ABSTRACT
Two-component signaling is a specialized mechanism that bacteria use to respond to changes in their environment. Nonpathogenic strains of Escherichia coli K-12 harbor 30 histidine kinases and 32 response regulators, which form a network of regulation that integrates many other global regulators that do not follow the two-component signaling mechanism, as well as signals from central metabolism. The output of this network is a multitude of phenotypic changes in response to changes in the environment. Among these phenotypic changes, many two-component systems control motility and/or the formation of biofilm, sessile communities of bacteria that form on surfaces. Motility is the first reversible attachment phase of biofilm development, followed by a so-called swim or stick switch toward surface organelles that aid in the subsequent phases. In the mature biofilm, motility heterogeneity is generated by a combination of evolutionary and gene regulatory events.
KEYWORDS: CheA, Escherichia coli, OmpR, RcsB, biofilm, fimbriae, flagella, motility, two-component signaling
INTRODUCTION
Bacteria colonize a wide variety of environmental niches and survive the challenges associated with them by tightly regulating gene expression. This happens in part by means of a phosphotransfer mechanism employing two-component signal transduction systems (2CSTSs) (for review articles, see references 1 to 5). The prototype 2CSTS consists of a sensor kinase and a response regulator. Most sensor kinases are membrane-bound proteins that autophosphorylate in the presence of ATP at a conserved histidine residue, from which the phosphoryl is transferred to a conserved aspartate in the response regulator (6). Nonpathogenic strains of Escherichia coli K-12 possess 30 histidine kinases and 32 response regulators (7), of which the majority control gene expression, primarily at the level of transcription. As specific examples of systems that follow the two-component mechanism and impact motility and biofilm, EnvZ/OmpR controls the response to changes in osmolarity (8), RscC/RcsD/RcsB (referred to as RcsCDB throughout this article) activates colanic acid synthesis (for a review, see reference 9), CheA/CheY/CheB controls the direction of flagellar motor rotation (for an early review, see reference 10), and QseC/QseB connects quorum sensing with motility, biofilm development, and virulence (11).
This minireview article focuses on the impact of 2CSTS signaling on the expression of motility and biofilm genes. Special emphasis will be on the involvement of motility in the early phases of biofilm development and the swim or stick switch that allows bacteria to transit from reversible to irreversible attachment. The concept of motility heterogeneity as one mechanism of niche adaptation in the mature biofilm will be developed. Specific examples of coupling mechanisms between motility and/or biofilm and pathogenesis are included for pathogenic E. coli and a small selection of other bacterial pathogens.
Motility and biofilm.
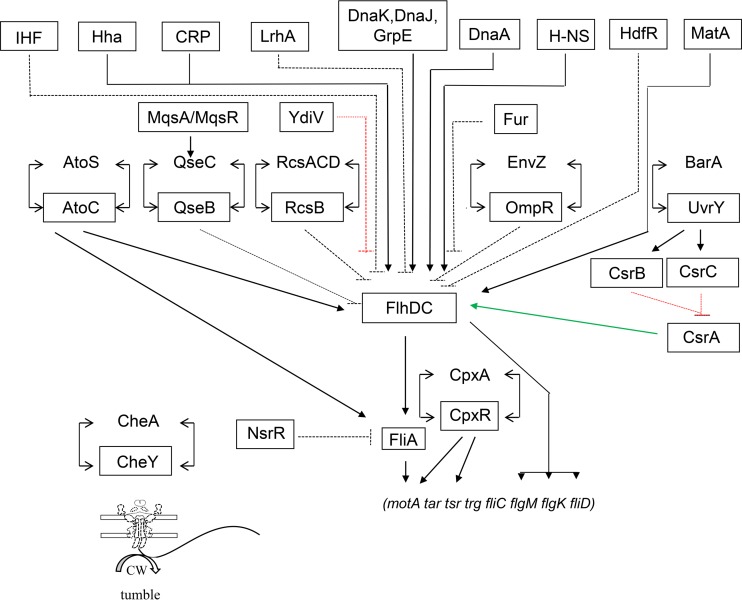
In E. coli, motility requires the synthesis of flagella, which is controlled in part by a 2CSTS. A three-tier hierarchy controls transcription of some 50 flagellar genes in 14 operons and includes numerous regulatory proteins; tier I is the flhDC operon that encodes FlhD and FlhC (12) and is required for the transcription of all other flagellar genes. Given this central role and the high energetic cost of flagellar synthesis, it does not come as a surprise that flhDC undergoes complex and precise regulation in response to a myriad of environmental conditions. Regulation of the flhDC operon by 2CSTSs and other regulators is depicted in Fig. 1.
FIG 1.
Global control of flagella. This figure has been modified from a previous review article (142). Positive effects on transcription are indicated by black solid lines and arrowheads; negative effects on transcription are indicated by black dashed lines with blunt ends. The green arrow indicates positive posttranscriptional regulation, and the red dashed lines with blunt ends indicate negative nontranscriptional regulation. Regulators that exert a direct effect on their targets are shown in boxes. The control of flagellar rotation by the CheA/CheY system is included in the figure. The flagellar motor complex is reprinted from reference 143 (CW, counterclockwise). All global regulators, as well as response regulators of 2CSTSs, are boxed. Sensor kinases are not boxed. (Adapted from reference 142.)
Motility is not just a way by which bacteria move from one location to another. It is also the first phase of biofilm development. The phasing model was first described for Pseudomonas aeruginosa. In this model, biofilm development starts with reversible attachment, followed by irreversible attachment, maturation, and dispersal (13). Flagella are recognized as the cell surface organelle for reversible attachment; type I fimbriae, pili, and curli were proposed to aid irreversible attachment, and the extracellular polymeric substance (EPS) facilitated maturation (for a review, see reference 14).
The phasing model for biofilm development includes the so-called swim or stick switch, where an individual bacterium can either swim (reversible attachment) or stick (irreversible attachment and maturation). Switching requires surface sensing (for a review, see reference 15), followed by a transition from the expression of flagella to the synthesis of fimbriae, pili, and/or curli and finally the production of EPS. Obviously, such a timely synthesis of cell surface organelles is dependent on precisely coordinated regulation of gene expression (16), some of which is mediated by 2CSTS signaling.
EnvZ/OmpR inversely regulates transcription of ompC and ompF, as well as flagellar genes and genes encoding type I fimbriae and curli.
The EnvZ/OmpR system was first discovered as part of the response to changes in osmolarity. Such changes are detected by the sensor kinase EnvZ, leading to autophosphorylation at a conserved His (17). This event is followed by a phosphotransfer to the response regulator OmpR at a conserved Asp (18). The resultant OmpR-P modulates the reciprocal synthesis of the outer membrane porins OmpF and OmpC (19–21). The first evidence that OmpR-P controls transcription of the flhDC operon came from Shin and Park (22) who found that the increase in the binding affinity for the flhD promoter by the phosphorylation of OmpR was approximately 10-fold. At the same time, research by Prüß and Wolfe (23) showed that acetyl phosphate, a metabolic signal that E. coli can use to phosphorylate OmpR (22), negatively affected flhDC transcription. In addition, a knockout mutation in ompR exhibited increased flhDC transcription in a growth phase-independent manner (24). An ompR mutant also exhibited a lack of temporal expression from the flhD promoter during biofilm development (25).
In addition to porin and flagellar genes, OmpR also impacts the transcription of the type I fimbria genes (26). The eight genes that are required for type I fimbriae form one large operon (27), including the fimA gene that encodes the major structural subunit (28) and the fimE and fimB genes that encode the recombinases for phase variation (29). These two proteins invert a 314-bp switch element designated fimS that is located upstream of fimA and flanked by inverted repeats. Whereas FimB facilitates switching in both directions, FimE preferentially switches from “phase-on” to “phase-off” (30). OmpR binds to the fimB promoter; a mutation in ompR exhibited increased fimB transcription relative to the parent, as well as a “phase-on” orientation of the switch element, in agreement with an increased level of type I fimbria synthesis (31).
The major regulator for curli, another irreversible attachment organelle, is CsgD that controls both of the curli-encoding operons csgAB and csgDEFG (32). Transcription of csgD is also activated by EnvZ/OmpR (33). Altogether, EnvZ/OmpR contribute to the transition from the reversible to the irreversible attachment phase in biofilm development by inhibiting flhDC and increasing the synthesis of type I fimbriae and curli.
RcsCDB is a positive regulator of colanic acid synthesis and a negative regulator of flagellar and curli genes.
RcsCDB was originally described as an activator of colanic acid (34), one of the extracellular polymeric substances that contribute to maturation of a developing biofilm. While RcsCDB follows the two-component signaling mechanism, it consists of more than two proteins. The hybrid kinase RcsC autophosphorylates at the conserved His. Phosphorelay ensues by transfer of the phosphoryl group to the conserved Asp in the receiver domain of the same molecule. The phosphoryl group is then transferred to the conserved His in RcsD, ultimately phosphorylating the response regulator RcsB at the conserved Asp (35–37). RcsB homodimer formation is required to activate transcription of the wza operon that encodes the colanic acid genes and was originally designated cps (38).
RcsCDB does not just contribute to the transition from reversible/irreversible attachment to the mature biofilm by activating colanic acid synthesis. It also downregulates csgD (39) and flhD (40). Regulation of flhD requires heterodimer formation of RcsB with the auxiliary protein RcsA, followed by binding to the conserved RcsAB box on the flhD promoter (40). Heterodimer formation can occur with a range of transcriptional regulators (41–43), often in a way that is unaffected by the phosphorylation state of RcsB (43, 44) and instead favored at high concentrations of the protein (45). Altogether, the RcsCDB system has a myriad of functions in E. coli and other members of the family Enterobacteriaceae (9, 46–49).
QseC/QseB connects quorum sensing with motility, biofilm, and virulence gene expression.
The first function that was attributed to QseC/QseB was its involvement in the regulation of flagellar genes by quorum sensing in E. coli K-12 and enterohemorrhagic E. coli (EHEC) (11). QseB was shown to bind to the flhD promoter directly at high- and low-affinity sites, and transcription initiation appeared dependent on the sigma factor FliA (50). Differences between mutations in qseB and qseC pointed toward the phosphatase activity of the system to be located on the sensor kinase QseC (51). In particular, deletion of QseC but not QseB attenuated the formation of intracellular communities and virulence by uropathogenic E. coli (UPEC). Intriguingly, the attachment organelles type I fimbriae and curli were downregulated alongside with flagella in the absence of QseC (51). This is different from EnvZ/OmpR and RcsCDB which regulate multiple biofilm-associated cell surface organelles in an inverse manner.
In addition to QseB, the sensor kinase QseC phosphorylates two other response regulators, QseF and KdpE, to control the Shiga toxin gene stx2 and the LEE (locus of enterocyte effacement) pathogenicity genes, respectively (52). Site-directed mutagenesis of the QseC periplasmic sensing domain yielded four mutants that all exhibited increased levels of phosphorylation but differed in their motility, LEE, and Shiga toxin expression phenotypes (53). The mutants also differed in their abilities to phosphorylate QseB, KdpE, and QseF. It was concluded that the mutations influenced the phosphotransfer preference of QseC (53).
In E. coli O157:H7, QseC/QseB constitutes a remarkable connection between the host stress response and bacterial environmental signaling, for which it has earned the term “adrenergic receptor” (54). When living in the intestine, E. coli responds to the autoinducer-3 that is produced by the gut flora and the human stress hormones epinephrine and norepinephrine (for a review, see reference 55). This trio of signals has been described as an “interkingdom chemical signaling system” (55). The signaling transduction cascade includes chemotaxis by using the serine receptor Tsr (56, 57) and activation of QseC (58). In the search for novel therapeutic targets and drugs, blocking the binding of epinephrine or norepinephrine to QseC with a proposed LED209 drug resulted in decreased QseC/QseB signaling and reduced motility and virulence (59), as well as attenuation of colitis in mice (60).
BarA/UvrY controls the expression of genes of metabolism, motility, biofilm, and virulence.
The BarA/UvrY 2CSTS regulates genes important for carbon storage and responds to acetate, formate, and carboxylates (61). UvrY synthesis is enhanced by polyamines (62). The signal is transmitted by activating the transcription of genes for small regulatory RNA (sRNA) molecules. sRNAs have gained increasing attention in recent years as powerful regulators of transcription (for a recent review, see reference 63). In the case of BarA/UvrY, strong interaction was observed between UvrY-P and csrB and csrC (64). These encoded sRNAs are part of the carbon storage system. Together with the RNA-binding protein and posttranscriptional regulator CsrA, they were initially described as modulators of gene expression when bacteria transit into stationary phase (for a review, see reference 65). CsrA is a repressor of stationary-phase genes, and it also activates flhDC (66). The fact that the activating effect of CsrA on flhDC is not on the transcriptional level but on the posttranscriptional level is indicated by a green arrow in Fig. 1. The inhibitory effect of the sRNA molecules CsrB and CsrC on CsrA is one of sequestration. Multiple binding sites for sRNA on CsrA enable CsrB and CsrC to act as a CsrA sink (65). This sequestration effect is distinguished from transcriptional regulation by red lines in Fig. 1. Likewise, the negative effect of the phosphodiesterase YdiV on FlhD/FlhC is indicated by the red dashed line with a blunt end. This effect is one of posttranslation, where YdiV binds to the FlhD/FlhC complex at high concentrations to inhibit its transcriptional activity (67).
As a final 2CSTS in the context of regulation of flhDC transcription, AtoS/AtoC facilitates the response to acetoacetate, spermidine, and histamine via enhancing transcription of flhD and fliA (68). This 2CSTS may expand the range of nutrients that E. coli is able to utilize.
Many regulatory proteins act upon flhDC transcription beyond regulation by 2CSTSs (Fig. 1). These regulatory proteins are LrhA (69), H-NS (70), HdfR (71), integration host factor IHF (72), Hha (73), the toxin/antitoxin system MqsA/MqsR (74) through QseB, MatA (75), and the iron response regulator Fur (76). Temperature regulation of flhDC transcription is complemented by DnaA (77) or the action of DnaK, DnaJ, and GrpE (78). Metabolic control of the flhDC operon includes catabolite repression through cyclic AMP (cAMP)-cAMP receptor protein (CRP) (79). Additional sRNAs that impact flhDC expression in a negative way are ArcZ, OmrA, OmrB, and OxyS; McaS exerts a positive effect (80). Note that these five regulatory molecules are not included in Fig. 1.
Additional downstream regulation of flagellar genes.
While it may make sense to control a complex hierarchy at the top level, fine tuning transcription can take place at other levels. In particular, the tier II gene fliA has emerged as a second center of regulation (Fig. 1). Specifically, the nitric oxide-sensitive repressor NsrR negatively regulates the fliA promoter, as well as attachment (81). A 2CSTS that acts upon the transcription of tier III genes is CpxA/CpxR. This 2CSTS responds to envelope stress, inhibits LEE gene transcription (82), and activates the synthesis pathway that leads to the production of an antimicrobial peptide that causes multidrug resistance (83). Direct binding of CpxR-P to the promoter regions of the motA and tsr operons has been shown (84) and so has transcriptional repression of csgD by CpxR (33, 85). This makes CpxA/CpxR another 2CSTS that inversely regulates flagellar and curli genes.
As a final 2CSTS to be discussed, CheA/CheY affects the direction of rotation of the flagellar motor (Fig. 1). This 2CSTS functions with a second response regulator, CheB, as well as many other chemotaxis proteins to control bacterial motility toward an attractant or away from a repellant by modulating the direction of flagellar rotation (for a recent review on chemotaxis, see reference 86). Since CheA lacks the classical transmembrane domain, it associates with a distinct set of transmembrane proteins known as methyl-accepting chemotaxis proteins (for a recent review, see reference 87) and an adaptor protein CheW (88–91). The response regulator CheY is a single-domain molecule, whose structure has been determined in its unphosphorylated (92) and phosphonated (93) form, in complex with CheA (94, 95), and bound to its FliM target (96). Phosphorylation of CheY enables its binding to FliM at the flagellar motor (96–98), where it causes clockwise rotation of the flagella, resulting in cell tumble and change in the direction of motility (99, 100).
An additional regulatory mechanism that may not involve the control of gene expression and is not included in Fig. 1 is the interaction of H-NS with the flagellar rotor protein FliG. Direct interaction of H-NS with FliG was first seen by using a yeast two-hybrid system (101). Later, it was shown that this interaction increased the flagellar rotational speed and caused hypermotility (102). More-recent research, however, indicates that the H-NS effect on motility happens via an indirect and complex route (103).
What does metabolism have to do with it all?
Some response regulators can receive signals not only from their cognate kinase but also from central metabolism. An example of such a metabolic intermediate is acetyl phosphate which stands between acetyl coenzyme A (acetyl-CoA) and acetate (104) and accumulates when the flux of carbon through glycolysis is high (105). Acetyl phosphate can phosphorylate OmpR (22) and other response regulators (106–108). This constitutes a mechanism of activation that links central metabolism with signal transduction (109). In one specific example of an in vivo effect of acetyl phosphate, a depletion of serine from a mixed amino acid growth medium by E. coli K-12 was linked to a reduction in flhDC transcription, presumably through the production of acetyl phosphate and increased levels of OmpR-P (110). Serine depletion simultaneously resulted in an increase in motility and a decrease in the cell division rate (110). With respect to biofilm development, acetyl phosphate (22, 23) acts at the swim or stick switch. In cases of low acetyl phosphate levels, OmpR-P is low, transcription from the flhD promoter is high, and bacteria are highly motile. This scenario resembles reversible attachment. In contrast, under conditions of high acetyl phosphate levels, OmpR-P is high, flhDC transcription is low, and motility is low (22, 23), but type I fimbriae are expressed (109). This scenario resembles irreversible attachment. The metabolic control of the swim or stick switch is in agreement with a model that was developed a few years after the phasing model and described the formation of biofilm as an output of the entire metabolic and gene regulatory network of the bacteria (111).
Interestingly, RcsB (112, 113) and CheY (114) can also be acetylated, using either acetyl-CoA or acetyl phosphate as donor of the acetyl group (115, 116). In the case of E. coli CheY, acetylation happens at two specific lysine residues, K91 and K109 (117). Acetylated CheY (CheY-ace) binds only to the low-affinity CheY-binding sites of FliM at the flagellar rotor. The cooperative binding of CheY-P to these sites and the synergistic effect of acetylation and phosphorylation on CheY activation suggest that binding of CheY-ace increases the affinity for CheY-P (O. Afanzar and M. Eisenbach, personal communication). For a recent review on protein acetylation, see reference 118.
A second messenger molecule that does not involve 2CSTS signaling is cyclic di-GMP (c-di-GMP). This molecule helps facilitate the swim or stick switch (119) by inhibiting flagellar synthesis and activating the synthesis of curli and cellulose (for reviews, see references 120 and 121). Cellular levels of c-di-GMP result from the interplay of two groups of enzymes, diguanylate cyclases (122) and phosphodiesterases (123). In the case of the flagellar system, the diguanylate cyclase YegE is under the control of the stationary-phase sigma factor σS and increases the level of c-di-GMP (124). The phosphodiesterase YhjH is under the control of the flagellar sigma factor FliA and decreases c-di-GMP levels. c-di-GMP itself is an inhibitor of flagellar synthesis and an activator of curli through their master regulator CsgD (124). Activation of cellulose synthase involves the diguanylate cyclase YaiC and the phosphodiesterase YoaD (125).
Motility and biofilm, a paradox or not?
There is an apparent paradox included in the previous paragraphs. On one hand, the proposal of flagella as the cell surface organelle for reversible attachment is consistent with the idea of motility being an advantage for bacteria forming a biofilm. However, the concept of the swim or stick switch makes motility and biofilm appear like two mutually exclusive processes. The question arises whether motility is an advantage or a disadvantage when bacteria form a biofilm.
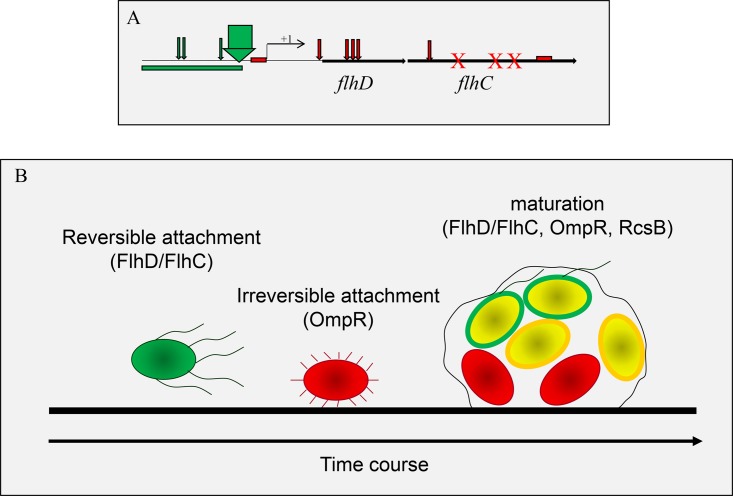
The solution to this apparent paradox may lie in the distinction between an advantage for the individual cell (swim or stick) and an advantage for the population (reversible attachment as a phase). What constitutes an advantage for an individual bacterium at a specific time and/or location may differ from what is beneficial for the entire population. To permit such niche adaptation, bacteria have evolved diverse mechanisms that generate motility heterogeneity within their population. E. coli couples evolutionary events with control of gene expression by multiple 2CSTSs. The evolutionary event is a selection for mutations in the flhDC operon. Among the many possible types of mutations, IS elements in particular are known to lead to changes in gene expression and phenotypes (for a review, see reference 126). The first such study regarding E. coli motility was by Barker and coworkers (127), who found an insertion of IS5 at bp −99 to −96 from the flhDC transcriptional start in an originally poorly motile version of E. coli MG1655 Fnr− that now exhibited a 2.7-fold increase in the rate of migration on motility plates. Interestingly, Wang and Wood (128) identified the same insertion (IS5 at −99 to −96) in the flhD promoter of strain BW25311 that increased motility and biofilm amounts. Lee and Park found IS elements up to 315 bp from the flhDC transcriptional start (71), and IS elements identified by Fahrner and Berg were identified up to bp −476 (129). These mutations are summarized in Fig. 2A and have the following characteristics in common. (i) They were selected from a previously nonmotile E. coli K-12 strain. (ii) They carried the IS element 96 or more bp upstream of the flhDC transcriptional start. (iii) They were recovered under conditions where motility was an advantage.
FIG 2.
Generation of motility heterogeneity by E. coli. (A) Demonstration of the mechanism by which E. coli generates motility heterogeneity by means of mutating the flhDC operon. The big green arrow depicts the IS5 at positions −99 to −96 (127, 128). Other IS elements that rendered E. coli hypermotile and were found by Lee and Park (71) are indicated by slim green arrows. The green bar indicates the region from positions −476 to −100 where Fahrner and Berg (129) found IS elements. The mutations found by Horne and coworkers (130) are indicated by red symbols. The red boxes indicate small deletions, the X letters depict point mutations, and the red arrows show insertion sites of IS elements. The mutations found by Horne et al. (130) rendered E. coli nonmotile. (B) Map of temporal and spatial expression patterns onto biofilm development. Transcription from the flhD promoter is indicated in green, ompR transcription is indicated in red, and rcsB transcription is indicated in yellow. For reversible and irreversible attachment, cell surface organelles are drawn in a slightly darker shade of the base color. For the mature biofilm, bacteria on the outermost edge of the biofilm are multicolored based on the spatial transcription data for flhD and rcsB. Note that the spatial distribution of transcription was determined across the biofilm and not across individual bacteria.
A fifth study on IS elements in the flhD promoter by Horne and coworkers (130) reported the isolation of nonmotile derivatives from biofilm of the originally highly motile MC1000 strain. The locations of these mutations are included in Fig. 2A and are all further downstream than the mutations that increased motility. The types of mutations include small deletions, point mutations causing frameshifts and truncations, and insertions of IS elements. Many of these mutations are in the open reading frames for flhD or flhC, others are in the ribosome-binding site or the −10/−35 sites for the RNA polymerase. The conclusion from this study was that mutations in the flhDC operon can occur in many places and that the location of the mutation determines whether the resulting E. coli is more or less motile than the original strain. The authors of this article (130) believe that mutagenesis of the flhDC operon may be a mechanism by which E. coli generates motility heterogeneity within their biofilm and that this heterogeneity constitutes a selective advantage for the total population of bacteria that form the biofilm.
This hypothesis is supported by a study by Samanta and coworkers, who performed temporal and spatial expression studies with the flhD promoter from the E. coli K-12 strain AJW678 that did not contain an IS element (25). The temporal transcription pattern of flhD was highest at 12 h and lowest at 35 h and increased again toward 51 h. The temporal profile for ompR was the inverse of that for flhD, and transcription of rcsB increased steadily throughout biofilm growth. Temporal expression patterns were used to map gene expression to biofilm phases, where flhD was an indicator of reversible attachment, ompR represented irreversible attachment, and rcsB was indicative of maturation (Fig. 2B). In the mature biofilm, flhDC transcription was highest at the outermost edge of the biofilm and lowest near the attachment surface. The spatial transcription of ompR was highest at the attachment surface and decreased with increasing distance from the surface, whereas transcription of rcsB increased toward the edge of the biofilm (131). Temporal and spatial patterns of flhD expression were abolished in ompR and rcsB mutants (25).
This minireview will conclude with two examples where motility heterogeneity has implications outside biofilm development. In one study, the authors deliberately generated heterogeneity at the level of phenotype and gene expression by exposing E. coli to several antibiotics, allowing them to perform a process they termed “mid-term adaptation”; among the genes that were described as “transcriptome-level signatures” were genes of the motility apparatus, the cps operon, and ompR (132). As a second example from Salmonella enterica, motility heterogeneity aids virulence. The fliZ gene encodes a posttranslational activator of FlhD/FlhC (133). The fliT (134) and ydiV (135) genes are tier III flagellar genes encoding anti-FlhD/FlhC factors. The competitive action of YdiV and FliZ generates subpopulations of motile fliC-ON and nonmotile fliC-OFF bacteria within the population (136). YdiV and FliZ constitute a nutrient-tunable bistable switch that allows bacteria to express different phenotypes under identical growth conditions (137). It promotes a selective advantage during the infection process because it permits a division of labor, where motile cells are pathogenic and nonmotile cells serve as reservoir to feed the infection (138).
Conclusion and outlook.
In summary, 2CSTSs provide the bacteria with a complex network of regulation that controls motility, biofilm development, and pathogenesis and is responsive to a large variety of signals from the environment. An intriguing unanswered question in this context is why motility and biofilm are so often reciprocally regulated by the same 2CSTS but sometimes appear coregulated. The answer to this question may feed into the growing recognition that the molecular response to environmental signals can take place at the level of a whole bacterial community, as well as at a single-cell level. The phenotypic heterogeneity that results from the latter is one mechanism by which bacteria can perform niche adaptation. Among the future aspects of this type of research is an investigation of phenotypic heterogeneity in natural communities, including symbiotic (139) and pathogenic (140) relationships with hosts and multispecies environmental communities (141). Such research takes the well-established approach of determining taxonomic relationships between multiple species of the same community (“who is there?”) to the level of functionality (“what are they doing?”). In this sense, studying heterogeneity of phenotypic traits within populations removes a current limit to the predictive power of the taxonomic approach (141).
ACKNOWLEDGMENTS
B.M.P. is funded by the North Dakota Agricultural Experiment Station and Hatch project 1009442 through USDA/NIFA.
I thank Preeti Sule (Texas A&M University) for helpful discussions and Shelley Horne and Meredith Schroeder (NDSU) for critically reading the manuscript.
REFERENCES
- 1.Gao R, Mack TR, Stock AM. 2007. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci 32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3:165–170. doi: 10.1016/S1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 3.Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev 70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 5.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitrophanov AY, Groisman EA. 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev 22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heermann R, Jung K (ed). 2010. Stimulus perception and signaling in histidine kinases, p 135–161. In Kramer R, Jung K (ed), Bacterial signaling. Wiley-VCH Verlag GmbH & KGaA, Weinheim, Germany. [Google Scholar]
- 8.Forst SA, Roberts DL. 1994. Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res Microbiol 145:363–373. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 9.Clarke DJ. 2010. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol 5:1173–1184. doi: 10.2217/fmb.10.83. [DOI] [PubMed] [Google Scholar]
- 10.Bourret RB, Hess JF, Borkovich KA, Pakula AA, Simon MI. 1989. Protein phosphorylation in chemotaxis and two-component regulatory systems of bacteria. J Biol Chem 264:7085–7088. [PubMed] [Google Scholar]
- 11.Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 12.Bartlett DH, Frantz BB, Matsumura P. 1988. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5′ consensus sequences of operons under FlbB and FlaI control. J Bacteriol 170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Houdt R, Michiels CW. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol 156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 15.O'Toole GA, Wong GC. 2016. Sensational biofilms: surface sensing in bacteria. Curr Opin Microbiol 30:139–146. doi: 10.1016/j.mib.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 17.Forst S, Delgado J, Inouye M. 1989. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci U S A 86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato M, Aiba H, Tate S, Nishimura Y, Mizuno T. 1989. Location of phosphorylation site and DNA-binding site of a positive regulator, OmpR, involved in activation of the osmoregulatory genes of Escherichia coli. FEBS Lett 249:168–172. doi: 10.1016/0014-5793(89)80617-9. [DOI] [PubMed] [Google Scholar]
- 19.Aiba H, Mizuno T. 1990. Phosphorylation of a bacterial activator protein, OmpR, by a protein kinase, EnvZ, stimulates the transcription of the ompF and ompC genes in Escherichia coli. FEBS Lett 261:19–22. doi: 10.1016/0014-5793(90)80626-T. [DOI] [PubMed] [Google Scholar]
- 20.Aiba H, Nakasai F, Mizushima S, Mizuno T. 1989. Phosphorylation of a bacterial activator protein, OmpR, by a protein kinase, EnvZ, results in stimulation of its DNA-binding ability. J Biochem 106:5–7. doi: 10.1093/oxfordjournals.jbchem.a122817. [DOI] [PubMed] [Google Scholar]
- 21.Hall MN, Silhavy TJ. 1981. Genetic analysis of the ompB locus in Escherichia coli K-12. J Mol Biol 151:1–15. doi: 10.1016/0022-2836(81)90218-7. [DOI] [PubMed] [Google Scholar]
- 22.Shin S, Park C. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol 177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prüß BM, Wolfe AJ. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol 12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 24.Prüß BM. 1998. Acetyl phosphate and the phosphorylation of OmpR are involved in the regulation of the cell division rate in Escherichia coli. Arch Microbiol 170:141–146. doi: 10.1007/s002030050626. [DOI] [PubMed] [Google Scholar]
- 25.Samanta P, Clark ER, Knutson K, Horne SM, Prüß BM. 2013. OmpR and RcsB abolish temporal and spatial changes in expression of flhD in Escherichia coli biofilm. BMC Microbiol 13:182. doi: 10.1186/1471-2180-13-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwan WR, Lee JL, Lenard FA, Matthews BT, Beck MT. 2002. Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infect Immun 70:1391–1402. doi: 10.1128/IAI.70.3.1391-1402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marc D, Dho-Moulin M. 1996. Analysis of the fim cluster of an avian O2 strain of Escherichia coli: serogroup-specific sites within fimA and nucleotide sequence of fimI. J Med Microbiol 44:444–452. doi: 10.1099/00222615-44-6-444. [DOI] [PubMed] [Google Scholar]
- 28.Klemm P. 1984. The fimA gene encoding the type-1 fimbrial subunit of Escherichia coli. Nucleotide sequence and primary structure of the protein. Eur J Biochem 143:395–399. [DOI] [PubMed] [Google Scholar]
- 29.Klemm P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J 5:1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClain MS, Blomfield IC, Eisenstein BI. 1991. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol 173:5308–5314. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rentschler AE, Lovrich SD, Fitton R, Enos-Berlage J, Schwan WR. 2013. OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology 159:316–327. doi: 10.1099/mic.0.059386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol 180:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P, Dorel C. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol 183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottesman S, Trisler P, Torres-Cabassa A. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol 162:1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stout V, Gottesman S. 1990. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol 172:659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brill JA, Quinlan-Walshe C, Gottesman S. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol 170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeda S, Fujisawa Y, Matsubara M, Aiba H, Mizuno T. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC → YojN → RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol Microbiol 40:440–450. doi: 10.1046/j.1365-2958.2001.02393.x. [DOI] [PubMed] [Google Scholar]
- 38.Trisler P, Gottesman S. 1984. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J Bacteriol 160:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrieres L, Clarke DJ. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol 50:1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- 40.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 49:823–832. doi: 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 41.Wehland M, Bernhard F. 2000. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J Biol Chem 275:7013–7020. [DOI] [PubMed] [Google Scholar]
- 42.Wehland M, Kiecker C, Coplin DL, Kelm O, Saenger W, Bernhard F. 1999. Identification of an RcsA/RcsB recognition motif in the promoters of exopolysaccharide biosynthetic operons from Erwinia amylovora and Pantoea stewartii subspecies stewartii. J Biol Chem 274:3300–3307. doi: 10.1074/jbc.274.6.3300. [DOI] [PubMed] [Google Scholar]
- 43.Pannen D, Fabisch M, Gausling L, Schnetz K. 2016. Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J Biol Chem 291:2357–2370. doi: 10.1074/jbc.M115.696815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhne C, Singer HM, Grabisch E, Codutti L, Carlomagno T, Scrima A, Erhardt M. 2016. RflM mediates target specificity of the RcsCDB phosphorelay system for transcriptional repression of flagellar synthesis in Salmonella enterica. Mol Microbiol 101:841–855. doi: 10.1111/mmi.13427. [DOI] [PubMed] [Google Scholar]
- 45.Filippova EV, Wawrzak Z, Ruan J, Pshenychnyi S, Schultz RM, Wolfe AJ, Anderson WF. 2016. Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli. Protein Sci 25:2216–2224. doi: 10.1002/pro.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang YH, Ferrieres L, Clarke DJ. 2006. The role of the Rcs phosphorelay in Enterobacteriaceae. Res Microbiol 157:206–212. doi: 10.1016/j.resmic.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Krin E, Danchin A, Soutourina O. 2010. RcsB plays a central role in H-NS-dependent regulation of motility and acid stress resistance in Escherichia coli. Res Microbiol 161:363–371. doi: 10.1016/j.resmic.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Sharma VK, Bayles DO, Alt DP, Looft T, Brunelle BW, Stasko JA. 2017. Disruption of rcsB by a duplicated sequence in a curli-producing Escherichia coli O157:H7 results in differential gene expression in relation to biofilm formation, stress responses and metabolism. BMC Microbiol 17:56. doi: 10.1186/s12866-017-0966-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oropeza R, Salgado-Bravo R, Calva E. 2015. Deletion analysis of RcsC reveals a novel signalling pathway controlling poly-N-acetylglucosamine synthesis and biofilm formation in Escherichia coli. Microbiology 161:903–913. doi: 10.1099/mic.0.000050. [DOI] [PubMed] [Google Scholar]
- 50.Clarke MB, Sperandio V. 2005. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol 57:1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- 51.Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. 2009. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol 73:1020–1031. doi: 10.1111/j.1365-2958.2009.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog 5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker CT, Russell R, Njoroge JW, Jimenez AG, Taussig R, Sperandio V. 2017. Genetic and mechanistic analyses of the periplasmic domain of the enterohemorrhagic Escherichia coli QseC histidine sensor kinase. J Bacteriol 199:e00861-16. doi: 10.1128/JB.00861-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng J, Huang YC, Huang J, Yang K. 2016. The role of the sensor kinase, QseC, an adrenergic receptor of Escherichia coli, in bacterial translocation during hemorrhagic shock. J Trauma Acute Care Surg 80:972–976. doi: 10.1097/TA.0000000000001007. [DOI] [PubMed] [Google Scholar]
- 55.Moreira CG, Sperandio V. 2016. The epinephrine/norepinephrine/autoinducer-3 interkingdom signaling system in Escherichia coli O157:H7. Adv Exp Med Biol 874:247–261. [DOI] [PubMed] [Google Scholar]
- 56.Pasupuleti S, Sule N, Cohn WB, MacKenzie DS, Jayaraman A, Manson MD. 2014. Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3,4-dihydroxymandelic acid. J Bacteriol 196:3992–4000. doi: 10.1128/JB.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bansal T, Englert D, Lee J, Hegde M, Wood TK, Jayaraman A. 2007. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun 75:4597–4607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasko DA, Moreira CG, Li de R, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rooks MG, Veiga P, Reeves AZ, Lavoie S, Yasuda K, Asano Y, Yoshihara K, Michaud M, Wardwell-Scott L, Gallini CA, Glickman JN, Sudo N, Huttenhower C, Lesser CF, Garrett WS. 2017. QseC inhibition as an antivirulence approach for colitis-associated bacteria. Proc Natl Acad Sci U S A 114:142–147. doi: 10.1073/pnas.1612836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chavez RG, Alvarez AF, Romeo T, Georgellis D. 2010. The physiological stimulus for the BarA sensor kinase. J Bacteriol 192:2009–2012. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakamoto A, Terui Y, Yamamoto T, Kasahara T, Nakamura M, Tomitori H, Yamamoto K, Ishihama A, Michael AJ, Igarashi K, Kashiwagi K. 2012. Enhanced biofilm formation and/or cell viability by polyamines through stimulation of response regulators UvrY and CpxR in the two-component signal transducing systems, and ribosome recycling factor. Int J Biochem Cell Biol 44:1877–1886. doi: 10.1016/j.biocel.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 63.Jagodnik J, Brosse A, Le Lam TN, Chiaruttini C, Guillier M. 2017. Mechanistic study of base-pairing small regulatory RNAs in bacteria. Methods 117:67–76. doi: 10.1016/j.ymeth.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 64.Zere TR, Vakulskas CA, Leng Y, Pannuri A, Potts AH, Dias R, Tang D, Kolaczkowski B, Georgellis D, Ahmer BM, Romeo T. 2015. Genomic targets and features of BarA-UvrY (-SirA) signal transduction systems. PLoS One 10:e0145035. doi: 10.1371/journal.pone.0145035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romeo T, Vakulskas CA, Babitzke P. 2013. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol 15:313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei BL, Brun-Zinkernagel AM, Simecka JW, Prüß BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 67.Li B, Li N, Wang F, Guo L, Huang Y, Liu X, Wei T, Zhu D, Liu C, Pan H, Xu S, Wang HW, Gu L. 2012. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res 40:11073–11085. doi: 10.1093/nar/gks869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Theodorou MC, Theodorou EC, Kyriakidis DA. 2012. Involvement of AtoSC two-component system in Escherichia coli flagellar regulon. Amino Acids 43:833–844. doi: 10.1007/s00726-011-1140-7. [DOI] [PubMed] [Google Scholar]
- 69.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol 45:521–532. doi: 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- 70.Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, Collatz E. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol 176:5537–5540. doi: 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee C, Park C. 2013. Mutations upregulating the flhDC operon of Escherichia coli K-12. J Microbiol 51:140–144. doi: 10.1007/s12275-013-2212-z. [DOI] [PubMed] [Google Scholar]
- 72.Yona-Nadler C, Umanski T, Aizawa S, Friedberg D, Rosenshine I. 2003. Integration host factor (IHF) mediates repression of flagella in enteropathogenic and enterohaemorrhagic Escherichia coli. Microbiology 149:877–884. doi: 10.1099/mic.0.25970-0. [DOI] [PubMed] [Google Scholar]
- 73.Sharma VK, Bearson BL. 2013. Hha controls Escherichia coli O157:H7 biofilm formation by differential regulation of global transcriptional regulators FlhDC and CsgD. Appl Environ Microbiol 79:2384–2396. doi: 10.1128/AEM.02998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gonzalez Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J Bacteriol 188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lehti TA, Bauchart P, Dobrindt U, Korhonen TK, Westerlund-Wikstrom B. 2012. The fimbriae activator MatA switches off motility in Escherichia coli by repression of the flagellar master operon flhDC. Microbiology 158:1444–1455. doi: 10.1099/mic.0.056499-0. [DOI] [PubMed] [Google Scholar]
- 76.Kurabayashi K, Agata T, Asano H, Tomita H, Hirakawa H. 2016. Fur represses adhesion to, invasion of, and intracellular bacterial community formation within bladder epithelial cells and motility in uropathogenic Escherichia coli. Infect Immun 84:3220–3231. doi: 10.1128/IAI.00369-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mizushima T, Koyanagi R, Katayama T, Miki T, Sekimizu K. 1997. Decrease in expression of the master operon of flagellin synthesis in a dnaA46 mutant of Escherichia coli. Biol Pharm Bull 20:327–331. doi: 10.1248/bpb.20.327. [DOI] [PubMed] [Google Scholar]
- 78.Shi W, Zhou Y, Wild J, Adler J, Gross CA. 1992. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol 174:6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Lay N, Gottesman S. 2012. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol 86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Partridge JD, Bodenmiller DM, Humphrys MS, Spiro S. 2009. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol Microbiol 73:680–694. doi: 10.1111/j.1365-2958.2009.06799.x. [DOI] [PubMed] [Google Scholar]
- 82.De la Cruz MA, Morgan JK, Ares MA, Yanez-Santos JA, Riordan JT, Giron JA. 2016. The two-component system CpxRA negatively regulates the locus of enterocyte effacement of enterohemorrhagic Escherichia coli involving sigma(32) and Lon protease. Front Cell Infect Microbiol 6:11. doi: 10.3389/fcimb.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weatherspoon-Griffin N, Yang D, Kong W, Hua Z, Shi Y. 2014. The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J Biol Chem 289:32571–32582. doi: 10.1074/jbc.M114.565762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Wulf P, Kwon O, Lin EC. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J Bacteriol 181:6772–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol 187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parkinson JS, Hazelbauer GL, Falke JJ. 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gegner JA, Graham DR, Roth AF, Dahlquist FW. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975–982. doi: 10.1016/0092-8674(92)90247-A. [DOI] [PubMed] [Google Scholar]
- 89.Conley MP, Wolfe AJ, Blair DF, Berg HC. 1989. Both CheA and CheW are required for reconstitution of chemotactic signaling in Escherichia coli. J Bacteriol 171:5190–5193. doi: 10.1128/jb.171.9.5190-5193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bourret RB, Davagnino J, Simon MI. 1993. The carboxy-terminal portion of the CheA kinase mediates regulation of autophosphorylation by transducer and CheW. J Bacteriol 175:2097–2101. doi: 10.1128/jb.175.7.2097-2101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McNally DF, Matsumura P. 1991. Bacterial chemotaxis signaling complexes: formation of a CheA/CheW complex enhances autophosphorylation and affinity for CheY. Proc Natl Acad Sci U S A 88:6269–6273. doi: 10.1073/pnas.88.14.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Volz K, Matsumura P. 1991. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem 266:15511–15519. [DOI] [PubMed] [Google Scholar]
- 93.Halkides CJ, McEvoy MM, Casper E, Matsumura P, Volz K, Dahlquist FW. 2000. The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY. Biochemistry 39:5280–5286. doi: 10.1021/bi9925524. [DOI] [PubMed] [Google Scholar]
- 94.Welch M, Chinardet N, Mourey L, Birck C, Samama JP. 1998. Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nat Struct Biol 5:25–29. doi: 10.1038/nsb0198-25. [DOI] [PubMed] [Google Scholar]
- 95.Mo G, Zhou H, Kawamura T, Dahlquist FW. 2012. Solution structure of a complex of the histidine autokinase CheA with its substrate CheY. Biochemistry 51:3786–3798. doi: 10.1021/bi300147m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee SY, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, Huang L, Kustu S, Berry EA, Wemmer DE. 2001. Crystal structure of an activated response regulator bound to its target. Nat Struct Biol 8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- 97.Bren A, Eisenbach M. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol 278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 98.Welch M, Oosawa K, Aizawa S, Eisenbach M. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci U S A 90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barak R, Eisenbach M. 1992. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry 31:1821–1826. doi: 10.1021/bi00121a034. [DOI] [PubMed] [Google Scholar]
- 100.Sagi Y, Khan S, Eisenbach M. 2003. Binding of the chemotaxis response regulator CheY to the isolated, intact switch complex of the bacterial flagellar motor: lack of cooperativity. J Biol Chem 278:25867–25871. doi: 10.1074/jbc.M303201200. [DOI] [PubMed] [Google Scholar]
- 101.Marykwas DL, Schmidt SA, Berg HC. 1996. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol 256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 102.Donato GM, Kawula TH. 1998. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J Biol Chem 273:24030–24036. doi: 10.1074/jbc.273.37.24030. [DOI] [PubMed] [Google Scholar]
- 103.Kim EA, Blair DF. 2015. Function of the histone-like protein H-NS in motility of Escherichia coli: multiple regulatory roles rather than direct action at the flagellar motor. J Bacteriol 197:3110–3120. doi: 10.1128/JB.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown TD, Jones-Mortimer MC, Kornberg HL. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol 102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 105.Holms WH. 1986. The central metabolic pathways of Escherichia coli: relationship between flux and control at a branch point, efficiency of conversion to biomass, and excretion of acetate. Curr Top Cell Regul 28:69–105. doi: 10.1016/B978-0-12-152828-7.50004-4. [DOI] [PubMed] [Google Scholar]
- 106.McCleary WR, Stock JB, Ninfa AJ. 1993. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol 175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol 189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lima BP, Lennon CW, Ross W, Gourse RL, Wolfe AJ. 2016. In vitro evidence that RNA polymerase acetylation and acetyl phosphate-dependent CpxR phosphorylation affect cpxP transcription regulation. FEMS Microbiol Lett 363:fnw011. doi: 10.1093/femsle/fnw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolfe AJ, Chang DE, Walker JD, Seitz-Partridge JE, Vidaurri MD, Lange CF, Prüß BM, Henk MC, Larkin JC, Conway T. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol Microbiol 48:977–988. doi: 10.1046/j.1365-2958.2003.03457.x. [DOI] [PubMed] [Google Scholar]
- 110.Prüß BM, Matsumura P. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J Bacteriol 178:668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Monds RD, O'Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol 17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 112.Hu LI, Chi BK, Kuhn ML, Filippova EV, Walker-Peddakotla AJ, Basell K, Becher D, Anderson WF, Antelmann H, Wolfe AJ. 2013. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J Bacteriol 195:4174–4186. doi: 10.1128/JB.00383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Castano-Cerezo S, Bernal V, Post H, Fuhrer T, Cappadona S, Sanchez-Diaz NC, Sauer U, Heck AJ, Altelaar AF, Canovas M. 2014. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol Syst Biol 10:762. doi: 10.15252/msb.20145227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yan J, Barak R, Liarzi O, Shainskaya A, Eisenbach M. 2008. In vivo acetylation of CheY, a response regulator in chemotaxis of Escherichia coli. J Mol Biol 376:1260–1271. doi: 10.1016/j.jmb.2007.12.070. [DOI] [PubMed] [Google Scholar]
- 115.Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G, Lima BP, Scholle M, Mrksich M, Anderson WF, Gibson BW, Schilling B, Wolfe AJ. 2014. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9:e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, Walker-Peddakotla A, Sorensen DJ, Zemaitaitis B, Gibson BW, Wolfe AJ. 2015. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol Microbiol 98:847–863. doi: 10.1111/mmi.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fraiberg M, Afanzar O, Cassidy CK, Gabashvili A, Schulten K, Levin Y, Eisenbach M. 2015. CheY's acetylation sites responsible for generating clockwise flagellar rotation in Escherichia coli. Mol Microbiol 95:231–244. doi: 10.1111/mmi.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wolfe AJ. 2016. Bacterial protein acetylation: new discoveries unanswered questions. Curr Genet 62:335–341. doi: 10.1007/s00294-015-0552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 120.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 121.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Romling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 126.Zhang Z, Saier MH Jr. 2011. Transposon-mediated adaptive and directed mutations and their potential evolutionary benefits. J Mol Microbiol Biotechnol 21:59–70. doi: 10.1159/000333108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Barker CS, Prüß BM, Matsumura P. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J Bacteriol 186:7529–7537. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang X, Wood TK. 2011. IS5 inserts upstream of the master motility operon flhDC in a quasi-Lamarckian way. ISME J 5:1517–1525. doi: 10.1038/ismej.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fahrner KA, Berg HC. 2015. Mutations that stimulate flhDC expression in Escherichia coli K-12. J Bacteriol 197:3087–3096. doi: 10.1128/JB.00455-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Horne SM, Sayler J, Scarberry N, Schroeder M, Lynnes T, Prüß BM. 2016. Spontaneous mutations in the flhD operon generate motility heterogeneity in Escherichia coli biofilm. BMC Microbiol 16:262. doi: 10.1186/s12866-016-0878-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Samanta P. 2014. Gene expression and evolution in Escherichia coli biofilm. PhD dissertation. North Dakota State University, Fargo, ND. [Google Scholar]
- 132.Erickson KE, Otoupal PB, Chatterjee A. 2017. Transcriptome-level signatures in gene expression and gene expression variability during bacterial adaptive evolution. mSphere 2:e00009-17. doi: 10.1128/mSphere.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Saini S, Brown JD, Aldridge PD, Rao CV. 2008. FliZ is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J Bacteriol 190:4979–4988. doi: 10.1128/JB.01996-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yamamoto S, Kutsukake K. 2006. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar typhimurium. J Bacteriol 188:6703–6708. doi: 10.1128/JB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wada T, Hatamoto Y, Kutsukake K. 2012. Functional and expressional analyses of the anti-FlhD4C2 factor gene ydiV in Escherichia coli. Microbiology 158:1533–1542. doi: 10.1099/mic.0.056036-0. [DOI] [PubMed] [Google Scholar]
- 136.Stewart MK, Cookson BT. 2014. Mutually repressing repressor functions and multi-layered cellular heterogeneity regulate the bistable Salmonella fliC census. Mol Microbiol 94:1272–1284. doi: 10.1111/mmi.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koirala S, Mears P, Sim M, Golding I, Chemla YR, Aldridge PD, Rao CV. 2014. A nutrient-tunable bistable switch controls motility in Salmonella enterica serovar Typhimurium. mBio 5:e01611-14. doi: 10.1128/mBio.01611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stewart MK, Cookson BT. 2012. Non-genetic diversity shapes infectious capacity and host resistance. Trends Microbiol 20:461–466. doi: 10.1016/j.tim.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Givaudan A, Lanois A. 28 December 2016. Flagellar regulation and virulence in the entomopathogenic bacteria—Xenorhabdus nematophila and Photorhabdus luminescens. Curr Top Microbiol Immunol doi: 10.1007/82_2016_53. [DOI] [PubMed] [Google Scholar]
- 140.Woo TE, Duong J, Jervis NM, Rabin HR, Parkins MD, Storey DG. 8 November 2016. Virulence adaptations of Pseudomonas aeruginosa isolated from patients with non-cystic fibrosis bronchiectasis. Microbiology doi: 10.1099/mic.0.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rodriguez-Torres MD, Islas-Robles A, Gomez-Lunar Z, Delaye L, Hernandez-Gonzalez I, Souza V, Travisano M, Olmedo-Alvarez G. 2017. Phenotypic microdiversity and phylogenetic signal analysis of traits related to social interaction in Bacillus spp. from sediment communities. Front Microbiol 8:29. doi: 10.3389/fmicb.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Prüß BM, Besemann C, Denton A, Wolfe AJ. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J Bacteriol 188:3731–3739. doi: 10.1128/JB.01780-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Prüß BM, Liu J, Higgs PI, Thompson LK. 2015. Lessons in fundamental mechanisms and diverse adaptations from the 2015 Bacterial Locomotion and Signal Transduction Meeting. J Bacteriol 197:3028–3040. doi: 10.1128/JB.00384-15. [DOI] [PMC free article] [PubMed] [Google Scholar]