes: Transcripts abundance levels of selected genes in joint tissue samples were assessed by quantitative polymerase chain reaction. Total RNA samples were collected 2, 7, 14, 21 and 28 days after osteoarthritis induction. The CT values are presented as a mean and SEM (n = 8–10).
Abbreviations: CT, cycle treshold; EC, endocannabinoid; ECS, endocannabinoid system; MMP, matrix metalloproteinase; SEM, standard error of the mean; TIMP, tissue inhibitors of metalloproteinase.
Table S2.
Correlation matrix for DWB data and expression levels
| Δ weight (%m) vs. Sox9 | Δ weight (%m) vs. Comp | Δ weight (%m) vs. Mmp3 | Δ weight (%m) vs. Mmp9 | Δ weight (%m) vs. Mmp13 | Δ weight (%m) vs. Timp1 | ||
|---|---|---|---|---|---|---|---|
 weight (%m) vs gene expression weight (%m) vs gene expression |
r | 0.05944 | −0.04753 | −0.09938 | −0.09954 | 0.4122 | 0.2161 |
| 95% confidence interval | −0.8685 to 0.8949 | −0.8924 to 0.8714 | −0.9026 to 0.8582 | −0.9026 to 0.8582 | −0.7389 to 0.9493 | −0.8232 to 0.9225 | |
| R square | 0.003533 | 0.002259 | 0.009877 | 0.009908 | 0.1699 | 0.04668 | |
| P (one-tailed) | 0.4622 | 0.4698 | 0.4368 | 0.4367 | 0.2452 | 0.3635 | |
| p-value summary | ns | ns | ns | ns | ns | ns | |
| Significant? (α = 0.05) | No | No | No | No | No | No | |
| Number of XY pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
|
| |||||||
| Δ weight (%m) vs. Cnr1 | Δ weight (%m) vs. Cnr2 | Δ weight (%m) vs. Faah | Δ weight (%m) vs. Napepld | Δ weight (%m) vs. Alox15 | Δ weight (%m) vs. Ptsg2 | ||
|
| |||||||
| r | −0.5224 | −0.03812 | −0.1887 | 0.005075 | 0.3837 | 0.3332 | |
| 95% confidence interval | −0.9615 to 0.6677 | −0.8905 to 0.8736 | −0.9182 to 0.8322 | −0.8812 to 0.8835 | −0.7539 to 0.9458 | −0.7778 to 0.9394 | |
| R square | 0.2729 | 0.001453 | 0.03561 | 2.58E-05 | 0.1472 | 0.111 | |
| P (one-tailed) | 0.1832 | 0.4757 | 0.3806 | 0.4968 | 0.2619 | 0.2919 | |
| p-value summary | ns | ns | ns | ns | ns | ns | |
| Significant? (α = 0.05) | No | No | No | No | No | No | |
| Number of XY pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
|
| |||||||
 weight (%m) vs gene expression weight (%m) vs gene expression |
Δ weight (%m) vs. Timp2 | Δ weight (%m) vs. Gde1 | Δ weight (%m) vs. Plcb1 | Δ weight (%m) vs. Alox12 | Δ weight (%m) vs. Pla2g2a | Δ weight (%m) vs. TIMP1 | |
|
| |||||||
| r | 0.04022 | 0.5007 | 0.3764 | 0.399 | 0.2813 | 0.2321 | |
| 95% confidence interval | −0.8731 to 0.8909 | −0.6837 to 0.9593 | −0.7575 to 0.9449 | −0.7460 to 0.9477 | −0.7995 to 0.9322 | −0.8177 to 0.9250 | |
| R square | 0.001618 | 0.2507 | 0.1417 | 0.1592 | 0.07911 | 0.05387 | |
| P (one-tailed) | 0.4744 | 0.1951 | 0.2662 | 0.2529 | 0.3233 | 0.3536 | |
| p-value summary | ns | ns | ns | ns | ns | ns | |
| Significant? (α = 0.05) | No | No | No | No | No | No | |
| Number of XY pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
|
| |||||||
| Δ weight (%m) vs. SOX9 | Δ weight (%m) vs. COMP | Δ weight (%m) vs. MMP3 | Δ weight (%m) vs. MMP2 | Δ weight (%m) vs. MMP9 | Δ weight (%m) vs. MMP13 | ||
|
| |||||||
| r | 0.5237 | 0.6089 | 0.07722 | 0.9253 | −0.4425 | 0.3316 | |
| 95% confidence interval | −0.6667 to 0.9617 | −0.5909 to 0.9701 | −0.8640 to 0.8984 | 0.2340 to 0.9952 | −0.9528 to 0.7216 | −0.7785 to 0.9392 | |
| R square | 0.2743 | 0.3707 | 0.005963 | 0.8562 | 0.1958 | 0.11 | |
| P (one-tailed) | 0.1825 | 0.1379 | 0.4509 | 0.0121 | 0.2278 | 0.2928 | |
| p-value summary | ns | ns | ns | * | ns | ns | |
| Significant? (α = 0.05) | No | No | No | Yes | No | No | |
| Number of XY pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
|
| |||||||
| Δ weight (%m) vs. CB1 | Δ weight (%m) vs. CB2 | Δ weight (%m) vs. FAAH | Δ weight (%m) vs. NAPE- PLD | Δ weight (%m) vs. 15LOX2 | Δ weight (%m) vs. COX2 | ||
|
| |||||||
| r | 0.8844 | 0.5222 | 0.8038 | −0.8895 | 0.364 | 0.7891 | |
| 95% confidence interval | 0.009190 to 0.9924 | −0.6679 to 0.9615 | −0.2701 to 0.9865 | −0.9927 to -0.03346 | −0.7636 to 0.9434 | −0.3071 to 0.9854 | |
| R square | 0.7821 | 0.2726 | 0.6461 | 7.91E-01 | 0.1325 | 0.6226 | |
| P (one-tailed) | 0.0232 | 0.1834 | 0.0506 | 0.0217 | 0.2735 | 0.0563 | |
| p-value summary | * | ns | ns | * | ns | ns | |
| Significant? (alpha = 0.05) | Yes | No | No | Yes | No | No | |
| Number of XY Pairs | 5 | 5 | 5 | 5 | 5 | 5 | |
Notes: Correlation matrix for DWB data and expression levels. Pearson correlation analysis has been performed: 1) between behavioral effects and alterations in gene expression levels, and 2) between behavioral effects and alterations in protein expression levels (grey shading); p<0.05 has been considered significant.
Abbreviations: COMP, cartilage oligomeric matrix protein; MMP, matrix metalloproteinase; TIMP, tissue inhibitors of metalloproteinase; FAAH, fatty acid amide hydrolase; NAPE-PLD, N-acyl phosphatidylethanolamine phospholipase D.