ABSTRACT
The Aer2 chemoreceptor from Pseudomonas aeruginosa contains a PAS sensing domain that coordinates b-type heme and signals in response to the binding of O2, CO, or NO. PAS-heme structures suggest that Aer2 uniquely coordinates heme via a His residue on a 310 helix (H234 on Eη), stabilizes O2 binding via a Trp residue (W283), and signals via both W283 and an adjacent Leu residue (L264). Ligand binding may displace L264 and reorient W283 for hydrogen bonding to the ligand. Here, we clarified the mechanisms by which Aer2-PAS binds heme, regulates ligand binding, and initiates conformational signaling. H234 coordinated heme, but additional hydrophobic residues in the heme cleft were also critical for stable heme binding. O2 appeared to be the native Aer2 ligand (dissociation constant [Kd] of 16 μM). With one exception, mutants that bound O2 could signal, whereas many mutants that bound CO could not. W283 stabilized O2 binding but not CO binding, and it was required for signal initiation; W283 mutants that could not stabilize O2 were rapidly oxidized to Fe(III). W283F was the only Trp mutant that bound O2 with wild-type affinity. The size and nature of residue 264 was important for gas binding and signaling: L264W blocked O2 binding, L264A and L264G caused O2-mediated oxidation, and L264K formed a hexacoordinate heme. Our data suggest that when O2 binds to Aer2, L264 moves concomitantly with W283 to initiate the conformational signal. The signal then propagates from the PAS domain to regulate the C-terminal HAMP and kinase control domains, ultimately modulating a cellular response.
IMPORTANCE Pseudomonas aeruginosa is a ubiquitous environmental bacterium and opportunistic pathogen that infects multiple body sites, including the lungs of cystic fibrosis patients. P. aeruginosa senses and responds to its environment via four chemosensory systems. Three of these systems regulate biofilm formation, twitching motility, and chemotaxis. The role of the fourth system, Che2, is unclear but has been implicated in virulence. The Che2 system contains a chemoreceptor called Aer2, which contains a PAS sensing domain that binds heme and senses oxygen. Here, we show that Aer2 uses unprecedented mechanisms to bind O2 and initiate signaling. These studies provide both the first functional corroboration of the Aer2-PAS signaling mechanism previously proposed from structure as well as a signaling model for Aer2-PAS receptors.
KEYWORDS: chemoreceptor, PAS domain, signal transduction, Pseudomonas aeruginosa, heme, oxygen
INTRODUCTION
Pseudomonas aeruginosa is a common environmental bacterium and a significant cause of opportunistic human disease. It survives in complex environments with the aid of 26 chemoreceptors and four chemosensory systems that collectively sense environmental conditions and modify bacterial behavior. The roles of three of these chemosensory systems are known: one modulates type IV pili production and twitching motility (Pil-Chp system), another controls biofilm formation (Wsp system), and a third regulates flagellum-mediated chemotaxis (Che system) (1, 2). The role of the fourth chemosensory system, Che2 (PA0173-PA0179), is currently unknown. Che2 expresses a complete set of chemosensory proteins (CheY2, CheA2, CheW2, CheR2, CheD, and CheB2), including a chemoreceptor (PA0176) called Aer2 (previously called McpB). Aer2 was so named because it, along with classical Aer, was observed to mediate aerotaxis by P. aeruginosa (3). However, we and others have not observed Aer2-mediated chemotaxis or aerotaxis in P. aeruginosa (4, 5). Moreover, it is now understood that the response regulators (CheY proteins) of Che2-like systems do not bind to the bacterial flagellar motor protein, FliM, to modulate swimming behavior (6–8; K. J. Watts and E. Orillard, unpublished data). This suggests that the primary role of Che2 is something other than the control of chemotaxis or aerotaxis. Notably, a role for Che2 in virulence has been suggested (9, 10).
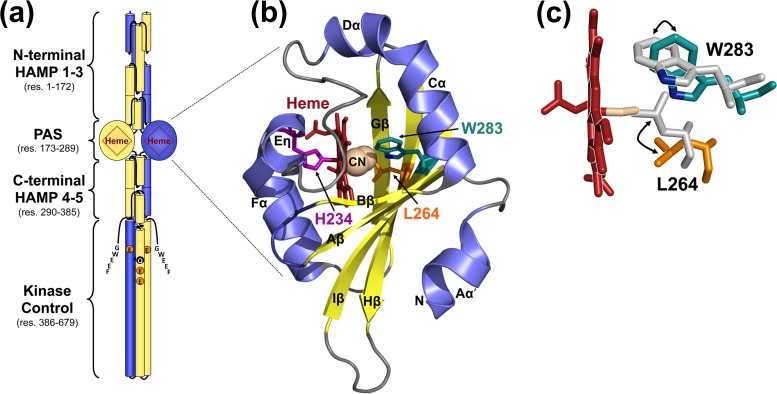
The Che2 chemoreceptor Aer2 has no membrane-spanning segments. However, during the early stationary phase of P. aeruginosa growth, Che2 proteins form a cluster at the cell pole that is held together solely by Aer2 (5, 10). Importantly, Che2 proteins do not colocalize with Che (chemotaxis system) proteins (5). Aer2 has an unusual architecture, with a PAS sensory domain sandwiched between three N-terminal and two C-terminal HAMP domains (Fig. 1a). These domains precede a kinase control module that is typical of methyl-accepting chemoreceptors. The kinase control module has four predicted methylation sites (QEEE) and a C-terminal pentapeptide (GWEEF) for binding the adaptation enzymes CheR2, CheB2, and CheD (Fig. 1a) (11). In P. aeruginosa, receptor deamidation/demethylation by CheB2 (and possibly CheD), as well as methylation by CheR2, is expected to fine-tune Aer2-mediated responses. The kinase control module of Aer2 shares significant sequence identity with the kinase control modules of the major Escherichia coli chemoreceptors. Thus, Aer2 is able to control the E. coli chemotaxis pathway through direct interactions with the E. coli adapter protein, CheW, and the histidine kinase, CheA (4). When Aer2 is expressed in otherwise chemoreceptor-less E. coli, it mediates repellent tumbling (signal-on) responses to O2, CO, and NO (4). Gas-bound Aer2 causes rapid autophosphorylation of bound CheA with subsequent phosphotransfer to CheY. Phospho-CheY in turn binds to the E. coli flagellar switch protein, FliM, causing a directional change in flagellar rotation from counterclockwise to clockwise, resulting in E. coli tumbling.
FIG 1.
P. aeruginosa Aer2 and the structure of its PAS domain. (a) Model of an Aer2 dimer showing the PAS domain sandwiched between three N-terminal and two C-terminal HAMP domains. The C-terminal kinase control module has four predicted methylation sites (QEEE) and a C-terminal pentapeptide (GWEEF) for binding adaptation enzymes. (b) Crystal structure of the Aer2 PAS domain in cartoon form with heme cofactor (shown as red sticks) and bound cyanide (shown as spheres) (PDB entry 3VOL) (13). The Fe-CN bond angle is 137° (13). The side chains of three amino acids relevant to this study, H234, L264, and W283, are shown as sticks. For clarity, the PAS structure is shown rotated 180° around the x axis compared with the orientation shown in panel a. (c) Cyanide-bound heme and a structural overlay showing the locations of the L264 and W283 side chains in both the unliganded (Fe3+ heme, gray side chains; PDB entry 4HI4) (14) and liganded (Fe3+-CN heme; colored side chains) (13) Aer2 PAS domain. The position of the W283 nitrogen, which is predicted to bond with O2, is shown in blue. Abbreviations: res, residue; CN, cyanide.
The Aer2 gas response is initiated in the PAS (Per-ARNT-Sim) domain, which itself binds pentacoordinate b-type heme (4). PAS domains are common sensing and signaling domains in nature. They have a broadly conserved structure that consists of a central antiparallel β-sheet with five β-strands (Aβ, Bβ, Gβ, Hβ, and Iβ) flanked by several α-helices (Cα, Dα, Eα, and Fα) (12). There are currently two structures for the Aer2 PAS domain: one contains cyanide bound to ferric heme (cyanomet, or Fe3+-CN; PDB entry 3VOL) (13) (Fig. 1b), and the other contains unliganded ferric heme (Fe3+; PDB entry 4HI4) (14). These structures revealed several unusual PAS features, including an extended Cα/Dα helix, a short 310 helix, called Eη (that replaces Eα), heme coordination via a His residue on Eη, and potential O2 stabilization via the indole group of a Trp residue on Iβ (Fig. 1b). In contrast, other PAS domains with b-type heme, like those in E. coli DOS (EcDOS) or Sinorhizobium meliloti FixL (RmFixL), coordinate heme with a His residue on the Fα helix and stabilize O2 binding via an Arg residue on Gβ (15). The two Aer2 PAS structures represent nonphysiological heme states (cyanomet and ferric heme), but they do represent structures with and without ligand, and overlaying these two structures highlights several residues that may be important for conformational signaling. In the absence of ligand, the Iβ Trp residue W283 appears to rotate ∼90°, and an adjacent Leu residue, L264 on Hβ, contracts toward the heme iron center to occupy the position where CN− was bound (Fig. 1c) (14). The heme itself appears to shift ∼2.0 Å upon ligand binding, and the heme pocket adjusts accordingly (14).
The Aer2 PAS domain is flanked on either side by poly-HAMP units (Fig. 1a). Individual HAMP domains form parallel four-helix bundles that are commonly found in prokaryotic proteins as signal-transducing modules (16). In Aer2, there appears to be minimal PAS-HAMP interactions, and overall, Aer2 assumes a linear domain arrangement (Fig. 1a) (14). This contrasts with the aerotaxis receptor, Aer, where side-on PAS-HAMP interactions allow PAS to control the HAMP signaling state through direct interactions (17). For Aer2, several structures have been solved for the N-terminal HAMP domains (18, 19). Those structures show that HAMP1 is separated from HAMP2-3 by a helical extension (18, 19). HAMP1 is also largely dispensable for Aer2 function (4). In contrast, N-terminal HAMP2-3, and C-terminal HAMP4-5, each form integrated di-HAMP units that are indispensable for Aer2 function (4, 19). The HAMP1 and HAMP2 structures represent signal-on and signal-off states, respectively (18), lending support to the hypothesis that poly-HAMP chains relay signals by interconverting HAMP signaling states along the HAMP chain.
Based on experimental evidence, our current signaling model for Aer2 includes the following features: Aer2 PAS-heme binds O2, generating a conformational signal that is transmitted to the PAS Iβ strand (4, 13, 14). N-terminal HAMP2-3 structures do not transmit signals but function to stabilize the PAS signaling state by altering their conformations in response to PAS ligand binding (4, 18). The PAS conformational signal is transmitted to C-terminal HAMP4-5, which together function as a unit to inhibit signaling from the kinase control module (4). Therefore, without a PAS ligand, the kinase control module conveys the signal-off state, but in the presence of PAS ligand, HAMP4-5 no longer inhibits the kinase control module, resulting in a signal-on output and the autophosphorylation of bound CheA2. Without HAMP4-5, the default state of the isolated kinase control module is signal-on (4). The purpose of the current study is to clarify the mechanisms used by the Aer2 PAS domain to bind heme, regulate ligand binding, and initiate conformational signaling. We provide evidence that (i) the Eη His coordinates heme binding, (ii) the hydrophobic heme pocket is crucial for stable heme binding, (iii) O2 is the native ligand of the Aer2 PAS domain, (iv) the unprecedented Iβ Trp stabilizes O2-binding but not CO-binding and plays a pivotal role in signal initiation, and (v) the Hβ Leu and other conserved PAS residues are important for heme binding, stable gas binding, and signal transduction.
RESULTS
Aer2 PAS coordinates heme with a uniquely positioned histidine residue.
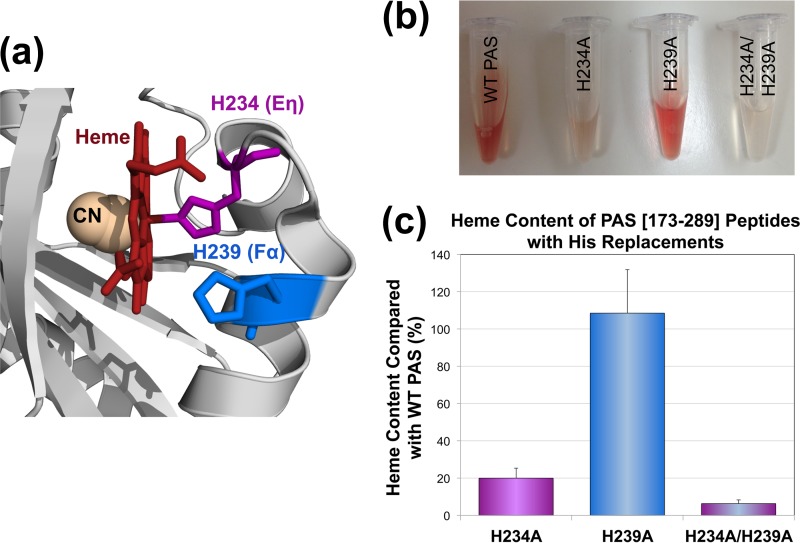
Structural studies suggest that the Aer2 PAS domain from P. aeruginosa coordinates b-type heme via a His residue (H234) that resides on a short Eη helix (Fig. 1b) (13, 14). In contrast, other PAS domains coordinate b-type heme with a His residue on the PAS Fα helix (15, 20). Notably, Eη and Fα His residues are both highly conserved in Aer2-PAS homologs (Fig. 2a; see also Fig. S1 in the supplemental material). To test the contributions of each histidine to heme binding in P. aeruginosa Aer2, H234A (Eη His), H239A (Fα His), and H234A/H239A were introduced into the PAS peptide Aer2[173-289]. Aer2[173-289] is expressed with an N-terminal 6× His tag and contains all necessary PAS heme-binding components (4). The purified PAS-H234A peptide showed a significant heme-binding defect, whereas PAS-H239A retained wild-type (WT) heme content (Fig. 2b and c). This confirms that the Eη His is the predominant means of coordinating heme in Aer2. However, 20% of PAS-H234A molecules retained heme, and PAS-H234A/H239A exhibited a significant decrease in heme content versus H234A alone (6% heme, P < 0.05) (Fig. 2c). Thus, the dual His replacement peptide has a lower heme affinity, suggesting that H239 contributes to heme coordination in the absence of H234.
FIG 2.
Heme coordination in the Aer2 PAS domain. (a) Location of the Eη (H234) and Fα His (H239) side chains in the cyanomet structure of the Aer2 PAS domain (13). Aer2-PAS structures indicate that the Eη His coordinates heme (13, 14), whereas the Fα His coordinates heme in other PAS-heme proteins (15, 20). Both His residues are highly conserved in Aer2-PAS homologs (see Fig. S1). (b) Purified Aer2 PAS[173-289] peptides (imidazole bound; 2.6 to 4 mg ml−1) showing less red color in Aer2-H234A and Aer2-H234A/H239A than in WT Aer2 and Aer2-H239A. (c) Heme content of PAS peptides with Eη and Fα His replacements, given as a percentage of WT PAS heme content, corrected for peptide concentration (see Materials and Methods). Error bars represent the standard deviations from multiple experiments. Abbreviations: CN, cyanide; WT, wild type.
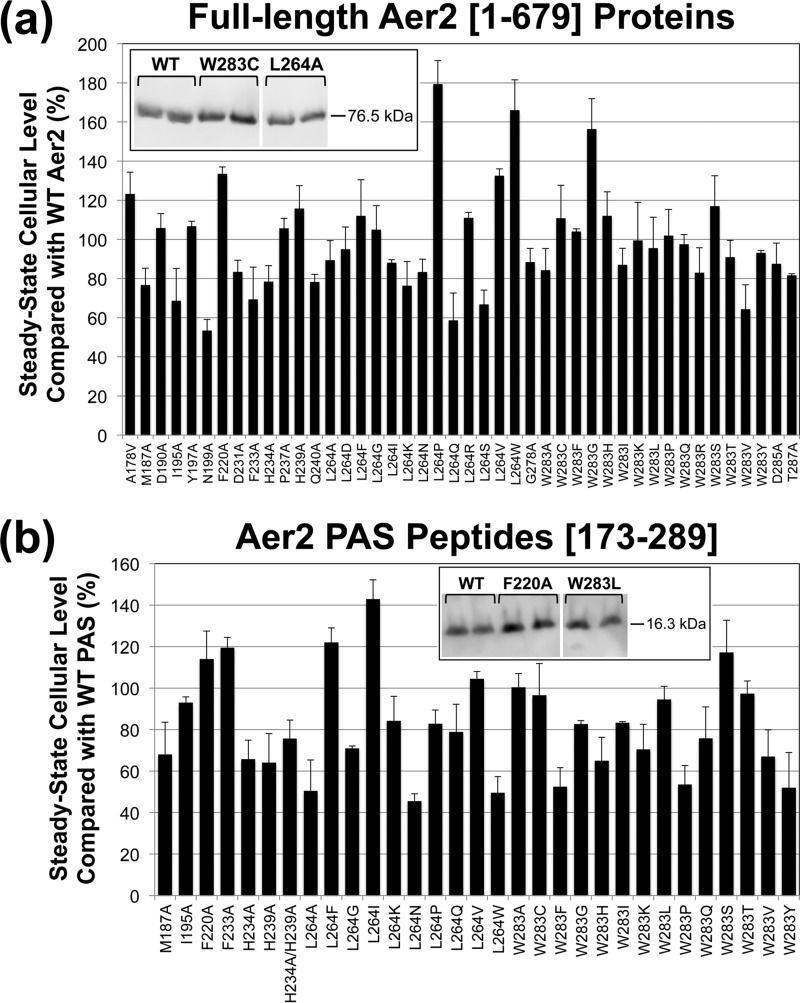
To determine the effect of the His substitutions on Aer2 signaling, mutations encoding H234A and H239A were introduced separately into full-length aer2 in an E. coli expression plasmid. Both Aer2 mutants had steady-state expression levels comparable to that of WT Aer2 (Fig. 3a). When WT Aer2[1-679] is expressed in chemoreceptorless E. coli BT3388, it directs E. coli to tumble in the presence of O2 because Aer2 signaling activates the E. coli chemotaxis cascade (4). When O2 is replaced with N2, Aer2 no longer signals, and after 5 to 10 s, BT3388 cells resume smooth-swimming behavior (∼2% of the cells tumble at any time) (4). BT3388 cells expressing Aer2-H239A behaved like cells expressing WT Aer2: cells tumbled in air (20.9% O2) and had smooth-swimming behavior in N2. In contrast, Aer2-H234A orchestrated tumbling in air like that of WT Aer2, but cells remained tumbling biased in N2 (∼60% of cells tumbled in N2 versus ∼2% for WT Aer2). Aer2-H234A therefore has a signal-on bias (Fig. 4a). The partial response may reflect the ability of a small proportion of heme-retaining molecules to respond to O2 changes.
FIG 3.
Steady-state cellular levels of full-length Aer2 proteins and PAS peptides in E. coli. (a) Steady-state levels of full-length Aer2 proteins compared with WT Aer2[1-679] in E. coli BT3388. Aer2 expression was induced with 50 μM IPTG, and protein levels were determined from Western blots, an example of which is shown in the inset box. Lanes are from the same gel; intervening lanes are represented by white space. (b) Steady-state levels of PAS peptides compared with WT PAS[173-289] in E. coli BL21(DE3). Aer2 peptide expression was induced with 100 μM IPTG, and protein levels were determined from Western blots. (See the example in the inset box. Lanes are from the same gel; intervening lanes are represented by white space.) Error bars represent the standard deviations from multiple experiments.
FIG 4.
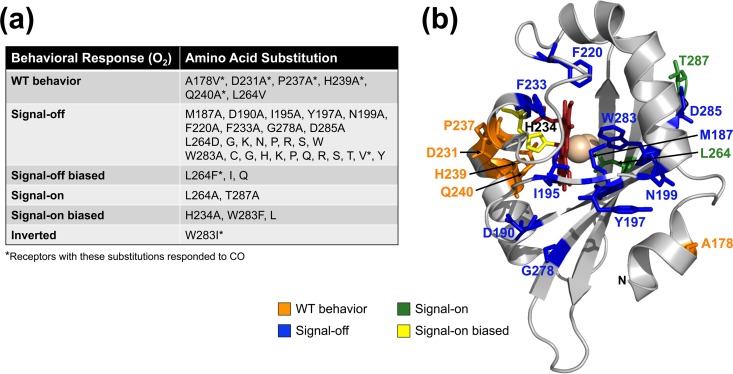
Aer2 mutant phenotypes in temporal assays. (a) Effects of amino acid substitutions on Aer2-mediated behavior in E. coli BT3388. Signal-off mutants exhibited smooth-swimming behavior (∼2% tumbling) in both air and N2, whereas signal-on mutants tumbled constantly in both air and N2. Neither signal-off nor signal-on mutants responded to the introduction or removal of O2. Signal-off-biased mutants responded to the introduction of O2 but adapted, unlike WT Aer2 in BT3388, which remained signal-on in the presence of O2. Signal-on-biased mutants responded to the removal of O2, but at least 50% of the cells continued to tumble in N2. Residue substitutions marked by an asterisk resulted in receptors that could respond to CO, i.e., they directed cell tumbling in the presence of CO. CO responses could not be determined for signal-on-biased and signal-on mutants. (b) Alanine mutants mapped onto the cyanomet structure of Aer2. Original side chains are shown as color-coded sticks based on the O2 responses listed in panel a.
When WT Aer2 is expressed in E. coli, cells respond to both O2 and CO (4). To test for a CO response, E. coli BT3388 cells expressing Aer2 are monitored in CO temporal assays. In these assays, cells are perfused with N2 (to remove O2) until they resume smooth swimming, after which CO is perfused for 10 s. If the receptor can respond to CO, cells tumble and continue to tumble for up to 30 s after CO has been removed (4). To determine if Aer2-H239A can respond to CO, cells expressing Aer2-H239A were perfused with CO under anaerobic conditions. Similar to WT Aer2, cells expressing Aer2-H239A responded to CO by tumbling, and the tumbling persisted for ∼30 s after CO was removed. However, a CO response could not be determined for Aer2-H234A, because cells expressing Aer2-H234A tumbled too extensively in the absence of O2.
PAS structures suggest a signaling mechanism.
Two PAS domain structures currently exist for Aer2, one with cyanide bound to ferric heme (cyanomet [Fe3+-CN]) (13) and another containing ferric heme without ligand (ferric heme [Fe3+]) (14). The ligand-bound structure suggests that Aer2 stabilizes O2 binding via a Trp residue on the PAS Iβ strand (W283) (Fig. 1) (13). Moreover, an overlay of the two PAS structures suggests that both the Iβ Trp and an adjacent Leu residue on Hβ (L264) reorient in response to ligand binding. In the absence of ligand, the indole group of the Iβ Trp may rotate ∼90°, whereas the adjacent Hβ Leu residue contracts toward the heme iron center to occupy the position where CN− was bound (Fig. 1c) (14). To determine the importance of W283 and L264 for Aer2 sensing and signaling, we performed site-directed random mutagenesis on each residue and analyzed the effects on receptor function, heme binding, and ligand binding.
The Iβ Trp is important for gas binding and signal initiation.
The PAS Iβ Trp is 100% conserved in 100 Aer2 PAS-like sequences (Fig. S1) and may be essential for stabilizing heme-O2 binding. To determine the role of the Iβ Trp in P. aeruginosa Aer2, we performed site-directed random mutagenesis on the W283 codon in the construct that expresses full-length Aer2. Expression was induced in E. coli BT3388 with 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and individual mutants were screened under the microscope for behavioral defects. Mutants with non-WT behavior were sequenced to determine the amino acid substitution at W283. After several rounds of mutagenesis and screening, 12 amino acid changes were identified at W283 that altered behavior (Fig. 4a). W283H and W283Y were not identified during the screen, but they were specifically engineered since these amino acids stabilize O2-binding in other heme proteins (21–23). W283A was similarly not identified during screening but was created as part of the PAS alanine mutagenesis described below.
All 15 of the W283 mutant proteins were stably expressed in E. coli BT3388 (Fig. 3a), but 12 of the receptors were signal-off receptors that did not respond to the addition or removal of O2 (Fig. 4a). Cells expressing these receptors swam smoothly in both the presence and absence of O2, even after induction with 1 mM IPTG to produce high cellular levels of Aer2. In contrast, cells expressing Aer2-W283F, L, or I retained some functionality (Fig. 4a). Aer2-W283F and Aer2-W283L were signal-on-biased mutants that orchestrated tumbling in air like cells expressing WT Aer2, but when air was removed, 50 to 80% of the cells continued to tumble (Fig. 4a). Cells expressing Aer2-W283I had an inverted phenotype where ∼50% of the cells tumbled in N2 but became smooth swimming after ∼30 s in air.
Heme-CO binding does not require amino acid stabilization and should not require W283. However, only two of the 15 W283 mutants, Aer2-W283I and Aer2-W283V, responded to CO (Fig. 4a). Cells expressing these mutants tumbled when CO was added in either N2 or air. It was not possible to determine whether cells expressing Aer2-W283F or Aer2-W283L responded to CO, because cells expressing these mutants tumbled too extensively to determine a CO response.
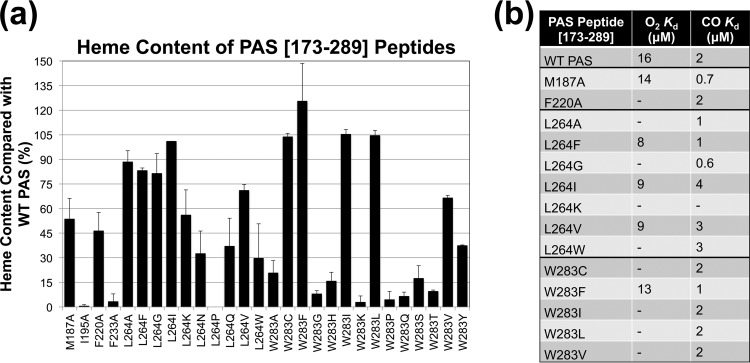
To determine the O2 and CO binding affinities of W283 mutants, W283-encoding mutations were transferred into the Aer2-PAS expression construct, Aer2[173-289], and the PAS peptides were purified on nickel-nitrilotriacetic acid (Ni-NTA)–agarose. W283 mutants that were analyzed included those that responded to O2 or CO and 11 of the signal-off mutants; these were compared with WT Aer2 (which was determined to have an O2 dissociation constant [Kd] of 16 μM and a CO Kd of 2 μM) (Fig. S2 shows WT O2 and CO titrations). Unexpectedly, PAS peptides for nine of the 11 W283 signal-off mutants exhibited very low heme content when purified (3 to 37.5% of WT heme levels) (Fig. 5a), and gas-binding affinities could not be determined. To test whether these heme-binding defects were also present in full-length receptors, we purified full-length Aer2-W283Y and Aer2-L264N (see below), both of which had PAS peptides with heme-binding defects (Fig. 5a). Neither of the purified full-length receptors showed heme binding (data not shown). The other signal-off mutants, W283C and W283V, had sufficient heme content to analyze (Fig. 5a). Aer2-W283C and Aer2-W283V did not respond to O2 and did not bind it; during O2 titrations, both mutants exhibited Met-heme spectra with rapid oxidation from Fe(II) to Fe(III) heme (Met-heme spectra were independently verified by oxidizing proteins with potassium ferricyanide and comparing with the spectra from O2 titrations) (Fig. S2c). However, both mutants bound CO with WT affinity, and Aer2-W283V was able to respond to it (Fig. 4a and 5b).
FIG 5.
PAS peptide heme content and gas-binding affinities. (a) Heme content of PAS peptides with amino acid substitutions, given as a percentage of WT PAS heme content, corrected for peptide concentration (see Materials and Methods). Values below 40% indicate a substantial heme-binding defect. Aer2[173-289]-L264P contained no measureable heme. Error bars represent the standard deviations from multiple experiments. (b) PAS peptide O2 and CO binding affinities. A dash indicates that O2- or CO-bound spectra were not observed, so binding affinities could not be determined.
Of the three W283 mutants that responded to O2 in the behavioral assay (Aer2-W283F, -L, and -I), only Aer2-W283F appeared to bind O2, and its O2 affinity was similar to that of WT Aer2-PAS (Fig. 5b). In contrast, Aer2-W283L and Aer2-W283I both responded to O2, but purified PAS peptides with these substitutions did not bind O2 (Fig. 4 and 5b) and were rapidly oxidized from Fe(II) to Fe(III) heme. Sawai et al. similarly reported that purified full-length Aer2-W283L does not bind O2 (13). It is possible that O2 binding to these mutants is too transient to observe during in vitro O2 titrations but is sufficiently stable in vivo to generate a behavioral response. All of the W283 mutants tested bound CO with affinities similar to that of WT Aer2 (Fig. 5b), irrespective of whether the corresponding full-length receptors responded to CO or not (Fig. 4). Overall, these data indicate that the Iβ Trp is important for stable heme and O2 binding but not CO binding and is important for signal initiation in the Aer2 PAS domain.
Substitutions at the Hβ Leu alter gas binding and signaling.
Aer2 PAS structures suggest that the Hβ Leu residue, L264, is involved in initiating PAS signaling (14). The L264 side chain appears to occupy the PAS ligand-binding site but swings out of the site when ligand binds (Fig. 1c) (14). Hβ Leu is well conserved in Aer2 PAS-like sequences, but other hydrophobic amino acids are also found at the same position, primarily Val and, to a lesser extent, Ile (Fig. S1). To determine if Aer2 can function with Val at 264, Aer2-L264V was engineered by site-directed mutagenesis. Full-length Aer2-L264V mediated an O2 response but exhibited a 30-s-delayed smooth-swimming response in N2 and did not respond to CO. Therefore, Aer2 can function with Val at 264, but the behavioral response is restricted to O2. Notably, PAS-L264V bound O2 and CO with affinities that were similar to that of the WT (Fig. 5b).
To determine whether other replacements at L264 affect the O2 response, we performed site-specific random mutagenesis on the L264 codon and screened for defective mutants, as outlined above for W283. L264A was not identified during the screen but was created as part of the PAS alanine mutagenesis described below. After several rounds of mutagenesis and screening, 12 amino acid substitutions were identified at L264 that altered the O2 response (Fig. 4a). All of these mutants were stably expressed in E. coli BT3388 (Fig. 3a). Eight of the mutants were nonfunctional, signal-off mutants (Fig. 4a), even after induction with 1 mM IPTG. In contrast, Aer2-L264A was locked signal-on, causing cells to tumble constantly in air and in N2 (Fig. 4a). The remaining three mutants, Aer2-L264F, -I, and -Q, were signal-off-biased mutants that responded to O2 (Ile shows some conservation at this position) (Fig. 4a; Fig. S1). Cells expressing Aer2-L264I and Aer2-L264Q had WT O2 responses, whereas cells expressing Aer2-L264F had a reduced tumble response to O2 (∼60% of cells tumbled). However, all three mutants adapted to O2 (cells became less tumble biased) over the course of several minutes. Only one of the L264 mutants, Aer2-L264F, tumbled in response to the addition of CO (this mutant also responded to O2) (Fig. 4a). A CO response could not be determined for Aer2-L264A, because cells expressing this Aer2 variant tumbled constantly in the presence and absence of O2.
To determine the O2 and CO binding affinities of the L264 mutants, L264-encoding mutations were transferred to the Aer2 PAS peptide Aer2[173-289], and the peptides were purified on Ni-NTA–agarose. L264 mutants that were analyzed included those that responded to O2 or CO, the signal-on mutant Aer2-L264A, and five of the signal-off mutants; these were compared with WT Aer2-PAS (Fig. 5). Four of the mutants expressed PAS peptides that contained very low heme content when purified. This included three of the signal-off mutants, L264N, -P, and -W, and one of the functional mutants, L264Q (0 to 37% of WT heme levels) (Fig. 5a). Gas-binding affinities could not be determined for PAS-L264N, -P, and -Q but were determined for PAS-L264W due to higher peptide purity. PAS-L264P showed no detectable heme spectra, even after scanning concentrated (135 μM) protein. There was no defect in the steady-state cellular expression level of PAS-L264P (Fig. 3b). Of the remaining L264 mutants, three bound O2 and CO (L264F, -I, and -V), three bound CO but not O2 (L264A, -G, and -W), and one bound neither gas (L264K) (Fig. 5b). The three L264 mutants that bound O2 (L264F, -I, and -V) also responded to it. In contrast, PAS-L264G, -I, -V, and -W all bound CO but did not respond to it. Of the L264 mutants that did not stably bind O2, PAS-L264A and PAS-L264G were rapidly oxidized by O2 and PAS-L264K was slowly oxidized by O2. In contrast, PAS-L264W showed no shift in its Soret maxima during O2 titrations, suggesting the absence of heme-O2 interactions. This finding lends support to the hypothesis that L264 must move out of the ligand-binding site to allow for O2 binding. However, L264W still allowed CO binding, indicating that the CO-binding angle (perpendicular to the plane of the heme) was permitted (Fig. 5b).
Of all the mutants in this study, the signal-off mutant, Aer2-L264K, was the only mutant that did not bind either O2 or CO. Moreover, the deoxy spectra of PAS-L264K contained a β band and a prominent α band (Fig. S2d), suggesting the formation of a hexacoordinate heme. This would entail coordination to H234 on the proximal side of the heme as well as coordination on the distal side, quite possibly by the amino group of lysine (e.g., Lys can coordinate heme in place of Met in cytochrome c-550 [24]). This differs from WT deoxy-Aer2, which contains pentacoordinate heme (Fig. S2d) (4).
Aer2 signaling is disrupted by alanine replacements at conserved residues.
To complement the mutagenesis experiments on W283 and L264, 16 residues that are highly conserved in Aer2 homologs (Fig. S1, marked by asterisks) were selected for site-directed alanine mutagenesis (A178 was replaced with Val). Conserved Gly residues that are structural elements at turns were excluded. The results for L264A and W283A were discussed above. Most of the mutants exhibited stable steady-state expression levels in E. coli BT3388 (Fig. 3a). However, 10 of the 16 Ala mutants were signal-off mutants and did not respond to either O2 or CO (Fig. 4). This included the four heme cleft mutants that were tested: M187A, I195A, F220A, and F233A. In contrast, Aer2-A178V, Aer2-P237A, and Aer2-Q240A mediated WT responses to both O2 and CO (Fig. 4). Aer2-D231A similarly orchestrated WT-like responses to O2 and CO but had a 30-s-delayed smooth-swimming response in N2 after O2 was removed. Overall, the Ala mutants that had WT or signal-on-biased behavior congregated on the Eη and Fα helices (Fig. 4b, orange and yellow residues). Very few signal-on mutants were identified in this study. Aer2-T287A, like Aer2-L264A, was a signal-on mutant that caused cells to tumble constantly in both air and N2. Because of this, a CO response could not be determined for Aer2-T287A. L264 and T287 both reside on the PAS β-sheet, which is the signal output surface of the PAS domain (12, 14).
To determine gas-binding affinities for the four heme-pocket mutants (M187A, I195A, F220A, and F233A), relevant mutations were transferred to the construct expressing Aer2[173-289] and the PAS peptides were purified. PAS peptides with I195A and F233A had severe heme-binding defects (Fig. 5a), even though neither of these mutants had steady-state expression defects (Fig. 3b). In contrast, PAS peptides with M187A and F220A both bound heme (Fig. 5a) and could bind CO (Fig. 5b), yet neither mutant responded to CO. Aer2-F220A neither bound nor responded to O2, whereas Aer2-M187A bound O2 with WT affinity but was also rapidly oxidized by O2 (Fig. 5a). Sawai et al. similarly reported that full-length Aer2-M187A binds O2 (13). Aer2-M187A was the only mutant in this study that bound, but did not respond, to O2.
Signal-on behavior is independent of Aer2 methylation.
When WT Aer2 is expressed in E. coli, it does not adapt to O2. This is because Aer2 is methylated by the E. coli methyltransferase CheR but is not demethylated by the E. coli methylesterase CheB (4). When WT Aer2 is expressed in an E. coli strain lacking CheR and CheB, Aer2 remains unmethylated and the cells have a low tumbling frequency (∼5% of cells tumble in O2 [4]). Hence, robust signal-on behavior requires receptor methylation. In this study, two locked signal-on mutants (L264A and T287A) and three signal-on-biased mutants (H234A, W283F, and W283L) were identified. To determine if their phenotypes were dependent on receptor methylation, full-length receptors containing each of these amino acid substitutions were expressed in E. coli UU2610, which lacks all E. coli chemoreceptors, as well as CheR and CheB (25). In UU2610, the tumbling bias of Aer2-L264A decreased ∼20% in both air and in N2. However, the tumbling biases of the other four mutants were not diminished by the lack of receptor methylation in UU2610. This suggests that the signal-on biases of these receptors are due primarily to the amino acid changes in the PAS domain and not receptor methylation status.
DISCUSSION
The Eη histidine coordinates heme in the Aer2 PAS domain.
Crystal structures of the Aer2 PAS domain identify the Eη His (H234) as the proximal heme-coordinating ligand (13, 14). This differs from the PAS domains of RmFixL, EcDOS, Acetobacter xylinum PDEA-1 (AxPDEA-1), and Burkholderia xenovorans RcoM, where, in each case, an Fα His residue coordinates b-type heme (15, 20). Although the Fα His is well conserved in Aer2-PAS homologs (see Fig. S1 in the supplemental material), in P. aeruginosa Aer2 it resides ∼10 Å from the heme Fe, in contrast to the Eη His, which lies ∼2 Å away. In the current study, the Fα His substitution, H239A, did not affect PAS heme content or behavioral responses. In contrast, the Eη His substitution, H234A, imposed substantial heme-binding and behavioral defects (Fig. 2), confirming that it is the proximal coordinating His of Aer2. However, Aer2-H234A had a signal-on bias, unlike all other heme-binding mutants in this study, which were signal-off or signal-off biased. In addition, the 80% heme loss in PAS-H234A is less than that observed for other PAS-heme domains when the proximal coordinating His is replaced. For example, His substitutions in RcoM result in <1% heme (20), and a His-to-Ala alteration in the Aer2 PAS-2 domain from Vibrio cholerae results in 2% heme (K. J. Watts, unpublished data). In Aer2-PAS, it is unclear how heme might be coordinated in the absence of H234, particularly since H239 lies at a more remote location at the entrance of the heme cleft. A dual PAS-H234A/H239A mutant had significantly less heme than PAS-H234A (Fig. 2), suggesting that Fα-H239 contributes to heme coordination in the absence of Eη-H234. Alternatively, the dual His replacements might distort the heme pocket in a way that prevents heme retention. Heme might instead be retained by PAS-H234A through hydrophobic pocket interactions. Hydrophobic interactions are sufficient to bind b-type hemes in the YybT family proteins from Bacillus and Geobacillus, which have no natural proximal heme-coordinating residue in their PAS domains (26, 27).
The hydrophobic heme cleft is critical for stabilizing heme binding in Aer2.
In Aer2-PAS, the heme cleft is a hydrophobic pocket in which the imidazole ring of H234 coordinates the heme-Fe, and H251 hydrogen bonds to the heme-7-proprionate (13). In this study, replacing heme-coordinating H234 caused a substantial heme-binding defect (PAS-H234A retained 20% heme) (Fig. 2). However, other amino acid substitutions in both the proximal (I195A and F233A) and distal (L264N, -P, -Q, and -W and W283A, -G, -H, -K, -P, -Q, -S, -T, and -Y) heme cleft likewise caused substantial heme-binding defects (0 to 37.5% of WT heme content) (Fig. 5a). Remarkably, some of these defects were greater than that caused by H234A. This suggests that hydrophobic heme cleft interactions are critical for stabilizing heme binding in Aer2. On the proximal side of the cleft, I195 and F233 both reside ∼4 Å from, and parallel to, the heme; the severe defects caused by Ala substitutions at these residues (0.6 to 3% heme content) shows that moderate perturbations in the PAS heme pocket can alter heme binding. Moreover, perturbations in O2 stabilizing and signaling residues, W283 and L264, also affected heme binding. In Bradyrhizobium japonicum FixL, replacing the distal O2-stabilizing Arg residue with Ala relaxes heme-protein coupling (28), and a similar event could be responsible for heme loss in PAS-W283 mutants.
Oxygen is the native ligand of the Aer2 PAS domain.
In the absence of imidazole, P. aeruginosa Aer2 purifies in the oxy-bound state (13). However, Aer can signal in response to the binding of O2, CO, and NO (4). This is atypical for heme sensors; they often bind all three gases, but typically they respond to only one. For example, the histidine kinase FixL is inhibited by O2 but not by CO or NO, even though it binds these two more tightly (29). Protohemes generally bind O2, CO, and NO with relative affinities of 1, 103, and 106, respectively (30, 31). In this study, we determined that the O2 and CO affinities of the isolated Aer2 PAS domain are 16 μM and 2 μM, respectively (Fig. 5b). The O2 affinity of Aer2-PAS is comparable with the O2 affinities of the PAS-heme O2 sensors EcDOS (13 μM), AxPDEA-1 (∼10 μM), and RmFixL (31 μM) (15, 32). In the DOS, PDEA-1, and FixL PAS domains, a Gβ Arg residue interacts directly with bound O2: substitutions at this residue substantially lower O2 but not CO affinities and affect O2-regulated behavior (28, 33). In Aer2, substitutions at the Iβ Trp likewise prevented O2 but not CO binding (Fig. 5b, W283C, -I, -L, and -V). Because changes in highly conserved residues predominately affect O2 binding properties and responses, these data suggest that O2 is the native ligand of the Aer2 PAS domain. We did not test NO binding in this study. However, unlike Aer2, heme-NO sensors usually exclude O2 binding; e.g., H-NOX (heme-nitric oxide/oxygen) proteins that lack hydrogen bond donors bind NO instead of O2 (34, 35), the PAS-heme domain of YybT is rapidly oxidized by O2 (27), and nitrophorins exclude O2 by maintaining their heme in the Fe(III) state (36).
The role of Aer2 PAS residues in ligand binding and signal transduction.
The data from this study indicate that the Iβ Trp, W283, stabilizes O2 binding to the Aer2 PAS-heme domain. Twelve of 15 W283 mutants, including those with amino acids that commonly stabilize O2 binding to other heme proteins (His, Tyr, and Arg [37]), resulted in signal-off Aer2 receptors that did not respond to O2. For W283 mutants that did not stably bind O2 in vitro (W283C, -I, -L, and -V), the heme cofactor was rapidly oxidized upon exposure to O2 (Fig. S2c). This has been observed similarly for EcDOS Gβ Arg mutants (33). Our data also indicate that W283 is critical for initiating conformational signaling from the PAS domain. This perhaps is not surprising given that the PAS Iβ strand connects directly to the C-terminal HAMP4 domain, and W283 resides close to the PAS DXT motif, which has been proposed to be involved in conformational signaling (12, 14). None of the 15 W283 mutants in this study retained WT function (Fig. 4a), and PAS-W283F was the only Trp mutant that preserved a WT O2 affinity (Fig. 5b). The side chain of Phe is similar in size to the nitrogen-containing Trp pyrrole ring that is predicted to hydrogen bond with heme-bound O2. However, Phe lacks a hydrogen-bonding moiety. One possibility is that O2 binding is supported by a solvent molecule in the distal pocket that acts as a hydrogen bond donor; this scenario was observed in a DevS mutant from Mycobacterium tuberculosis when the O2-stabilizing Tyr residue was replaced with Phe (38). In this study, Aer2-W283F retained partial functionality (as a signal-on-biased mutant), but Aer2 is clearly fine-tuned to use Trp for O2 binding and signal initiation.
The hypothesis that the Hβ Leu, L264, moves out of the ligand-binding site when O2 binds to heme (Fig. 1c) (14) was supported by the results of this study. Notably, the bulky Trp substitution, L264W, appeared to block O2 binding, and L264K formed a hexacoordinate heme that did not bind O2. However, the Hβ Leu itself was not specifically required for function: Aer2-L264F and Aer2-L264Q retained some functionality, and L264 substitutions that occur in other Aer2 PAS-like homologs, Val and Ile (Fig. S1), resulted in Aer2 receptors that bound and responded to O2 (Fig. 4 and 5). Interestingly, smaller hydrophobic replacements, L264A and L264G, resulted in nonresponsive Aer2 receptors that did not bind O2 and furthermore were rapidly oxidized by it. This suggests that (i) the size of the amino acid at the Hβ Leu is important and (ii) Hβ Leu is choreographed to move concomitantly with the Iβ Trp so that Trp can rotate into place and bond with O2 (Fig. 1c). Notably, O2 binding alone was not sufficient for PAS signaling. Aer2-M187A bound O2 but was unable to respond to it. M187 resides on the Aβ strand, adjacent to W283 on Iβ, but it does not exhibit a significant conformational shift between the cyanomet and ferric PAS-heme structures. Still, the distortion caused by the Ala replacement at residue 187 could feasibly block PAS β-sheet signaling rearrangements that are required for downstream signaling.
Like Aer2-M187A, most of the mutants in this study were signal-off mutants that did not respond to O2 or CO (Fig. 4a). The amino acid changes in these mutants most likely disengage PAS control of the downstream HAMP4-5 unit so that HAMP4-5 continues to inhibit the activity of the kinase control module. D285A specifically disrupts the conserved DXT motif that has a proposed role in conformational signaling between the PAS and C-terminal HAMP domains (12, 14). In Aer2-N199A, the Ala substitution could also disrupt interactions between the PAS N-terminal cap (N-cap, Aα′ helix) (Fig. 1b) and the PAS core (Cα helix); these interactions are required for both structural stability (N199A was the least stable receptor in this study) (Fig. 3a) and for N-cap reorientation during signaling (14, 39).
All of the PAS peptides tested in this study (with the exception of PAS-L264K) bound CO with an affinity that was the same as that of the WT peptide or better (M187A and L264G bound CO in the nanomolar range) (Fig. 5b). This is not surprising, since heme-CO binding does not require amino acid stabilization due to its high inherent affinity for heme (30). However, heme-CO binding did not predict function. Mutants that responded to CO bound it with WT affinity (W283I and V and L264F), but so did many of the mutants that did not respond to CO (Fig. 4 and 5). In the latter instances, CO binding apparently could not induce the conformational changes required for signal transduction.
These studies provide insight into the mechanisms used by the Aer2 PAS domain to regulate heme binding and ligand binding and initiate conformational signaling. The results support the model based on differences between the cyanomet and ferric PAS-heme structures, corroborating the roles of W283 and L264 in O2 stabilization and PAS signaling (Fig. 1) (14). When O2 binds to Aer2-PAS, it generates a conformational signal that is transmitted via the PAS Iβ strand to modulate the activity of the C-terminal HAMP and kinase control domains. Future studies will test an expanded model whereby O2-mediated signaling evokes a PAS dimer-to-monomer transition (14), resulting in the signal-off conformation of HAMP4, the signal-on conformation of HAMP5 (18), and deinhibition of the kinase control module (4).
MATERIALS AND METHODS
Bacterial plasmids and strains.
Full-length P. aeruginosa PAO1 Aer2[1-679] (PA0176) was expressed from pLH1, a pProEX-derived plasmid that expresses Aer2 with an N-terminal His6 tag (4). The Aer2 PAS domain, Aer2[173-289], was likewise expressed from pProEX with an N-terminal His6 tag (4). Full-length Aer2 was expressed in chemoreceptorless E. coli strains BT3388 (tar tsr trg tap aer [40]) and UU2610 (tar tsr trg tap aer cheR cheB [25]). UU2610 also lacks the E. coli adaptation enzymes CheR and CheB. Aer2 PAS peptides were expressed in E. coli BL21(DE3).
Mutagenesis and cloning.
Site-directed mutagenesis was performed on pLH1 using site-specific primers and PfuUltra II Fusion DNA polymerase (Agilent Technologies, Santa Clara, CA). To replace native codons with Ala or Val codons, 30 amplification cycles were performed with an annealing temperature of 55°C. For site-directed random mutagenesis, primers containing an equimolar mix of all four nucleotides at the L264 or W283 codons were used; however, the amplification conditions described above consistently created DNA insertions following the primer site. To solve this problem, we tested stepwise annealing temperatures from 55°C to 68°C and analyzed the constructs created. The lowest proportion of DNA inserts occurred when 68°C was used as the annealing temperature and 20 amplification cycles were performed. These conditions yielded no obvious bias for codon replacements with one, two, or three nucleotide changes and subsequently were used to create most of the site-specific random mutants identified in this study. Site-specific mutagenesis products were treated with DpnI (New England BioLabs, Ipswich, MA) to remove template strands and then electroporated into E. coli. Aer2 expression was induced with 600 μM IPTG, and products of the correct size were confirmed by Western blotting with HisProbe-horseradish peroxidase (Thermo Scientific, Rockford, IL). All mutations were confirmed by sequencing the entire coding sequence of aer2.
To create Aer2-PAS peptides expressing specific amino acid changes in the PAS domain, the PAS coding region (residues 173 to 289) was PCR amplified from pLH1-derived plasmids using PfuUltra II fusion DNA polymerase. PCR products were ligated into the NcoI and SalI sites of pProEX. Peptide expression and DNA sequencing were performed as described above.
Steady-state cellular Aer2 levels.
The steady-state cellular levels of the full-length Aer2 mutants were compared with that of WT Aer2 after inducing BT3388 cells with 50 μM IPTG. In contrast, the cellular levels of the PAS peptides were compared with that of WT Aer2[173-289] after inducing BL21(DE3) cells with 100 μM IPTG. Samples were electrophoresed in duplicate, and experiments were repeated on two to four separate days. Bands were visualized on HisProbe Western blots and quantified on a BioSpectrum digital imager (UVP, Upland, CA).
Behavioral assays.
BT3388 cells were grown at 30°C in tryptone broth containing 0.5 μg ml−1 thiamine and induced with 200 μM IPTG. At this induction level, the number of Aer2 receptors in E. coli is comparable with the total number of chemoreceptors in WT E. coli cells (4). Cells were placed into a gas perfusion chamber where the gas was toggled between air (20.9% O2) and N2, and cell behavior was analyzed (41, 42). Mutants that were signal-off (smooth swimming in air and in N2) were retested after induction with 1 mM IPTG to produce higher cellular levels of Aer2. Behavioral responses to O2 were repeated two or more times on at least two separate days. To determine CO responses, BT3388 cells (induced with 200 μM IPTG) were perfused with N2 for 30 s prior to perfusing with CO gas (>99% purity; Sigma-Aldrich, St. Louis, MO), which was added through the open end of the chamber for 10 s. For Aer2-W283I and Aer2-W283V, CO responses were also tested while air was being perfused.
Protein purification.
WT Aer2[173-289]/BL21(DE3) and relevant mutants were grown in LB broth, Lennox, containing 25 μg ml−1 5-aminolevulinic acid (Sigma-Aldrich) to enhance heme synthesis and incorporation. After 3 to 5 h of induction with 600 μM IPTG, cells were centrifuged at 10,000 × g for 15 min and resuspended to 1% of their original volume in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, and 10 mM imidazole) containing 0.3 mg ml−1 lysozyme, 1 μg ml−1 DNase I, and 100 μl of protease inhibitor cocktail for His-tagged proteins (Sigma-Aldrich). The cells were lysed by freeze-thawing five times, followed by sonication. Soluble protein was acquired by removing cellular debris at low speed (10,000 × g for 20 min) and the membrane fraction at high speed (485,000 × g for 1 h). The high-speed supernatant was applied to an Ni-NTA–agarose column (Qiagen, Valencia, CA) and allowed to empty by gravity flow. The column was washed with 10 column volumes of wash buffer 1 (50 mM Tris, pH 7.5, 500 mM NaCl, and 20 mM imidazole), followed by 8 to 10 column volumes of wash buffer 2 (50 mM Tris, pH 7.5, 500 mM NaCl, and 50 mM imidazole). Aer2 peptides were eluted by adding 1 ml of elution buffer (50 mM Tris, pH 7.5, 500 mM NaCl, and 250 mM imidazole) to the column, but only the red-colored fraction was collected. For proteins with no obvious red color, two ∼0.4-ml elution fractions were collected. Aer2 peptides usually were most concentrated in the second eluted fraction. The concentration of eluted protein was determined using a bicinchoninic acid protein assay (Thermo Scientific), and the quality of the sample was determined by staining SDS-PAGE gels with Coomassie brilliant blue.
Heme binding.
The proportion of heme bound to the WT Aer2[173-289] PAS domain was determined using a pyridine hemochrome assay (43, with modifications communicated by M. Gilles-Gonzalez). Briefly, WT PAS peptide and hemin standards (10 to 50 μM) were added to an alkaline pyridine solution and scanned from 350 to 700 nm under both dithionite-reduced and ferricyanide-oxidized conditions. Heme concentrations were determined from the reduced minus oxidized spectra, using an extinction coefficient of 23.4 mM−1 cm−1 for the absorbance difference of A556 (reduced) minus A539 (oxidized). The heme content determined from the pyridine hemochrome assay was used to standardize PAS-heme concentrations used in ligand-binding assays.
To determine whether purified PAS peptides had heme-binding defects, 10 μM imidazole-bound PAS peptides were scanned from 300 nm to 700 nm in a BioMate 3S spectrophotometer (Thermo Scientific). Samples were overlaid by zeroing at 700 nm, and the maximum absorbance of each Soret peak was determined. Maximum Soret absorbances were divided by the maximum Soret absorbance of the WT PAS peptide. Peptide concentrations were determined by electrophoresing 2.5 μg of each purified protein in duplicate on SDS-PAGE as outlined above, staining gels with Coomassie brilliant blue, and quantifying the density of each PAS peptide on a BioSpectrum digital imager. The average density of each PAS peptide was divided by the average density of the WT PAS peptide (which itself was usually 85 to 90% pure). The heme content/peptide ratio then was calculated for each mutant and averaged from multiple purifications. Ratios below 40% indicated a substantial heme-binding defect from which gas affinity constants generally were not determined.
Gas-binding affinities.
Deoxy-heme was created by adding 0.5 mM dithionite to 4 to 10 μM anaerobic PAS-heme in an anaerobic hood (Coy Laboratory Products, Grass Lake, MI). Deoxy-PAS was added to a quartz septum-sealed cuvette (Starna Cells, Atascadero, CA) and used directly for CO binding. For O2 affinities, sufficient O2 was added to the cuvette to oxidize the dithionite (as determined spectrophotometrically). To create gas-saturated buffers, buffer (50 mM Tris, pH 8.0, 50 mM KCl, and 5%, vol/vol, ethylene glycol) was perfused with either CO (Sigma-Aldrich) or air. To create 50% CO-saturated buffer, a volume of N2-saturated buffer was added to an equal volume of CO-saturated buffer in a Reacti-Vial (Thermo Scientific) and used immediately. Gas solutions were transferred to gas-tight Hamilton syringes (Hamilton, Reno, NV) and titrated into the deoxy protein solution. Stepwise spectra were recorded on a Beckman DU 650 spectrophotometer (Beckman Coulter, Brea, CA) after each addition of buffer. The amount of bound gas was estimated from the UV-visible spectrum by linear interpolation of the unliganded (Fe2+) and liganded (Fe2+-O2, Fe2+-CO) spectra. For WT PAS and most of the PAS mutants, the Soret maxima occurred at 428 to 432 nm for deoxy-heme, 414 to 416 nm for oxy-heme, and 421 to 422 nm for carbonmonoxy-heme. After O2 titrations were complete, CO was perfused directly into the cuvette to differentiate O2-bound protein from Met-heme protein. O2-bound or ferrous protein, but not ferric protein, showed CO-bound spectra after the addition of CO.
Met-heme absorption spectra.
To create Met-heme, 50 μM purified PAS peptide was oxidized with 50 μM potassium ferricyanide at room temperature for 15 min. To evaluate Met-heme spectra, samples were purified on a Micro Bio-Spin column (Bio-Rad, Hercules, CA) and scanned spectrophotometrically.
Supplementary Material
ACKNOWLEDGMENTS
We thank Magi Ishak Gabra, Lana Haddad, Virginia Henry, and Vinicius Cabido for constructing and testing several of the mutants used in this study. We also thank Marie-Alda Gilles-Gonzalez for providing modifications to the pyridine hemochrome method, Paul Herrmann for helpful discussions, and Brian Crane for helpful comments and discussions on the manuscript.
This research was supported by laboratory start-up funds to K.J.W., the Loma Linda University Biomedical Undergraduate Research Program, the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health, award number R01GM108655 to K.J.W., and NIGMS award number 2R25GM060507 (for support of D.G.).
The content of this work is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00003-17.
REFERENCES
- 1.Kato J, Kim HE, Takiguchi N, Kuroda A, Ohtake H. 2008. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng 106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 2.Sampedro I, Parales RE, Krell T, Hill JE. 2014. Pseudomonas chemotaxis. FEMS Microbiol Rev 39:17–46. [DOI] [PubMed] [Google Scholar]
- 3.Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. 2004. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol Lett 231:247–252. doi: 10.1016/S0378-1097(04)00009-6. [DOI] [PubMed] [Google Scholar]
- 4.Watts KJ, Taylor BL, Johnson MS. 2011. PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor. Mol Microbiol 79:686–699. doi: 10.1111/j.1365-2958.2010.07477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guvener ZT, Tifrea DF, Harwood CS. 2006. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol 61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 6.Biswas M, Dey S, Khamrui S, Sen U, Dasgupta J. 2013. Conformational barrier of CheY3 and inability of CheY4 to bind FliM control the flagellar motor action in Vibrio cholerae. PLoS One 8:e73923. doi: 10.1371/journal.pone.0073923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyakutake A, Homma M, Austin MJ, Boin MA, Hase CC, Kawagishi I. 2005. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J Bacteriol 187:8403–8410. doi: 10.1128/JB.187.24.8403-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasgupta J, Dattagupta JK. 2008. Structural determinants of V. cholerae CheYs that discriminate them in FliM binding: comparative modeling and MD simulation studies. J Biomol Struct Dyn 25:495–503. doi: 10.1080/07391102.2008.10507196. [DOI] [PubMed] [Google Scholar]
- 9.Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, Tummler B, Filloux A. 2009. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog 5:e1000540. doi: 10.1371/journal.ppat.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol 51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Fontana C, Corral Lugo A, Krell T. 2014. Specificity of the CheR2 methyltransferase in Pseudomonas aeruginosa is directed by a C-terminal pentapeptide in the McpB chemoreceptor. Sci Signal 7:ra34. doi: 10.1126/scisignal.2004849. [DOI] [PubMed] [Google Scholar]
- 12.Moglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawai H, Sugimoto H, Shiro Y, Ishikawa H, Mizutani Y, Aono S. 2012. Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa. Chem Commun (Camb) 48:6523–6525. doi: 10.1039/c2cc32549g. [DOI] [PubMed] [Google Scholar]
- 14.Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ, Crane BR. 2013. Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol 425:886–901. doi: 10.1016/j.jmb.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilles-Gonzalez MA, Gonzalez G. 2005. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem 99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Dunin-Horkawicz S, Lupas AN. 2010. Comprehensive analysis of HAMP domains: Implications for transmembrane signal transduction. J Mol Biol 397:1156–1174. doi: 10.1016/j.jmb.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 17.Garcia D, Watts KJ, Johnson MS, Taylor BL. 2016. Delineating PAS-HAMP interaction surfaces and signalling-associated changes in the aerotaxis receptor Aer. Mol Microbiol 100:156–172. doi: 10.1111/mmi.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Airola MV, Sukomon N, Samanta D, Borbat PP, Freed JH, Watts KJ, Crane BR. 2013. HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol 11:e1001479. doi: 10.1371/journal.pbio.1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Airola MV, Watts KJ, Bilwes AM, Crane BR. 2010. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure 18:436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerby RL, Youn H, Roberts GP. 2008. RcoM: a new single-component transcriptional regulator of CO metabolism in bacteria. J Bacteriol 190:3336–3343. doi: 10.1128/JB.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podust LM, Ioanoviciu A, Ortiz de Montellano PR. 2008. 2.3 A X-ray structure of the heme-bound GAF domain of sensory histidine kinase DosT of Mycobacterium tuberculosis. Biochemistry 47:12523–12531. doi: 10.1021/bi8012356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloek AP, Yang J, Mathews FS, Frieden C, Goldberg DE. 1994. The tyrosine B10 hydroxyl is crucial for oxygen avidity of Ascaris hemoglobin. J Biol Chem 269:2377–2379. [PubMed] [Google Scholar]
- 23.Olson JS, Mathews AJ, Rohlfs RJ, Springer BA, Egeberg KD, Sligar SG, Tame J, Renaud JP, Nagai K. 1988. The role of the distal histidine in myoglobin and haemoglobin. Nature 336:265–266. doi: 10.1038/336265a0. [DOI] [PubMed] [Google Scholar]
- 24.Worrall JA, van Roon AM, Ubbink M, Canters GW. 2005. The effect of replacing the axial methionine ligand with a lysine residue in cytochrome c-550 from Paracoccus versutus assessed by X-ray crystallography and unfolding. FEBS J 272:2441–2455. doi: 10.1111/j.1742-4658.2005.04664.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Q, Ames P, Parkinson JS. 2011. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol 80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan E, Rao F, Pasunooti S, Pham TH, Soehano I, Turner MS, Liew CW, Lescar J, Pervushin K, Liang ZX. 2013. Solution structure of the PAS domain of a thermophilic YybT protein homolog reveals a potential ligand-binding site. J Biol Chem 288:11949–11959. doi: 10.1074/jbc.M112.437764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao F, Ji Q, Soehano I, Liang ZX. 2011. Unusual heme-binding PAS domain from YybT family proteins. J Bacteriol 193:1543–1551. doi: 10.1128/JB.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunham CM, Dioum EM, Tuckerman JR, Gonzalez G, Scott WG, Gilles-Gonzalez MA. 2003. A distal arginine in oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure. Biochemistry 42:7701–7708. doi: 10.1021/bi0343370. [DOI] [PubMed] [Google Scholar]
- 29.Gilles-Gonzalez MA, Gonzalez G, Sousa EH, Tuckerman J. 2008. Oxygen-sensing histidine-protein kinases: assays of ligand binding and turnover of response-regulator substrates. Methods Enzymol 437:173–189. doi: 10.1016/S0076-6879(07)37010-9. [DOI] [PubMed] [Google Scholar]
- 30.Tsai AL, Berka V, Martin E, Olson JS. 2012. A “sliding scale rule” for selectivity among NO, CO, and O(2) by heme protein sensors. Biochemistry 51:172–186. doi: 10.1021/bi2015629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai AL, Martin E, Berka V, Olson JS. 2012. How do heme-protein sensors exclude oxygen? Lessons learned from cytochrome c′, Nostoc puntiforme heme nitric oxide/oxygen-binding domain, and soluble guanylyl cyclase. Antioxid Redox Signal 17:1246–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. 2000. DOS, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka A, Takahashi H, Shimizu T. 2007. Critical role of the heme axial ligand, Met95, in locking catalysis of the phosphodiesterase from Escherichia coli (Ec DOS) toward cyclic diGMP. J Biol Chem 282:21301–21307. doi: 10.1074/jbc.M701920200. [DOI] [PubMed] [Google Scholar]
- 34.Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. 2004. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochemistry 43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 35.Kosowicz JG, Boon EM. 2013. Insights into the distal heme pocket of H-NOX using fluoride as a probe for H-bonding interactions. J Inorg Biochem 126:91–95. doi: 10.1016/j.jinorgbio.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 36.Jain R, Chan MK. 2003. Mechanisms of ligand discrimination by heme proteins. J Biol Inorg Chem 8:1–11. doi: 10.1007/s00775-002-0405-8. [DOI] [PubMed] [Google Scholar]
- 37.Martinkova M, Kitanishi K, Shimizu T. 2013. Heme-based globin-coupled oxygen sensors: linking oxygen binding to functional regulation of diguanylate cyclase, histidine kinase, and methyl-accepting chemotaxis. J Biol Chem 288:27702–27711. doi: 10.1074/jbc.R113.473249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yukl ET, Ioanoviciu A, Nakano MM, de Montellano PR, Moenne-Loccoz P. 2008. A distal tyrosine residue is required for ligand discrimination in DevS from Mycobacterium tuberculosis. Biochemistry 47:12532–12539. doi: 10.1021/bi801234w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Key J, Hefti M, Purcell EB, Moffat K. 2007. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry 46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 40.Yu HS, Saw JH, Hou S, Larsen RW, Watts KJ, Johnson MS, Zimmer MA, Ordal GW, Taylor BL, Alam M. 2002. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol Lett 217:237–242. doi: 10.1111/j.1574-6968.2002.tb11481.x. [DOI] [PubMed] [Google Scholar]
- 41.Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci U S A 94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor BL, Watts KJ, Johnson MS. 2007. Oxygen and redox sensing by two-component systems that regulate behavioral responses: behavioral assays and structural studies of Aer using in vivo disulfide cross-linking. Methods Enzymol 422:190–232. doi: 10.1016/S0076-6879(06)22010-X. [DOI] [PubMed] [Google Scholar]
- 43.Appleby CA, Bergersen FJ. 1980. Preparation and experimental use of leghaemoglobin. John Wiley & Sons Ltd, Chichester, United Kingdom. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.