ABSTRACT
Fluctuations in nutrient availability often result in recurrent exposures to the same stimulus conditions. The ability to memorize the past event and use the “memory” to make adjustments to current behaviors can lead to a more efficient adaptation to the recurring stimulus. A short-term phenotypic memory can be conferred via carryover of the response proteins to facilitate the recurrent response, but the additional accumulation of response proteins can lead to a deviation from response homeostasis. We used the Escherichia coli PhoB/PhoR two-component system (TCS) as a model system to study how cells cope with the recurrence of environmental phosphate (Pi) starvation conditions. We discovered that “memory” of prior Pi starvation can exert distinct effects through two regulatory pathways, the TCS signaling pathway and the stress response pathway. Although carryover of TCS proteins can lead to higher initial levels of transcription factor PhoB and a faster initial response in prestarved cells than in cells not starved, the response enhancement can be overcome by an earlier and greater repression of promoter activity in prestarved cells due to the memory of the stress response. The repression counterbalances the carryover of the response proteins, leading to a homeostatic response whether or not cells are prestimulated. A computational model based on sigma factor competition was developed to understand the memory of stress response and to predict the homeostasis of other PhoB-regulated response proteins. Our insight into the history-dependent PhoBR response may provide a general understanding of how TCSs respond to recurring stimuli and adapt to fluctuating environmental conditions.
IMPORTANCE Bacterial cells in their natural environments experience scenarios that are far more complex than are typically replicated in laboratory experiments. The architectures of signaling systems and the integration of multiple adaptive pathways have evolved to deal with such complexity. In this study, we examined the molecular “memory” that is generated by previous exposure to stimulus. Under our experimental conditions, activating effects of autoregulated two-component signaling and inhibitory effects of the stress response counterbalanced the transcriptional output to approach response homeostasis whether or not cells had been preexposed to stimulus. Modeling allows prediction of response behavior in different scenarios and demonstrates both the robustness of the system output and its sensitivity to historical parameters such as timing and levels of exposure to stimuli.
KEYWORDS: PhoBR, autoregulation, computer modeling, homeostasis, memory, sigma factors, stress response, transcriptional regulation, transcriptional repression, two-component regulatory systems
INTRODUCTION
Cellular adaptations to environmental perturbations are often mediated by modulation of gene expression. Balancing the benefits and costs of gene expression frequently requires cells to evolve different optimal protein expression levels under different environmental conditions (1–3). The ever-changing environments constantly challenge cells to optimize strategies to prevail under fluctuating conditions. It has been shown that cells respond faster to a change of environmental condition when they have experienced similar conditions in the recent past (4–6). These “memories” of past events, or history-dependent behaviors, are generally believed to originate from either response hysteresis, usually involving bistability of positive-feedback loops, or a slow purge of the induced protein after stimulus removal, providing carryover of the functional protein for the recurring environmental perturbation (6–10). For the latter, the duration of memory depends on the rate of decrease in the levels of the response proteins that confer memory. Recurrent responses in addition to the residual response from previous induction can result in a significant increase in the levels of response proteins, exceeding the optimal level. Less is known about whether or not cells maintain a homeostatic optimal expression level during repeated exposures to environmental perturbations.
In order to understand the apparent conflicts between memory and homeostasis, we focused on the activation response of the Escherichia coli PhoB/PhoR (PhoBR) two-component system (TCS) that occurs when cells are stimulated a second time. TCSs are widely used in prokaryotes to respond to environmental cues, and PhoBR is one of the archetype TCSs (11, 12). The sensor histidine kinase (HK) PhoR responds to limitation of environmental phosphate (Pi) concentrations and modulates its activities to adjust the phosphorylation level of the response regulator (RR) protein PhoB (13, 14). Phosphorylated PhoB (PhoB∼P) regulates transcription of genes responsible for assimilation of different phosphorus sources as well as expression of the phoBR operon encoding the PhoB and PhoR proteins themselves to adapt to different Pi conditions. It has been demonstrated that different levels of PhoBR under different Pi conditions provide near-maximal fitness under the respective conditions. The importance of this was underscored by the finding that challenging cells with unfavorable PhoBR levels led to diverse homeostatic efforts to mediate optimal expression levels through acquisition of mutations (3). Maintaining expression optimality appears to be a strong evolutionary force to select the positive autoregulatory scheme for adaptation to different Pi conditions.
Preexposure to Pi limitation was shown to cause a faster and greater response when cells were starved of Pi again (4). This memory effect, or “learning” behavior, is suggested to result from the stability of the PhoB and PhoR proteins that were synthesized during the prior Pi starvation phase and that remained at high levels during the recurring stimulation. Understanding the effects of prestarvation on the PhoBR-regulated response and unraveling the molecular principles behind such history-dependent behaviors rely on elucidation of a few central issues as follows.
(i) It has been shown that the steady-state output of some TCSs can be robust with respect to variations of TCS levels (15, 16). How are the activation kinetics (initial responses to stimulus) influenced by variations in PhoBR levels? Can high initial PhoBR levels elicit a response that is sufficiently fast to account for the memory effect?
(ii) PhoBR levels at the start of a recurring starvation process depend on the rate of protein concentration decrease during the span of Pi-replete growth between the two periods of Pi limitation. How do the PhoBR levels change during the Pi-replete growth period? How do the magnitude and duration of memory correlate with the growth time?
(iii) If cells experience only a short period of Pi-replete growth, significant amounts of PhoBR and PhoBR-regulated proteins from the prestarvation period would remain. A faster and greater response for the recurring stimulation would produce even more of these response proteins. The combination of these two effects would result in a protein concentration much higher than the optimal levels observed previously. Would this confer significant fitness cost? Or does there exist some degree of protein homeostasis that maintains protein levels close to the optimum?
(iv) If cells can achieve some degree of response homeostasis, what is the counterbalancing mechanism?
In order to answer the questions given above, we evaluated impacts of PhoBR levels on activation kinetics and investigated the response dynamics for cells with or without prestarvation of Pi. Measurements of reporter activity, protein expression level, and PhoB phosphorylation revealed a complex dependence of the Pi limitation response on cellular history. A computational model has been developed to enhance understanding of the effects of multiple regulatory pathways on response dynamics and protein homeostasis in a fluctuating environment with recurrent exposures to stimulus.
RESULTS
Dependence of activation kinetics on PhoBR levels.
The E. coli PhoBR system is a typical positively autoregulated TCS. Expression of PhoB increased shortly after cells encountered the activating Pi limitation condition and gradually reached an optimal level (3) ∼40 min later (Fig. 1A). RU1616, a strain with an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter replacing the wild-type (WT) promoter of phoBR, was engineered to constitutively produce different PhoBR levels at different IPTG concentrations (16). PhoB levels were used to track the expression levels of the entire phoBR operon, which was possible because the ratio of PhoB to PhoR remains equal to that seen with the WT strain across different IPTG concentrations (16). An IPTG concentration of 150 μM has been shown to produce a PhoB level comparable to the WT level induced under Pi-depleted conditions (16).
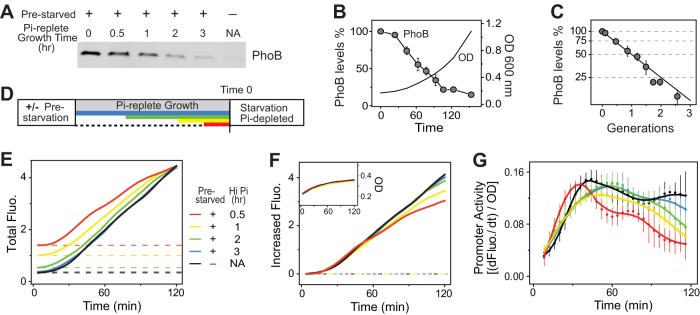
FIG 1.
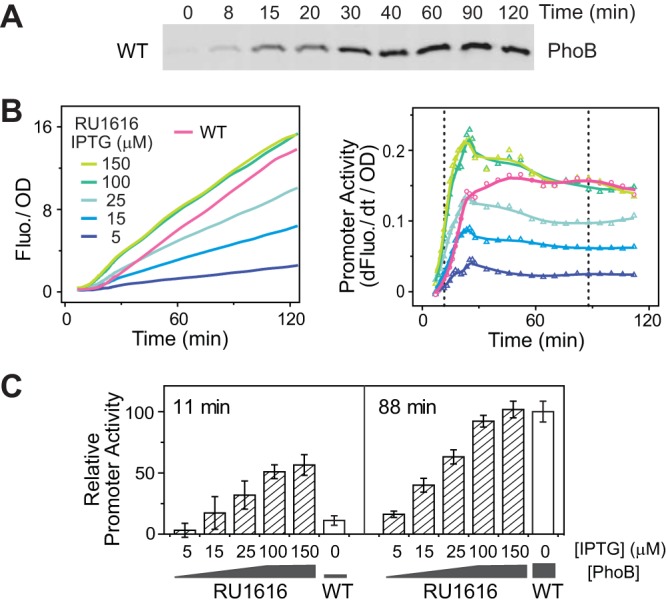
Expression levels of phoBR affect activation kinetics of phoA-yfp. (A) Time-dependent expression of PhoB upon Pi starvation. One immunoblot representative of four replicates is shown. (B) Temporal dynamics of phoA-yfp activation upon Pi starvation. Data representing OD-normalized fluorescence (Fluo.) are shown in the left panel, and data representing the first derivative of fluorescence (dFluo./dt) are shown in the right panel. The first derivative of fluorescence measures the rate of YFP synthesis, reflecting the promoter activities. BW25113 (WT) and RU1616 (a strain containing a phoBR operon under the control of an IPTG-inducible promoter) carrying the pRG161 reporter plasmid were assayed for Pi starvation response in MOPS medium with a starting concentration of Pi of 2 μM. Dotted lines represent two time points at which the relative promoter activities were compared in panel C. (C) IPTG concentrations of 5, 15, 25, 100, and 150 μM resulted in PhoB expression levels of 0.93, 1.7, 2.4, 8.1, and 8.2 μM in RU1616, while Pi limitation raised the PhoB level in the WT strain from 0.43 to 9.3 μM (28). Data are shown as means ± standard deviations (SD) of results from 11 individual wells assayed in a single microplate.
The activation kinetics upon Pi depletion were monitored using a plasmid-carried yellow fluorescent protein (YFP) reporter gene placed after the promoter of phoA, a PhoB-regulated gene encoding an alkaline phosphatase (AP). Higher PhoBR levels resulted in a faster increase of YFP fluorescence when bacteria were resuspended in Pi-depleted medium (Fig. 1B). The time derivative of YFP fluorescence, corresponding to the rate of YFP synthesis, is used to indicate the real-time promoter activities. The promoter activity quickly rose upon Pi starvation and reached a steady state thereafter, recapitulating the kinetics of PhoB phosphorylation (17). PhoBR levels correlate well with the steady-state level as well as with the rising rate of promoter activity (Fig. 1B and C). An IPTG concentration of 5 μM yielded a low expression level of PhoB; thus, a low level of promoter activity was observed for both the early rising phase (11 min) and the later steady phase (88 min). Higher constitutive PhoBR levels clearly enhanced the starvation response. For the WT strain, in which the expression of phoBR is autoregulated, the promoter activity reached a high steady-state level comparable to that of RU1616 with 150 μM IPTG when PhoB was fully induced, but it was initially low due to the low starting level of PhoB, causing a 10- to 15-min delay in the overall kinetics. Thus, for cells with recurring exposures to stimulation, a high initial PhoB level resulting from a prior period of Pi starvation may expedite the response by a maximal time of 10 to 15 min.
Effects of prestarvation and Pi-replete growth on the recurrent response.
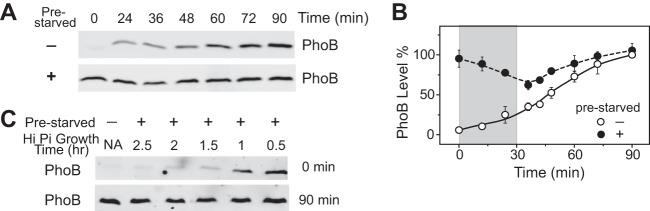
Cells prestarved of Pi for 50 min were grown in Pi-replete medium to examine the stability of the PhoB protein. As the Pi-replete growth continued, PhoB levels kept decreasing (Fig. 2A to C). The rate of decrease agreed with a growth dilution effect, except for a minor deviation for growth times longer than 2 h (Fig. 2C). The doubling time was approximately 55 min in Pi-replete MOPS (morpholinepropanesulfonic acid) medium. The PhoB level was halved for every doubling of bacterial growth, and it took longer than 3 h for the PhoB concentration to return to approximately the prestimulus level. The levels of PhoA and the phoA-yfp operon reporter have also been discovered to be growth diluted upon stimulus removal (17). An instantaneous dephosphorylation of PhoB∼P has been observed to shut off the pathway, following which there was no new synthesis either of PhoB or of proteins whose expression is regulated by PhoB.
FIG 2.
Activation kinetics is dependent on growth dilution time for prestarved WT cultures. (A to C) Growth dilution of PhoB levels. Cells prestarved of Pi for 50 min were grown in Pi-replete medium (1 mM Pi) for the indicated times. One representative of six immunoblots is shown in panel A (NA, not applicable). Other immunoblots with more time points were used for the PhoB quantifications shown in panels B (time) and C (generations). Growth ODs were converted to values representing doubling generations of bacteria by exponential fitting of the growth curve to replot the data in panel C. The solid line represents the theoretical growth dilution curve. (D to G) Activation kinetics of phoA-yfp for prestarved WT cultures. The experimental timeline is shown in panel D. Different times of growth dilution in Pi-replete medium are represented in colors. The time of cells reencountering Pi-depleted medium is set as time zero. Total fluorescence (E), the increased fluorescence upon stimulation (F) and bacterial OD (inset in panel F), and promoter activities (G) are illustrated with smoothed solid lines. Dashed lines in panel E indicate different initial levels of fluorescence after the growth dilution, and these values were subsequently subtracted to give the newly increased fluorescence shown in panel F. Error bars represent SD of results from 11 individual wells.
Different growth dilution times in Pi-replete medium resulted in different starting concentrations of PhoB and YFP for similarly prestarved cells. The second Pi starvation activated the phoA promoter and increased the reporter fluorescence (Fig. 2D to G). Surprisingly, the reporter fluorescence levels eventually converged, in spite of the differences in growth dilution times and initial reporter levels (Fig. 2E). Cells with shorter periods of growth dilution had higher levels of initial reporter fluorescence but displayed a counterbalancing smaller increase of fluorescence (Fig. 2F), leading to the convergence of reporter levels. Close examination of the promoter activities revealed that prestarved cells with 0.5 h of growth dilution indeed showed slightly stronger initial promoter activity than those with longer times of growth dilution or those that had not been prestarved (Fig. 2G). However, as Pi starvation continued, the promoter activity of prestarved cells was quickly repressed whereas the promoter activity of cells without prestarvation remained high. The time when the promoter activity became repressed correlated with the growth dilution time. The shorter the growth dilution time, the earlier the repression occurred. It appears that cells memorize the prior event of Pi starvation but that the memory has two opposing effects on the response. One allows cells to activate phoA more quickly in response to subsequent starvation, while the other has an adverse effect on promoter activity that offsets the residual enhanced response from prior stimulation and eventually leads toward response homeostasis.
The early enhancing and later repressing effects of prestarvation were reaffirmed in a similar growth dilution experiment with a longer assay time (see Fig. S1 in the supplemental material). Cells with prestarvation displayed earlier repression than cells without prestarvation. The fluorescence levels seen with cultures with different growth dilution times remained similar after converging. However, in this experiment, the time required for fluorescence convergence was ∼90 min, much shorter than the time of 120 min shown in Fig. 2E. It appears that there were substantial differences between the experiments with respect to the response homeostasis profiles, although the counterbalancing effect, or the smaller increase of fluorescence for prestarved cells, was consistently present.
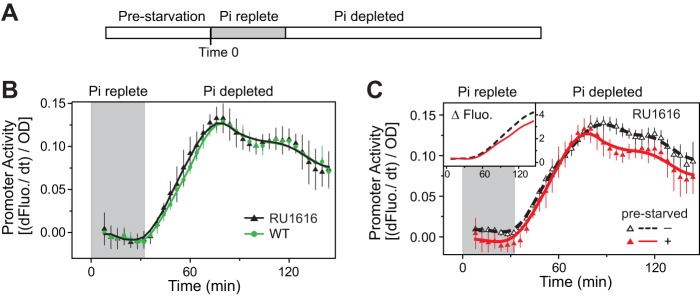
The observed counterbalancing regulation suggests a history-dependent repression of promoter activity that contrasts with the previously reported “learning” behavior in the PhoBR system, i.e., a faster and greater response to starvation recurrence (4). To examine the cause of such a discrepancy or the reason behind the experimental variations in the two studies, reporter outputs were assayed under experimental conditions parallel to those of the previous study (4). Cells were prestarved for 50 min and then were immediately resuspended in MOPS medium containing 50 μM Pi (Fig. 3; see also Fig. S2A). A Pi concentration of 50 μM is still above the activation threshold of 4 μM (14), indicating a Pi-replete condition. Cells consumed Pi and maintained fast growth during the first ∼30 min of growth until Pi became depleted (Fig. S2, gray-shaded area). At that time, a second exposure to Pi depletion activated the response and the level of the response was again observed to be lower than that of cells without prestarvation.
FIG 3.
Prestarvation decreases PhoB-regulated promoter activity in both the WT and RU1616 strains. (A) Experimental timeline. Cells were resuspended in MOPS medium (50 μM Pi) with a starting OD of ∼0.2 at time zero, and Pi became limited approximately 30 min later, indicating an ∼30-min period of growth dilution under Pi-replete conditions (area shaded in gray). (B) Comparison of phoA promoter activities in strains containing autoregulated phoBR (WT) and constitutively expressed phoBR (RU1616) when cells were prestarved. (C) Activation kinetics of the phoA promoter in RU1616 with or without prestarvation of Pi. The increased fluorescence upon stimulation is shown in the inset. An IPTG concentration of 150 μM was present in the medium for RU1616 to maintain a steady PhoB level approximately equal to that of fully induced WT. Data are shown as means ± SD of results from 11 individual wells.
With identical growth conditions, response profiles were similar, with little variation for cultures from individual wells in a single microplate assay (Fig. S2A). In contrast, substantial variation was observed among assays performed with different preparations or compositions of nutrients (Fig. S2B to E). Any slight difference in the medium composition, the starting optical density (OD) of bacteria, or the efficiency of removal of excess Pi from Pi-replete conditions to initiate starvation could result in different growth rates and consumption rates of Pi that could lead to different activation profiles. The extent of response homeostasis appears extremely sensitive to growth conditions. Complete convergence of response outputs, or true homeostasis, was not always observed. However, the counterbalancing effect, or the attenuation of the response of prestarved cells, was consistently present (Fig. S2).
Effects of autoregulation on response repression.
To investigate whether the autoregulation of phoBR plays any role in the counterbalancing mechanism, responses to recurrent Pi starvation were examined in RU1616, a strain in which PhoBR is not autoregulated (Fig. 3). For the WT cells, an ∼30-min period of growth dilution time resulted in >50% of the response proteins from the prior stimulation being carried over to the second stimulation. With a high initial level of PhoBR, prestarved WT cells displayed kinetics of promoter activity indistinguishable from that of RU1616 (Fig. 3B). A decrease in promoter activity during the later stage of the starvation recurrence was observed for both the WT and the nonautoregulated strain (Fig. 3B). The repression of promoter activity for RU1616 cells with prestarvation was earlier and greater than that seen with those without prestarvation (Fig. 3C). The existence of repression in the nonautoregulated RU1616 strain resulted in a smaller increase of fluorescence for prestarved cells (Fig. 3C, inset), suggesting a counterbalancing mechanism that is not dependent on the positive autoregulation of phoBR.
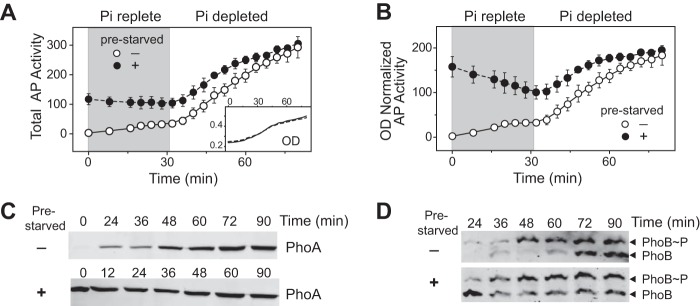
To confirm that the repression of output is not an artifact of the YFP reporter plasmid, we examined the expression dynamics of the endogenous phoA gene, which encodes an alkaline phosphatase (AP). During the initial 30 min of growth under Pi-replete conditions, cells that had not been prestarved displayed only a slight increase of AP activity and the total AP levels remained low (Fig. 4A). In contrast, total AP activity for prestarved cells was relatively high due to the carryover of PhoA proteins from the prior activation. When Pi is replete, total AP levels remain constant because there is no new production of PhoA. Cell growth during the period when Pi was replete, monitored as an increase in OD, resulted in a “dilution” or a decrease in the OD-normalized AP activity as well as in the PhoA protein level for prestarved cells (Fig. 4B and C). Once Pi became limited at ∼30 min, Pi starvation resulted in increases in the total amount of AP activity, the OD-normalized AP activity, and the PhoA expression level, all approaching homeostatic levels independent of prestarvation (Fig. 4A to C). A reduced rate of PhoA production at the later stage of starvation, reflecting a history-dependent repression of the promoter activity, appears to counterbalance the high initial PhoA level in prestarved cells.
FIG 4.
AP activity and PhoA protein levels approach homeostasis. WT cells were grown in MOPS medium (50 μM Pi). (A) Equal volumes of cultures were removed at the indicated times to assay the total AP activity. (B) Total AP activities were normalized to bacterial ODs to derive AP activities per 0.3 OD of cells. Gray-shaded areas indicate growth under the Pi-replete condition. (C and D) OD-normalized samples were probed for PhoA levels by Western blotting with an anti-AP antibody (C) or analyzed for PhoB phosphorylation using Phos-tag gels (D). Data are shown as means ± SD of results from duplicate experiments, and one representative of two immunoblots is shown.
Repression of the promoter activity was not due to repression of PhoB phosphorylation, because a reduction in PhoB∼P levels was not observed (Fig. 4D). At early time points, cells that had been prestarved showed slightly higher PhoB∼P levels than those that had not, reflecting the memory effect of high PhoB levels carried over from prior stimulation. At the later stage of the time course, PhoB∼P levels were comparable for cells with or without prestarvation (Fig. 4D). Therefore, the history-dependent repression occurs downstream of PhoB/PhoR TCS signaling, possibly at the level of transcription or translation.
Involvement of stress response in the history-dependent repression.
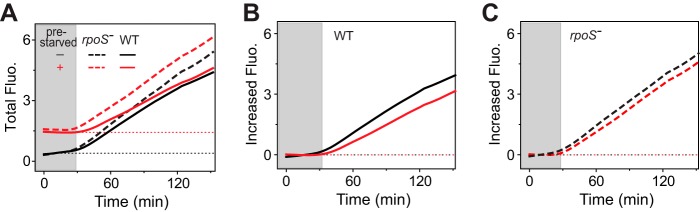
It has been well documented that Pi starvation can elicit a developmental reprogramming of gene expression for a general stress response and adaptation to stationary phase (18–22). Under nutrient-depleted conditions, E. coli cells increase expression of the rpoS gene encoding the stress sigma factor σS and the level of a small molecule alarmone (p)ppGpp and of other factors to globally repress or activate different sets of genes to resist the stress condition. It has been shown that several PhoB-regulated genes, including phoA, are negatively regulated by increased levels of σS and ppGpp (23, 24). To investigate the role of the stress response in the history-dependent repression, effects of prestarvation on phoA-yfp expression were examined in an rpoS deletion strain (Fig. 5). Prior exposure to Pi-depleted conditions caused a slight repression of transcription output in the rpoS deletion strain that was much lower than that in WT cells (Fig. 5B and C). The extent of repression was attenuated in the rpoS deletion strain, suggesting a role for σS in the counterbalancing mechanism.
FIG 5.
Deletion of rpoS attenuates the inhibitory effect of prestarvation. Activation of the phoA-yfp reporter in plasmid pRG161 was assayed in the WT strain and rpoS deletion strain RU1646 (rpoS−) with or without prestarvation. Gray-shaded areas indicate growth under the Pi-replete condition. Total fluorescence (A) and the increased fluorescence for the WT (B) and RU1646 (C) strains are shown as averages of results from 10 individual wells from one microplate assay.
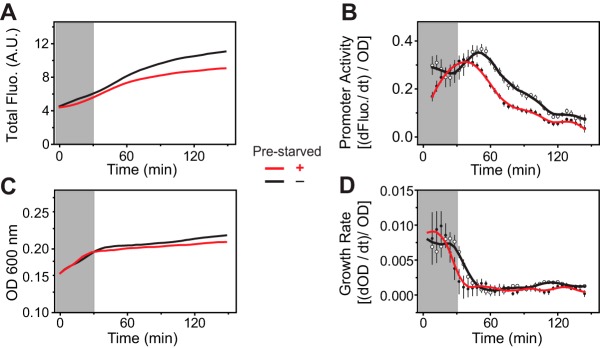
We reasoned that the observed history-dependent repression of phoA could have been a result of negative regulation by the general stress response if the general stress response to the second Pi starvation had been altered by prior exposures to Pi-depleted conditions. If this were true, history-dependent repression of promoter activity would not be restricted to genes regulated by PhoB and should be observed independently of PhoBR signaling. We examined how prestarvation of Pi affects the expression dynamics of a YFP reporter driven by a constitutive Ptet promoter in a phoBR deletion strain (Fig. 6). Repression of the promoter activity was observed after Pi depletion for all cells with or without prior exposure to Pi limitation (Fig. 6B), reflecting the inhibitory effect of the general stress response. However, cells prestarved of Pi displayed earlier reduction of the promoter activity than cells not previously stressed. Moreover, the growth rate for prestarved cells, indicated by the normalized first derivatives of the OD, displayed an earlier decrease than was seen with cells without prestarvation (Fig. 6C and D), suggesting an earlier entry into the stationary phase. It appears that a prior exposure to Pi starvation can accelerate the general stress response and lead to expedited repression of gene expression during a recurrence of starvation.
FIG 6.
Prestarvation reduces activity of the constitutive Ptet promoter. (A and B) Plasmid pRG278 carrying Ptet-yfp was placed in phoBR deletion strain BW25142 for analyses of total fluorescence (A.U., arbitrary units) (A) and promoter activity (B) during Pi starvation as described in Materials and Methods. Gray-shaded areas indicate growth under the Pi-replete condition. (C and D) ODs of bacterial cultures and the OD-normalized first derivatives, indicating the real-time growth rates, are illustrated in panels C and D, respectively. Data from cells without prestarvation of Pi are shown with black lines, while red lines indicate data from prestarved cells. The average of results from 8 individual wells from one microplate assay is shown for each condition, and error bars indicate SD. Similar patterns of reporter activity were observed for independent assays.
Modeling the effects of stress response on protein homeostasis.
Our data suggest that expedited repression of gene expression occurs during the response to a recurrence of stress. “Memory” of prior stress conditions results in inhibitory effects that offset the boosted learning behavior caused by the “memory” of prior PhoBR activation. We developed a simplified transcription regulation model to understand the history-dependent repression of the phoA reporter and to explain the counterbalancing mechanism (Fig. 7; see also Fig. S3).
FIG 7.
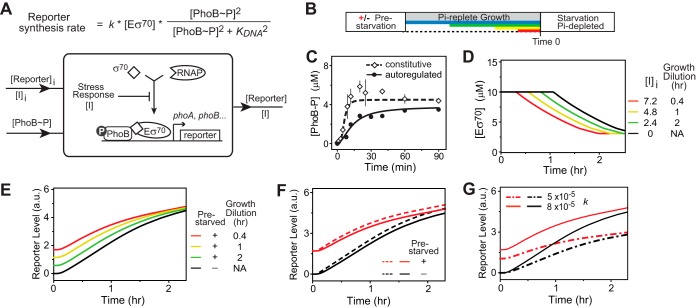
A model of stress response explains the homeostasis of reporter levels. (A) Model scheme. PhoB-regulated reporter activity is a function of the concentration of PhoB∼P and of the effective concentration of RNAP-σ70 complex (Eσ70), with the rate equation shown. Parameter k describes the gene-specific factors, such as promoter strength and translation efficiency, that determine the protein expression level. Stress response is represented with an arbitrary stress factor, I, that decreases the concentration of Eσ70 (see Modeling Details in the supplemental material). Reporter output is dependent on the input of [PhoB∼P] as well as on the history-related initial levels of I and reporter expression. (B) Illustration of timeline. Different growth dilution times are shown in different colors. (C) Profiles of PhoB∼P input. In vivo phosphorylation kinetics of PhoB have been quantified based on previous Phos-tag analyses of intracellular PhoB∼P (17, 28) for constitutively expressed phoBR in strain RU1616 and autoregulated phoBR in the WT strain. Fitted Hill curves were used for PhoB∼P inputs and represent the boundary of PhoB phosphorylation kinetics with the initial concentration of PhoB at the high level (dashed line) and the low level (solid line). (D) Modeling the dependence of [Eσ70] on prestarvation. Prestarvation increases the concentration of I, and different growth dilution times in Pi-replete medium result in different initial concentrations of I ([I]i). Higher [I]i levels cause an earlier entry into the stationary phase that decreases [Eσ70]. (E) Homeostasis of reporter output. Different Eσ70 dynamics (shown in panel D) result in converging levels of reporter output regardless of the time spent in Pi-replete medium. (F) Simulation of reporter output affected by PhoB∼P profiles. Dashed lines represent data modeled with the PhoB∼P kinetic profile from RU1616, while solid lines indicate data modeled with the WT PhoB∼P profile described in the panel B legend. (G) Effect of gene-specific parameter k on output homeostasis for cells with (red) or without (black) prestarvation.
One major mechanism for inhibition of gene expression during the stationary phase is through the sigma factor competition that reduces the amount of functional RNA polymerase (RNAP) bound to the promoters (25). During exponential growth of E. coli cells, housekeeping sigma factor σ70 is required for transcription of most genes to recognize promoters and bind to the core RNAP. During the stationary phase, increased levels of σS compete with σ70 for interactions with a limited number of core RNAPs, while other factors, such as ppGpp and the anti-σ factor Rsd, further reduce the availability of σ70 for RNAP interaction, leading to reduced transcription activity (19, 25–27). In our model, expression of phoA is described with a rate function that is dependent on the binding of PhoB∼P to promoter DNA and the concentration of the functional RNAP-σ70 complex (Eσ70) (Fig. 7A). Kinetics of PhoB phosphorylation were derived from previous Phos-tag analyses (17, 28) and used as system inputs (Fig. 7C). Based on a sigma factor competition model reported previously (29), the concentration of Eσ70 was determined by the concentrations of σS and σ70 (Fig. S3A). Development of the stress response is modeled with an arbitrary factor, I, that changes the effective concentrations of σS and σ70 (see Modeling Details in the supplemental material), leading to a reduction in [Eσ70] and thus to repression of transcription activity (Fig. 7D; see also Fig. S3B).
Memory effects are modeled as different initial levels of reporter expression and stress factor I that feed into the system (Fig. 7A). It is assumed that the memory of the stress response is conferred through a similar mechanism, with carryover of some “memory” molecules, modeled by I, to the second stress response. The level of I is assumed to follow the same growth dilution rule as the reporter response (Fig. S3C and E). Shorter times of growth dilution between the two starvations led to higher initial levels of I and thus to a quicker entry into the stationary phase and an earlier reduction of the Eσ70 concentration (Fig. 7D and S3D), which results in a smaller increase in reporter expression for cells with high initial levels of reporter, thus approaching a homeostasis of reporter responses (Fig. 7E). If the stress response is not considered, the counterbalancing effect was absent and response homeostasis was unable to be reached (Fig. S3G).
It is apparent that the extent of counterbalancing effects is sensitive to the kinetics of the stress response. Different rates of σS accumulation and σ70 sequestration mediated by other stress factors enable different degrees of inhibition of gene expression; thus, a complete convergence of transcription output may not always be achieved (Fig. S4). The progress of the stress response is known to be tightly correlated with the growth rates of cells. Small differences in nutrient conditions or metabolic states of cells may potentially alter the dynamics of development of the stress response, thus leading to different levels of repression and different extents of homeostasis. This may have contributed to the experimental variations represented in Fig. S2.
Prestarvation also has a positive memory effect on the activation kinetics due to high initial concentrations of PhoB and PhoR proteins. High starting levels of PhoBR give rise to fast phosphorylation kinetics of PhoB, as demonstrated in Fig. 7C. The kinetic PhoB∼P profile from RU1616, which expresses PhoBR at a constant level close to that of induced WT cells, is used as the upper boundary, or the fastest kinetics that a WT strain can achieve with a high initial level of residual PhoB from prior Pi starvation. The fast kinetics in PhoB phosphorylation are not readily reflected in transcription dynamics, because transcription of PhoB-regulated genes also depends on the binding affinity of PhoB∼P to promoter DNA (Fig. S5). High affinity, or low Kd (dissociation constant), causes saturated occupancy of promoter at a very low concentration of PhoB∼P, leading to almost indistinguishable transcription dynamics between cells with different PhoB∼P profiles (Fig. S5). The affinity of PhoB∼P for the phoA promoter has been previously determined using activities of the same phoA-yfp reporter at different PhoBR expression levels (28). Modeling with that affinity level revealed a slight increase in transcription for the nonautoregulated PhoB∼P profile over the WT profile, but the increase was not sufficient to overcome the homeostatic effects caused by the stress response (Fig. 7F).
As the expression rate function in Fig. 7A shows, expression of a PhoB-regulated gene is also dependent on parameter k, which denotes gene-specific factors determining the protein expression efficiency. The model predicts that response homeostasis can be reached for a different value of gene-specific parameter k (Fig. 7G). This suggests that response homeostasis may not be restricted to one particular gene. Indeed, we observed homeostatic kinetics of PhoB expression for cells with or without prestarvation similar to that of the PhoA response (Fig. 8A and B). Cells with different growth dilution times displayed different initial levels of PhoB, but all reached similar levels for the second Pi starvation independently of cellular history (Fig. 8C). This is consistent with our previous discovery of a strong evolutionary force maintaining an optimal level of PhoB expression for maximal fitness (3).
FIG 8.
PhoB protein levels reach similar levels independently of growth history. (A and B) Dynamics of PhoB expression. WT cells with or without prestarvation of Pi were shifted to MOPS medium (50 μM Pi) at time zero. Equal ODs of bacteria were lysed and analyzed by Western blotting (A), and data were quantified (B). Phosphate became limited after ∼30 min (area shaded in gray). Data are shown as means ± SD of results from four immunoblots. (C) PhoB expression levels for cells with different growth histories. Prestarvation of Pi and growth dilution in Pi-replete medium were performed as described in Materials and Methods.
DISCUSSION
It has long been known that even simple microbes can generate history-dependent responses to recurrences of environmental perturbations, appearing as if they had memorized the past experience of a similar condition and made the adjustments necessary to respond more efficiently to the current one (30). One extensively characterized example of bacterial memory is adaptation in chemotaxis. A short-term memory is provided by the receptor methylation system that allows cells to compare concentrations of attractants and repellants along the swimming track and adjust behavior accordingly to achieve directed movement (31). In addition, two distinct memory mechanisms, referred to as response memory and phenotypic memory, have been described in bacterial cells (6). Response memory involves a certain form of regulatory circuitry, usually positive feedback, to generate bi- or multistable behaviors that are dependent on prior states of the cell. Such memories have been studied for engineered or mutated TCS proteins (32, 33). The WT PhoB/PhoR system, albeit positively autoregulated, does not display any bistable response to the natural stimulus (3). Its memory-like learning behavior (4) is not necessarily dependent on autoregulation but is best understood as phenotypic memory that results from transmission of stable proteins through a limited number of bacterial generations for an altered response to the recurrence of stimulus. Such short-term memory could be beneficial, as suggested in other systems (6, 9), or only incidental, due to the fact that the time scale of protein dilution is much greater than the time scale of the fast kinetics of shutting off the pathway through PhoB dephosphorylation (17).
Moreover, because a single stimulus may elicit multiple responses through different pathways, phenotypic memories conferred by different regulators may have distinct or even conflicting effects on the recurrent response. Pi limitation can induce the PhoBR-regulated PHO response that is accelerated by past memory as well as the stress response that has an inhibitory effect as a consequence of prior exposures. Under the experimental conditions used in this study, the counterbalancing effects exerted by the memory of the stress response seemed to dominate the response enhancement caused by high initial PhoBR levels. The stress response in bacteria is under sophisticated control; thus slight differences in the strains examined or nutrient contents or growth conditions may give rise to dramatic differences in memory effects. This may explain the difference between our data corresponding to counterbalancing regulation and the previous report of the “learning” behavior in the PhoB/PhoR TCS (4). Because it has been demonstrated that the total levels of PhoA remain constant during Pi-replete growth, we were able to follow the change in total PhoA levels and quantify PhoA proteins synthesized from different starvation events for WT strain BW25113. In contrast, in the previous study, a temperature-sensitive PhoA allele was utilized to eliminate PhoA interference from prestarvation and to facilitate tracking solely of the PhoA protein produced from the second starvation. Cells were starved of Pi at 30°C instead of 37°C, and the Pi-replete growth was performed at 42°C to allow PhoA degradation. Temperature increases, or heat shocks, are known to induce expression of alternative sigma factors, rpoH and rpoS, and to elicit stress responses (34, 35) that can potentially influence the memory effects. The mutagenesis process used for the temperature-sensitive PhoA mutant isolation may also generate alleles with an altered stress response profile, because rpoS is susceptible to mutation under nutrient-limited conditions (36, 37). The altered stress profiles could prevent the repressing effects of stress memory.
“Memory” of stress responses has been reported for different aspects of cellular adaptations to stress conditions (38–40). The timing of growth resumption from stationary phase has been discovered to correlate with the timing of entry into stationary phase, although the molecular mechanism of this phenomenon has not been elucidated (40). In our model, we assumed a phenotypic memory mechanism to explain the observed early entry into the stationary phase for the recurrence of Pi starvation. An arbitrary memory factor, I, was assumed to track with the concentration of σS, but the molecule that confers memory is not likely to be σS. σS is short-lived during exponential growth, and the stability of σS is tightly controlled (19). More studies are required to understand the mechanism of stress memory. Any protein or other molecule that can alter the accumulation rate of σS (19, 41) can potentially display some history-dependent behavior.
Cells usually show strong selection for optimal expression of proteins that have a high cost (1–3, 42). The homeostatic response to fluctuating environments is an attempt to maintain a stable and optimal state of the cell. Response homeostasis often involves negative regulation to provide stability to variations (7, 43). Here we show that the history-dependent repression of transcription provides a negative regulatory mechanism to approach output homeostasis for the positively autoregulated PhoBR system for cases in which there is insufficient time for the response proteins to return to the basal level before a recurrent stimulation takes place. In other TCSs, additional negative regulation can be observed for many positively autoregulated TCSs. Cell wall homeostasis requires a fine-tuned response from the CpxRA system (44), and both positive-feedback and negative-feedback loops have been discovered (45). Interestingly, several positively autoregulated TCSs have been shown to couple with stress responses (46–48); a potential common role for stress responses in protein homeostasis awaits further investigation.
MATERIALS AND METHODS
Strains and plasmids.
Reporter plasmid pRG161 containing phoA-yfp was used for assaying output responses. E. coli BW25113 is the WT strain, while RU1616 contains a phoBR operon with the autoregulated WT promoter replaced with an IPTG-inducible promoter (16). An rpoS deletion strain, RU1646, was constructed by the λ red recombination technique (49) via replacing the rpoS gene with a kanamycin resistance cassette. To investigate general transcription regulation, phoBR deletion strain BW25142 carrying pRG278 was used to examine YFP expression from the constitutive Ptet promoter (3).
Bacterial growth conditions and phosphate starvation.
Bacteria were grown at 37°C in MOPS minimal medium with appropriate antibiotics, 0.4% (wt/vol) glucose, and amino acid mix (40 μg/ml) as described previously (28). Specifically, MOPS minimal medium containing 40 mM MOPS, 4 mM Tricine, 50 mM NaCl, 5 mM NH4Cl, 0.276 mM K2SO4, 0.523 mM MgCl2, 0.01 mM FeSO4, and other micronutrients was prepared as described previously (50). Glucose and an amino acid mix solution were later added to the indicated concentrations. Different amounts of a stock solution of 100 mM KH2PO4 were added for each assay to achieve the different indicated Pi concentrations.
Cells from overnight MOPS cultures were used to inoculate fresh Pi-replete (1 mM KH2PO4) MOPS medium. Once the OD (at 600 nm) reached 0.3 to 0.5, bacteria were harvested and resuspended in MOPS medium (0 μM KH2PO4) with a starting OD of ∼0.2 for prestarvation. After 50 min of Pi starvation, bacteria were pelleted and resuspended in Pi-replete MOPS medium again, with different starting ODs based on the times of expected growth dilution. A low starting OD of ∼0.08 was used for 3-h growth, while an OD of 0.35 was typically used for 0.5-h growth. Final ODs of all cultures were usually within the range of 0.4 to 0.7. Bacteria suspensions were kept on ice until ∼10 min before the start of the growth experiment. All growth experiments were started in a chronological order so that all cultures with different growth times could be harvested at the same time. Cell pellets were then washed twice with MOPS medium (50 μM Pi [nonactivating]) and directly resuspended in MOPS medium with the indicated concentration of Pi (2 or 50 μM Pi) for subsequent assays.
Measurement of alkaline phosphatase, protein expression, and phosphorylation levels.
Cells prepared as described above were grown again in MOPS medium (50 μM Pi) with a starting OD of ∼0.2, and aliquots were removed at the indicated time points and pelleted. Protein expression, in vivo phosphorylation, and AP activity levels were measured as described previously (16, 51). Briefly, AP activities were measured using 7 mM p-nitrophenylphosphate as the substrate and the relative rate of absorbance change at 420 nm was calculated to represent the AP activities. Protein expression levels were examined with Western blots using anti-AP (Sigma-Aldrich) (1:5,000) antibody or anti-PhoB (1:1,500) antisera followed by Cy3- or Cy5-conjugated secondary antibodies (GE Healthcare) (1:5,000). In vivo phosphorylation levels of PhoB were analyzed using Phos-tag gels to separate unphosphorylated and phosphorylated PhoB proteins as previously described (16). Cells were lysed in 55 μl 1× BugBuster reagent (Novagen) followed by denaturation with 18 μl 4× SDS loading buffer. All samples were frozen immediately in a dry ice-ethanol bath and later analyzed using Phos-tag gels and quantitative Western blots. Images of immunoblots were quantified by ImageJ (52).
Fluorescence reporter assays.
Prestarved bacteria and cells from Pi-replete cultures were inoculated in prewarmed MOPs medium with a starting OD of ∼0.15 to 0.2. Bacterial cultures were assayed in a Varioskan plate reader (Thermo Scientific) at 37°C with constant shaking (3 mm orbital, 240 rpm). For each individual strain or condition, 8 to 11 replicates were typically assayed simultaneously. YFP fluorescence (excitation [ex.], 488 nm; emission [em.], 530 nm) and absorbance (OD at 600 nm) levels were measured repeatedly (every 4 min). Fluorescence and OD readings were smoothed with a moving average of five time points, except for the data in Fig. 1, in which an adjacent average of three time points was used for smoothing. Variations between individual wells in a single microplate assay were small (see Fig. S2A in the supplemental material); thus, the mean values of total fluorescence or of increased fluorescence are shown as smooth lines in figures without error bars. First derivatives of fluorescence and OD values were calculated numerically as previously described by differentiating the second-order Lagrange interpolating polynomial values (17). Promoter activity was calculated as (dFluo./dt)/OD, where “dFluo./dt” represents first derivative of fluorescence, while the real-time growth rate was derived as (dOD/dt)/OD.
Model of PhoB-regulated transcription.
Expression of the PhoB-regulated reporter was modeled with the equation shown in Fig. 7. Mathematic modeling was performed using the Simbiology tool of Matlab (see Modeling Details in the supplemental material). The rate of reporter synthesis is dependent on the binding of PhoB∼P to promoter DNA and on the effective concentration of an open complex of RNA polymerase (RNAP) and sigma factor Eσ70. PhoB∼P binds DNA as a dimer (53, 54); thus, a Hill equation with a coefficient of 2 was used to model the binding. The binding constant was set at 1 μM, close to the values derived from transcription reporter assays as well as in vitro experiments (28). Kinetics of PhoB∼P were derived from previous Phos-tag analyses (17, 28), and the fitted Hill curves served as inputs for transcription modeling. The stress response is described with an arbitrary factor, S, that models the inhibitory effects of σS, anti-σ factor Rsd, ppGpp, or other factors on formation of Eσ70 (see Modeling Details in the supplemental material). Briefly, an increase in [S] upon Pi starvation is modeled to raise the concentration of σS for RNAP competition and reduce the concentration of Eσ70 through inhibition by other factors, e.g., σ70 sequestration by anti-σ factor Rsd. The effective concentration of Eσ70 is described with the following equation based on a simple competition model (29):
| (1) |
in which [ET] is the total concentration of RNAP. The concentration of S and the level of reporter after 1 h of prestarvation were calculated and multiplied by the growth dilution factor to serve as initial values for the second round of starvation response modeling.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a grant from the National Institutes of Health (R01GM047958).
Footnotes
For a commentary on this article, see ">https://doi.org/10.1128/JB.00420-17.
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00390-17.
REFERENCES
- 1.Dekel E, Alon U. 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature 436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 2.Foucault ML, Depardieu F, Courvalin P, Grillot-Courvalin C. 2010. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci U S A 107:16964–16969. doi: 10.1073/pnas.1006855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao R, Stock AM. 2013. Evolutionary tuning of protein expression levels of a positively autoregulated two-component system. PLoS Genet 9:e1003927. doi: 10.1371/journal.pgen.1003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffer SM, Westerhoff HV, Hellingwerf KJ, Postma PW, Tommassen J. 2001. Autoamplification of a two-component regulatory system results in “learning” behavior. J Bacteriol 183:4914–4917. doi: 10.1128/JB.183.16.4914-4917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zacharioudakis I, Gligoris T, Tzamarias D. 2007. A yeast catabolic enzyme controls transcriptional memory. Curr Biol 17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 6.Lambert G, Kussell E. 2014. Memory and fitness optimization of bacteria under fluctuating environments. PLoS Genet 10:e1004556. doi: 10.1371/journal.pgen.1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alon U. 2007. Network motifs: theory and experimental approaches. Nat Rev Genet 8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 8.Burrill DR, Silver PA. 2010. Making cellular memories. Cell 140:13–18. doi: 10.1016/j.cell.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jablonka E, Oborny B, Molnar I, Kisdi E, Hofbauer J, Czaran T. 1995. The adaptive advantage of phenotypic memory in changing environments. Philos Trans R Soc Lond B Biol Sci 350:133–141. doi: 10.1098/rstb.1995.0147. [DOI] [PubMed] [Google Scholar]
- 10.Xiong W, Ferrell JE Jr. 2003. A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature 426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 11.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capra EJ, Laub MT. 2012. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanner BL. 1996. Phosphorus assimilation and control of the phosphate regulon, p 1357–1381. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB Jr, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella. American Society for Microbiology Press, Washington, DC. [Google Scholar]
- 15.Batchelor E, Goulian M. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc Natl Acad Sci U S A 100:691–696. doi: 10.1073/pnas.0234782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao R, Stock AM. 2013. Probing kinase and phosphatase activities of two-component systems in vivo with concentration-dependent phosphorylation profiling. Proc Natl Acad Sci U S A 110:672–677. doi: 10.1073/pnas.1214587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao R, Stock AM. 2017. Quantitative kinetic analyses of shutting off a two-component system. mBio 8:e00412-17. doi: 10.1128/mBio.00412-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentry DR, Hernandez VJ, Nguyen LH, Jensen DB, Cashel M. 1993. Synthesis of the stationary-phase sigma factor σs is positively regulated by ppGpp. J Bacteriol 175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 21.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz N, Silhavy TJ. 2003. Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J Bacteriol 185:5984–5992. doi: 10.1128/JB.185.20.5984-5992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spira B, Yagil E. 1998. The relation between ppGpp and the PHO regulon in Escherichia coli. Mol Gen Genet 257:469–477. doi: 10.1007/s004380050671. [DOI] [PubMed] [Google Scholar]
- 24.Taschner NP, Yagil E, Spira B. 2004. A differential effect of σs on the expression of the PHO regulon genes of Escherichia coli. Microbiology 150:2985–2992. doi: 10.1099/mic.0.27124-0. [DOI] [PubMed] [Google Scholar]
- 25.Nyström T. 2004. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol 54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- 26.Österberg S, del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu Rev Microbiol 65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 27.Feklístov A, Sharon BD, Darst SA, Gross CA. 2014. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 28.Gao R, Stock AM. 2015. Temporal hierarchy of gene expression mediated by transcription factor binding affinity and activation dynamics. mBio 6:e00686-15. doi: 10.1128/mBio.00686-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauri M, Klumpp S. 2014. A model for sigma factor competition in bacterial cells. PLoS Comput Biol 10:e1003845. doi: 10.1371/journal.pcbi.1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf DM, Fontaine-Bodin L, Bischofs I, Price G, Keasling J, Arkin AP. 2008. Memory in microbes: quantifying history-dependent behavior in a bacterium. PLoS One 3:e1700. doi: 10.1371/journal.pone.0001700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Block SM, Segall JE, Berg HC. 1982. Impulse responses in bacterial chemotaxis. Cell 31:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 32.Chang DE, Leung S, Atkinson MR, Reifler A, Forger D, Ninfa AJ. 2010. Building biological memory by linking positive feedback loops. Proc Natl Acad Sci U S A 107:175–180. doi: 10.1073/pnas.0908314107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ram S, Goulian M. 2013. The architecture of a prototypical bacterial signaling circuit enables a single point mutation to confer novel network properties. PLoS Genet 9:e1003706. doi: 10.1371/journal.pgen.1003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arsène F, Tomoyasu T, Bukau B. 2000. The heat shock response of Escherichia coli. Int J Food Microbiol 55:3–9. doi: 10.1016/S0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 35.Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev 66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spira B, de Almeida Toledo R, Maharjan RP, Ferenci T. 2011. The uncertain consequences of transferring bacterial strains between laboratories—rpoS instability as an example. BMC Microbiol 11:248. doi: 10.1186/1471-2180-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Notley-McRobb L, King T, Ferenci T. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol 184:806–811. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathis R, Ackermann M. 2016. Response of single bacterial cells to stress gives rise to complex history dependence at the population level. Proc Natl Acad Sci U S A 113:4224–4229. doi: 10.1073/pnas.1511509113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 40.Jõers A, Tenson T. 2016. Growth resumption from stationary phase reveals memory in Escherichia coli cultures. Sci Rep 6:24055. doi: 10.1038/srep24055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bougdour A, Gottesman S. 2007. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci U S A 104:12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eames M, Kortemme T. 2012. Cost-benefit tradeoffs in engineered lac operons. Science 336:911–915. doi: 10.1126/science.1219083. [DOI] [PubMed] [Google Scholar]
- 43.Becskei A, Serrano L. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 44.Delhaye A, Collet JF, Laloux G. 2016. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio 7:e00047-16. doi: 10.1128/mBio.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raivio TL, Popkin DL, Silhavy TJ. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol 181:5263–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown DR, Barton G, Pan Z, Buck M, Wigneshweraraj S. 2014. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Commun 5:4115. doi: 10.1038/ncomms5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atkinson MR, Blauwkamp TA, Bondarenko V, Studitsky V, Ninfa AJ. 2002. Activation of the glnA, glnK, and nac promoters as Escherichia coli undergoes the transition from nitrogen excess growth to nitrogen starvation. J Bacteriol 184:5358–5363. doi: 10.1128/JB.184.19.5358-5363.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. 2006. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc Natl Acad Sci U S A 103:13503–13508. doi: 10.1073/pnas.0606026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao R, Tao Y, Stock AM. 2008. System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol Microbiol 69:1358–1372. doi: 10.1111/j.1365-2958.2008.06355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco AG, Canals A, Coll M. 2012. PhoB transcriptional activator binds hierarchically to pho box promoters. Biol Chem 393:1165–1171. doi: 10.1515/hsz-2012-0230. [DOI] [PubMed] [Google Scholar]
- 54.Ritzefeld M, Walhorn V, Kleineberg C, Bieker A, Kock K, Herrmann C, Anselmetti D, Sewald N. 2013. Cooperative binding of PhoBDBD to its cognate DNA sequence—a combined application of single-molecule and ensemble methods. Biochemistry 52:8177–8186. doi: 10.1021/bi400718r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.