ABSTRACT
Two-component signal transduction systems (TCSs), typically composed of a sensor histidine kinase (HK) and a response regulator (RR), are the primary mechanism by which pathogenic bacteria sense and respond to extracellular signals. The pathogenic bacterium Vibrio cholerae is no exception and harbors 52 RR genes. Using in-frame deletion mutants of each RR gene, we performed a systematic analysis of their role in V. cholerae biofilm formation. We determined that 7 RRs impacted the expression of an essential biofilm gene and found that the recently characterized RR, VxrB, regulates the expression of key structural and regulatory biofilm genes in V. cholerae. vxrB is part of a 5-gene operon, which contains the cognate HK vxrA and three genes of unknown function. Strains carrying ΔvxrA and ΔvxrB mutations are deficient in biofilm formation, while the ΔvxrC mutation enhances biofilm formation. The overexpression of VxrB led to a decrease in motility. We also observed a small but reproducible effect of the absence of VxrB on the levels of cyclic di-GMP (c-di-GMP). Our work reveals a new function for the Vxr TCS as a regulator of biofilm formation and suggests that this regulation may act through key biofilm regulators and the modulation of cellular c-di-GMP levels.
IMPORTANCE Biofilms play an important role in the Vibrio cholerae life cycle, providing protection from environmental stresses and contributing to the transmission of V. cholerae to the human host. V. cholerae can utilize two-component systems (TCS), composed of a histidine kinase (HK) and a response regulator (RR), to regulate biofilm formation in response to external cues. We performed a systematic analysis of V. cholerae RRs and identified a new regulator of biofilm formation, VxrB. We demonstrated that the VxrAB TCS is essential for robust biofilm formation and that this system may regulate biofilm formation via its regulation of key biofilm regulators and cyclic di-GMP levels. This research furthers our understanding of the role that TCSs play in the regulation of V. cholerae biofilm formation.
KEYWORDS: Vibrio cholerae, VxrAB, biofilms, c-di-GMP, motility
INTRODUCTION
Vibrio cholerae is the causative agent of the gastrointestinal disease cholera, responsible for approximately 3 to 5 million cases of severe diarrhea and 120,000 deaths annually (1, 2). A resident of aquatic reservoirs, V. cholerae can be found as free-swimming planktonic cells or in matrix-protected cellular aggregates, known as biofilms (2–4). Evidence suggests that biofilms form during the aquatic and intestinal phases of the V. cholerae life cycle and play an important role in environmental survival, as well as in the intestinal and transmission stages of infection (5–9). V. cholerae biofilm formation requires the production and secretion of an extracellular matrix composed of matrix proteins, nucleic acids, and Vibrio polysaccharide (VPS), a glycoconjugate that is essential for the formation of three-dimensional biofilm structures (10–16). A complex regulatory network governs this process, tightly controlling V. cholerae biofilm production. While important biofilm regulators and their genetic interactions have been examined, relatively little is known about how environmental signals are integrated into the biofilm regulatory network (8, 17–19).
Like most pathogenic bacteria, V. cholerae utilizes two-component signal transduction systems (TCSs) as a means for sensing and responding to different environment stimuli, such as nutrient availability, pH, oxygen, osmolarity, quorum sensing signals, and numerous host factors (19–23). The genome of V. cholerae is predicted to encode 43 histidine kinases (HKs) and 49 response regulators (RRs), according to the reference genome of O1 EL Tor strain N16961 (http://www.ncbi.nlm.nih.gov/Complete_Genomes/RRcensus.html and http://www.p2cs.org). An additional 3 RRs (VpsT, VpsR, and VC0396) were identified based on an analysis of the genome for REC domains; thus, it is predicted that the V. cholerae O1 EL Tor N16961 genome encodes 53 putative RRs. Only 9 RRs have been previously shown to impact biofilm formation. VpsR, VpsT, and LuxO are activators of biofilm formation, while PhoB, VarA, VieA, and CarR are repressors of biofilm formation (22, 24–31). VC1348 and VCA0210 are RRs that contain HD-GYP domains with predicted cyclic di-GMP (c-di-GMP) phosphodiesterase activity. Overexpression of these RRs led to a significant decrease in biofilm formation (32). To date, there is no systematic analysis reporting on the contribution of each TCS to biofilm formation in Vibrio cholerae.
Here, we report our results for the analysis of vpsL expression, as a representative biofilm gene, in strains with in-frame deletions of each RR. We found that VxrB, an RR that we recently characterized for its roles in intestinal colonization and in regulation of the type 6 secretion system (T6SS) (33), was also a positive regulator of biofilm formation.
RESULTS
The VxrB RR is a newly identified positive regulator of vps expression in V. cholerae.
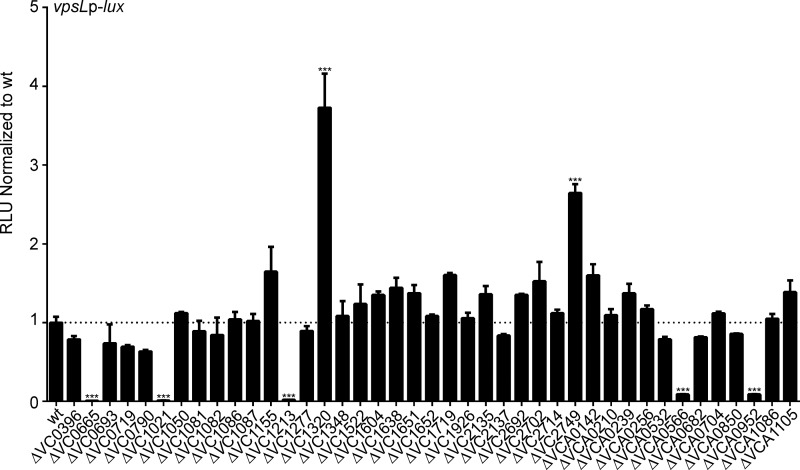
To assess the role of V. cholerae TCSs in biofilm formation, we utilized an in-frame deletion mutant library of 41 response regulators (RR). In this study, the 11 RRs that were predicted to be involved in chemotaxis (CheY, CheV, and CheB proteins) and VC2368 (which we were unable to generate an in-frame deletion for) were not included (34, 35). VPS is required for biofilm formation, and vps transcription is a useful readout of potential biofilm forming capacity. Therefore, we analyzed the expression of vpsL, the first gene in the vps-II cluster, which, along with the vps-I cluster genes, encodes components that are required for VPS production and biofilm formation. We used a transcriptional fusion of the regulatory region of vpsL and the luciferase transcriptional reporter luxCADBE (PvpsL-lux). Our results revealed 7 RR-null mutants with significant changes in vpsL expression compared with that in the wild type (Fig. 1). Consistent with previous studies, we observed a 122-fold decrease in vpsL expression in the ΔvpsR (VC0665) strain, an 81-fold decrease in vpsL expression in the ΔluxO (VC1021) strain, an 11-fold decrease in vpsL expression in the ΔvpsT (VCA0952) strain, and a 4-fold increase in vpsL expression in the ΔcarR (VC1320) strain (24, 27, 28). A 3-fold increase in vpsL expression was observed in the ΔntrC (VC2749) strain, indicating that this RR may be a repressor of biofilm formation. Additionally, we observed a 47-fold decrease in vpsL expression in a ΔvarA (VC1213) mutant. Furthermore, we found that the ΔvxrB (VCA0566) strain had an 11-fold decrease in vpsL expression, indicating that this RR may be a positive regulator of biofilm formation (Fig. 1). Since VxrB was not previously reported to be a regulator of biofilm, we focused this work on characterizing how this TCS influences biofilm formation.
FIG 1.
Analysis of biofilm gene expression in the wild type and in response regulator (RR) deletion mutants. A transcriptional reporter harboring the regulatory region of vpsL upstream of a promoterless lux reporter (PvpsL-lux) was used to analyze the expression of biofilm genes in 41 ΔRR mutants, including ΔVC0396 (qstR), ΔVC0665 (vpsR), ΔVC0693, ΔVC0719 (phoB), ΔVC0790, ΔVC1021 (luxO), ΔVC1050, ΔVC1081, ΔVC1082, ΔVC1086, ΔVC1087, ΔVC1155, ΔVC1213 (varA), ΔVC1277, ΔVC1320 (carR), ΔVC1348, ΔVC1522, ΔVC1604, ΔVC1638, ΔVC1651 (vieB), ΔVC1652 (vieA), ΔVC1719 (torR), ΔVC1926 (dctD1), ΔVC2135 (flrC), ΔVC2137 (flrA), ΔVC2692 (cpxR), ΔVC2702 (cbrR), ΔVC2714 (ompR), ΔVC2749 (ntrC), ΔVCA0142 (dctD2), ΔVCA0210, ΔVCA0239, ΔVCA0256, ΔVCA0532, ΔVCA0566 (vxrB), ΔVCA0682 (uhpA), ΔVCA0704 (pgtA), ΔVCA0850, ΔVCA0952 (vpsT), ΔVCA1086, and ΔVCA1105 strains (names included in parentheticals where relevant). Cultures of the wild type and ΔRR mutants were grown to exponential phase (OD600 of ∼0.3) and luminescence was measured. The graph represents the averages and standard deviations of relative light units (RLU) obtained from at least three technical replicates from two biological replicates, normalized to wild-type levels. RLU are reported in luminescence counts · min−1 · ml−1 · OD600−1. These values were then normalized to the wild-type RLU, and data are shown as fold changes above or below a wild-type value of 1. Data were analyzed using a one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. ***, P < 0.001.
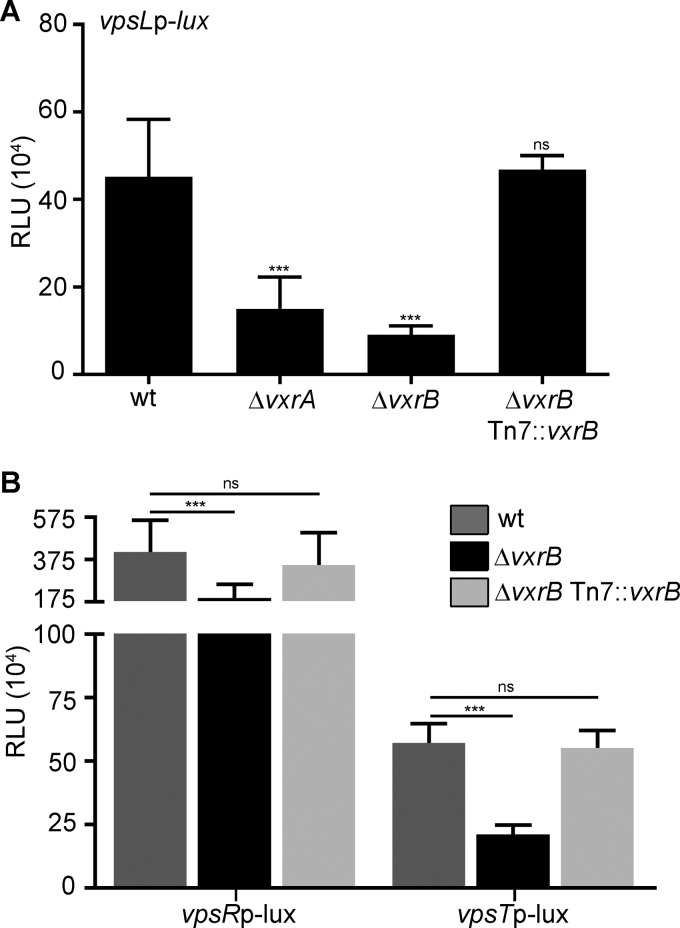
We recently determined that VxrB, along with its cognate sensor histidine kinase (HK), VxrA, regulates the type VI secretion system (T6SS) and virulence (33). Another recent study additionally showed that the VxrAB TCS plays a role in cell wall homeostasis in response to antibiotic treatment (in that study, the authors renamed VxrAB as WigKR) (36). However, this is the first report of its role in the regulation of biofilm-gene expression. To further characterize the role of the VxrAB TCS in vpsL regulation, we analyzed vpsL expression in the ΔvxrA strain. We observed that, similar to that with ΔvxrB deletion, a ΔvxrA mutant downregulates expression of vpsL by 3-fold. A ΔvxrB strain harboring vxrB under the control of its own promoter (PvxrA) in the Tn7 site was complemented for vpsL expression (Fig. 2A). These results demonstrate that the VxrAB TCS positively regulates vpsL gene expression.
FIG 2.
Analysis of biofilm gene expression in the wild type and in ΔvxrA and ΔvxrB mutants. (A) Cultures of wild-type, ΔvxrA, ΔvxrB, and ΔvxrB Tn7::vxrB strains containing PvpsL-lux were grown to exponential phase (OD600 of ∼0.3) and luminescence was measured. The graph represents the averages and standard deviations of RLU obtained from three technical replicates from three biological replicates. RLU are reported in luminescence counts · min−1 · ml−1 · OD600−1. Data were analyzed using a one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. ***, P < 0.001; ns, not significant. (B) Cultures of wild-type and ΔvxrB strains containing PvpsR-lux or PvpsT-lux were grown to exponential phase (OD600 of∼0.3) and luminescence was measured. The graph represents the averages and standard deviations of RLU obtained from three technical replicates from three biological replicates. RLU are reported in luminescence counts · min−1 · ml−1 · OD600−1. Data were analyzed using two-tailed Student's t tests. ***, P < 0.0001.
VxrB acts upstream of the major biofilm regulators, VpsR and VpsT.
The two major positive regulators of vps genes are VpsR and VpsT. To determine if VxrB affects the regulators of vps gene expression, we measured the expression of each regulator in the ΔvxrB strain using the transcriptional fusions PvpsR-lux and PvpsT-lux. Expression of vpsL, vpsR, and vpsT was decreased by 4-fold, 2.5-fold, and 3-fold, respectively, in the ΔvxrB strain compared with that in the wild type, suggesting that VxrB could regulate vpsL expression by activating the expression of vpsR and vpsT (Fig. 2B). A ΔvxrB strain harboring vxrB under the control of its own promoter (PvxrA) in the Tn7 site was complemented for vpsR and vpsT expression (Fig. 2B).
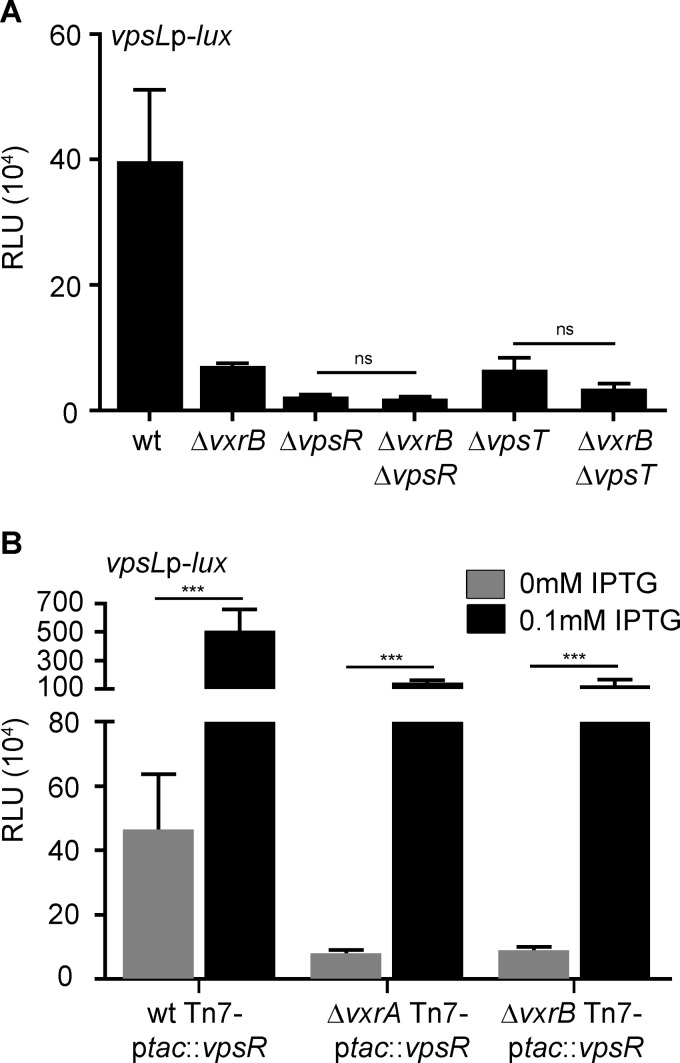
We performed an epistasis analysis with vpsR, vpsT and vxrB. Our results revealed that the deletion of vxrB in the ΔvpsR or ΔvpsT strain had no significant effect on vpsL expression compared with that in the single mutants (ΔvpsR and ΔvpsT strains) (Fig. 3A). These data suggest that VpsR and VpsT function downstream of VxrB for vpsL expression. Given that VpsR is at the bottom of the vps regulatory cascade, we reasoned that VxrB-dependent regulation of vpsL might ultimately be due to increased expression of vpsR. To test this, we expressed vpsR (inserted in the Tn7 site) from an isopropyl-β-d-1-thiogalactopyranoside (IPTG) inducible promoter and analyzed vpsL expression in wild-type (WT), ΔvxrA, and ΔvxrB strains. We observed that vpsL expression was increased by 11-fold when vpsR was overexpressed in the wild type. The overexpression of vpsR in the ΔvxrA or ΔvxrB strain also resulted in increased vpsL expression, although ∼5-fold less than in the wild type. Taken together, these results suggest that VxrB can regulate vpsL expression independently of VpsR.
FIG 3.
Epistasis analysis of regulators of vps gene expression. (A) Cultures of wild-type, ΔvxrB, ΔvpsR, ΔvxrB ΔvpsR, ΔvpsT, and ΔvxrB ΔvpsT strains containing PvpsL-lux were grown to exponential phase (OD600 of ∼0.3) and luminescence was measured. The graph represents the averages and standard deviations of RLU obtained from three technical replicates from three biological replicates. RLU are reported in luminescence counts · min−1 · ml−1 · OD600−1. Data were analyzed using a one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. ns, not significant. (B) Cultures of wild-type, ΔvxrA, and ΔvxrB strains containing PvpsL-lux and an IPTG-inducible copy of VpsR in the Tn7 site were grown to exponential phase (OD600 of ∼0.3) and luminescence was measured. The graph represents the averages and standard deviations of RLU obtained from three technical replicates from three biological replicates. RLU are reported in luminescence counts · min−1 · ml−1 · OD600−1. Data were analyzed using two-tailed Student's t tests. ***, P < 0.0001.
Members of the vxr operon affect vps expression and biofilm formation.
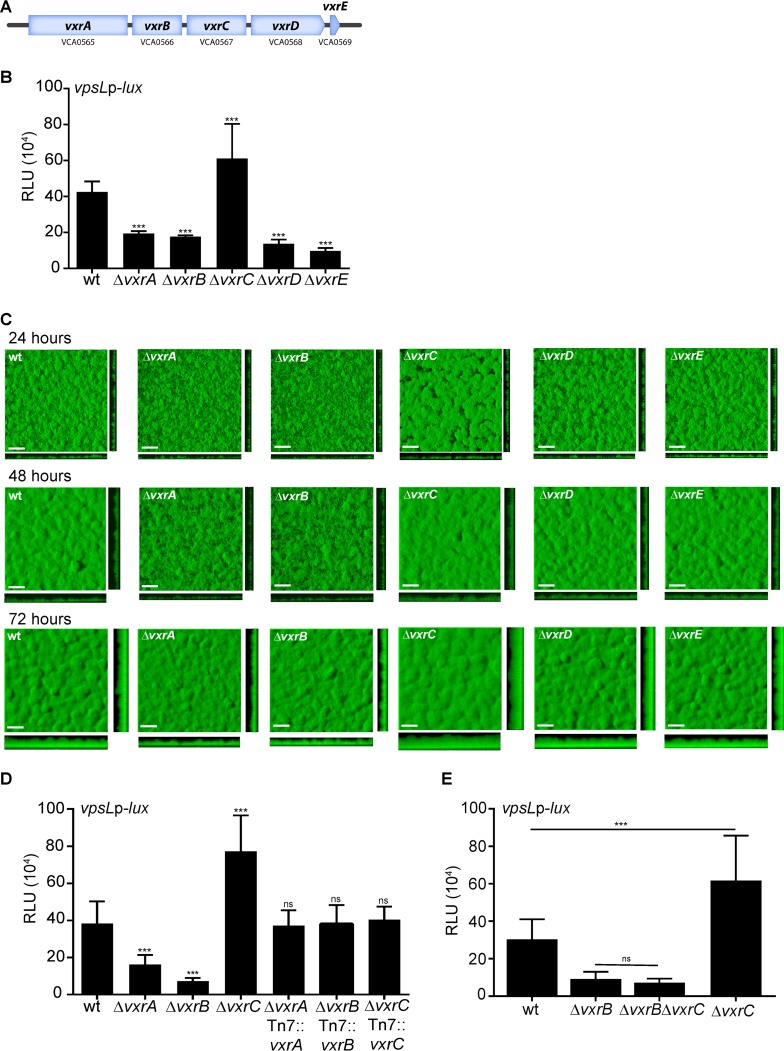
The VxrAB TCS is encoded as part of a 5-gene operon, composed of the HK vxrA, the RR vxrB, and three genes of unknown function, vxrC, vxrD, and vxrE (Fig. 4A). We previously demonstrated that the vxrABCDE genes are cotranscribed (33). To characterize the role of vxr genes in biofilm formation, we analyzed vpsL expression normalized to total protein in mutants lacking vxr operon genes (Fig. 4B). For this study, we analyzed PvpsL-lux in biofilm-grown cells. As demonstrated above, the ΔvxrA and ΔvxrB strains both had decreased vpsL expression compared with that in the wild type. The ΔvxrC mutant showed a 1.5-fold increase in vpsL expression compared with the wild type, while the ΔvxrD and ΔvxrE strains showed a 3-fold and 4-fold decrease in vpsL expression, respectively.
FIG 4.
Analysis of biofilm gene expression and biofilm formation in vxr operon mutants. (A) The 5-gene vxr operon structure, containing the TCS vxrAB and three genes of unknown function, vxrCDE. (B) Biofilms of the wild type and the ΔvxrA, ΔvxrB, ΔvxrC, ΔvxrD, and ΔvxrE strains containing PvpsL-lux were grown in flow cells for 24 h, and luminescence was measured from harvested biofilm cells. The graph represents the averages and standard deviations of RLU obtained from three technical replicates from three independent biological samples. RLU are reported in luminescence counts · min−1 · ml−1 · μg−1 total protein. Data were analyzed using a one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. ***, P < 0.001. (C) Top and orthogonal views of biofilms formed in flow cells by wild-type, ΔvxrA, ΔvxrB, ΔvxrC, ΔvxrD, and ΔvxrE strains 72 h after inoculation. Bars, 30 μm. (D) Cultures of wild-type, ΔvxrA, ΔvxrB, ΔvxrC, ΔvxrA Tn7::vxrA, ΔvxrB Tn7::vxrB, and ΔvxrC Tn7::vxrC strains containing PvpsL-lux were grown to exponential phase (OD600 of ∼0.3) and luminescence was measured. The graph represents the averages and standard deviations of RLU obtained from three technical replicates from three independent biological replicates. RLU are reported in luminescence counts · min−1 · ml−1 · OD600−1. Data were analyzed using a one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. ***, P < 0.001; ns, not significant. (E) Cultures of wild-type, ΔvxrB, ΔvxrC, and ΔvxrB ΔvxrC strains containing PvpsL-lux were grown to exponential phase (OD600 of ∼0.3) and luminescence was measured. The graph represents the averages and standard deviations of RLU obtained from three technical replicates from three independent biological replicates. RLU are reported in luminescence counts · min−1 · ml−1 · OD600−1. Data were analyzed using oa ne-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. ***, P < 0.001.
We analyzed the biofilm formation of isogenic strains expressing green fluorescent protein (GFP) from the chromosome. These experiments were done in flow cells, and the biofilms were imaged using confocal laser scanning microscopy at 24, 48, and 72 h postinoculation (Fig. 4C). We used the quantitative analysis software COMSTAT (37) to calculate biomass, the average and maximum thickness, substratum coverage, and roughness of the biofilms at 72 h, when the differences in biofilm formation were most defined. The ΔvxrA and ΔvxrB strains formed visibly thinner biofilms compared with that of the wild type, with 1.5- to 1.6-fold decreases in biomass and average and maximum thickness compared with that of the wild type (Table 1). By contrast, the ΔvxrC strain showed significantly increased biofilm formation compared with that of the wild type, and biomass and average thickness were both increased by 1.4-fold, while the maximum thickness increased by 1.3-fold (Table 1). The ΔvxrD and ΔvxrE strains formed biofilms similar to that of the wild type (Fig. 4B and C; Table 1). Since the ΔvxrA, ΔvxrB, and ΔvxrC strains formed biofilms that were significantly different from that of the wild type, we generated ΔvxrA, ΔvxrB, and ΔvxrC strains harboring vxrA, vxrB, and vxrC, respectively, under the control of their own promoters (PvxrA) in the Tn7 site and found that in these strains, vpsL expression was similar to that in the wild type (Fig. 4D). We next analyzed the genetic interaction between vxrC and vxrB with respect to the regulation of biofilm formation. The ΔvxrB and ΔvxrC strains showed decreased and increased vpsL expression, respectively, compared to that in the wild type. The ΔvxrB ΔvxrC strain phenocopied the vpsL expression level of the ΔvxrB strain, indicating that the ΔvxrB mutation is epistatic to the ΔvxrC mutation (Fig. 4E). Altogether, our results show that VxrA and VxrB both positively regulate vpsL expression and biofilm formation, while VxrC may act as a repressor of biofilm formation. The roles of VxrD and VxrE in regulating biofilm formation are unclear, as both the ΔvxrD and ΔvxrE strains showed decreased vpsL expression but formed biofilms similar to that of the wild type.
TABLE 1.
COMSTAT analysis of biofilms of wild type, ΔvxrA, ΔvxrB, ΔvxrC, ΔvxrD, and ΔvxrE strainsa
| Time postinoculation and strain | Biomass (μm3/μm2 [SD]) | Mean thickness (μm [SD]) |
Substrate coverageb (SD) | Roughness coefficient (SD) | |
|---|---|---|---|---|---|
| Avg | Maximum | ||||
| 24 h | |||||
| Wild type | 7.77 (0.70) | 8.53 (0.99) | 14.00 (1.30) | 0.90 (0.05) | 0.23 (0.04) |
| ΔvxrA mutant | 6.00 (0.79)d | 6.38 (1.03) | 12.67 (1.74) | 0.87 (0.09) | 0.28 (0.07) |
| ΔvxrB mutant | 5.94 (0.73)d | 6.25 (0.86)c | 13.41 (1.30) | 0.92 (0.05) | 0.30 (0.08) |
| ΔvxrC mutant | 8.73 (0.63) | 9.43 (2.23) | 14.44 (2.14) | 0.91 (0.07) | 0.23 (0.02) |
| ΔvxrD mutant | 6.66 (0.84) | 6.40 (0.94) | 11.79 (1.55) | 0.87 (0.19) | 0.33 (0.16) |
| ΔvxrE mutant | 7.087 (0.85) | 7.88 (1.68) | 13.26 (1.48) | 0.87 (0.08) | 0.25 (0.06) |
| 48 h | |||||
| Wild type | 10.94 (0.27) | 12.12 (0.88) | 22.84 (2.20) | 0.91 (0.10) | 0.14 (0.03) |
| ΔvxrA mutant | 7.49 (1.02)e | 8.31 (1.38)c | 17.39 (2.89) | 0.87 (0.21) | 0.18 (0.05) |
| ΔvxrB mutant | 8.35 (1.16)d | 9.59 (1.59)c | 20.18 (3.18) | 0.91 (0.13) | 0.20 (0.06) |
| ΔvxrC mutant | 12.79 (2.35) | 13.86 (2.94) | 28.73 (7.43) | 0.98 (0.03) | 0.13 (0.04) |
| ΔvxrD mutant | 11.06 (1.03) | 12.49 (0.83) | 22.10 (4.15) | 0.93 (0.05) | 0.15 (0.03) |
| ΔvxrE mutant | 9.76 (1.30) | 10.99 (0.59) | 21.66 (3.78) | 0.96 (0.06) | 0.17 (0.03) |
| 72 h | |||||
| Wild type | 19.42 (1.49) | 20.29 (1.25) | 31.09 (2.20) | 1.00 (0.00) | 0.14 (0.03) |
| ΔvxrA mutant | 11.58 (2.31)e | 12.36 (2.18)e | 21.22 (3.31)d | 0.91 (0.07)c | 0.19 (0.06) |
| ΔvxrB mutant | 12.79 (2.08)d | 14.61 (1.82)d | 24.75 (2.44) | 0.91 (0.07)c | 0.21 (0.07)c |
| ΔvxrC mutant | 26.34 (3.52)e | 27.82 (3.29)e | 40.07 (5.86)d | 0.98 (0.04) | 0.11 (0.03) |
| ΔvxrD mutant | 18.95 (3.30) | 19.61 (3.74) | 30.35 (6.51) | 0.98 (0.04) | 0.13 (0.03) |
| ΔvxrE mutant | 19.69 (3.15) | 20.75 (4.13) | 31.09 (4.79) | 0.96 (0.07) | 0.13 (0.03) |
Total biomass, the average and maximum thicknesses, substrate coverage, and roughness coefficient were calculated using COMSTAT. The values are the means (standard deviations) of data from at least six z-series image stacks. Significance was determined by ANOVA. Dunnett's multiple comparison test identified samples that differ significantly from biofilms formed by the wild-type strain.
A value of 0 indicates no coverage (equivalent to 0%), while a value of 1 indicates full coverage (equivalent to 100%).
P ≤ 0.05.
P ≤ 0.001.
P ≤ 0.0001.
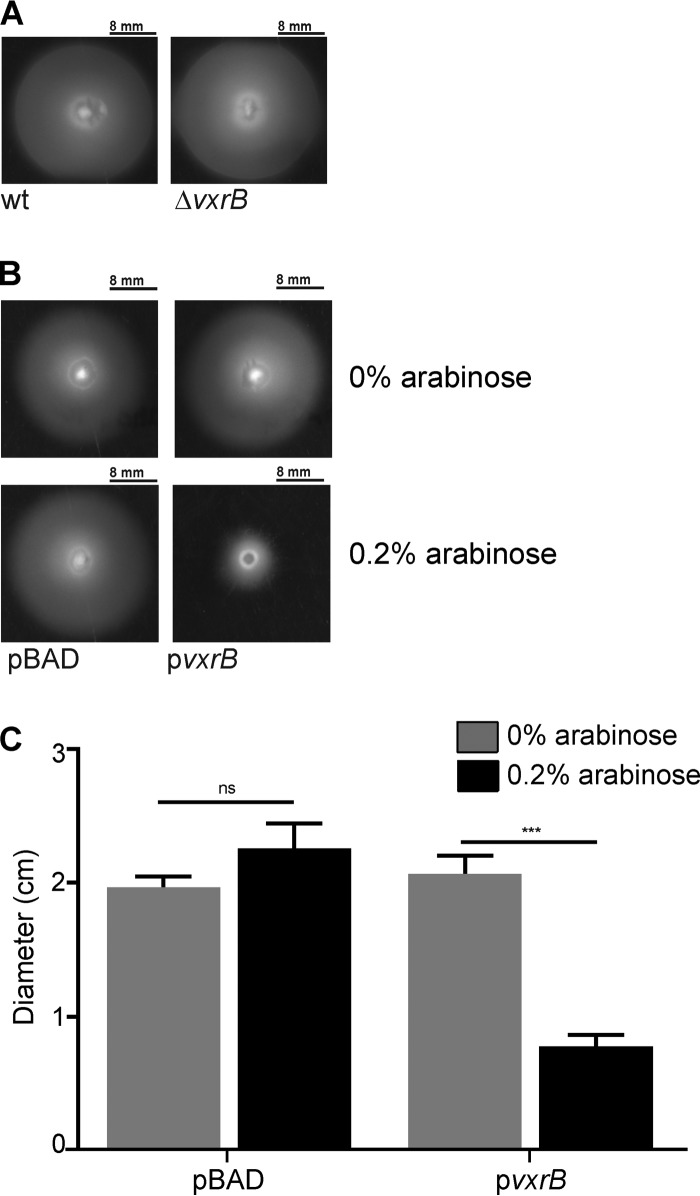
Overexpression of wild-type VxrB negatively impacts motility.
Biofilm formation and motility are inversely regulated in V. cholerae. Therefore, we analyzed the role of VxrB in the regulation of motility. We measured swimming motility of the ΔvxrB strain by performing a motility assay on soft agar plates. We observed no difference in the average migration zone diameters between the ΔvxrB and wild-type strains (Fig. 5A). To determine the effect that vxrB overexpression could have on motility, we cloned this gene in an expression plasmid under the control of an arabinose-inducible promoter (pBAD-vxrB). In the absence of arabinose, the migration zones of the wild type harboring the empty plasmid and the wild type containing pBAD-vxrB were similar (Fig. 5B). In the presence of 0.2% arabinose, the wild type containing pBAD-vxrB showed an ∼50% decrease in migration compared with that of the wild type with the empty plasmid (Fig. 5B and C). These results suggest that overexpression of VxrB can impact motility.
FIG 5.
Analysis of swimming motility in ΔvxrB and VxrB overexpression strains. (A) Migration zones of wild-type and ΔvxrB strains after 16 h of growth on 0.3% agar plates. (B) Migration zones of the wild type expressing empty vector (pBAD) and pBAD-vxrB (pvxrB) after 16 h of growth on 0.3% agar plates containing 0% or 0.2% arabinose. (C) Strains were grown on LB agar plates containing 0.3% agar at 30°C for 16 h before migration zones were measured. The error bars indicate the standard deviations of the results from 6 biological replicates. Data were analyzed using two-tailed Student's t tests. ***, P < 0.0001.
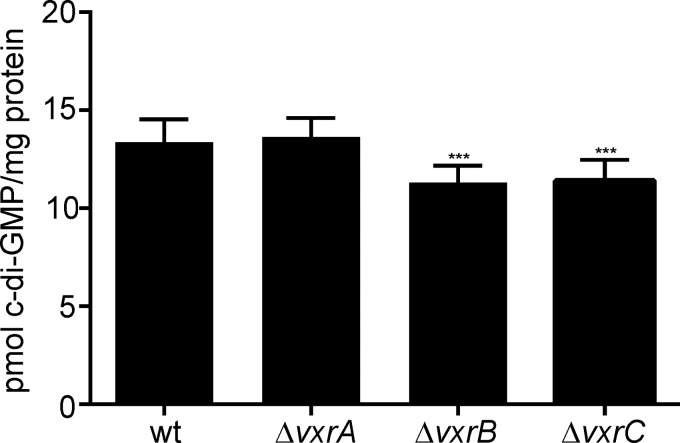
VxrB increases levels of the second messenger signaling molecule c-di-GMP.
Given VxrB's role as a positive regulator of biofilms and a negative regulator of motility, we asked if VxrB could be affecting cellular c-di-GMP levels, as this second messenger signaling molecule inversely regulates biofilms and motility. We measured intracellular c-di-GMP levels in wild-type, ΔvxrA, ΔvxrB, and ΔvxrC strains and found that ΔvxrB and ΔvxrC strains had small but statistically significant decreases in levels of c-di-GMP compared with that in the wild type, while the ΔvxrA strain had c-di-GMP levels similar to that of the wild type (Fig. 6). Thus, cellular c-di-GMP levels are altered in the ΔvxrB and ΔvxrC strains.
FIG 6.
Analysis of c-di-GMP levels in the wild type and in ΔvxrA, ΔvxrB, and ΔvxrC mutants. Strains were grown aerobically to an OD600 of ∼0.3 before c-di-GMP was extracted from whole-cell protein and quantified by high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS). The error bars indicate the standard deviations of the results from 8 biological replicates. Data were analyzed using a one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. ***, P < 0.001.
The T6SS does not impact vpsL expression or biofilm formation.
Our previous work indicated that VxrB positively regulates the T6SS (33). As biofilms provide an environment where cell-to-cell contact is frequent and where high activity of the T6SS might be expected, we next analyzed the role of the T6SS in biofilm formation and determined if VxrB's influence on biofilm formation could act partially through the regulation of, or activation by, the T6SS.
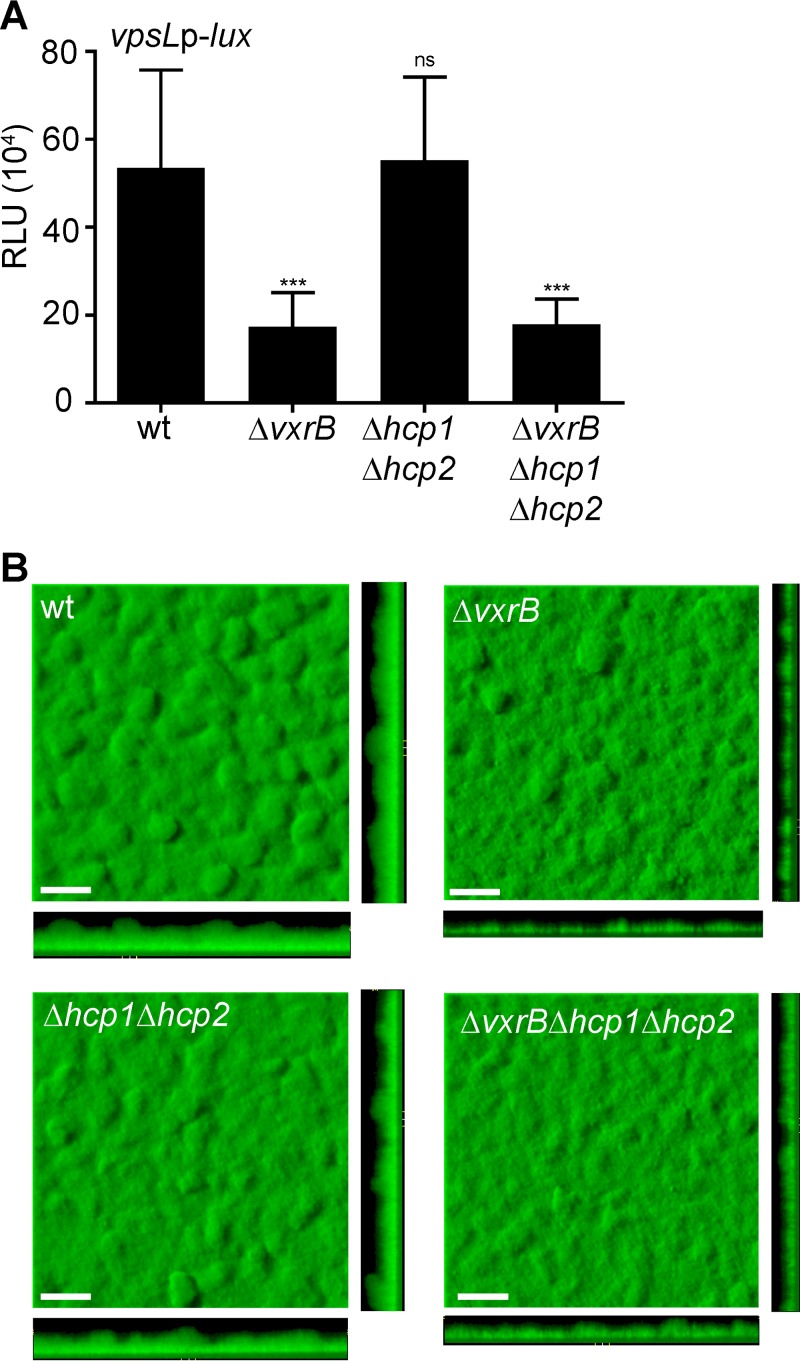
A strain lacking hcp1 and hcp2 (Δhcp1 Δhcp2), the genes required to produce the major T6SS structural component, Hcp, was used to assess the role of the T6SS in biofilm formation (38). We also evaluated vpsL expression in cells grown to exponential phase in wild-type, ΔvxrB, Δhcp1 Δhcp2, and ΔvxrB Δhcp1 Δhcp2 strains (Fig. 7A). As expected, the ΔvxrB strain demonstrated decreased vpsL expression, as did the ΔvxrB Δhcp1 Δhcp2 strain. The Δhcp1 Δhcp2 strain showed no significant difference in vpsL expression compared with that of the wild type. Biofilm formation was also examined by growing strains expressing GFP in flow cells and imaging using confocal laser scanning microscopy at 72 h postinoculation (Fig. 7B). The ΔvxrB and the ΔvxrB Δhcp1 Δhcp2 strains showed decreased biofilm formation, while the Δhcp1 Δhcp2 strain showed no significant difference in biofilm formation compared with that of the wild type. These observations were supported by COMSTAT analysis, which was used to analyze biomass, the average and maximum thickness, substratum coverage, and roughness of biofilms (Table 2). Altogether, these findings indicate that the T6SS does not contribute to the regulation of biofilm formation by VxrB under the conditions we analyzed.
FIG 7.
Analysis of the impact of the type 6 secretion genes on biofilm gene expression and biofilm formation. (A) Cultures of wild-type, ΔvxrB, Δhcp1 Δhcp2, and ΔvxrB Δhcp1 Δhcp2 strains containing PvpsL-lux were grown to exponential phase (OD600 of ∼0.3) and luminescence was measured. The graph represents the averages and standard deviations of RLU obtained from three technical replicates from three independent biological samples. RLU are reported in luminescence counts · min−1 · ml−1 · OD600l−1. Data were analyzed using a one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. ***, P < 0.001; ns, not significant. (B) Top and orthogonal views of biofilms formed in flow cells by wild-type, ΔvxrB, Δhcp1 Δhcp2, and ΔvxrB Δhcp1 Δhcp2 strains after 72 h. Bars, 30 μm.
TABLE 2.
COMSTAT analysis of biofilms formed after 72 h by wild type, ΔvxrB, Δhcp12, and ΔvxrB Δhcp1 Δhcp2 strainsa
| Strain | Biomass (μm3/μm2 [SD]) | Mean thickness (μm [SD]) |
Substrate coverageb (SD) | Roughness coefficient (SD) | |
|---|---|---|---|---|---|
| Avg | Maximum | ||||
| Wild type | 20.41 (2.63) | 22 (3.34) | 33.74 (6.084) | 1.00 (00) | 0.11 (0.02) |
| ΔvxrB mutant | 11.68 (2.34)e | 13.58 (2.12)e | 23.48 (2.76)e | 0.95 (0.05)c | 0.21 (0.06)e |
| Δhcp1 Δhcp2 mutant | 21.77 (0.73) | 22.97 (0.88) | 36.1 (2.65) | 1.00 (00) | 0.11 (0.02) |
| ΔvxrB Δhcp1 Δhcp2 mutant | 18.57 (2.82) | 17.26 (1.68)d | 27.70 (2.832)c | 0.99 (0.02) | 0.14 (0.01) |
Total biomass, the average and maximum thicknesses, substrate coverage, and roughness coefficient were calculated using COMSTAT. The values are the means (standard deviations) of data from at least six z-series image stacks. Significance was determined by ANOVA. Bonferroni's multiple-comparison test identified samples that differ significantly from biofilms formed by the wild-type strain.
A value of 0 indicates no coverage (equivalent to 0%), while a value of 1 indicates full coverage (equivalent to 100%).
P ≤ 0.05.
P ≤ 0.001.
P ≤ 0.0001.
DISCUSSION
V. cholerae biofilms enhance survival in the aquatic environment and facilitate transmission to a human host. This study serves to evaluate the role of TCSs, typically utilized to regulate cellular processes in response to extracellular signals, in biofilm formation. We systematically analyzed the role of V. cholerae RRs in biofilm formation and identified several that play roles in biofilm regulation (Fig. 8). Consistent with previous studies, we observed a decrease in vpsL expression in a ΔvpsR (VC0665) mutant, a ΔluxO (VC1021) mutant, and a ΔvpsT (VCA0952) mutant and an increase in vpsL expression in a ΔcarR (VC1320) mutant (22, 24, 27, 28, 30). We did not observe a phenotype for a ΔphoB (VC0719) mutant, which is only expected to act as repressor of biofilm formation under phosphate-limiting conditions (25, 26). As expected for the El Tor biotype, a ΔvieA (VC1652) mutant did not have altered vpsL levels. In the classical biotype of V. cholerae, VieA negatively regulates biofilms via an EAL domain, independently of its phosphorylation status and DNA-binding activity. The EAL domain functions as a phosphodiesterase (PDE) that degrades the secondary messenger c-di-GMP, an important positive regulator of biofilm formation (29). This phenotype is not observed in the El Tor biotype, as the transcription of vieSAB is subject to H-NS silencing (39, 40). We also did not observe a phenotype for the ΔVC1348 or ΔVCA0210 strain; however, this was also consistent with previously published findings, which demonstrated a biofilm phenotype only when these proteins were overexpressed (32).
FIG 8.
Model of all known TCSs that regulate V. cholerae biofilm formation. The RRs VpsR (VC0665) and VpsT (VCA0952) are the major positive regulators of biofilm formation. Their cognate TCS partners have yet to be identified. The PhoB (VC0719-20) TCS acts as a repressor of biofilm formation under phosphate-limited conditions. LuxO (VC1021) acts as an activator of biofilm formation by positively regulating the expression of small regulatory RNAs responsible for repressing translation of hapR, the master quorum sensing regulator and the major negative regulator of biofilm formation. VarSA (VC2453/VC1213) represses biofilm formation by interfering with the LuxO-mediated activation of Qrr sRNAs through its activation of the inhibitory regulatory small RNAs CsrBCD and their subsequent inhibition of the global regulator CsrA. Repression of CsrA reduces Qrr sRNA levels, leading to increased HapR levels and decreased vps gene expression. CarSR (VC1319-20) negatively regulates biofilm formation. VieA (VC1652) represses biofilm formation independently of its cognate histidine kinase via an EAL domain responsible for degrading the secondary messenger c-di-GMP, an important positive regulator of biofilm formation. The NtrBC (VC2748-49) TCS was identified as a potential repressor of biofilms in this study. Although described in the text, VC1348 and VCA0210 are not pictured. These RRs contain HD-GYP domains responsible for degrading c-di-GMP. Although the deletion of these genes does not impact biofilm formation, their overexpression leads to a significant decrease in biofilm formation, indicating that they may act as repressors of biofilm formation. Finally, we have included the newly identified VxrAB (VCA0565-66) TCS, which positively regulates biofilm formation.
Interestingly, we observed a 47-fold decrease in vpsL expression in the ΔvarA (VC1213) strain, which is contradictory to results from previously published work (31). The VarSA TCS is thought to impact biofilm formation through its control of levels of CsrA, a small RNA binding protein that regulates a number of cellular processes. Specifically, VarA is predicted to repress biofilm formation by interfering with the LuxO-mediated activation of Qrr sRNAs through its positive regulation of the inhibitory regulatory small RNAs CsrBCD. These small RNAs sequester CsrA, which in turn reduces Qrr sRNA levels, leading to increased HapR levels and decreased vps gene expression (41–44). However, a recent study demonstrated that the loss of VarA results in excessive levels of active CsrA, which negatively impacts cell growth and leads to single-amino-acid suppressor mutations in CsrA (45). These mutations were shown to alter the regulatory function of CsrA. Our sequence analysis of csrA in the ΔvarA PvpsL-lux strain revealed that this strain had an amino acid substitution of T11P. It was speculated by Mey et al. that the amino acid substitutions clustered in the N-terminal half of CsrA (R6H, R6L, T11P, I14T, and T19P) result in the accumulation of a “less active” form of CsrA (45). Thus, our finding is consistent with the observation that the loss of VarA results in suppressor mutations in csrA and that these mutations can alter CsrA activity, including its positive regulation of biofilm formation. Collectively, these observations explain why we did not observe the expected increase in vpsL expression in our ΔvarA strain. Although varA mutant strains do not produce the CsrA-sequestering sRNAs and have increased levels of the positive biofilm regulator, CsrA, we hypothesize that CsrA's regulatory activity was altered due to the accumulation of a suppressor mutation. Finally, we found an increase in vpsL expression in a ΔntrC (VC2749) mutant, indicating that this RR may be a repressor of biofilm formation.
We focused our studies mainly on a new positive regulator of vps and biofilm formation, VxrB. The observations reported here demonstrate that VxrAB TCS activates biofilm formation and vpsL expression and represses motility. We showed that VxrB positively regulates vpsR and vpsT, the two major regulators of V. cholerae biofilms, and that VxrB regulates vpsL expression upstream of VpsR and VpsT. Finally, we demonstrated that VxrB positively regulates cellular levels of the second messenger signaling molecule c-di-GMP, which acts as a positive regulator of biofilm formation and a negative regulator of motility (46). While we demonstrated that VxrB may act through major biofilm regulators to exert its regulatory control on biofilm formation, further investigation of where VxrB is integrated into the vps regulatory network is necessary. It is possible that VxrB controls the activity of vps genes through its modulation of cellular c-di-GMP levels, as increased c-di-GMP levels have been shown to lead to increased vpsL, vpsR, and vpsT expression (39, 47). Alternatively, VpsR and VpsT may be the direct regulatory targets of VxrB, as both regulators positively regulate diguanylate cyclase (DGC) genes, which are responsible for producing c-di-GMP (48). Given that a VxrA mutant did not demonstrate a decrease in c-di-GMP levels and that a VxrC mutant had a decrease in c-di-GMP levels despite positively regulating biofilm formation, further investigation into the regulatory role of the Vxr TCS on c-di-GMP levels is merited. Determining the direct regulatory targets of VxrB would provide additional insight into our understanding of the role of TCS in the regulation and control of biofilm formation.
Our previous work demonstrated that vxrABCDE are cotranscribed and that vxrCDE have minor but significant roles in intestinal colonization (33). Here, we demonstrated that only the ΔvxrA, ΔvxrB, and ΔvxrC strains produced biofilms that were different from that of the wild type, though ΔvxrD and ΔvxrE strains did demonstrate decreased vpsL expression. It is possible that VxrD and VxrE play minor roles in the activation of vpsL expression, potentially through the VxrAB pathway, but do not significantly impact biofilm formation. VxrC was shown to act as a repressor of vpsL expression and biofilm formation, and this phenotype was shown to be dependent on the presence of VxrB. This indicates that VxrC is in a pathway with the VxrAB TCS and that VxrC may interact with VxrA or VxrB to inhibit its regulation of biofilm formation. Further analysis of the Vxr operon members and their role in activating or repressing the VxrAB TCS will be essential for fully understanding the mechanism of action of this system.
c-di-GMP positively regulates biofilm formation and negatively regulates motility. Given the role of VxrAB in positively regulating biofilm formation and c-di-GMP levels, we investigated the impact of VxrB on motility. Our results demonstrated that deletion of vxrB did not impact motility but that overexpression of VxrB led to a decrease in motility. This is consistent with results from a previous study that demonstrated that overexpression of wild-type VxrB or constitutively active VxrB resulted in a loss of motility (36).
We have previously shown that VxrB is a positive regulator of the T6SS, a protein delivery system that requires cell-to-cell contact to translocate toxic effector proteins into a diverse group of target cells, including other bacteria, phagocytic amoebas, and human macrophages (33, 38, 49–51). In a biofilm, where cells are already in close contact, the T6SS could contribute to localized death and biofilm remodeling. The role of the T6SS in V. cholerae biofilms is unknown, and very few studies have been done in other species linking the T6SS to biofilm formation. In Pseudomonas aeruginosa strain PAO1, the structural component that makes up the inner sheath of the T6SS, Hcp, was shown to take part in the biofilm maturation stage (52). Additionally, a structural component of the outer sheath of the T6SS, tssC1, was demonstrated to promote antibiotic resistance in P. aeruginosa biofilms (53). In Agrobacterium tumefaciens, the ExoR regulator, an important regulator of biofilm formation, was shown to also regulate the T6SS, and in Burkholderia cenocepacia, a novel hybrid HK was identified that controls biofilm formation and the T6SS (54, 55). Thus, we wanted to determine the contribution of the T6SS on biofilm formation in V. cholerae and whether the decreased biofilm formation phenotype of the vxrB mutant is due to decreased T6SS activity. Our initial results indicate that the T6SS does not impact V. cholerae biofilm formation under the conditions tested. However, it is possible that under alternative growth conditions, the T6SS plays a structural or functional role within biofilms.
Determining the activating signals sensed by the VxrAB TCS will provide valuable insight into its mechanism of action. A recent study indicated that the VxrAB system may sense signals generated in response to β-lactam exposure, potentially either directly sensing the antibiotics themselves or sensing cell wall stress or degradation products (36). A VxrA homolog in Vibrio parahaemolyticus, VbrK, was shown to directly bind β-lactams to activate its cognate RR and stimulate the expression of a β-lactamase and additional β-lactam antibiotic resistance genes (56). Given that V. cholerae does not encode β-lactamase genes, it is interesting to consider the conditions under which VxrAB might encounter these potential signals and to speculate about the role they might play in directing the activity of this TCS. Subinhibitory concentrations of aminoglycoside antibiotics have been demonstrated to induce biofilm formation in P. aeruginosa and Escherichia coli, and it is possible that VxrAB mediates a similar response to β-lactam antibiotics in V. cholerae (57). Further investigation of the signals sensed by VxrAB is needed to fully elucidate the mechanism by which this TCS regulates diverse processes in V. cholerae.
It is evident that the VxrAB TCS plays an important regulatory role in V. cholerae and governs the T6SS, virulence, and cell wall homeostasis. This work identifies a new role for this system in biofilm formation and provides a better understanding of how VxrAB regulates important phases in the V. cholerae life cycle.
MATERIALS AND METHODS
Strains and growth conditions.
All response regulator deletion strains used in Fig. 1 are listed in reference 33. Additional strains used in this study are listed in Table 3. V. cholerae and Escherichia coli strains were grown aerobically in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.5) at 30°C and 37°C, respectively. LB agar contained granulated agar (Difco) at 1.5% (wt/vol). Medium additives were used when necessary at the following concentrations: rifampin, 100 μg/μl; ampicillin, 100 μg/μl; and chloramphenicol, 20 μg/μl for E. coli and 5 μg/μl or 2.5 μg/μl for V. cholerae.
TABLE 3.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| CC118λpir | Δ(ara-leu)araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 λpir | 60 |
| S17-1λpir | Tpr Smr recA thi pro rK− mK+ RP4::2-Tc::MuKm Tn7 λpir | 61 |
| V. cholerae strains | ||
| FY_VC_0001 | O1 El Tor A1552, wild type, Rifr | 62 |
| FY_VC_0237 | FY_VC_0001 mTn7 gfp Rifr Gmr | 46 |
| FY_VC_2272 | ΔVC0665 (vpsR) | 24 |
| FY_VC_9332 | ΔVCA0565 (vxrA) | 33 |
| FY_VC_8758 | ΔVCA0566 (vxrB) | 33 |
| FY_VC_0099 | ΔVCA0952 (vpsT) | 27 |
| FY_VC_9369 | ΔVCA0567 (vxrC) | 33 |
| FY_VC_9417 | ΔVCA0568 (vxrD) | 33 |
| FY_VC_9394 | ΔVCA0569 (vxrE) | 33 |
| FY_VC_9469 | ΔvxrB Tn7::vxrB | 33 |
| FY_VC_9952 | ΔvxrB Δhcp1 Δhcp2 | 33 |
| FY_VC_9569 | ΔVC1415 ΔVCA0017 (Δhcp1 Δhcp2) | 33 |
| FY_VC_9390 | ΔvxrA mTn7 gfp Rifr Gmr | This study |
| FY_VC_8764 | ΔvxrB mTn7 gfp Rifr Gmr | This study |
| FY_VC_9392 | ΔvxrC mTn7 gfp Rifr Gmr | This study |
| FY_VC_9437 | ΔvxrD mTn7 gfp Rifr Gmr | This study |
| FY_VC_9439 | ΔvxrE mTn7 gfp Rifr Gmr | This study |
| FY_VC_9234 | ΔvpsR ΔvxrB Rifr | This study |
| FY_VC_9237 | ΔvpsT ΔvxrB Rifr | This study |
| FY_VC_11356 | FY_VC_0001 mTn7 PTAC-vpsR Rifr Gmr | This study |
| FY_VC_11357 | ΔvxrA mTn7 PTAC-vpsR Rifr Gmr | This study |
| FY_VC_11358 | ΔvxrB mTn7 PTAC-vpsR Rifr Gmr | This study |
| FY_VC_11355 | ΔvxrB ΔvxrC | This study |
| FY_VC_12056 | ΔvxrA-Tn7::vxrA | This study |
| FY_VC_12057 | ΔvxrC-Tn7::vxrC | This study |
| Plasmids | ||
| pGP704sacB28 | pGP704 derivative, mob/oriT sacB Apr | 27 |
| pUX-BF13 | oriR6K helper plasmid, mob/oriT, provides the Tn7 transposition function in trans, Apr | 63 |
| pMCM11 | pGP704::mTn7 gfp Gmr Apr | M. Miller and G. Schoolnik |
| pBBRlux | luxCDABE-based promoter fusion vector, Cmr | 64 |
| pFY-0950 | pBBRlux vpsL promoter, Cmr | 65 |
| pFY-0989 | pBBRlux vpsR promoter, Cmr | This study |
| pFY-0988 | pBBRlux vpsT promoter, Cmr | This study |
| pBAD/myc | Arabinose-inducible expression vector with C-terminal myc epitope and six-His tags | |
| His-B | ||
| pFY-2074 | pBAD-vxrB-noTag Ampr | This study |
| pFY-3071 | pBAD-vxrBD78A-noTag Ampr | This study |
| pFY-3073 | pBAD-vxrBD78E-noTag Ampr | This study |
Strain and plasmid construction.
Plasmids were constructed using standard cloning methods or the Gibson Assembly recombinant DNA technique (New England BioLabs, Ipswich, MA). Gene deletions were carried out using allelic exchange of the native open reading frame (ORF) with the truncated ORF, as previously described (58). Complementation of ΔvxrB was carried out using a Tn7-based system, as previously described (33). Briefly, triparental matings with donor E. coli S17λpir carrying pGP704-Tn7 with the gene of interest, helper E. coli S17λpir harboring pUX-BF13, and V. cholerae deletion strains were carried out by mixing all three strains and incubating mating mixtures on LB agar plates for 18 h at 30°C. Transconjugants were selected on thiosulfate-citrate-bile salts-sucrose (TCBS; BD Difco, Franklin Lakes, NJ) agar medium containing gentamicin (15 μg/μl) at 30°C. Insertion of the complementation construct to the Tn7 site was verified by PCR. V. cholerae wild-type and mutant strains were tagged with the green fluorescent protein gene (gfp) according to a previously described procedure (14). The gfp-tagged V. cholerae strains were verified by PCR and used in biofilm analyses. Transcriptional fusions were constructed by cloning the upstream regulatory regions of selected genes into the pBBR-lux plasmid, as previously described (59).
Luminescence assays from planktonically grown cells.
Overnight cultures of V. cholerae cells were diluted 1:500 in appropriate medium containing chloramphenicol (5 μg/ml). Cells were then grown aerobically at 30°C to an optical density at 600 nm (OD600) of 0.3 to 0.4, and then the luminescence of cells was measured using a PerkinElmer Victor3 multilabel counter (PerkinElmer, Waltham, MA). Lux expression is reported as counts · min−1 · ml−1 · OD600−1, shown as relative light units (RLU). Assays were repeated with three biological replicates. Three technical replicates were measured for all assays. Statistical analysis was performed using one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test.
Luminescence assays from biofilm-grown cells.
Flow cells were inoculated by diluting overnight-grown cultures of V. cholerae cells harboring PvpsL-lux 1:200 and injecting cells into an Ibidi m-Slide VI0.4 (Ibidi 80601; Ibidi LLC, Verona, WI). After inoculation, the bacteria were allowed to adhere at room temperature for 1 h with no flow. Next, the flow of 2% (vol/vol) LB (0.2 g/liter tryptone, 0.1 g/liter yeast extract, 1% NaCl) containing chloramphenicol (2.5 μg/ml) was initiated at a rate of 7.5 ml/h and continued for up to 24 h at 25°C. After 24 h, biofilms were harvested in 1 ml phosphate-buffered saline (PBS) for luminescence reading. The luminescence of cells was read using a PerkinElmer Victor3 multilabel counter (PerkinElmer, Waltham, MA) and is reported as counts · min−1 · ml−1 · μg−1 protein concentration, as calculated by the bicinchoninic acid (BCA) assay (Thermo Fisher, Waltham, MA) using bovine serum albumin (BSA) as standards. Lux expression is reported as relative light units (RLU). Assays were repeated with two biological replicates, and three technical replicates were measured for all assays. Statistical analysis was performed using ANOVA and Bonferroni's multiple-comparison test.
Biofilm assays.
Flow cells were inoculated by diluting overnight-grown cultures of gfp-tagged V. cholerae strains 1:200 (OD600 of 0.02) and injecting cells into an Ibidi m-Slide VI0.4 (Ibidi 80601; Ibidi LLC, Verona, WI). After inoculation, the bacteria were allowed to adhere at room temperature for 1 h with no flow. Next, the flow of 2% (vol/vol) LB (0.2 g/liter tryptone, 0.1 g/liter yeast extract, 1% NaCl) was initiated at a rate of 7.5 ml/h and continued for up to 72 h. Confocal laser scanning microscopy (CLSM) images of the biofilms were captured with an LSM 5 PASCAL system (Zeiss) using an excitation wavelength of 488 nm and an emission wavelength of 543 nm. Three-dimensional images of the biofilms were reconstructed using Imaris software (Bitplane) and quantified using COMSTAT 2 (37). Statistical analysis of COMSTAT data was performed using ANOVA and Bonferroni's multiple-comparison test.
Motility assays.
Soft agar motility plates were made using LB medium with 0.3% (wt/vol) agar supplemented with 100 μg/μl ampicillin or 100 μg/μl ampicillin and 0.1 mM IPTG. The plates were inoculated by stabbing the agar from an overnight colony of the strains to be tested. The plates were then incubated at 30°C. Diameters of the migration zones were measured after 16 h. Statistical analysis was performed using ANOVA and Bonferroni's multiple-comparison test.
Determination of intracellular c-di-GMP levels.
c-di-GMP extraction was performed as previously described (58). Briefly, V. cholerae wild-type, ΔvxrB, ΔvxrC, and ΔvxrB Tn7::vxrB-complemented strains were grown in LB broth to an OD600 of 0.4 before 40 ml of culture was harvested at 4,000 × g for 30 min. Cell pellets were allowed to dry briefly and then resuspended in 1 ml extraction solution (40% acetonitrile, 40% methanol, 0.1% formic acid, 19.9% high-pressure liquid chromatography [HPLC]-grade water), and incubated on ice for 15 min. Samples were then centrifuged at 16,000 × g for 5 min and 800 μl of supernatant was dried under vacuum and then lyophilized. Samples were resuspended in 50 μl of 184 mM NaCl and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Thermo-Electron Finnigan LTQ mass spectrometer coupled to a surveyor HPLC. The amount of c-di-GMP in samples was calculated with a standard curve generated from pure c-di-GMP suspended in 184 mM NaCl (Biolog Life Science Institute, Bremen, Germany). The concentrations used for the standard curve generation were 50 nM, 100 nM, 500 nM, 2 μM, 3.5 μM, 5 μM, 7.5 μM, and 10 μM. The assay is linear from 50 nM to 10 μM, with an R2 of 0.999. The c-di-GMP levels were normalized to total protein per ml of culture.
To determine protein concentration, 4 ml from each culture was harvested, the supernatant was removed, and cells were lysed in 1 ml of 2% sodium dodecyl sulfate (SDS). Total protein in the samples was determined with a bicinchoninic acid (BCA) assay (Thermo Fisher, Waltham, MA) using bovine serum albumin (BSA) as the standard. Each c-di-GMP quantification experiment was performed with four biological replicates. Statistical analysis was performed using ANOVA and Bonferroni's multiple-comparison test.
ACKNOWLEDGMENTS
We thank Benjamin Abrams from the UCSC Life Sciences Microscopy Center for his technical support, Qiangli Zhang for help with the high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) experiments and analysis, Ates Gurcan for his assistance with Fig. 8, and David Zamorano-Sánchez for his comments on the manuscript.
This work was supported by the NIH grant R01AI055987 to F.H.Y.
REFERENCES
- 1.Kaper JB, Morris JG, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faruque SM, Albert MJ, Mekalanos JJ. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev 62:1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charles RC, Ryan ET. 2011. Cholera in the 21st century. Curr Opin Infect Dis 24:472–477. doi: 10.1097/QCO.0b013e32834a88af. [DOI] [PubMed] [Google Scholar]
- 4.Teschler JK, Zamorano-Sánchez D, Utada AS, Warner CJA, Wong GCL, Linington RG, Yildiz FH. 2015. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol 13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islam MS, Jahid MIK, Rahman MM, Rahman MZ, Islam MS, Kabir MS, Sack DA, Schoolnik GK. 2007. Biofilm acts as a microenvironment for plankton-associated Vibrio cholerae in the aquatic environment of Bangladesh. Microbiol Immunol 51:369–379. doi: 10.1111/j.1348-0421.2007.tb03924.x. [DOI] [PubMed] [Google Scholar]
- 6.Faruque SM, Biswas K, Udden SMN, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc Natl Acad Sci U S A 103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alam M, Sultana M, Nair GB, Siddique AK, Hasan NA, Sack RB, Sack DA, Ahmed KU, Sadique A, Watanabe H, Grim CJ, Huq A, Colwell RR. 2007. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci U S A 104:17801–17806. doi: 10.1073/pnas.0705599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conner JG, Teschler JK, Jones CJ, Yildiz FH. 2016. Staying alive: Vibrio cholerae's cycle of environmental survival, transmission, and dissemination. Microbiol Spectr 4:VMBF-0015-2015. doi: 10.1128/microbiolspec.VMBF-0015-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA, Torreilles S, Schoolnik GK. 2010. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog 6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berk V, Fong JCN, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. 2012. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Absalon C, Van Dellen K, Watnick PI. 2011. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog 7:e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong JCN, Syed KA, Klose KE, Yildiz FH. 2010. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 156:2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A 96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J Bacteriol 188:1049–1059. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong JCN, Yildiz FH. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol 189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol Microbiol 57:1623–1635. doi: 10.1111/j.1365-2958.2005.04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol 17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Townsley L, Sison Mangus MP, Mehic S, Yildiz FH. 2016. Response of Vibrio cholerae to low-temperature shift: CpsV regulates type VI secretion, biofilm formation, and association with zooplankton. Appl Environ Microbiol 82:e00807-16. doi: 10.1128/AEM.00807-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shikuma NJ, Yildiz FH. 2009. Identification and characterization of OscR, a transcriptional regulator involved in osmolarity adaptation in Vibrio cholerae. J Bacteriol 191:4082–4096. doi: 10.1128/JB.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beier D, Gross R. 2006. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol 9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Calva E, Oropeza R. 2006. Two-component signal transduction systems, environmental signals, and virulence. Microb Ecol 51:166–176. doi: 10.1007/s00248-005-0087-1. [DOI] [PubMed] [Google Scholar]
- 22.Bilecen K, Yildiz FH. 2009. Identification of a calcium-controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ Microbiol 11:2015–2029. doi: 10.1111/j.1462-2920.2009.01923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shikuma NJ, Davis KR, Fong JNC, Yildiz FH. 2013. The transcriptional regulator, CosR, controls compatible solute biosynthesis and transport, motility and biofilm formation in Vibrio cholerae. Environ Microbiol 15:1387–1399. doi: 10.1111/j.1462-2920.2012.02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yildiz FH, Dolganov NA, Schoolnik GK. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol 183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratt JT, McDonough E, Camilli A. 2009. PhoB regulates motility, biofilms, and cyclic di-GMP in Vibrio cholerae. J Bacteriol 191:6632–6642. doi: 10.1128/JB.00708-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sultan SZ, Silva AJ, Benitez JA. 2010. The PhoB regulatory system modulates biofilm formation and stress response in El Tor biotype Vibrio cholerae. FEMS Microbiol Lett 302:22–31. doi: 10.1111/j.1574-6968.2009.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casper-Lindley C, Yildiz FH. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol 186:1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 29.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilecen K, Fong JCN, Cheng A, Jones CJ, Zamorano-Sánchez D, Yildiz FH. 2015. Polymyxin B resistance and biofilm formation in Vibrio cholerae is controlled by the response regulator CarR. Infect Immun 83:1199–1209. doi: 10.1128/IAI.02700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol 58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 32.McKee RW, Kariisa A, Mudrak B, Whitaker C, Tamayo R. 2014. A systematic analysis of the in vitro and in vivo functions of the HD-GYP domain proteins of Vibrio cholerae. BMC Microbiol 14:272. doi: 10.1186/s12866-014-0272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng AT, Ottemann KM, Yildiz FH. 2015. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog 11:e1004933. doi: 10.1371/journal.ppat.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler SM, Camilli A. 2005. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat Rev Microbiol 3:611–620. doi: 10.1038/nrmicro1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler SM, Camilli A. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci U S A 101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dörr T, Alvarez L, Delgado F, Davis BM, Cava F, Waldor MK. 2016. A cell wall damage response mediated by a sensor kinase/response regulator pair enables beta-lactam tolerance. Proc Natl Acad Sci U S A 113:404–409. doi: 10.1073/pnas.1520333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146 (Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 38.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect Immun 74:3633–3642. doi: 10.1128/IAI.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Ayala JC, Benitez JA, Silva AJ. 2015. RNA-Seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. PLoS One 10:e0118295. doi: 10.1371/journal.pone.0118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Tsou AM, Liu Z, Cai T, Zhu J. 2011. The VarS/VarA two-component system modulates the activity of the Vibrio cholerae quorum-sensing transcriptional regulator HapR. Microbiology 157:1620–1628. doi: 10.1099/mic.0.046235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang J, Jung K-T, Yoo C-K, Rhie G-E. 2010. Regulation of hemagglutinin/protease expression by the VarS/VarA-CsrA/B/C/D system in Vibrio cholerae. Microb Pathog 48:245–250. doi: 10.1016/j.micpath.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Jang J, Jung K-T, Park J, Yoo C-K, Rhie G-E. 2011. The Vibrio cholerae VarS/VarA two-component system controls the expression of virulence proteins through ToxT regulation. Microbiology 157:1466–1473. doi: 10.1099/mic.0.043737-0. [DOI] [PubMed] [Google Scholar]
- 45.Mey AR, Butz HA, Payne SM. 2015. Vibrio cholerae CsrA regulates ToxR levels in response to amino acids and is essential for virulence. mBio 6:e01064. doi: 10.1128/mBio.01064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol 188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. 2007. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J Bacteriol 189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng J, Ho B, Mekalanos JJ. 2011. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A 107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Southey-Pillig CJ, Davies DG, Sauer K. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol 187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Hinz AJ, Nadeau J-P, Mah T-F. 2011. Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J Bacteriol 193:5510–5513. doi: 10.1128/JB.00268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heckel BC, Tomlinson AD, Morton ER, Choi J-H, Fuqua C. 2014. Agrobacterium tumefaciens exoR controls acid response genes and impacts exopolysaccharide synthesis, horizontal gene transfer, and virulence gene expression. J Bacteriol 196:3221–3233. doi: 10.1128/JB.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aubert DF, Flannagan RS, Valvano MA. 2008. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect Immun 76:1979–1991. doi: 10.1128/IAI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, Wang Q, Zhang H, Yang M, Khan MI, Zhou X. 2016. Sensor histidine kinase is a β-lactam receptor and induces resistance to β-lactam antibiotics. Proc Natl Acad Sci U S A 113:1648–1653. doi: 10.1073/pnas.1520300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Beyhan S, Lim B, Linington RG, Yildiz FH. 2010. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J Bacteriol 192:4541–4552. doi: 10.1128/JB.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zamorano-Sánchez D, Fong JCN, Kilic S, Erill I, Yildiz FH. 2015. Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J Bacteriol 197:1221–1235. doi: 10.1128/JB.02439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol 172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Lorenzo V, Timmis KN. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 62.Yildiz FH, Schoolnik GK. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol 180:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene 109:167–168. doi: 10.1016/0378-1119(91)90604-A. [DOI] [PubMed] [Google Scholar]
- 64.Hammer BK, Bassler BL. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 104:11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shikuma NJ, Fong JCN, Yildiz FH. 2012. Cellular levels and binding of c-di-GMP control subcellular localization and activity of the Vibrio cholerae transcriptional regulator VpsT. PLoS Pathog 8:e1002719. doi: 10.1371/journal.ppat.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]