Abstract
Colletotrichum truncatum, a major fungal phytopathogen, causes the anthracnose disease on an economically important spice crop chilli (Capsicum annuum), resulting in huge economic losses in tropical and sub-tropical countries. It follows a subcuticular intramural infection strategy on chilli with a short, asymptomatic, endophytic phase, which contrasts with the intracellular hemibiotrophic lifestyle adopted by most of the Colletotrichum species. However, little is known about the molecular determinants and the mechanism of pathogenicity in this fungus. A high quality whole genome sequence and gene annotation based on transcriptome data of an Indian isolate of C. truncatum from chilli has been obtained. Analysis of the genome sequence revealed a rich repertoire of pathogenicity genes in C. truncatum encoding secreted proteins, effectors, plant cell wall degrading enzymes, secondary metabolism associated proteins, with potential roles in the host-specific infection strategy, placing it next only to the Fusarium species. The size of genome assembly, number of predicted genes and some of the functional categories were similar to other sequenced Colletotrichum species. The comparative genomic analyses with other species and related fungi identified some unique genes and certain highly expanded gene families of CAZymes, proteases and secondary metabolism associated genes in the genome of C. truncatum. The draft genome assembly and functional annotation of potential pathogenicity genes of C. truncatum provide an important genomic resource for understanding the biology and lifestyle of this important phytopathogen and will pave the way for designing efficient disease control regimens.
Introduction
Capsicum annuum (chilli or hot pepper), a native of Mexico [1], is an important vegetable crop and an indispensable spice used in everyday cuisine throughout the world. A serious limiting factor and major constraint to chilli cultivation has been fruit rot or anthracnose disease caused by Colletotrichum species [2], a large genus belonging to the family Glomerellaceae (Glomerellales, Sordariomycetes) of Ascomycota. It is one of the most common and important genera of plant-pathogenic fungi [3,4], with some species being endophytic on symptomless plants [5] or saprotrophic [4]. C. acutatum and C. gloeosporioides are often associated with anthracnose of chilli [6], but C. truncatum (previously known as C. capsici [7]) is the most predominant causative species in major chilli growing countries in South-East Asia, including India [8,9]. The disease is characterized by very dark, sunken necrotic lesions with concentric rings of acervuli containing characteristic, strongly curved conidia [7]; causing both pre- and post-harvest fruit rots and reducing their quality and marketability [10]. C. truncatum is a devastating pathogen of many other tropical crops and has a wide host range including Solanaceae, Brassicaceae, Fabaceae, Malvaceae, etc. [11] and is even reported to infect humans [12]. At present, there are 11 major species complexes/clades and 23 singleton species in the genus Colletotrichum [11]. Thorough characterization of the species causing anthracnose and knowledge of the molecular mechanism of its progression is critical to devise strategies to efficiently control the spread of the disease.
The Colletotrichum species use melanized appressoria to penetrate host tissues and follow host-specific infection strategy of either intracellular hemibiotrophy or subcuticular intramural necrotrophy [13][14]. Some Colletotrichum species adopt both the strategies on same or different hosts; for example, members of the acutatum and gloeosporioides species complexes [14]. The initial stages of infection are similar for both groups of pathogens; conidia adhere to, and germinate on plant surfaces, produce germ tubes and form appressoria which penetrate the cuticle directly. Following penetration, in intracellular hemibiotrophic infection the primary biotrophic hyphae grow intracellularly within the cell lumen without killing the host protoplast and subsequently give rise to secondary necrotrophic hyphae, as exemplified by most of the Colletotrichum species like C. lindemuthianum on Phaseolus vulgaris, C. graminicola on maize etc. [14]. In contrast, the subcuticular intramural pathogens form an intramural network of hyphae within the walls of epidermal and cortical cells beneath the cuticle and kill the host cells prior to the rapid necrotrophic mycelial spread [11] as exemplified by C. truncatum (C. capsici) on cowpea (Vigna unguiculata) [13] and cotton (Gossypium hirsutum L.) [14]. Recent studies suggest that C. truncatum colonises chilli through subcuticular intramural necrotrophy with a brief asymptomatic, endophytic phase after initial infection and prior to necrotrophic development [15,16]. Still, it contrasts with the hemibiotrophic lifestyle that is followed by most of the Colletotrichum species. Despite extensive efforts to devise measures for the management and control of anthracnose, it has been difficult to control in the field because of increasing resistance in the pathogen populations to common fungicides and non-availability of cultivated varieties of C. annuum showing satisfying levels of resistance against the fungus. The lack of information on the genome content and organization has been a limiting factor in the study of this important phytopathogen and for developing strategies aimed at limiting its spread.
In the post-genomics era, an increasing number of pathogenic fungi are being sequenced, leading to the discovery of several genes and virulence factors that play key roles in host infection and disease progression. A number of secreted fungal effector molecules have been identified that suppress plant defense responses and modulate plant physiology to facilitate the host colonization [17]. These rapidly evolving effectors help fungi adapt to highly diverse lifestyles, and act as virulence factors governing the compatible interactions with the host. The whole genome sequence of many Colletotrichum species belonging to different species complexes, viz., C. graminicola [18], C. higginsianum [18,19], C. orbiculare [20], C. fructicola (previously mis-identified as C. gloeosporioides Nara gc5 [21]) [20], C. fiorineae [22], C. sublineola [23], C. scovillei (previously C. acutatum, host chilli) [24], etc., are publicly available, giving an impetus to the Colletotrichum research. The comparative genomic analyses of important gene classes among different Colletotrichum species and related fungi had helped in the discovery of the core genes conserved in the genus Colletotrichum, and the expansion and contraction of certain gene classes that could be attributed to their specific lifestyles [18,20,25,26].
The genome of an Indian isolate of C. truncatum was sequenced, and a repertoire of putative pathogenicity genes like secretory proteases and cell wall degrading enzymes, candidate effectors, secondary metabolite (SM) biosynthesis genes, etc., were identified, which gave an insight into different aspects of its biology, lifestyle and host specificity. The comparative genomic analyses were carried out between all Colletotrichum species and other fungi with diverse lifestyles using both genome and proteome data that was publicly available. With the whole genome sequencing of this major member of the truncatum species complex, 9 out of 11 clades of the phylogenetic tree of the Colletotrichum genus presently have the genome information available, facilitating further genus-wide evolutionary and comparative studies.
Materials and methods
Fungal culture conditions and plant infection assay
Six Colletotrichum capsici cultures that were deposited in Microbial Type Culture Collection and Gene Bank (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh, India with MTCC no. 2071, 3414, 8473, 9691, 10147 and 10327 were procured and were propagated on potato dextrose agar (PDA, Diffco Laboratories, France). To confirm the species identity of all the fungal cultures sourced from MTCC, internal transcribed spacer region (ITS) of nuclear ribosomal cistron and 28S nuclear ribosomal large subunit rRNA gene (LSU) were PCR amplified using the conserved universal primer pairs [27] and sequenced.
The conidial suspension was prepared by adding sterile distilled water to the 7 day old colonies on PDA and scrapping the spores off the surface of the colonies using inoculation loops. The conidia were counted using haemocytometer at different dilutions and the concentrations were adjusted to 1 x 106 conidia/ml. The pathogenicity assay on C. annuum fruits was carried out through wound/drop method [2] to inoculate conidia from different fungal cultures. Koch’s postulates were tested by placing parts of lesions from inoculated chillies on PDA.
A highly virulent isolate of C. truncatum (MTCC no. 3414) was selected for all the subsequent experiments. The molecular phylogenetic analysis was carried out for the Colletotrichum species complexes based on multilocus alignment of five genes used in a previous study [18] (internal transcribed spacer of rRNA (ITS), chitin synthase-1 (CHS-1), histone3 (HIS3), actin (ACT) and tubulin (TUB2)) in C. truncatum (MTCC no. 3414) and 27 other Colletotrichum species using MEGA6 [28] by neighbour joining (NJ) method with 1000 bootstrap replicates. Monilochaetes infuscans was taken as an outgroup for the analysis.
Mycelia were harvested from liquid cultures grown in potato dextrose broth (PDB) and Czapeck’s medium at 28°C with shaking at 180 rpm for 3 days for DNA and RNA extractions, respectively. The fruits of chilli were surface sterilized with 1% sodium hypochlorite and inoculated with 1 x 106 conidia/ml of conidial suspension using non-wound/drop method [2], covered with nylon mesh, sealed in a plastic box and incubated at room temperature for different time points. The inoculated portions of the fruits were excised, ground in liquid nitrogen and stored at -80°C until RNA extraction.
Genome sequencing and assembly
The genomic DNA was isolated from C. truncatum (MTCC no. 3414) culture using DNeasy Plant Minikit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Two short insert paired-end (PE, average insert size of 300 bp and 500 bp), and two long insert mate-pair (average insert size of 3000 bp and 5000 bp) genomic DNA libraries were constructed and sequenced on the Illumina HiSeq 2000 platform (2x100 bp reads) at Cofactor Genomics (St. Louis, MO, USA). The demultiplexed reads, filtered for low quality and adapter sequences, were assembled using SOAPdenovo (v 1.05) [29]. The gene space coverage was estimated with Core Eukaryotic Genes Mapping Approach (CEGMA v2.5) [30] using default parameters and Benchmarking Universal Single-Copy Orthologues (BUSCO v1) [31] using fungal gene set and Fusarium graminearium as a species parameter in long mode.
RNA-sequencing (RNA-Seq) and transcriptome assembly
Non-strand-specific RNA-Seq was carried out for three in vitro samples, including 7 day old cultures on potato dextrose agar (PDA) supported by dialysis membrane for easy collection of fungal mass along with un-germinated conidia, 3 day old liquid cultures in Czapeck’s medium (CZ) and in vitro appresorial assay (APR). Three in planta samples which included chilli fruits inoculated with C. truncatum conidia and sampled at 0, 24 and 72 hours post inoculation (0 hpi as control, 24 hpi and 72 hpi, respectively) were also used for RNA-Seq. For in vitro appresorial assay, conidial suspension was placed on polystyrene petri dishes (Tarsons, India; 140 x 16 mm) in the form of droplets covering the whole surface, incubated overnight in a plastic box lined with moist tissue paper to maintain the humidity and observed under light microscope for the formation of appresoria. For RNA extraction, the petri dishes were washed with RNase-free water, appresoria were scraped off the surface using a cell scraper (Corning Inc., USA) adding Trizol reagent (Invitrogen, USA). For the rest of the samples too, Trizol method was used for RNA extraction followed by purification with RNeasy minispin columns (Qiagen, Hilden, Germany) according to the manufacturers’ instructions. The RNAs from all samples were treated with Turbo DNA-free DNase (Ambion, Texas, USA) to remove any genomic DNA contamination. RNA-seq libraries were prepared using the Illumina TruSeq 3 kit and were sequenced on Illumina HiSeq 2500 platform (2x100 bp reads) at SciGenom Labs Pvt. Ltd. (Cochin, Kerala, India). The quality check of sequenced reads (S1 Table) was performed using the FastQC suite (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The raw reads were quality filtered using the program Trimmomatic (v0.36) [32] and rRNA sequences were removed through SortMeRNA (v2.1) [33]. PCR duplicates were removed using PRINSEQ-lite (v0.20.4) [34] and PE reads were assembled into longer super-reads through MaSuRCA (v 3.13) [35]. All the in vitro and in planta reads were aligned to C. truncatum genome (intron sizes: 10–5000 bp) and the annotated genome of chilli (C. annuum cv. Zunla-1) [36] (intron sizes: 50–20,000), respectively using HISAT2 [37]. The in planta reads which did not map to chilli genome were aligned to C. truncatum genome assembly. Both genome-guided and de novo transcript assemblies of C. truncatum were obtained using Trinity (v2.2.0) [38] with jaccard-clip option to reduce the fusion genes. The non-redundant transcripts were retained using CD-HIT-EST [39] (95% identity threshold). Program to Assemble Spliced Alignments (PASA) [40] was used to align transcripts to genome and reconstruct the refined transcriptome. A set of likely coding regions was extracted for training ab initio gene prediction softwares through TransDecoder module bundled with PASA.
Gene prediction
Genes were predicted through a combination of evidence-based (transcriptome data and homology to known proteins) and ab initio methods through MAKER2 (v2.31.7) [41]. Repetitive elements in the genome assembly were masked using library of all fungal repeat elements in the Repbase database (v 20140131) through RepeatMasker (v4.0.5, http://www.repeatmasker.org/) as well as the de novo repeat library identified using RepeatModeler (v1.0.8, http://www.repeatmasker.org/RepeatModeler.html). Genes were predicted using SNAP (v2013-02-16) [42] trained on CEGMA output, AUGUSTUS (3.0.3) [43] trained on BUSCO output, auto-trained GeneMark-ES (v4.21) [44], and GeneId (v1.4) [45]. The training set of transcripts was supplied as organism-specific ESTs to MAKER2 along with C. lentis ESTs from NCBI as alternate ESTs and the known proteins from C. higginsianum [19], C. graminicola [18], C. fructicola [20] and SwissProt database. The process was reiterated using SNAP and AUGUSTUS trained on the output of the previous run including the transcriptome assembly from PASA. The standard set of protein coding genes was compiled following the recommendations of Campbell and coworkers [46], retaining high confidence ab initio gene models. Some of the scaffolds were randomly inspected through Apollo genome browser [47] to check for any annotation biases. Transfer RNAs (tRNAs) were predicted using tRNAScan-SE (v1.23) [48].
Differential gene expression analysis
RNA-Seq reads from in vitro and in planta samples were mapped onto the annotated genomes of C. truncatum and chilli, respectively, using Tophat2 [49]. The unmapped reads from in planta samples were retrieved through bamUtils (v 1.0.13, http://genome.sph.umich.edu/wiki/BamUtil) and mapped to C. truncatum genome through Tophat2. The differential gene expression between the samples was estimated by GFOLD (V1.1.4) [50].
Genome synteny and phylogenetic analysis
Synteny Mapping and Analysis Program (SyMAP) v4.2 [51] was used for synteny analysis of C. truncatum genome with that of C. higginsianum with minimum size of sequences set to 10 kb and masking sequences other than the protein coding genes. For phylogenetic analysis and comparative genomics, the genomes and/or proteomes of 10 Colletotrichum species and 12 other related plant pathogenic fungi with diverse lifestyles (S1 Fig) were downloaded from NCBI (Table 1). A phylogenetic tree was constructed for all the fungi with Aspergillus nidulans as outgroup. Briefly, the tightly clustered single copy orthologues present in all the fungi were identified through Proteinortho (v5.13) [52] and were aligned to each other using MUSCLE (v3.8.31) [53]. The trimmed alignment in PHYLIP format was generated through trimAl (v1.4.rev15) [54] and concatenated to generate a supermatrix using FASconCAT (v1.0) [55]. A maximum likelihood tree with 1000 bootstraps was inferred using RAxML (v 8.2.9) [56] based on the ProtTest (v3.0) [57] estimated models (Gamma distribution of rate heterogeneity on invariant sites with JTT model and empirical amino acid frequency) and was visualized through Figtree (http://tree.bio.ed.ac.uk/software/figtree/).
Table 1. The list of fungal genomes used for comparative analysis with C. truncatum sequenced in the present study (in bold).
| Organism | Strain | Host | Origin | Accession no. | Ref. |
|---|---|---|---|---|---|
| C. truncatum | MTCC #3414 | Capsicum annuum | India | NBAU00000000 | - |
| C. graminicola | M1.001 | Zea mays | USA | ACOD00000000.1 | [18] |
| C. higginsianum | IMI 349063 | Brassica rapa | Trinidad & Tobago | LTAN00000000.1 | [19] |
| C. orbiculare | MAFF 240422 | Cucumis sativus | Japan | AMCV00000000.1 | [20] |
| C. fructicola (deposited as C. gloeosporioides) | Nara gc5 | Fragaria x ananassa | Japan | ANPB00000000.1 | [20] |
| C. fioriniae | IMI 504882 | Fragaria x ananassa | New Zealand | JARH00000000.1 | [22] |
| C. sublineola | TX430BB | Sorghum bicolor | USA | JMSE00000000.1 | [23] |
| C. simmondsii | CBS 122122 | Carica papaya | Australia | JFBX00000000.1 | [26] |
| C. salicis | CBS 607.94 | Salix species | Netherlands | JFFI00000000.1 | [26] |
| C. nymphaeae | IMI 504889 | Fragaria x ananassa | Denmark | JEMN00000000.1 | [26] |
| C. incanum | MAFF 238704 | Raphanus sativus | Japan | LFIW00000000.1 | [5] |
| C. scovillei* | 1 | Capsicum annuum | South Korea | LUXP00000000.1 | [24] |
| Verticillium alfalfa | VaMs.102 | Medicago sativa | USA | ABPE00000000.1 | [58] |
| V. dahliae | VdLs.17 | Lactuca sativa | USA | ABJE00000000.1 | [58] |
| Fusarium graminearum | PH1 | Triticum species | USA | AACM00000000.1 | [59] |
| F. oxysporum | FOL 4287 | Solanum lycopersicum | Spain | AAXH00000000.1 | [59] |
| F. verticillioides | 7600 | Zea mays | USA | AAIM00000000.2 | [59] |
| Magnaporthe oryzae# | 70–15 | Oryza sativa | French Guiana | AACU00000000.3 | [60] |
| M. oryzae | P131 | Oryza sativa | Japan | AHZT00000000.1 | [61] |
| M. oryzae | Y34 | Oryza sativa | China | AHZS00000000.1 | [61] |
| N. crassa | OR74A | - | USA | AABX00000000.3 | [62] |
| Trichoderma reesei | QM6a | - | Solomon Islands | AAIL00000000.2 | [63] |
| Botrytis cinerea | B05.10 | Unknown | Germany | ASM83294v1 | [64] |
| Sclerotinia sclerotiorum | 1980 | Phaseolus vulgaris | USA | AAGT00000000.1 | [64] |
*Gene annotations were not available for further analyses at the time of writing this manuscript.
# The strain used for phylogenetic analysis.
Functional annotation
Putative functions were assigned to the predicted proteins that had at least one homologue identified through BLASTP (v2.2.26) against the SwissProt database (E-value threshold 1e-05) and/or a known protein domain identified through InterProScan 5 (v5.20–59.0) [65]. Secretome was predicted using a battery of tools; Signal-P (v4.1) [66], Phobius [67] and WoLF PSORT (v0.2) [68] to identify signal peptides or extracellular localization, and TMHMM (v2.0) [69], PS-SCAN (ftp://ftp.expasy.org/databases/prosite/ps_scan/ps_scan.pl) and PredGPI (http://gpcr.biocomp.unibo.it/predgpi/pred.htm) to exclude sequences with transmembrane domains, ER-retention signal and GPI anchors, respectively. The secretory proteins with nuclear localization signal were identified using NLStradamus (v1.8) [70], while putative effector proteins were predicted through EffectorP (v1.0) [71]. Carbohydrate Active enZymes (CAZymes) were identified by HMMER (v3.1b1, http://hmmer.janelia.org/) scan against the family-specific HMM profiles of dbCAN release 2.0 [72]. Putative proteases and protease inhibitors were predicted through online batch BLAST of proteins against the MEROPS database [73], excluding the sequences with incomplete domains and mutated active sites or metal ligands. Secondary Metabolite Unique Regions Finder (SMURF) [74] was used to identify potential secondary metabolite (SM) gene clusters. In addition to SMURF, another online tool, antibiotics and Secondary Metabolite Analysis SHell (antiSMASH fungal version 4.0.0rc1) [75] was also used with default parameters for the prediction of SM gene clusters in C. truncatum. The putative pathogenicity genes, transporters and cytochrome P450 genes were identified by retaining the best hits through BLASTP (E-value threshold 1e-10) against pathogen-host interactions database (PHI-base) [76], transporter classification database (TCDB) [77] and cytochrome P450 database [78], respectively (all accessed in October 2016). For comparative genomics, the same pipeline was run on the predicted proteomes of other Colletotrichum species and related ascomycete fungi downloaded from NCBI.
Results
Fungal culture authentication
The six C. capsici (C. truncatum) cultures procured from MTCC were authenticated by sequencing internal transcribed spacer region of nuclear rRNA (ITS), a universal DNA barcode marker, and 28S nuclear ribosomal large subunit rRNA gene (LSU), which sometimes discriminates species on its own or when combined with ITS. Conserved universal primer pairs for ITS and LSU were used to PCR amplify and sequence these genes. BLAST analysis against the NCBI non-redundant database showed 100% homology with C. truncatum. Pathogenicity and virulence of the cultures showing conidiation were established by inoculating chilli fruits and proving Koch’s postulates. A highly virulent fungal strain (MTCC no. 3414), isolated from chilli in Puducherry, India was selected for the subsequent experiments and genome sequencing. The molecular phylogenetic analysis based on five conserved genes of Colletotrichum species from different clades showed that C. truncatum (MTCC no. 3414), together with the other isolate of C. truncatum that infects Phaseolus lunatus, forms the truncatum clade, which diverged from the gloeosporioides clade (S2 Fig). The position of C. truncatum in the cladogram confirmed its identity in the Colletotrichum species complexes.
Genome and transcriptome assembly of C. truncatum
The genome of an Indian isolate of C. truncatum was sequenced on Illumina HiSeq 2000 platform, generating a high quality draft genome assembly consisting of 6,193 contigs arranged in 81 scaffolds with a total size of 55.3 Mb and N50 scaffold length of 1.66 Mb (Table 2). The estimated genome size and G-C content were comparable to most of the sequenced species of Colletotrichum genus (S1 Fig). A single scaffold (scaffold_70_2.0) of ~900 bp was excluded from assembly since it had adapter contamination. None of the sequences in the C. truncatum assembly showed any homology to complete mitochondrial genome sequences of C. acutatum and C. graminicola. The completeness of the genome assembly of C. truncatum was assessed using CEGMA that showed 235 complete genes out of 248 orthologues of core eukaryotic genes (CEGs) conserved in all eukaryotes in the assembled genome (94.76% coverage). However, all the partial and missing CEGs were identified through TBLASTN comparison of orthologues from other Colletotrichum species to C. truncatum genome (E-value < 1e-10, >70% identity and query coverage) representing 100% coverage of the gene space. Another tool BUSCO, based on 1,438 single-copy genes present in at least 90% of the fungi, reported 99% coverage of complete BUSCOs with 1,325 single-copy orthologues detected in the C. truncatum genome assembly.
Table 2. Assembly statistics of the C. truncatum genome assembly.
| Total number of contigs | 6,193 |
| Total number of scaffolds | 81 |
| Total length (bp) | 55,368,957 |
| Mean length (bp) | 683,567 |
| Max length (bp) | 3,859,631 |
| Min length (bp) | 906 |
| Largest 10 Total (bp) | 26,251,402 |
| Scaffold N50 (bp) | 1,667,354 |
| Number of Gaps | 6,793 |
| Total Gap Length (bp) | 2,630,941 |
| Mean Gap Length (bp) | 387 |
| Percent N's | 4.75% |
| GC content | 49.61% |
| Repetitive DNA (homology based/ de novo method, S2 and S3 Tables) | 0.44–3% |
| tRNA | 257 |
| Refined transcripts | 32,566 |
| Predicted protein coding genes | 13,724 |
| Coding region in genome | 51% |
| Genes with SwissProt homologues | 66% |
| Genes with IPR/Pfam annotations | 74% |
| Genes with GO annotations | 46% |
| Genes with AED scores between 0–0.25 | 82% |
RNA-Seq was carried out with three in vitro samples of C. truncatum and three in planta samples of inoculated chillies at different time points giving 13.7–39.5 million processed PE reads (S1 Table). ~90% of in vitro processed reads (super-reads, PEs and singletons) mapped to the C. truncatum genome, while >88% of reads from each in planta sample mapped to the chilli genome. The reads from in planta samples which failed to map on the chilli genome were aligned to the C. truncatum genome. Few reads (0.0048%) from 0 hpi mapped to the fungal genome, while a total of 3–6% reads from 24 and 72 hpi mapped to the fungal genome. The processed reads from three in vitro samples and the in planta reads mapping to C. truncatum genome were used for de novo transcriptome assembly using Trinity and comprising of 57,383 transcripts, while genome-guided assembly resulted in 38,425 transcripts. 94,309 non-redundant transcripts from both the assemblies were retained. The protein coding ORFs were predicted by aligning these transcripts to C. truncatum genome through PASA, generating a refined transcriptome with 32,566 sequences. 25,208 longest full length ORFs identified by TransDecoder module of PASA were taken as training set for ab initio gene model prediction.
Genome annotation
The genome annotation was performed using MAKER2 pipeline by integrating the species-specific RNA-Seq evidence, protein homology from SwissProt database and other Colletotrichum species, and ab initio gene predictions to get consensus gene models. A total of 13,724 protein coding genes were predicted in C. truncatum assembly. Annotation Edit Distance (AED), a quantitative measure that shows the high congruency of each annotated transcript with its supporting evidence if it is close to 0, was used to assess the annotated genes [46,79]. 11,315 genes had AED scores between 0 and 0.25, while 10,807 genes had AED of <0.2. In contrast, only 364 genes had AED between 0.75 and 1. Some of the gene models were randomly inspected for any annotation bias. 9,074 predicted proteins had known homologues in the SwissProt database. A functional annotation could be assigned through InterProScan to 1,522 out of the 4,650 proteins which had no homologues in SwissProt database. 10,189 proteins with a recognizable InterPro (IPR) and/or Pfam domain were identified in C. truncatum with gene ontology (GO) terms assigned to 6,385 predicted proteins (S4 Table). Interestingly, only 65% of genes with AED scores of <0.25 had a Pfam domain, whereas ~80% of genes with AED scores of >0.75 had a known domain. 31 of the 37 Pfam domains associated with fungal transcription factors (TFs) [80] were observed in 493 proteins in C. truncatum proteome. The most abundant domains, Zn(2)-Cys(6) binuclear cluster domains (PF00172) and fungal specific transcription factor domain (PF04082) were found in 169 and 107 proteins, respectively, whereas 125 proteins shared both the domains (S4 Table).
Genome synteny
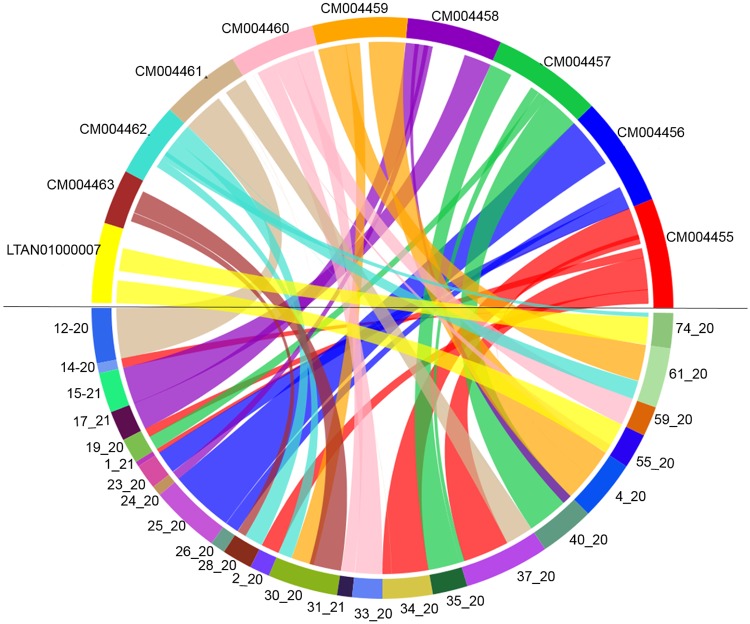
The comparison of C. truncatum genome assembly and gene annotation with one of the well annotated species, C. higginsianum revealed 65 syntenic blocks with conserved gene order among the two genomes (Fig 1). 44 scaffolds of C. truncatum could be aligned with 9 chromosomes and a contig of C. higginsianum with 9,255 syntenic hits between the two species. The size of conserved syntenic blocks ranged from 14 kb to 3.1 Mb with 18 blocks of sizes above 1 Mb. Most of the syntenic regions were inverted in C. truncatum and fragmented synteny was observed in many regions. The synteny plots showed many translocations, like 1.7 Mb of proximal and 0.9 Mb of distal regions of scaffold 61_20 of C. truncatum were in synteny with regions on chromosome 9 (CM004462) and chromosome 5 (CM004459) of C. higginsianum, respectively (S5 Table).
Fig 1. The synteny plot between C. higginsianum and C. truncatum genomes obtained using SyMAP.
The circular plot shows some of the syntenic blocks between the scaffolds of C. truncatum in lower half of the circle mapping to the reference chromosomes of C. higginsianum in upper half joined with the lines of same colour as the reference.
Orthologue analysis and phylogenetic relationship of C. truncatum with other fungi
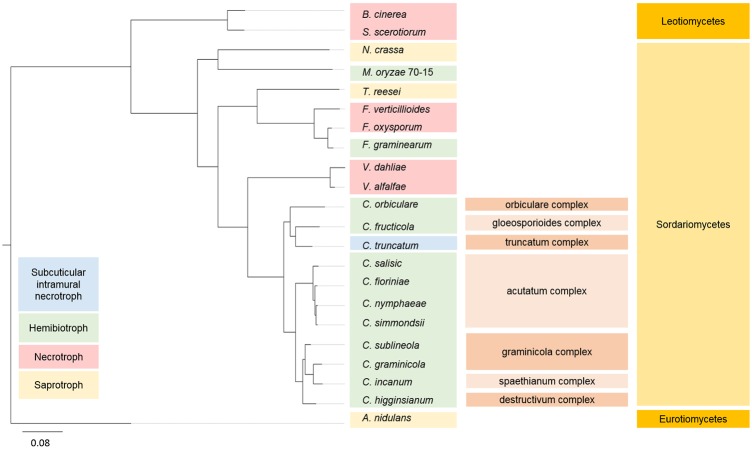
A phylogenetic tree was constructed based on the concatenated alignment of 515 highly conserved, single-copy orthologues of C. truncatum with Colletotrichum species and other ascomycete fungi with diverse lifestyles (Table 1). The inferred phylogeny showed that the genus Colletotrichum was most closely related to the Verticillium species and the two genera diverged from Fusarium, while C. truncatum and C. fructicola diverged from C. orbiculare (Fig 2). Interestingly, C. truncatum had more homologues in Fusarium species, specifically in F. oxysporum than in any other fungus outside the genus Colletotrichum. Orthologue analysis of Colletotrichum genus using Proteinortho showed that 88.8% of the C. truncatum genes (12,186) had an orthologue in at least one of the ten Colletotrichum species. There were 6,081 orthogroups consisting of proteins from all the Colletotrichum species, out of which, 6,010 contained single-copy orthologues. 1,409 genes of C. truncatum that did not cluster in orthogroups were subjected to BLASTP against the proteomes of other Colletotrichum species. The gene annotation and proteome of C. scovillei, which follows a subcuticular necrotrophic lifestyle on chilli, were not in the public domain at the time of writing this manuscript. Hence, only orthologue analysis was carried out through genome-wide comparison of C. truncatum genes. TBLASTX analysis showed that 12,792 of the total C. truncatum genes had homology in C. scovillei genome, of which 26 genes had a homologue only in C. scovillei among all Colletotrichum species. 377 genes had no homologues in any other species used in this study or SwissProt database, and hence represented the species-specific genes that are unique to C. truncatum.
Fig 2. A maximum likelihood tree of Colletotrichum species and other fungi with diverse lifestyles.
Bootstrap support values (1000 replicates) of 100% were obtained at the nodes. A. nidulans was taken as an outgroup for the analysis. The Colletotrichum species complexes and the families are shown in parallel. C. truncatum, C. gloeosporioides and C. orbiculare form a separate clade in the Colletotrichum genus suggesting their origin from a common ancestor.
Secretome and effector prediction
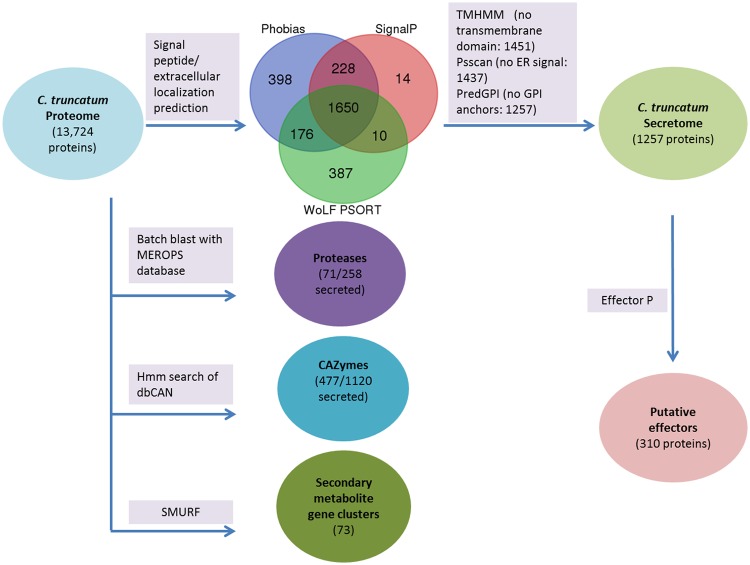
The secretome of a pathogen includes extracellular secreted proteins that are deployed to the host–pathogen interface during infection and includes important virulence factors such as effector proteins for manipulation of host cell dynamics and cell wall degrading enzymes. A highly stringent pipeline for secretome prediction in fungi, as described in FunSecKB2 [81] (Fig 3), was implemented to predict a refined secretome in C. truncatum (1,257 proteins) that represented 9.16% of the total proteome. The analysis of known domains in the secretome showed that it is rich in the plant cell wall degrading enzymes like proteases and CAZymes. 793 secretory proteins had Pfam annotations, most of which were associated with subtilisin-like serine proteases (40), oxidoreductases with FAD domain (28) and carbohydrate-binding modules (28). 310 putative effector proteins were predicted by EffectorP in the secretome. The homologues of some of the known effectors from other fungi, which were detected in other Colletotrichum species as well, were also identified in C. truncatum; like ChEC6 of C. higginsianum, an effector expressed specifically during appresorial penetration [82], necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins (NLPs), etc. (S6 Table). 59 proteins in the predicted secretome contained nuclear localization signal, 6 of which were specific to C. truncatum with no homology in Colletrotrichum species, while one had a homologue in C. scovillei only (CTRU_006472) and 15 of these were putative effectors, which may modulate the activity of host genes by localization into the host nucleus during infection. Most of the putative effectors in C. truncatum and other fungi were cysteine-rich, small, secreted proteins (SSPs, <300 amino acids, >3% cysteine) (S3 and S4 Figs) and lacked homology to any known protein in the SwissProt database, which are the hallmarks of effectors. The sizes of all the putative effectors in C. truncatum were below 358 amino acids (aa) except for CTRU_010949 (508 aa). 254 effectors of C. truncatum had orthologues in other fungi used in phylogenetic analysis, while 2 effectors had homologues only in C. scovillei. 75 of the 310 putative effectors had Pfam annotations, some of which were associated with enzymatic domains like pectin lyases (7), cutinase (6) and GDSL-like lipase/acylhydrolase family (5). Hence, 219 secreted proteins lacking homologues in the SwissProt database with no known function or Pfam domains were considered as candidate effectors for further functional studies (S4 Table). 34 secreted proteins of C. truncatum were specific to this species, of which 23 proteins lacking homology to any of the protein in SwissProt database were considered as species-specific effectors (S7 Table).
Fig 3. Work flow for functional annotation of some of the gene categories relevant to pathogenicity.
A stringent pipeline of tools was used to predict 1257 proteins that are highly likely to be secreted and other important categories like carbohydrate active enzymes (CAZymes), proteases and secondary metabolism (SM) gene clusters. 310 of the secreted proteins were predicted to be putative effectors, 477 were secreted CAZymes and 71 were secreted proteases. Secondary metabolite backbone genes were not detected in the secretome.
CAZymes
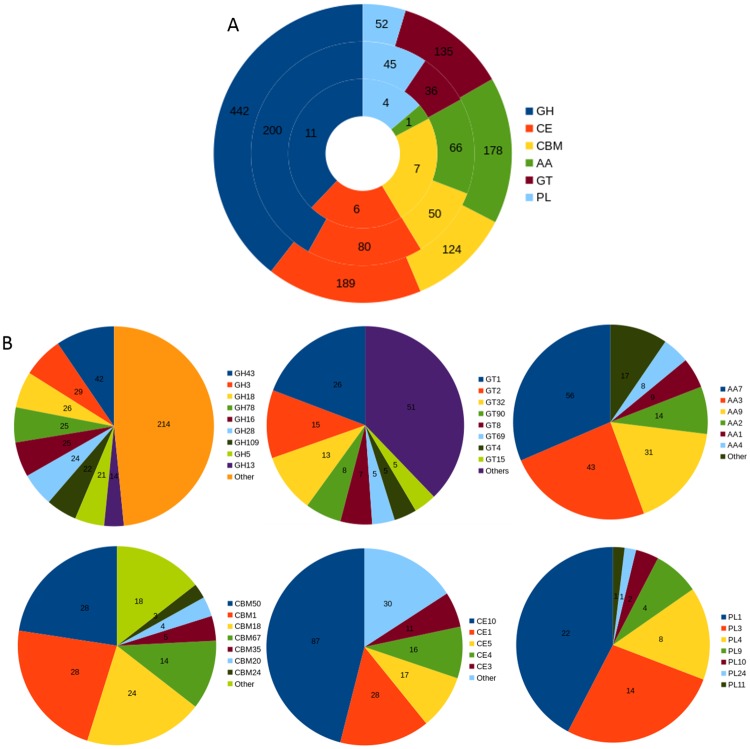
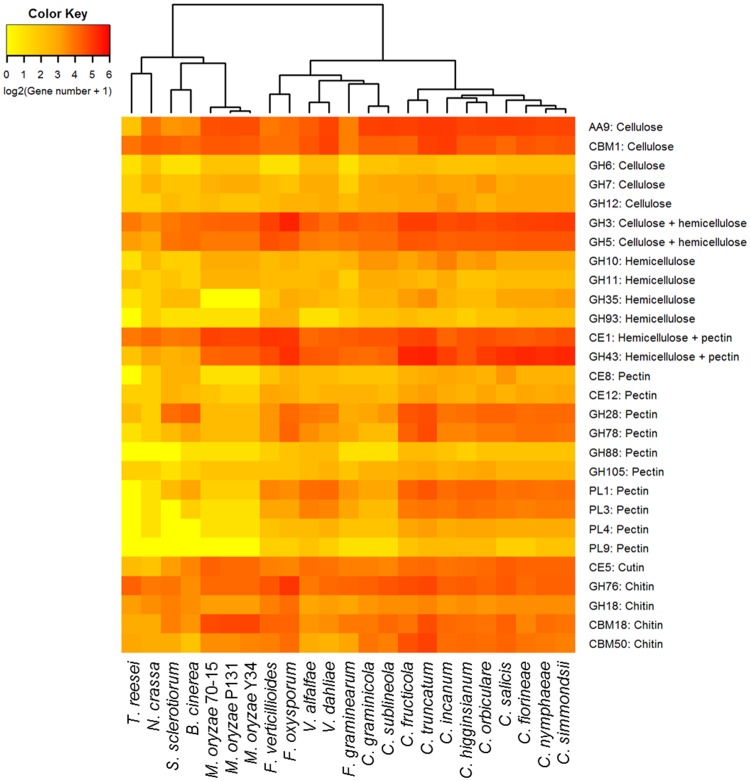
The carbohydrate metabolizing enzymes play an important role in the degradation of fungal and plant cell wall components (chitin, cellulose, hemicellulose, pectin, etc.) and utilization of plant polysaccharides during the host colonization by pathogenic fungi [83–85]. The C. truncatum genome had 1,036 genes encoding 147 different CAZyme families (Fig 4). 449 of C. truncatum genes associated with 88 CAZyme families were predicted to be secreted. The important classes of CAZymes like glycoside hydrolases (GH), carbohydrate esterases (CE), polysaccharide lyases (PL), auxillary activities (AA) and carbohydrate-binding modules (CBMs, lack catalytic activities but are included due to their association with catalytic modules) in C. truncatum were compared to other Colletotrichum species and other fungi. 84 CAZyme families were common in all the fungal species analysed (S8 Table), with choline esterases (CE10) representing the most abundant family (though most of the members of this family have non-carbohydrate substrates) followed by chitooligosaccharide oxidase (AA7). Cellobiose dehydrogenases (AA3, flavoproteins containing a flavin-adenine dinucleotide (FAD)-binding domain) was the third largest CAZyme family in all Colletotrichum species as well as in Leotiomycetes (Botrytis and Sclerotinia). The family AA9 with cellulase activity, which was previously designated as GH60, was also highly expanded in Colletotrichum species. The family GH43, with both hemicellulase and pectinase activities, was expanded in C. truncatum (42), C. fructicola (40) and acutatum species complex (35–39) as compared to other species in the genus (16–30). Species-specific expansion of one glycosyl transferase domain GT1 in C. truncatum (26) was also observed among all the fungi analysed.
Fig 4. Analysis of different CAZyme families in C. truncatum.
(A) The total number and category of CAZymes in predicted proteome (outer circle), secretome (middle circle) and putative effectors (inner circle) are shown. (B) The fraction of genes of major families of all the CAZyme classes, viz., glycoside hydrolases (GHs), glycosyltransferase (GTs), auxillary activities (AAs), carbohydrate-binding modules (CBMs), carbohydrate esterases (CEs) and polysaccharide lyases (PLs) are shown.
The heatmap of plant and fungal cell wall degrading enzymes (PCWDEs and FCWDEs) showed graminicola clade members, viz., C. graminicola and C. sublineola that specifically infect graminaceous monocot host, clustering away from the rest of the Colletotrichum species owing to the contraction of certain CAZyme families; while C. fructicola and C. truncatum with a broad host range formed a separate cluster due to the expansion of many families (Fig 5). 17 cutinases (CE5 family) were detected, 11 of which were homologous to CtCut1, a cutinase gene described previously for C. truncatum [86]. The chitin-binding domains belonging to the class CBM50 harboring 1–6 lysine motifs (LysM) and CBM18 were more abundant in C. truncatum genome as compared to other species. 7 genes had 3 CAZyme signatures; a chitin-binding CBM18 domain and up to three CBM50 domains as well as a GH18 catalytic domain with chitinase activity. 4 of these genes along with 10 other LysM genes (143–762 amino acid residues) were predicted to be secreted, while 3 were putative effectors (S9 Table). CTRU_001009 had 9 orthologues that were specific to genus Colletotrichum, and CTRU_002865 and CTRU_002908 had homologues in other fungi, while 11 of these proteins were unique to C. truncatum.
Fig 5. The heatmap showing important fungal and plant cell wall degrading CAZyme families in the proteomes of different fungi.
The members of graminicola species complex that specifically infect monocot hosts clustered away from the rest of the Colletotrichum species while C. fructicola and C. truncatum with broad host range showed the maximum expansion of pectin degrading CAZyme families within the genus.
The chitin- and cellulose-binding CBM1 family was specifically expanded in C. incanum (30) and C. truncatum (28) among the members of the Colletotrichum genus, with almost all genes predicted to be secreted. The genes containing CBM42 binding to arabinofuranose, were found in all fungi except C. truncatum and C. incanum. CBM67 binding to L-rhamnose showed high expansion specifically in C. truncatum (14) and F. oxysporum (15), whereas this family was absent in the members of graminicolous clade of Colletotrichum and also in the non-pathogenic fungi, N. crassa and T. reesei. Pectin-degrading enzymes (PL1 and PL3) showed the maximum expansion in Colletotrichum species when compared to other fungi. A pectin lyase family PL1 was observed in all fungi except T. reesei, and had highest expansion in C. truncatum (22).
Proteases
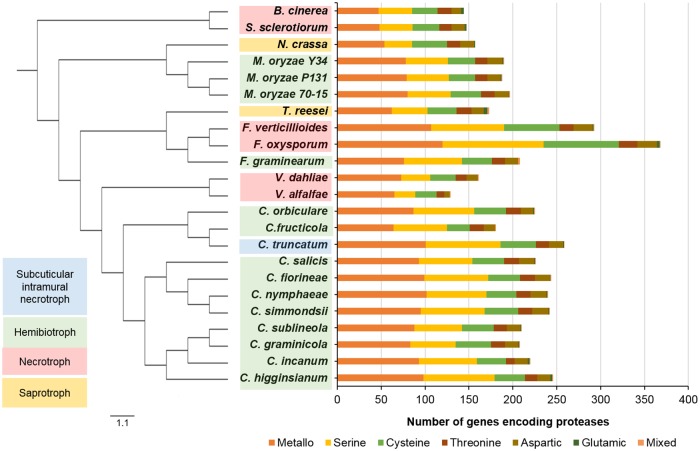
Proteases complement the plant cell wall degrading enzymes and protect the fungi from host pathogenicity-related proteins released during fungal invasion. Batch BLAST search against MEROPS protease database [73] identified 258 genes in C. truncatum belonging to 76 families of proteases and 10 genes belonging to 4 protease inhibitor families. The genes from S09 family were screened since they were likely to be alpha/beta hydrolases (carboxylesterases and lipases, but not proteases). The metalloproteases (101) comprised the largest category of proteases in C. truncatum, followed by serine (85) and cysteine proteases (40), with similar trend in other fungi (Fig 6). C. truncatum had 71 secretory proteases that included 35 serine and 29 metalloproteases. 13 out of 20 subtilisins (S08A) were predicted to be secreted (S10 and S12 Tables). The two chitin-degrading, secreted fungalysins (M36) in C. truncatum, CTRU_012332 and CTRU_004392, shared only 50% identity with each other and the former showed 91% identity to a C. fructicola fungalysin.
Fig 6. The comparative analysis of protease families in different fungal genomes.
Phylogenetic tree of fungi with diverse lifestyles is shown with corresponding protease families in each fungal genome. C. truncatum had the largest protease component among Colletotrichum species due to expansion of metallo- and serine proteases.
Interestingly, C. fructicola had the least number of proteases (185) among Colletotrichum species despite its wide host range. C. graminicola (213) and C. subliniola (215) also showed less diversity in proteases, similar to the trend observed with CAZymes (S11 Table). A total of 110 protease families were detected in all the fungi analysed, of which 46 families were common to all. The largest protease family in all the fungi studied was prolyl aminopeptidase (S33) followed by subtilisins (S08). The largest expansion of protease families was observed in Fusarium species, followed by Colletotrichum and Magnaporthe species. On the other hand, V. alfalfae (137) showed the least number of proteases among all the fungi. T. reesei (182) showed relatively large protease component than necrotrophic fungi like Verticillium spp, Sclerotinia and Botrytis; and non-pathogenic fungus Neurospora. The rare families included A11A that was observed only in F. oxysporum (7), S. sclerotiorum (6) and C. incanum (1).
Secondary metabolite (SM) gene clusters
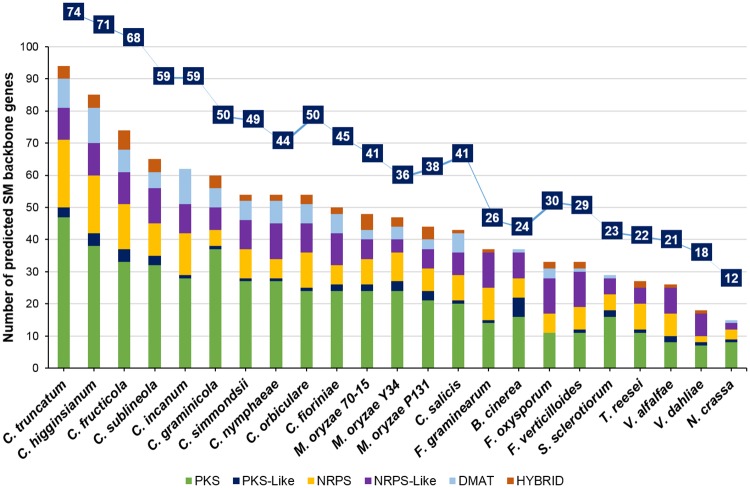
The SMs produced by the fungal phytopathogens are known to be associated with their pathogenicity and host range. The genes encoding the enzymes for the production of the SMs are located in the form of a cluster in the genome and their expression is transcriptionally co-regulated [87]. The comparative analysis of the SM gene clusters, predicted through SMURF, showed the highest number of such clusters in Colletotrichum species, ranging from 41 in C. salicis to 71 in C. higginsianum and 73 in C. truncatum (S13 Table), 2–4 fold higher than their close relatives in Verticillium (18–21 clusters) and Fusarium (26–30 clusters) (Fig 7). The SM backbone genes were mainly represented by 50 polyketide synthases (47 PKS and 3 PKS-like genes), 27 non-ribosomal protein kinases (17 NRPS and 10 NRPS-like genes), 9 dimethylallyl transferases (DMAT) and 4 PKS-NRPS hybrid genes in C. truncatum. The orthologue analysis identified 18 core SM backbone genes that were present in all the Colletotrichum species, including 6 PKS, 8 NRPS and 4 DMAT genes. The highest number of orthologues of SM backbone genes of C. truncatum was observed in C. higginsianum (59) and C. fructicola (58), respectively, most of which were PKS genes.
Fig 7. The comparative analysis of secondary metabolite (SM) gene clusters (line graph) and SM backbone genes (bar graph) predicted in different fungi using SMURF.
Colletotrichum species had maximum expansion of SM backbone genes among all the fungi analysed, while Verticillium species showed contraction of these genes.
Another tool, antiSMASH predicted 73 clusters in C. truncatum genome, including 29 type1 PKS (T1PKS), 12 NRPS, 7 each of t1PKS-NRPS hybrids, terpene and indole type, 1 each of T3PKS-T1PKS, indole-T1PKS and indole-T1PKS-NRPS hybrid type of SM clusters along with 8 other clusters (S14 Table). Some of the genes in 9 of the 73 antiSMASH predicted SM clusters showed homology to the known clusters in other fungi. One of the T1PKS clusters (cluster 71) showed 100% similarity with an alternapyrone biosynthetic gene cluster of Alternaria solani (S5 Fig). 51–68 SM gene clusters were predicted in C. scovillei, C. graminicola, C. fructicola and C. higginsianum, which were much less than those predicted in a previous study [26].
Cytochrome P450 monooxygenases (P450s) and transporters
P450s are the diverse, heme-thiolate proteins, which play an important role in primary and secondary metabolism, and fungal pathogenicity, and are often associated with the SM gene clusters [88]. 1,345 genes in C. truncatum had homologues in fungal cytochrome P450 database. 860 genes had more than 30% sequence identity to 347 proteins from the database with a minimum bit score of 60, showing a significant match. Comparative analysis of P450 proteins among all the fungi showed that many genes of C. higginsianum and C. graminicola, which were included in the database, were the core genes in the Colletotrichum genus (S15 Table).
Apart from P450s, transporters are also reported to be associated with the SM gene clusters and export of toxic SMs during host invasion and detoxification of fungal cells. 1,374 genes of C. truncatum had homologues (with >30% identity) in Transporter Classification Database (TCDB) with 340 genes belonging to the Major Facilitator Superfamily (MFS, 2.A.1), the largest category of secondary carriers involved in nutrient uptake and transport of toxins and drugs [89]. The second most expanded transporter family in C. truncatum was nuclear pore complex (NPC, 1.I.1) with 82 genes, involved in transport of mRNA and proteins across the nuclear envelope, followed by ABC transporter family (3.A.1) with 56 genes, which contains both uptake and efflux transport systems driven by ATP hydrolysis. The comparative analysis with other fungi showed the highest expansion of transporters in F. oxysporum and F. graminicola followed by C. fructicola and C. truncatum (S6 Fig). The electrochemical potential-driven transporters, which include a subclass of uniporters, symporters, antiporters (2.A) containing MFS (2.A.1), formed the largest class of transporters in all the fungi analysed.
Putative pathogenicity genes
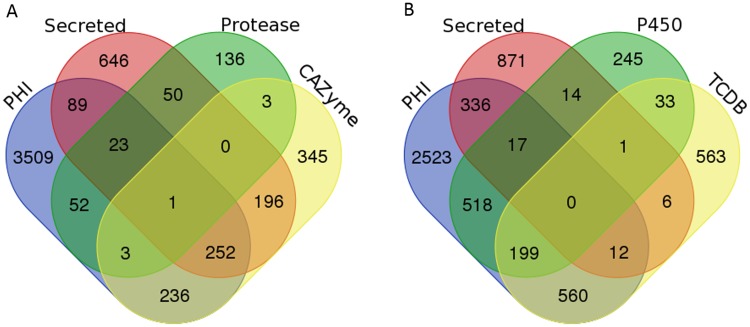
Pathogen-Host Interactions database (PHI-base) comprises of the genes associated with pathogenicity that are experimentally validated in different pathogens [76]. A total of 4,165 genes with homologues in the PHI-base were identified in C. truncatum, many of which were important for the fungal pathogenicity and virulence (S16 Table). The majority of these genes belonged to the category that show no effect on pathogenicity on mutagenesis, while others belonged to different functional categories relevant for pathogenicity including secreted proteases, CAZymes, P450s and transporters (Fig 8). 15.7% (2,156) of the predicted C. truncatum genes had homologues in PHI-base which were reported to affect the host-pathogen interactions during infection. Out of 4,165 genes, 1,470 genes were associated with reduced virulence, 263 with loss of pathogenicity, 287 with mixed phenotype, 59 with hypervirulence and 38 with effector functions. The comparative analysis of PHI-base homologues from other fungi revealed the maximum number of homologous genes from all categories in F. oxysporum, F. verticillioides, C. fructicola and C. truncatum, while M. oryzae had the highest number of effectors (S7 Fig).
Fig 8. Venn diagram showing the overlap of different gene categories relevant to fungal pathogenicity.
Overlap of secretory proteins and PHI-homologues with CAZymes and proteases (A), and cytochrome P450 and transporters with homologues in TCDB (B) representing the putative pathogenicity related genes with diverse functions.
Differential gene expression analysis
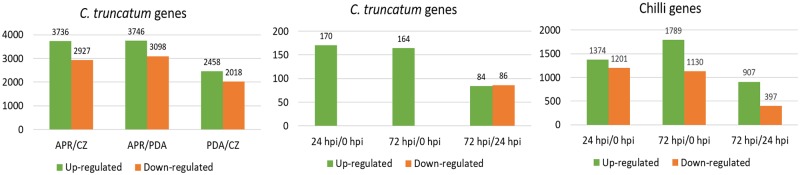
The RNA-Seq data from the limited number of samples was undertaken to generate evidence for gene annotation and to get clues regarding differentially expressed genes among in vitro and in planta samples, generating pilot data before scaling up the experiments. The GFOLD (or generalized fold change) algorithm was employed for this purpose since it was specifically designed for single samples without biological replicates and gives stable and biologically meaningful rankings of differentially expressed genes [50]. The normalized gene expression levels in each sample were used to estimate the fold changes (log2ratio) and those with at least 2-fold change (log2fdc ≥1 and Gfold (0.01) > 1) were considered as differentially expressed (Fig 9).
Fig 9. Differentially expressed genes under different conditions and infection stages in vitro and in planta.
(A) Number of differentially regulated genes of C. truncatum in APR and PDA compared to CZ and APR compared to PDA. (B) Number of differentially regulated genes of C. truncatum in planta at 24 and 72 hpi compared to 0 hpi (control, uninfected chilli) and at 72 hpi in comparison to 24 hpi. (C) Number of differentially regulated genes of chilli for 24 and 72 hpi in comparison to 0 hpi.
65–88% of the C. truncatum genes were expressed at different stages of development (in planta or in vitro). A total of 8,442 genes were differentially regulated in APR (in vitro appresorial assay) and PDA (hyphae and conidia grown on solid medium) compared to CZ (hyphae grown in liquid culture), 746 of which were predicted to be secreted, including 146 putative effectors, 274 CAZymes and 46 proteases. The majority of genes up-regulated in APR and PDA were associated with oxidation-reduction processes, transmembrane transport, Zn ion binding, ATP binding and nuclear transport, and included MFS-1, heterokaryon incompatibility protein (HET), fungal transcriptional regulatory proteins with Zn(2)-Cys(6) binuclear cluster domains, etc. Analysis of in planta samples showed 174 up-regulated genes in 24 hpi and 72 hpi compared to 0 hpi, including 13 CAZymes. 61 common genes were up-regulated in all in vitro and in planta samples compared to their respective controls (S17 Table). The fungal genes up-regulated in planta at 24 and 72 hpi included protein kinases, MFS-1, ring finger proteins with a role in the ubiquitanation pathway (E3 ubiquitin-protein ligase). More than 2,500 chilli genes were differentially regulated in early infection stages (24 and 72 hpi) as compared to control (0 hpi).
Discussion
The chilli anthracnose fungus, C. truncatum has been a serious threat for the major chilli producing countries for many decades. The wide host range and subcuticular intramural necrotrophic lifestyle make it an interesting candidate to study fungal pathogenicity and host specificity. With the development of tools for fungal transformation and availability of chilli genome sequences [36,90], chilli-C. truncatum pathosystem provides an excellent model to study host-pathogen interactions in which both the partners are amenable to genetic manipulation. The present study was undertaken with an aim to fill the lacunae in chilli-Colletotrichum interaction studies by providing a high quality reference genome sequence and annotation of this pathogen of a commercially important crop plant. The genome assembly of ~55 Mb was comparable to other sequenced Colletotrichum species except C. orbiculare (~90 Mb). The 6,793 contigs were assembled into 80 scaffolds, a low number for a genome sequenced on Illumina platform, due to the inclusion of two mate-pair libraries along with short insert paired-end reads for the assembly. This draft assembly represented the nuclear genome of C. truncatum, since no homology to the mitochondrial genome of other Colletotrichum species was observed.
Consensus gene models were predicted in C. truncatum through MAKER2 pipeline, taking into consideration the RNA-Seq data, homology with Colletotrichum species and SwissProt database as well as ab initio gene predictions from some of the best gene callers presently available for fungal genome annotation to enable accurate annotation of protein-coding genes. 13,724 genes were predicted with high confidence in C. truncatum as indicated by Annotation Edit Distance (AED) integrated in MAKER2. AED is a measure of the goodness of fit of an annotation to the evidence supporting it. AED of zero denotes perfect concordance with the available evidence (EST alignments, RNA-Seq and protein homology data), whereas a value of one indicates a complete absence of support for the annotated gene model [79]. In general, a well annotated genome is expected to have AED of less than 0.5 for 90% of the annotations and known domains for over 50% of its proteome [46]. >80% genes of the ‘gold-standard’ annotations of maize had AED scores of <0.2 [41]. In C. truncatum, 79% of the annotations had AED values of <0.2, which was very close to that of a well-annotated genome suggesting the reliability of these predictions. 74% of the total predicted proteins had a known IPR or Pfam domain and 66% had a homologue in SwissProt database. In contrast, only 2.6% genes had AED of 0.75–1, indicating little or no evidence support for these annotations. Interestingly, ~79% of the genes with AED of >0.75 had a Pfam domain, suggesting that they are not likely to be false positives. The other 21% genes with no known domain may include some unique genes and/or genes with undetectable expression or brief activation at certain stages, and can be prioritized for manual review. The C. truncatum genome assembly with small number of scaffolds, high N50 (1.6 Mb), detection of >99% of conserved eukaryotic/fungal genes, prediction of highly reliable gene models with less AED and presence of recognizable domains in a large number of predicted proteins point towards the high quality of the draft genome sequence of C. truncatum achieved in this study.
Synteny analysis of C. truncatum with C. higginsianum genome revealed a large fraction of genome which seems to be translocated or inverted in C. truncatum. Synteny analysis of C. graminicola with C. higginsianum and C. fructicola with C. orbiculare in earlier studies showed only 35–40% synteny [18,20], while >80% of the genes of the two species from graminicola species complex, C. graminicola and C. sublineola were syntenous [91]. Only 3% of the C. truncatum genome comprised of repetitive DNA elements (S2 and S3 Tables), which could be an under-estimation, likely due to the screening or collapse of the heterochromatin and other repetitive regions by the sequence assembly algorithm. The 4.75% of gaps observed in the assembly could also be attributed to the repetitive DNA, since some of the gaps that were inspected randomly had repetitive sequences flanking either the 5’ or 3’ ends. The completion of genome assembly and closure of the gaps in the existing sequence to get more contiguous assembly with fewer scaffolds will provide greater insights into the contribution of repetitive elements in the genome expansion and diversification of effectors. The predicted collinearity of existing scaffolds based on synteny mapping with C. higginsianum chromosomes can be used in future to get a nearly complete genome assembly.
The genome-wide comparative analyses of C. truncatum were carried out with 11 other Colletotrichum species (the proteomes for which are available in NCBI) and 8 other phylogenetically related fungi with diverse lifestyles including hemibiotrophs, necrotrophs and saprotrophs. The inferred phylogeny was consistent with the previous studies with Colletotrichum and Verticillium species diverging from Fusarium, pointing at their origin from a common ancestor. Hence, these genera are expected to share more homologues among each other and show similar expansion or contraction of certain gene families. ~89% of predicted genes had homologues in other Colletotrichum species reflecting the reliability of predicted gene models. Analysis of orthologues revealed the presence of 377 unique genes in C. truncatum, which had no homology to known proteins in SwissProt database and could be associated with its host-specific lifestyle. However, there is a possibility that predicted gene numbers may be inflated with incorrect or truncated gene structures which require manual curation.
The results of the comparative genomics were consistent with the previous findings that show the expansion of certain gene classes potentially involved in pathogenesis, in particular secreted proteins and effectors, CAZymes, proteases, SM synthesis and associated genes in Colletotrichum species when compared to other phytopathogenic fungi [18,20,25,92]. Though Colletotrichum genus is closely related to Verticillium species, the pathogenicity gene categories in all species including C. truncatum were similar to Fusarium and Magnaporthe species. C. truncatum had a diverse arsenal of such genes similar to the hemibiotrophic fungi when compared to necrotrophic and saptrotrophic fungi (except Fusarium species).
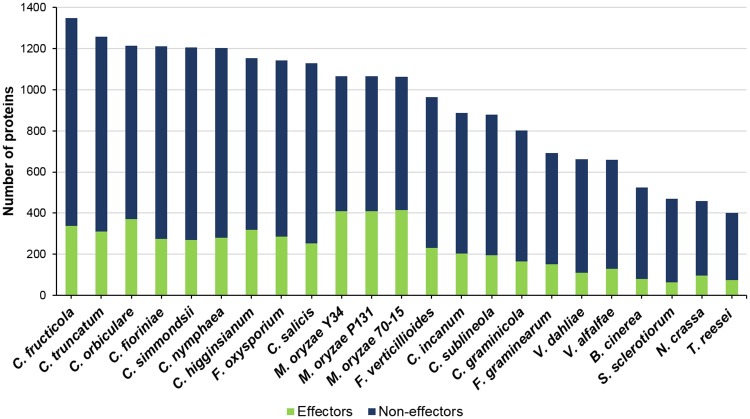
Pathogenic fungi overcome the host immune response by secreting effector molecules that manipulate cellular dynamics and help them evade the host defence responses and establish successful infection [93]. Effectors are typically lineage-specific, small, secreted, cysteine-rich proteins, most of which lack homology to known proteins, and have no recognizable domains [71,93]. However, some of the known effectors have homologues in related fungi as well as functional domains [94–96], though fungal effector proteins do not share a conserved host targeting signal like RxLR motifs of Oomycetes. A stringent strategy based on the tools described for fungal secretome prediction [81] was used in this study for the prediction of proteins that are highly likely to be secreted in C. truncatum as well as in other fungi. Though it resulted in reduction in the number of predicted secreted proteins reported in the previous studies through different tools (Fig 10), but reduced the probability of finding false positives in the secretomes of various fungi. However, use of such stringent strategies may result in some false negatives, similar reductions in the number of proteins in the refined secretomes of other fungi like F. graminearum [97], S. sclerotiorum [98] and B. cinerea [98] were observed using other stringent methods.
Fig 10. The number of secreted proteins and putative effectors, predicted using EffectorP, in all the fungal species analysed.
C. fructicola and C. truncatum had the largest number of secreted proteins while Magnaporthe species had the largest effector component.
The C. truncatum secretome was rich in proteins like FAD oxidases, subtilisins, pectin lyases and putative effectors, which have been reported to play a role in host infection. The presence of the nuclear localization signal in many of the effectors implied their crucial role in the manipulation of host genes, probably related to defence, through translocation into the host nucleus and transcriptional reprogramming. 219 candidate effectors were shortlisted based on their lack of homology to known proteins or domains. 36 species-specific proteins with no known domains and no homologues in the SwissProt database were predicted to be secreted in C. truncatum, 25 of which were putative effectors and were expressed in planta and/or in vitro. The homologues of some of the known fungal effectors that are present in all pathogenic fungi like LysM, NLPs etc. were identified in the C. truncatum genome as well. The homologues of C. higginsianum extracellular LysM protein effectors (ChELPs) [99] that were shown to be essential for appresorial function and suppressing chitin-triggered plant immunity, and another effector that is secreted from appresorial pore during early penetration (ChEC6) [82] were observed in C. truncatum as well and possibly might play an important role during host infection. Interestingly, CtNudix, a nudix hydrolase effector expressed during transition to necrotrophy in the cowpea anthracnose pathogen C. lentis [100], had no homologue in C. truncatum which might be in tune with its host specific necrotrophic lifestyle which contrasts hemibiotrophy. It was reported to have homologues only in hemibiotrophic fungi, but not in biotrophic or necrotrophic fungi [92,101].
Analysis of the major polysaccharide degrading enzyme (CAZyme) families with a wide range of substrates revealed lineage specific expansion of pectin degrading CAZymes (GH28, GH78, PL1 and PL3), cutinases (CE5) in C. truncatum and C. fructicola, both of which cause fruit rots in a number of hosts. These CAZyme families probably aid in the degradation of thick cuticle and complex cell walls of fruits rich in pectins and provide an evolutionary advantage to these fruit rot fungi. On the other hand, contraction of these families along with AA class in C. graminicola and C. sublineola points towards their dispensability for monocot infecting graminicolous clade as reported earlier [26]. Some Colletotrichum species like the members of gloeosporioides and acutatum clade, employ a stealth strategy, wherein they were reported to be either physiologically inactive on fruits until the ripening process starts, thus enabling them to avoid the host defence mechanisms within unripe, pre-climacteric fruits, or by endophytic species colonizing the living host tissues asymptomatically [102]. The abundance of CBM50 (LysM effectors) along with CBM18, the other chitin-binding module mainly associated with CE4 chitin deacetylase, and GH18 chitinase (apart from CBM50), and chitin- and cellulose-binding CBM1 family was a prominent feature of C. truncatum CAZyme and secretome components. These secreted CAZymes may play an important role during early infection by reducing the release of chitooligosaccharides by action of plant chitinases or sequestering the released chitin oligomers that act as Microbe Associated Molecular Patterns (MAMPs) and thereby preventing their recognition by plant immune system [20,64,94,99]. The expression of these CAZymes after host invasion may be responsible for the stealth strategy employed by C. truncatum during endophytic or quiescent phase of infection that has been observed even in a susceptible host like chilli [16,103].
Apart from CAZymes, proteases are the major plant cell wall degrading enzymes, of which serine- and metallo-proteases are the most important classes present in pathogenic fungi including Colletotrichum species. The aspartic protease family was also expanded in the Colletotrichum species that encounter acidic environment while infecting fruits. Interestingly, Trichoderma had larger protease component than non-pathogenic fungi like Neurospora and some of the necrotrophs. The expansion of proteases and CAZymes for lignocellulosic substrates in Trichoderma may be attributed to its saprophytic lifestyle that involves the digestion of dead plant material and cell wall components. C. truncatum showed a variety of metalloproteases, which possibly reflects a link between certain metalloproteases and necrotrophic lifestyle. The two fungalysins in C. truncatum (CTRU_004392 and CTRU_012332), the evolutionarily conserved chitin degrading enzymes present in all fungi analysed, had characteristic features of zinc metalloproteases, viz; a fungalysin/thermolysin propeptide (FTP) domain and M36 peptidase domain with catalytic activity and HEXXH motif [104]. Most of the fungi in family Sordariomycetes contain a single fungalysin gene. In a previous study, the phylogeny based on fungalysins from different fungi suggested that a gene duplication event probably might have occurred prior to the appearance of the Sordariomycetes, followed by numerous gene loss events of one or two copies in most of the members of Sordariomycetes [104]. This may explain the presence of two copies of this gene in C. truncatum and Verticillium species.
Subtilisin (S08A) was the most expanded family of alkaline proteases in C. truncatum as well as other species of the genus. Local alkalinization of the host tissue due to fungal secretion of ammonia was reported in many Colletotrichum species which provides an optimum pH to secreted hydrolases such as pectin-degrading enzymes [18,20]. The expansion of acidic and alkaline proteases in Colletotrichum species gives these fungi the ability to withstand a wide pH range encountered at different stages of infection in a number of hosts. There were reports of horizontal gene transfer (HGT) of subtilisin genes from plants to Colletotrichum fungi and hence such genes are named as Colletotrichum plant-like subtilisins (CPLS) [105]. C. truncatum had 4 proteins homologous to CPLSs that were similar to subtilisin-like protease (SBT1.7) of Arabidopsis thaliana based on SwissProt database search. CTRU_000179 and CTRU_000180 were found to be truncated and hence were fused and named as CTRU_000179A. All three CPLSs (CTRU_000179A, CTRU_007447 and CTRU_008341) had different sequences from each other, but retained the three characteristic domains, viz., peptidase inhibitor I9 domain (PF05922), the PA domain (PF02225) and the peptidase S8 domain (PF00082) with conserved catalytic triad at active site. When these three proteins were subjected to BLASTP analysis against the NCBI non-redundant database, among the top hits were subtilisin-like proteases from plants, including many Brassica species, Gossypium species, lotus, tobacco etc. and the proteins from two other distantly related ascomycetes, Rhynchosporium and Diaporthe species. This finding further supported the theory of HGT of some of the subtilisin genes from plants into an ancestor of Colletotrichum species. Apart from the subtilisins, there were 268 more proteins in C. truncatum proteome that had their best hits in A. thaliana in SwissProt database, while many proteins had hits in other plants like rice, maize, tobacco, tomato, chilli, cucurbits, etc. Additionally, many genes appeared to be of bacterial, rather than fungal, origin. This finding suggests that with additional analyses, an evidence for the HGT in many of these genes could be obtained in the future.
During host invasion, the fungal pathogens produce secondary metabolites that affect their pathogenicity and virulence, while many host plants produce antimicrobial compounds, SMs and toxins. Fungi counteract these toxins by enzymatic degradation or export them through membrane transporters. The comparative genomic analysis showed a genus-specific expansion of SM gene clusters in Colletotrichum species, including P450 genes, transporters and TFs suggesting their transcriptional co-regulation that may affect their virulence and help in nutrient uptake and export of toxins and antimicrobial compounds produced by the host. The expansion in SM gene clusters predicted through SMURF in C. truncatum (73), C. higginsianum (71) and C. fructicola (68) may also reflect their broad host range. An expansion of PKS and NRPS backbone genes, including some unique genes in C. truncatum, shows greater capacity of production of these SMs, which may play a significant role in host invasion by enabling appresorial penetration and pathogenicity. Another tool used for SM gene cluster prediction, antiSMASH predicted 73 clusters in C. truncatum, some of which had homologous genes from known SM clusters of other fungi (S5 Fig). >80 clusters were predicted for other Colletotrichum species in a previous study using an earlier version of antiSMASH (v1.2.2) [26]. The reduction in number of predicted clusters for some of these fungi (<68) could be attributed to the advanced version of the online tool (fungiSMASH) and the parameters used for the analysis.
The Pathogen-Host Interactions database (PHI-base) serves as a multi-species catalogue of experimentally validated pathogenicity genes (through gene/transcript disruption experiments) from plant and animal pathogens (bacteria, fungi, protists and nematodes) that are manually curated from peer-reviewed publications [76,106]. 15% of C. truncatum proteome had homologues in PHI-base associated with fungal pathogenicity including effectors, CAZymes, proteases, transporters and P450 genes. Most of the C. truncatum genes had homology to genes from F. oxysporum and M. oryzae due to a plethora of research carried out on these model fungi making them the highest represented pathogens in PHI-base. The majority of these genes was associated with reduced virulence. These genes, along with the genes associated with loss of pathogenicity and homologues of characterized effectors could be good candidates for further research. The comparative analysis of PHI-base homologues revealed a rich repertoire of pathogenicity genes in C. truncatum and C. fructicola among all fungi except the Fusarium species.
The RNA-Seq data was primarily used to support the annotation of protein coding genes and to get a glimpse of the differential gene expression at various developmental stages. GFOLD, which is one of the best available algorithms for differential expression analysis from single samples without biological replicates, was used to analyse RNA-Seq data. More fungal genes were expressed in vitro than in planta probably due to lesser representation of the fungal partner in the latter. In vitro appresoria (APR) showed the highest number of differentially regulated genes compared to the mycelial and conidial biomass grown in rich medium (PDA) and mycelia grown in minimal medium (CZ). Many secreted CAZymes, proteases, TFs and PHI-base homologues were up-regulated in appresoria as well as in planta, which may be needed to penetrate the host cell wall during initial infection. 80% of the species-specific genes in C. truncatum were expressed in vitro and/or in planta, some of which had at least one of the IPR or Pfam annotations indicating that these represent the genuine genes and not artifacts. Though the in vitro samples included about half of the total predicted genes that showed differential expression, the in planta samples had only a small fraction of such fungal genes, possibly due to the infection assay adopted, which avoided wounding the chilli surface that might have resulted in less representation of fungal transcripts. Even though the differential gene expression studies were performed without the biological replicates, the genes expressed during appresorial development and initial infection of chilli provide the candidates for future studies to understand the molecular mechanism of fungal pathogenicity.
Deciphering the genetic basis of the infection mechanism and host-specific lifestyle of fungal pathogens is crucial for a better understanding of host-fungal interactions and to develop effective and novel strategies to combat the pathogen. With the availability of whole genome sequences of chilli [90][36] and its major pathogens, C. scovillei (deposited as C. acutatum) [24] and C. truncatum (present study) in the public domain, the chilli-Colletotrichum pathosystem provides an attractive model to study host-pathogen interactions. Due to the unavailability of annotated genes of C. scovillei in public domain, direct comparison of pathogenicity genes among these two species, showing necrotrophy on the same host, was not possible. However, 26 genes of C. truncatum, which have homologs only in C. scovillei, could be candidates for further studies to delineate the host-specific infection strategy of both the fungi. The existence of conserved fungal genes and core genes from the genus Colletotrichum in C. truncatum genome reflects the good quality of assembly and gene predictions that were achieved in the present study. RNA-Seq analysis not only provided evidence for the gene structure prediction, but also provided the preliminary cues to design high throughput experiments to study gene regulation and a list of species-specific candidate genes for experimental validation and functional characterization. The comparative genomic analyses of the gene categories relevant for fungal pathogenicity revealed the conserved genes among Colletotrichum species with a broad host range and unique genes, including effectors; SM associated genes, PHI homologues, expanded CAZymes and proteases, and the absence of hemibiotrophic effector CtNudix in C. truncatum. Many of the genes belonging to these pathogenicity relevant classes could be associated with its host specific subcuticular intramural necrotrophic lifestyle and need to be functionally characterized. Summarily, this study provides a high quality reference genome sequence and annotation of genes with putative roles in pathogenicity and host interaction of an important Colletotrichum species, which would serve as a genomic resource to facilitate further functional and evolutionary studies of this agronomically important fungal pathogen and to develop novel disease control measures.
Supporting information
C. truncatum genome and proteome sizes were comparable to other Colletotrichum species, except C. orbiculare and F. oxysporum which had the largest genome and proteome among the fungi analysed, respectively.
(TIF)
Monilochaetes infuscans was taken as an outgroup. The species with genome sequence available at the time of sequencing of C. truncatum (MTCC no. 3414) are marked with red solid circles. Bootstrap support values (1000 replicates) above 50% are shown at the nodes.
(TIF)
The small (below 300 amino acids), secreted, cysteine-rich proteins can be considered as candidate effectors if they lack homology to known proteins and functional domains.
(TIF)
The median sizes of all secreted proteins in Colletotrichum species were below 300 amino acids (aa), except for M. oryzae strains which had median at 200 aa and encode maximum number of reported effectors among all the fungi analysed. The sizes of all the putative effectors in C. truncatum were below 358 aa except for CTRU_010949 (508 aa). The outliers were removed from the plot.
(TIF)
The corresponding homologous genes are shown in red, green and blue colours. MFS: Major Facilitator Superfamily (transporter).
(TIF)
Fusarium species had exceptionally high number of transporters among all fungi followed by Colletotrichum species. The electrochemical potential-driven transporters, which include a subclass of uniporters, symporters, antiporters (2.A) containing Major Facilitator Superfamily (2.A.1), formed the largest class of transporters in all the fungi analysed, followed by uptake and efflux transport systems driven by ATP hydrolysis, containing ABC transporter family (3.A.1).
(TIF)
(A) The homologues from some of the most abundant categories of genes in PHI-base. The category of genes with unaffected pathogenicity phenotype (validated through mutagenesis) were the largest among PHI-base homologues in all fungi followed by the category of genes with reduced pathogenicity. (B) The homologues from some of the least represented categories of genes in PHI-base. The category of genes with hypervirulence phenotype was the largest among all fungi except M. oryzae that had the largest effector component. F. oxysporum and F. verticillioides had the maximum number of PHI-base homologues reflecting the amount of experimental data available for this genus.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank P. Sairam Reddy, JK AgriGenetics Ltd, Hyderabad, India for the kind gift of C. annuum (var. RP16) fruits and seeds; Vineesha Oddi for her assistance with RNA extractions and online analyses, and Archana Tomar and Rohan Mishra for their help in computational analyses.
Data Availability
The genome sequence of Colletotrichum truncatum was deposited at the NCBI Genome Database under the accession number NBAU00000000. All the raw data used for genome and transcriptome assemblies can be accessed from the figshare repository using the following links: https://figshare.com/s/f8d4908e91600eebc7c3, https://figshare.com/s/09f44d16fc8966daedba, https://figshare.com/s/9374f90804c944c5399c, https://figshare.com/s/35c459e9f4ac27127902, https://figshare.com/s/e81941f2d19afa090c45, https://figshare.com/s/7bfd5631ac659c8f2b34, https://figshare.com/s/4c68fb7aaa33c4db1445, https://figshare.com/s/17ee5ef5ff3490eeb42f, https://figshare.com/s/6140645e2f22350ff824.
Funding Statement
This study was supported by the core grants of CDFD. No additional funding was received for this study. SR was the recipient of Junior and Senior Research Fellowships of the University Grants Commission (UGC), India towards the pursuit of a Ph.D. degree of the Manipal University. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Kraft KH, Brown CH, Nabhan GP, Luedeling E, Luna Ruiz JdJL, d’Eeckenbrugge GC, et al. Multiple lines of evidence for the origin of domesticated chili pepper, Capsicum annuum, in Mexico. Proc Natl Acad Sci. 2014;111: 6165–6170. doi: 10.1073/pnas.1308933111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Than PP, Jeewon R, Hyde KD, Pongsupasamit S, Mongkolporn O, Taylor PWJ. Characterization and pathogenicity of Colletotrichum species associated with anthracnose on chilli (Capsicum spp.) in Thailand. Plant Pathol. 2008;57: 562–572. [Google Scholar]
- 3.Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13: 414–430. doi: 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon PF, Damm U, Johnston PR, Weir BS. Colletotrichum—current status and future directions. Stud Mycol. 2012;73: 181–213. doi: 10.3114/sim0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacquard S, Kracher B, Hiruma K, Münch PC, Garrido-Oter R, Thon MR, et al. Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat Commun. 2016;7: 11362 doi: 10.1038/ncomms11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montri P, Taylor PWJ, Mongkolporn O. Pathotypes of Colletotrichum capsici, the causal agent of chili anthracnose, in Thailand. Plant Dis. 2009;93: 17–20. [DOI] [PubMed] [Google Scholar]
- 7.Damm U, Woudenberg JHC, Cannon PF, Crous PW, Damm U. Colletotrichum species with curved conidia from herbaceous hosts. 2009;39: 45–87. [Google Scholar]
- 8.Shenoy BD, Jeewon R, Lam WH, Bhat DJ, Than PP, Taylor PWJ, et al. Morpho-molecular characterisation and epitypification of Colletotrichum capsici (Glomerellaceae, Sordariomycetes), the causative agent of anthracnose in chilli. Fungal Divers. 2007;27: 197–211. [Google Scholar]
- 9.Than PP, Prihastuti H, Phoulivong S, Taylor PWJ, Hyde KD. Chilli anthracnose disease caused by Colletotrichum species. J Zhejiang Univ Sci B. 2008;9: 764–78. doi: 10.1631/jzus.B0860007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manandhar J, Hartman G, Wang T. Anthracnose development on pepper fruits inoculated with Colletotrichum gloeosporioides. Plant Disease. 1995;79(4): 380–3. [Google Scholar]
- 11.Jayawardena RS, Hyde KD, Damm U, Cai L, Liu M, Xh L, et al. Notes on currently accepted species of Colletotrichum. 2016;7: 1192–1260. [Google Scholar]
- 12.Squissato V, Yucel YH, Richardson SE, Alkhotani A, Wong DT, Nijhawan N, et al. Colletotrichum truncatum species complex: Treatment considerations and review of the literature for an unusual pathogen causing fungal keratitis and endophthalmitis. Med Mycol Case Rep. 2015;9: 1–6. doi: 10.1016/j.mmcr.2015.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey JA, O’Connell RJ, Pring RJ, Nash C. Infection strategies of Colletotrichum species. Colletotrichum: Biology, Pathology, and Control. 1992; 88–120. [Google Scholar]
- 14.Perfect SE, Hughes HB, O’Connell RJ, Green JR. Colletotrichum: A Model Genus for Studies on Pathology and Fungal–Plant Interactions. Fungal Genet Biol. 1999;27: 186–198. doi: 10.1006/fgbi.1999.1143 [DOI] [PubMed] [Google Scholar]
- 15.Auyong ASM, Ford R, Taylor PWJ. Genetic transformation of Colletotrichum truncatum associated with anthracnose disease of chili by random insertional mutagenesis. J Basic Microbiol. 2012;52: 372–382. doi: 10.1002/jobm.201100250 [DOI] [PubMed] [Google Scholar]
- 16.Ranathunge NP, Bajjamage H, Sandani P. Deceptive behaviour of Colletotrichum truncatum: strategic survival as an asymptomatic endophyte on non-host species. J Plant Prot Res. 2016;56: 157–162. [Google Scholar]
- 17.Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, et al. Fungal Effectors and Plant Susceptibility. Annu Rev Plant Biol. 2015;66: 513–545. doi: 10.1146/annurev-arplant-043014-114623 [DOI] [PubMed] [Google Scholar]
- 18.O’Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet. 2012;44: 1060–5. doi: 10.1038/ng.2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zampounis A, Pigné S, Dallery J-F, Wittenberg AHJ, Zhou S, Schwartz DC, et al. Genome Sequence and Annotation of Colletotrichum higginsianum, a Causal Agent of Crucifer Anthracnose Disease. Genome Announc. 2016;4: e00821–16. doi: 10.1128/genomeA.00821-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan P, Ikeda K, Irieda H, Narusaka M, O’Connell RJ, Narusaka Y, et al. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 2013;197: 1236–1249. doi: 10.1111/nph.12085 [DOI] [PubMed] [Google Scholar]
- 21.Weir BS, Johnston PR, Damm U. The Colletotrichum gloeosporioides species complex. Stud Mycol. 2012;73: 115–80. doi: 10.3114/sim0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baroncelli R, Sreenivasaprasad S, Sukno S, Thon MR, Holub E. Draft Genome Sequence of Colletotrichum acutatum Sensu Lato (Colletotrichum fioriniae). Genome Announc. 2014;2: e00112–14. doi: 10.1128/genomeA.00112-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baroncelli R, Sanz-Martín JM, Rech GE, Sukno S a, Thon MR. Draft Genome Sequence of Colletotrichum sublineola, a Destructive Pathogen of Cultivated Sorghum. Genome Announc. 2014;2: 10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J-H, Chon J-K, Ahn J-H, Choi I-Y, Lee Y-H, Kim KS. Whole genome sequence and genome annotation of Colletotrichum acutatum, causal agent of anthracnose in pepper plants in South Korea. Genomics Data. 2016;8: 45–46. doi: 10.1016/j.gdata.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gan P, Narusaka M, Kumakura N, Tsushima A, Takano Y, Narusaka Y, et al. Genus-wide comparative genome analyses of Colletotrichum species reveal specific gene family losses and gains during adaptation to specific infection lifestyles. Genome Biol Evol. 2016;8: evw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baroncelli R, Amby DB, Zapparata A, Sarrocco S, Vannacci G, Le Floch G, et al. Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genomics. 2016;17: 555 doi: 10.1186/s12864-016-2917-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. 2012;109(16):6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6. 0. 2013;30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20: 265–72. doi: 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23: 1061–1067. doi: 10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- 31.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva E V, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31: 3210–3212. doi: 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 32.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30: 2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopylova E, Noe L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28: 3211–3217. doi: 10.1093/bioinformatics/bts611 [DOI] [PubMed] [Google Scholar]
- 34.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27: 863–4. doi: 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimin A V, Marcais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29: 2669–2677. doi: 10.1093/bioinformatics/btt476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. 2014;111(14): 5135–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. Nature Research; 2015;12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29: 644–652. doi: 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22: 1658–1659. doi: 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 40.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31: 5654–5666. doi: 10.1093/nar/gkg770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. 2011;12(1):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5: 59 doi: 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanke M, Tzvetkova A, Morgenstern B. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 2006;7: S11 doi: 10.1186/gb-2006-7-s1-s11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33: 6494–506. doi: 10.1093/nar/gki937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parra G, Blanco E, Guigó R. GeneID in Drosophila. Genome Res. 2000;10: 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell MS, Holt C, Moore B, Yandell M, Campbell MS, Holt C, et al. Genome Annotation and Curation Using MAKER and MAKER-P. Current Protocols in Bioinformatics. 2014. pp. 4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis SE, Searle SMJ, Harris N, Gibson M, Lyer V, Richter J, et al. Apollo: a sequence annotation editor. Genome Biol. 2002;3: RESEARCH0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997; 25(5):955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14: R36 doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics. 2012;28: 2782–2788. doi: 10.1093/bioinformatics/bts515 [DOI] [PubMed] [Google Scholar]
- 51.Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: A system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16: 1159–68. doi: 10.1101/gr.5396706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lechner M, Findeiß S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. 2011;12(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25: 1972–1973. doi: 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kück P, Meusemann K. FASconCAT: Convenient handling of data matrices. Mol Phylogenet Evol. 2010;56: 1115–1118. doi: 10.1016/j.ympev.2010.04.024 [DOI] [PubMed] [Google Scholar]
- 56.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22: 2688–2690. doi: 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 57.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27: 1164–1165. doi: 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klosterman SJ, Subbarao K V., Kang S, Veronese P, Gold SE, Thomma BPHJ, et al. Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens. PLoS Pathog. 2011;7: e1002137 doi: 10.1371/journal.ppat.1002137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi M-J, Di Pietro A, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464: 367–373. doi: 10.1038/nature08850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434: 980–986. doi: 10.1038/nature03449 [DOI] [PubMed] [Google Scholar]
- 61.Xue M, Yang J, Li Z, Hu S, Yao N, Dean RA, et al. Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus Magnaporthe oryzae. PLoS Genet. 2012;8(8): e1002869 doi: 10.1371/journal.pgen.1002869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422: 859–868. doi: 10.1038/nature01554 [DOI] [PubMed] [Google Scholar]
- 63.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol. 2008;26: 553–560. doi: 10.1038/nbt1403 [DOI] [PubMed] [Google Scholar]
- 64.Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7 (8):e1002230 doi: 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30: 1236–40. doi: 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8: 785–786. doi: 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 67.Käll L, Krogh A, Sonnhammer EL. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J Mol Biol. 2004;338: 1027–1036. doi: 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- 68.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35: 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305: 567–580. doi: 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 70.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10: 202 doi: 10.1186/1471-2105-10-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sperschneider J, Gardiner DM, Dodds PN, Tini F, Covarelli L, Singh KB, et al. Effector P: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 2016;210: 743–761. doi: 10.1111/nph.13794 [DOI] [PubMed] [Google Scholar]
- 72.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40: W445–W451. doi: 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40: D343–D350. doi: 10.1093/nar/gkr987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, et al. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47: 736–741. doi: 10.1016/j.fgb.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39: W339–W346. doi: 10.1093/nar/gkr466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winnenburg R, Baldwin TK, Urban M, Rawlings C, Köhler J, Hammond-Kosack KE. PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 2006;34: D459–D464. doi: 10.1093/nar/gkj047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saier MH, Tran C V, Barabote RD, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34: D181–6. doi: 10.1093/nar/gkj001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eilbeck K, Moore B, Holt C, Yandell M. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics. 2009;10: 67 doi: 10.1186/1471-2105-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Todd RB, Zhou M, Ohm RA, Leeggangers HACF, Visser L, de Vries RP. Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genomics. 2014;15: 214 doi: 10.1186/1471-2164-15-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meinken J, Asch DK, Neizer-Ashun KA, Chang G-H, R.Cooper C JR, Min XJ. FunSecKB2: a fungal protein subcellular location knowledgebase. Comput Mol Biol. 2014;4: 1–17. [Google Scholar]
- 82.Kleemann J, Rincon-Rivera LJ, Takahara H, Neumann U, Ver Loren van Themaat E, van Themaat EVL, et al. Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 2012;8: e1002643 doi: 10.1371/journal.ppat.1002643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kubicek CP, Starr TL, Glass NL. Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol. 2014;52: 427–451. doi: 10.1146/annurev-phyto-102313-045831 [DOI] [PubMed] [Google Scholar]
- 84.Zhao Z, Liu H, Wang C, Xu J-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14(1): 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lyu X, Shen C, Fu Y, Xie J, Jiang D, Li G, et al. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci Rep. 2015;5: 15565 doi: 10.1038/srep15565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Auyong SM, Ford R, Taylor WJ. The Role of Cutinase and its Impact on Pathogenicity of Colletotrichum truncatum. Journal of Plant Pathology & Microbiology. 2015;6: 259. [Google Scholar]
- 87.Palmer JM, Keller NP. Secondary metabolism in fungi: does chromosomal location matter? Curr Opin Microbiol. 2010;13: 431–436. doi: 10.1016/j.mib.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Črešnar B, Petrič Š. Cytochrome P450 enzymes in the fungal kingdom. Biochim Biophys Acta—Proteins Proteomics. 2011;1814: 29–35. [DOI] [PubMed] [Google Scholar]
- 89.Saier MH, Reddy VS, Tsu B V., Ahmed MS, Li C, Moreno-Hagelsieb G. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016;44: D372–D379. doi: 10.1093/nar/gkv1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim S, Park M, Yeom S-I, Kim Y-M, Lee JM, Lee H-A, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 2014;46: 270–278. doi: 10.1038/ng.2877 [DOI] [PubMed] [Google Scholar]
- 91.Buiate EAS, Xavier K V, Moore N, Torres MF, Farman ML, Schardl CL, et al. A comparative genomic analysis of putative pathogenicity genes in the host-specific sibling species Colletotrichum graminicola and Colletotrichum sublineola. BMC Genomics. BMC Genomics; 2017; 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crouch J, O’Connell R, Gan P, Buiate E, Torres MF, Beirn L, Shirasu K, Vaillancourt L. The genomics of Colletotrichum Genomics of plant-associated fungi: monocot pathogens. 2014. pp. 69–102. [Google Scholar]
- 93.Selin C, de Kievit TR, Belmonte MF, Fernando WGD. Elucidating the role of effectors in plant-fungal interactions: Progress and challenges. Front Microbiol. 2016;7: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kombrink A, Thomma BPHJ, Kombrink A, Berg van den G, Yadeta K. LysM Effectors: Secreted Proteins Supporting Fungal Life. Heitman J. PLoS Pathog. 2013;9: e1003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guyon K, Balagué C, Roby D, Raffaele S. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics. 2014;15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shiller J, Van de Wouw AP, Taranto AP, Bowen JK, Dubois D, Robinson A, et al. A Large Family of AvrLm6-like Genes in the Apple and Pear Scab Pathogens, Venturia inaequalis and Venturia pirina. Front Plant Sci. Frontiers; 2015;6: 980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown NA, Antoniw J, Hammond-Kosack KE, Cho S, Trail F. The Predicted Secretome of the Plant Pathogenic Fungus Fusarium graminearum: A Refined Comparative Analysis. PLoS One. 2012;7: e33731 doi: 10.1371/journal.pone.0033731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Heard S, Brown NA, Hammond-Kosack K, Levis C, Boccara M. An Interspecies Comparative Analysis of the Predicted Secretomes of the Necrotrophic Plant Pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS One. 2015;10: e0130534 doi: 10.1371/journal.pone.0130534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takahara H, Hacquard S, Kombrink A, Hughes HB, Halder V, Robin GP, et al. Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol. 2016;211: 1323–1337. doi: 10.1111/nph.13994 [DOI] [PubMed] [Google Scholar]
- 100.Bhadauria V, Banniza S, Vandenberg A, Selvaraj G, Wei Y. Overexpression of a novel biotrophy-specific Colletotrichum truncatum effector, CtNUDIX, in hemibiotrophic fungal phytopathogens causes incompatibility with their host plants. Eukaryot Cell. 2013;12: 2–11. doi: 10.1128/EC.00192-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dong S, Wang Y, Sun D, Ojcius D, Zhao J. Nudix Effectors: A Common Weapon in the Arsenal of Plant Pathogens. PLOS Pathog. 2016;12: e1005704 doi: 10.1371/journal.ppat.1005704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Latunde-Dada AO. Colletotrichum: tales of forcible entry, stealth, transient confinement and breakout. Mol Plant Pathol. 2001;2: 187–198. doi: 10.1046/j.1464-6722.2001.00069.x [DOI] [PubMed] [Google Scholar]
- 103.Ranathunge NP, Mongkolporn O, Ford R, Taylor PWJ. Colletotrichum truncatum Pathosystem on Capsicum spp: infection, colonization and defence mechanisms. Australas Plant Pathol. 2012;41: 463–473. [Google Scholar]
- 104.Sanz-Martin JM, Pacheco-Arjona JR, Bello-Rico V, Vargas WA, Monod M, Diaz-Minguez JM, et al. A highly conserved metalloprotease effector enhances virulence in the maize anthracnose fungus Colletotrichum graminicola. Mol Plant Pathol. 2016;17: 1048–1062. doi: 10.1111/mpp.12347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jaramillo VDA, Vargas WA, Sukno SA, Thon MR. Horizontal Transfer of a Subtilisin Gene from Plants into an Ancestor of the Plant Pathogenic Fungal Genus Colletotrichum. PLoS One. 2013;8(3):e59078 doi: 10.1371/journal.pone.0059078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Urban M, Pant R, Raghunath A, Irvine AG, Pedro H, Hammond-Kosack KE. The Pathogen-Host Interactions database (PHI-base): additions and future developments. Nucleic Acids Res. 2015;43: D645–D655. doi: 10.1093/nar/gku1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C. truncatum genome and proteome sizes were comparable to other Colletotrichum species, except C. orbiculare and F. oxysporum which had the largest genome and proteome among the fungi analysed, respectively.
(TIF)
Monilochaetes infuscans was taken as an outgroup. The species with genome sequence available at the time of sequencing of C. truncatum (MTCC no. 3414) are marked with red solid circles. Bootstrap support values (1000 replicates) above 50% are shown at the nodes.
(TIF)
The small (below 300 amino acids), secreted, cysteine-rich proteins can be considered as candidate effectors if they lack homology to known proteins and functional domains.
(TIF)
The median sizes of all secreted proteins in Colletotrichum species were below 300 amino acids (aa), except for M. oryzae strains which had median at 200 aa and encode maximum number of reported effectors among all the fungi analysed. The sizes of all the putative effectors in C. truncatum were below 358 aa except for CTRU_010949 (508 aa). The outliers were removed from the plot.
(TIF)
The corresponding homologous genes are shown in red, green and blue colours. MFS: Major Facilitator Superfamily (transporter).
(TIF)
Fusarium species had exceptionally high number of transporters among all fungi followed by Colletotrichum species. The electrochemical potential-driven transporters, which include a subclass of uniporters, symporters, antiporters (2.A) containing Major Facilitator Superfamily (2.A.1), formed the largest class of transporters in all the fungi analysed, followed by uptake and efflux transport systems driven by ATP hydrolysis, containing ABC transporter family (3.A.1).
(TIF)
(A) The homologues from some of the most abundant categories of genes in PHI-base. The category of genes with unaffected pathogenicity phenotype (validated through mutagenesis) were the largest among PHI-base homologues in all fungi followed by the category of genes with reduced pathogenicity. (B) The homologues from some of the least represented categories of genes in PHI-base. The category of genes with hypervirulence phenotype was the largest among all fungi except M. oryzae that had the largest effector component. F. oxysporum and F. verticillioides had the maximum number of PHI-base homologues reflecting the amount of experimental data available for this genus.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The genome sequence of Colletotrichum truncatum was deposited at the NCBI Genome Database under the accession number NBAU00000000. All the raw data used for genome and transcriptome assemblies can be accessed from the figshare repository using the following links: https://figshare.com/s/f8d4908e91600eebc7c3, https://figshare.com/s/09f44d16fc8966daedba, https://figshare.com/s/9374f90804c944c5399c, https://figshare.com/s/35c459e9f4ac27127902, https://figshare.com/s/e81941f2d19afa090c45, https://figshare.com/s/7bfd5631ac659c8f2b34, https://figshare.com/s/4c68fb7aaa33c4db1445, https://figshare.com/s/17ee5ef5ff3490eeb42f, https://figshare.com/s/6140645e2f22350ff824.