Abstract
Fungi are increasingly being recognized as common members of the microbiomes found on nearly all mucosal surfaces, and interest is growing in understanding how these organisms may contribute to health and disease. In this review, we investigate recent developments in our understanding of the fungal microbiota or “mycobiota” including challenges faced in characterizing it, where these organisms are found, their diversity, and how they interact with host immunity. Growing evidence indicates that like the bacterial microbiota, the fungal microbiota is often altered in disease states, and increasingly studies are being designed to probe the functional consequences of such fungal dysbiosis on health and disease.
INTRODUCTION
The mammalian microbiota, the sum of microorganisms associated with body surfaces, is a key factor in host health and disease. The microbiota influences diverse functions including gut permeability and barrier function, vitamin synthesis, metabolism, neurologic activity, metabolism of pharmaceuticals, and inflammation and immunity (Thomas et al., 2017b). While characterization of microbiota has been hindered in previous decades by limitations in culture methods, targeted and shotgun high-throughput sequencing technologies have more recently revolutionized our understanding of microbial diversity at various body sites. While most research and interest has focused on bacterial microbiota, current evidence documents that nearly anywhere there is a bacterial microbiota, there are also fungal, viral, archae, and perhaps protozoan community members. Here we focus our discussion on the fungal microbiota or “mycobiota”, where these organisms are found, their diversity, and what is currently known about their functional roles in health and disease. We have included a brief list of the fungi mentioned in this review (Table 1), and refer the reader to other materials for specific discussions of pathogenic fungi (Kohler et al., 2014).
Table 1.
Fungi mentioned in this review
| Fungal organism | Scientific Classification (Phylum, class) | Morphologies | Interaction with mammalian host | Principal Habitat |
|---|---|---|---|---|
| Aspergillus spp. | Ascomycota, Eurotiomycetes | Mold, hyphae | Pathogen, Opportunistic pathogen, Commensal | Soil dwelling, ubiquitous in the environment |
| Aureobasidium spp. | Ascomycota, Dothideomycetes | Mold, hyphae, Yeast or yeast-like | Opportunistic pathogen, Commensal | Soil dwelling, plant material |
| Candida albicans | Ascomycota, Saccharomycetes | Yeast and hyphae | Opportunistic pathogen, Commensal | Gastrointestinal tract, mucosa, skin |
| Candida parapsilosis | Ascomycota, Saccharomycetes | Yeast or yeast-like | Opportunistic pathogen, Commensal | Gastrointestinal tract, mucosa, skin, soil dwelling |
| Candida pseudotropicalis | Ascomycota, Saccharomycetes | Yeast or yeast-like | Commensal | Dairy products, fruit juices |
| Candida tropicalis | Ascomycota, Saccharomycetes | Yeast or yeast-like | Opportunistic pathogen, Commensal | Gastrointestinal tract, mucosa, skin, soil dwelling |
| Cladosporium spp. | Ascomycota, Dothideomycetes | Mold, hyphae | Commensal | Soil dwelling, plant material |
| Clavispora lusitaniae | Ascomycota, Saccharomycetes | Yeast or yeast-like | Opportunistic pathogen, Commensal | Gastrointestinal tract, plant material |
| Cryptococcus spp. | Basidiomycota, Tremellomycetes | Yeast or yeast-like | Pathogen | Soil dwelling |
| Cyberlindnera jadinii | Ascomycota, Saccharomycetes | Yeast or yeast-like | Commensal | Gastrointestinal tract, food sources |
| Epicoccum spp. | Ascomycota, Dothideomycetes | Mold, hyphae | Commensal | Soil dwelling, ubiquitous in the environment |
| Fusarium spp. | Ascomycota, Sordariomycetes | Hyphae | Opportunistic pathogen, Commensal | Soil dwelling, plant material |
| Malassezia furfur | Basidiomycota, Malasseziomycetes | Yeast or yeast-like (mainly), hyphae | Opportunistic pathogen, Commensal | Sebaceous skin |
| Malassezia globosa | Basidiomycota, Malasseziomycetes | Yeast or yeast-like (mainly), hyphae | Opportunistic pathogen, Commensal | Sebaceous skin |
| Malassezia restricta | Basidiomycota, Malasseziomycetes | Yeast or yeast-like (mainly), hyphae | Opportunistic pathogen, Commensal | Sebaceous skin |
| Malassezia sympodialis | Basidiomycota, Malasseziomycetes | Yeast or yeast-like (mainly), hyphae | Opportunistic pathogen, Commensal | Sebaceous skin |
| Mucor spp. | Zygomycota, Zygomycetes | Mold, hyphae | Opportunistic pathogen, Commensal | Gastrointestinal tract, soil dwelling, plant material |
| Penicillium spp. | Ascomycota, Eurotiomycetes | Mold, hyphae | Commensal | Soil dwelling, ubiquitous in the environment |
| Pichia spp. | Ascomycota, Saccharomycetes | Yeast or yeast-like | Commensal | Gastrointestinal tract, mucosa, skin, soil dwelling, plant material |
| Rhodotorula spp. | Basidiomycota, Microbotryomycetes | Yeast or yeast-like | Opportunistic pathogen, Commensal | Soil dwelling, water sources, Gastrointestinal tract |
| Saccharomyces boulardii | Ascomycota, Saccharomycetes | Yeast or yeast-like | Commensal | Gastrointestinal tract, food sources |
| Saccharomyces cerevisiae | Ascomycota, Saccharomycetes | Yeast or yeast-like | Commensal | Gastrointestinal tract, food sources |
| Wallemia spp. | Basidiomycota, Wallemiomycetes | Mold, hyphae | Commensal | Soil dwelling, house dust |
Diverse fungi are found in the gut, on the skin, in the mouth, and other mucosal surfaces. Since microbial diversity measurements are typically derived from sequencing data that can be influenced by PCR primer usage, depth of metagenomics sequencing, and efficiency of microbial collection and DNA preparation, among other factors, good estimates of relative bacterial and fungal numbers can be difficult to obtain. However, in most cases, it can be assumed that bacteria vastly outnumber fungi, although when considering physiological and immunological functions, one should note that fungal cells are typically a hundred fold larger than bacterial cells, and that many fungal metabolic functions are likely unique to fungi.
Growing evidence indicates that like the bacterial microbiota, the fungal microbiota is often altered in disease states, and increasingly studies are being designed to probe the functional consequences of such fungal dysbiosis on health and disease. This is a rapidly growing area of research: a 2013 review noted that the term “mycobiome” appeared in only 10 papers indexed in PubMed (Cui et al., 2013), while today (2017) there are over a hundred and we can expect this to continue to grow.
CHALLENGES IN DEFINING THE MYCOBIOTA
Characterization of microbiota by current culture-independent sequencing methods requires efficient preparation of microbial DNA from anatomical sites. As with analyses of bacterial communities, analyses of fungal communities are influenced by methods of DNA preparation. Fungi are encased in thick cell walls, and methodology originally developed for isolating bacterial genomic DNA is not necessarily ideal for recovery of fungal DNA. One recent study on matched oral samples directly compared the fungal DNA signatures detected when using a variety of different DNA preparation methods. Fungal signatures proved to be more sensitive to DNA isolation methods than bacterial signatures (Vesty et al., 2017). To directly assess the relative efficiency of recovering DNA from different fungi and how this might skew signatures, we tested isolation and analysis approaches using model libraries of known ratios of 4 fungi commonly found in intestinal samples (Tang et al., 2015).
Once recovered, different methods of evaluating fungal content of DNA will yield different results. Shotgun sequencing of total genomic DNA is perhaps the least biased approach, but it requires enormously deep sequencing to distill the fungal signature from all of the other sequences (i.e. bacteria) typically also acquired in a sample. For comparison, using this approach Lewis et al. identified 5 fungi in human stool samples and 6.5 × 1011 bases of sequence (Lewis et al., 2015) whereas other fungal-specific approaches typically identify 50–60 genera (Hoarau et al., 2016; Liguori et al., 2016). Such fungal-specific approaches typically involve PCR amplification and sequencing either of two fungal ribosomal DNA (rDNA) “internal transcribed spacer regions” (ITS1 or ITS2). Similar to the fact that PCR primers targeting different variable regions of bacterial rDNA in 16S sequencing can more or less efficiently isolate and identify different groups of bacteria, so too can different primers targeting different ITS target regions prove more or less effective at isolating different groups of fungi (Bokulich and Mills, 2013). Compounding this challenge, unlike bacterial 16S regions targeted for bacterial community characterization, fungal ITS regions vary in length between fungi which adds an additional potential source of bias during PCR amplification and sequencing approaches (Schoch et al., 2012). In mixed samples, length dependent sequence recovery bias can lead to overrepresentation of the relative abundance of fungi with shorter ITS sequences when using common sequencing platforms such as the Illumina MiSeq and Ion Torrent PGM (Motooka et al., 2017).
After acquiring fungal ITS sequence information, investigators are faced with the challenge of identifying the organisms represented by these sequences. This effort is frustrated by a lack of quality-controlled reference databases, evolving recognition and definition of new fungal species/complexes, and confusion as to the taxonomic lineage of many species as well as common use of very different names for sexual and asexual forms of many frequently-encountered fungi. (Halwachs et al., 2017) Because GenBank acts primarily as an archive, many sequences submitted have been annotated with incorrect or poorly-defined species names. It is estimated that more than 10% of the publicly-available fungal ITS sequences are annotated incorrectly at the species level (Nilsson et al., 2006). Further, many fungal ITS sequences in GenBank are annotated as “unidentified fungus”. The most commonly-used and mature database of distilled fungal ITS sequences is the “UNITE” database (Koljalg et al., 2013). Some groups, including ourselves, have developed in-house manually-curated ITS databases for evaluation of specific types of samples (i.e. intestinal) (Tang et al., 2015) or for pathogenic fungi (Irinyi et al., 2015).
Once sequences are identified, a reader or investigator should be aware of what the identifications and numbers mean. Database alignments are good at finding the best matches, but further investigation is required to be certain as a user that any given sequence is correctly identified. Within some genera of fungi, sequence variability is high enough that species (and often sub-species) identifications are relatively strong. For example, ITS sequences can be pretty useful at discriminating important species of Candida and Aspergillus, genera containing many species that can be pathogens, especially in immunocompromised patients (Irinyi et al., 2015). In contrast, for other genera, variability between species is sufficiently low that it can be impossible to tell species apart even if the database alignment returns a match. For example, ITS variation is insufficient to discriminate between most species of Cladosporium (Schubert et al., 2007).
Together, these caveats make direct comparison of fungal microbiota profiles between different research groups and different sources problematic. Like any microbiome experiments, analyses of fungal communities require good experimental design, thoughtful interpretation, and internal controls.
FUNGAL COMMUNITIES IN HEALTHY CONDITIONS
Gastrointestinal tract
The gastrointestinal (GI) tract is tasked with nutrient acquisition from various food sources, but it must also maintain homeostasis with a multitude of microbial residents. Of mucosal surfaces, it is by far populated with the most and most different kinds of microbes. The human GI tract has an average length of 30 feet from the oral cavity to the anus with varying niches and microenvironments capable of sheltering diverse microbial tenants and travelers. In the healthy state, studies in both humans and mice have revealed a greater diversity of fungal organisms than previously described using culture-based methods (Iliev and Underhill, 2013).
A culture-independent sequencing-based approach to the healthy oral mycobiota was first reported by Ghannoum and co-workers (Ghannoum et al., 2010). The study involved a cohort of twenty healthy individuals from varying racial backgrounds and ages ranging from 21 to 60 years of age. In total they identified 85 fungal genera with enormous interpersonal diversity. The most commonly-encountered genera included Candida, Cladosporium, Aureobasidium, an unidentified Saccharomycetales, Aspergillus, and Saccharomyces. A followup study by a different group that used a limited number of study subjects (6), reported similar findings but added Malassezia and Epicoccum to the list of high-abundance organisms (Dupuy et al., 2014).
The intestines of mice harbor the common fungal genera Candida, Saccharomyces, and Cladosporium in addition to more than 50 other genera (Dollive et al., 2013; Iliev et al., 2012). The highest concentration of fungal organisms is found in the colon as measured by fungal rDNA (Iliev et al., 2012). At least one study in mice has suggested that intestinal (fecal) mycobiota may be prone to episodic fluctuations and less stable than bacterial microbiota (Dollive et al., 2013). In a study looking at human fecal material from 96 healthy individuals, a total of 66 fungal genera were reported (Hoffmann et al., 2013). Similar to the murine gut, the most common genera were Saccharomyces, Candida and Cladosporium, being present in 89%, 57% and 42% of samples respectively. Of interest, recent consumption of carbohydrates correlated with an increase in the Candida fecal burden (Hoffmann et al., 2013). Further subsequent studies examining the stool of healthy individuals reported Candida, Penicillium, Wallemia, Cladosporium, and Saccharomyces as the most prevalent genera (Chehoud et al., 2015; Lewis et al., 2015; Mar Rodriguez et al., 2015; Sokol et al., 2016). A healthy bacterial gut community may keep Candida and other fungal populations in check. Healthy mice are generally resistant to C. albicans colonization, but broad spectrum antibacterial treatment along with exposure to C. albicans can generate a high grade C. albicans colonization state. The mechanisms by which bacterial communities interfere with fungal colonization in the gut are largely unknown, but one study has demonstrated that selected commensal anaerobic bacteria (Bacteroides thetaiotamicron and Blautia producta) can induce secretion of anti-fungal peptides by colonic epithelial cells through mechanisms involving the hypoxia-responsive transcription factor, HIF-1α (Fan et al., 2015).
Human infants from 1–4 months of age harbor an intestinal mycobiota predominantly encompassed by the Saccharomycetales, an order containing many common yeasts, and Malasseziales, an order containing many common skin fungi, which eventually matures from 5–11 months to a diminished Malasseziales presence while retaining the Saccharomycetales (Fujimura et al., 2016). A recent study in mice suggests that fungi in early life may play an important role in the maturation of secondary lymphoid organs by promoting intestinal trafficking of dendritic cells expressing the retinoic acid-synthesizing enzyme RALDH to peripheral lymph nodes where, via a mechanism involving production of retinoic acid, they promote homing of lymphocytes to both gut-associated lymphoid tissues and peripheral lymph nodes (Zhang et al., 2016). Whether a similar mechanism is at work in developing humans is not yet known.
Genitourinary system
The fungal microbiota of the vagina has not been as thoroughly investigated as the bacterial microbiota, which is dominated by Lactobacillus spp. (Human Microbiome Project, 2012; Li et al., 2012). A study of 494 Caucasian Estonian women between 15 and 44 years of age with no history of vaginal candidiasis reported a prevalence of Candida sp. dominated by C. albicans (34.1%), Pichia kudriavzevii (2.3%) and C. parapsilosis (0.3%) (Drell et al., 2013). A study in very low birth weight (VLBW) infants looking at Candida albicans transmission from mother to baby reported that of the infants (n=46) born to C. albicans-positive mothers, 18 (39%) became colonized themselves. Vertical transmission from mother to baby accounted for 65% of Candida colonization in infants born to Candida-positive mothers, although the contributing sites could not be established (Bliss et al., 2008).
Respiratory tract
As recently as the 1990s, it was felt that the human lower respiratory tract was sterile except in states of disease (Cabello et al., 1997). With the use of non-culture-based sequencing techniques, it is now understood that microorganisms can be found at all levels of the respiratory tract. With the notable exception of cystic fibrosis patients, study of the lung microbiome is relatively young, with descriptions of non-culture-based profiling of respiratory microorganisms mostly appearing within the last decade.
Uniformly, when non-culture-based sequencing techniques are applied to look for fungi in respiratory specimens, a diverse array of fungal organisms is identified. This should not be surprising since human lungs continually exchange 5–8 L/min of air with an environment containing 102–104 or more fungal spores per cubic meter indoors and outdoors (Burge, 2002). Spores and fungal fragments smaller than 2–3 microns can be inhaled deep into the terminal airways and alveoli where, if immune defenses are intact, they are usually cleared (Latge, 1999). In healthy individuals, studies examining the lower respiratory tract generally find a low burden of fungal DNA consisting predominantly of environmental organisms such as Aspergillus and Cladosporium (Charlson et al., 2012; van Woerden et al., 2013). In healthy hosts with no structural lung disease and intact immune defenses, the extent to which these fungal organisms represent transient inhaled environmental or microaspirated upper airway organisms versus a self-renewing microbial community is not known. A recent well-designed study of the respiratory tract bacterial microbiome suggests that microaspiration is the primary source of lung bacteria in healthy humans, but this study did not examine fungi which may predominantly arrive in the lung via a different source such as inhalation of environmental spores (Dickson et al., 2017). It should be noted that the profile of fungal organisms in the healthy lower respiratory tract is likely distinct from the upper airway, as Candida is usually the dominant fungal genus identified in oral wash specimens from healthy individuals (Charlson et al., 2012; Ghannoum et al., 2010).
Skin
The skin also contains a community of commensal fungal organisms. The lipophilic fungus Malassezia, including several common species, is the dominant fungal skin organism on most adults (Findley et al., 2013; Paulino et al., 2006). Malassezia spp. require long chain fatty acids for optimal growth and are found on most skin areas, but are most closely associated with lipid-rich sebum secreted by sebaceous glands (Gaitanis et al., 2012). In children, who have less sebaceous gland activity on their skin than adults, fungal communities are more diverse and include organisms such as Aspergillus, Epicoccum, and Phoma in addition to Malassezia (Jo et al., 2016). It is unknown whether the presence of Malassezia provides any beneficial effects for the skin in healthy hosts, but Malassezia have been reported to produce potent aryl hydrocarbon receptor (Ahr) ligands that may promote epithelial cell health and protection from ultraviolet radiation (Velegraki et al., 2015). Still, use of antifungal medications that deplete Malassezia are generally not associated with adverse skin changes. Fungal pathogens can be asymptomatically carried on the skin of healthy individuals, and over 50% of health care workers may be carriers of Candida albicans on their skin, with several Candida outbreaks attributed to carriage by health care workers (Brunetti et al., 2008).
FUNGAL COMMUNITIES IN DISEASE
Fungi in the oral cavity and intestines
In the oral cavity, the pathogenic potential of Candida albicans was first reported in HIV+ individuals with a progressive CD4+ T cell loss who developed oral pharyngeal candidiasis (Klein et al., 1984). Pro-inflammatory TH17 CD4+ T cells are responsible for anti-Candida immune defense and help maintain a state of détente with the commensal organism (Cassone and Cauda, 2012). Interestingly, keeping Candida in a commensal state may also be promoted by fungal-fungal interactions as suggested by Mukherjee and co-workers (Mukherjee et al., 2014). In HIV+ individuals there is an inverse correlation between the abundance of Pichia and the level of Candida colonization. In healthy controls where Pichia was observed there was also an absence of Cryptococcus, Fusarium and Aspergillus, known fungal pathogens. Pichia-conditioned media was able to directly inhibit the growth of Candida, Aspergillus and Fusarium. The inhibitory effects of Pichia-conditioned media was attributed to a secreted protein, perhaps a mycotoxin (Mukherjee et al., 2014).
Inflammatory bowel disease (IBD) encompasses two gastrointestinal diseases: ulcerative colitis (affecting the colon) and Crohn’s disease (affecting the entire GI tract) with an affected population of about 2.5 million people (Xavier and Podolsky, 2007). Intestinal inflammation is believed to be attributed to an aberrant immune response against commensal gut microbes, but the exact pathogenesis remains to be elucidated. Genetic predisposing factors are also important, and in fact polymorphisms in CARD9 and IL23R, which respectively encode for a signaling adapter protein and the Interleukin 23 Receptor and are both important in anti-fungal defense, are strongly associated with IBD (Jostins et al., 2012). In addition, the mycobiota may exacerbate established disease since a polymorphic haplotype of CLEC7A (the gene for Dectin-1, an innate immune receptor essential for immunity to diverse types of fungi) has been linked to severe disease in patients with ulcerative colitis, and mice lacking Dectin-1 are more susceptible to colitis (Iliev et al., 2012). Finally, high serum titers of anti-Saccharomyces cerevisiae antibodies (ASCA), which are specific for fungal cell wall-associated mannan, is a clinical biomarker for identifying a large portion of Crohn’s disease patients (Joossens et al., 2002).
One of the first studies to investigate mycobiota differences between healthy controls and IBD patients used 18S rDNA-based denaturing gradient gel electrophoresis (DGGE) (Ott et al., 2008). DGGE is limited in providing in-depth microbial analysis, and the main differences reported in the mycobiota between the study groups was an increase in the fungal diversity of Crohn’s patients. No single fungal organism was positively correlated with either Crohn’s disease or ulcerative colitis (Ott et al., 2008). Two recent papers from the same investigative groups have reported on the mycobiota of pediatric IBD using high-throughput sequencing (Chehoud et al., 2015; Lewis et al., 2015). In the first paper IBD was associated with both a lower bacterial and fungal diversity and a higher abundance of Candida in IBD samples (Chehoud et al., 2015). In a close follow-up study, Crohn’s disease severity and antibiotic use correlated with greater fungal proportions of Saccharomyces cerevisiae, Clavispora lusitaniae, Cyberlindnera jadinii, Candida albicans and Kluyveromyces marxianus (Lewis et al., 2015). A related study in familial CD noted a positive correlation between Candida tropicalis and CD (Hoarau et al., 2016). Decreased intestinal fungal diversity was also reported in a study examining the stool of 235 adult IBD patients and 38 healthy controls (Sokol et al., 2016). A prominent feature of the IBD mycobiota in this study was a reduction in ascomycetes and a reciprocal increase in the abundance of basidiomycetes, with marked differences during IBD flare (Sokol et al., 2016). Ascomycetes and basidiomycetes are the two largest phyla in the fungal kingdom, and a change at this level suggests a fundamental change in the mycobiota. While Candida albicans abundance was increased in Crohn’s disease, it could not be statistically associated with disease in this study. Interestingly, a clear reduction in Saccharomyces and particularly Saccharomyces cerevisiae was observed in IBD and during active flare, which has also been reported in mouse model (Iliev et al., 2012; Sokol et al., 2016). How S. cerevisiae might function in the gut, perhaps as immune regulatory or directly antagonistic to Candida, remains to be determined.
A recent study suggests a role for the intestinal mycobiota in development of ethanol-induced liver disease in people and mice (Yang et al., 2017). The investigators noticed that alcohol feeding of mice increases the intestinal fungal burden and heightens the levels of β-glucan, a component of the fungal cell wall, in the circulation. In patients, alcohol-induced disease was associated with increased growth of Candida, increased ASCA in blood, and this increased ASCA correlated with mortality rates. In the mice, development of liver disease was found to be dependent on Dectin-1, probably on Kupffer cells detecting circulating β-glucan. It will be interesting to see if genetic variations in Dectin-1 (CLEC7A) or CARD9 are similarly associated with the risk of developing alcohol-induced liver disease.
Fungi in metabolic disease
Metabolic syndrome in humans is diagnosed by several clinical markers which include elevated blood pressure and fasting glucose levels, high levels of triglycerides in the serum, abdominal obesity and low level of “good” cholesterol in the blood. Obesity is the excessive accumulation of body fat often defined by a body mass index (BMI) of greater than 30 kg/m2 which predisposes individuals to develop metabolic syndrome (Garrow and Webster, 1985). Obesity-related changes in the microbiota were suggested by early studies in humans and obese mice that reported decreased ratios of Firmicutes and Bacteroidetes, two common phyla of bacteria (Devaraj et al., 2013; Ma et al., 2014; Turnbaugh et al., 2006). Additionally, the obese bacterial microbiome was more metabolically active and thus harvested more energy from the diet (Turnbaugh et al., 2006). One study has specifically characterized the mycobiota in obese subjects (Mar Rodriguez et al., 2015). The investigators noted no alteration of the overall richness of the mycobiota in obese subjects, although family-level biodiversity was lower and Zygomycota (a phylum of fungi much less common that ascomycetes or basidiomycetes) appeared depleted in obese subjects. Among the zygomycetes, Mucor spp. were specifically noted as more prevalent in non-obese controls compared to obese subjects, and the decreased relative abundance of Mucor in obese subjects was reversed when patients lost weight. It is uncertain whether these mycobiota changes are simply a marker for obesity or whether fungal dysbiosis can actually contribute to the pathogenesis of obesity. If the latter is true, this could suggest that the mycobiota may be a potential therapeutic target to combat obesity and related metabolic disorders.
Mycobiota and lung disease
The most well-studied lung microbiome has been in cystic fibrosis (CF) patients. Patients with CF have abnormal mucous production and undergo lung colonization with microorganisms shortly after birth (Stoltz et al., 2015). Fungal “colonizers” are commonly detected in lungs of CF patients, with Aspergillus and Candida found in culture in over 50% of patients, but their impact on disease progression is controversial (Sudfeld et al., 2010; Valenza et al., 2008). Several small studies have applied non-culture-based sequencing techniques to profile lower respiratory tract fungi in patients with CF. Although these studies generally identify DNA from over a dozen fungal species in respiratory samples, they have consistently identified Candida species as the most prevalent (Delhaes et al., 2012; Kramer et al., 2015; Willger et al., 2014).
Several groups have examined lung mycobiota in patients with systemic immunosuppression. In a study of 21 lung transplant recipients on lifelong immunosuppression, these patients were found to have higher fungal burdens in lower respiratory tract bronchoalveolar lavage specimens compared to healthy controls (Charlson et al., 2012). Depending on the individual, Aspergillus or Candida were usually the dominant organism identified. Patients with high levels of Candida also generally had high levels in oral wash specimens, whereas patients with high Aspergillus in the lungs had little detectable Aspergillus in oral washes. In another well-designed study that compared 32 patients with HIV and 24 healthy controls, all patients underwent bronchoalveolar lavage (BAL), oral wash, and induced sputum collection (Cui et al., 2015). Patients with HIV had a distinct profile of fungi in the BAL compared to healthy controls and their own oral washes, characterized by increased detection of Pneumocystis jirovecii, even though these patients did not have Pneumocystis pneumonia (Cui et al., 2015).
In addition to lung-resident fungi, fungal organisms in the gut may impact lung disease. In mouse models of asthma, alteration of gut fungi induced by antifungal medications or a Candida colonization protocol have both been shown to enhance airway inflammation (Noverr et al., 2005; Wheeler et al., 2016). One study suggests that prostaglandin E2 generated by Candida enzymes in the gut reaches the lungs and influences lung macrophages to alter allergic airway disease (Kim et al., 2014). In human infants the presence of Candida and Rhodotorula has been associated with an increased risk of developing asthma later in life, although the mechanisms that might lead to this are unknown (Fujimura et al., 2016).
Mycobiota and skin disease
Malassezia spp. have been implicated in a variety of skin diseases. Malassezia is a nearly ubiquitous commensal skin colonizer, but in selected individuals it can act as a pathogen. Pityriasis versicolor is the only common skin disease that is directly and unequivocally attributable to Malassezia. It is characterized by skin invasion of the hyphal form of Malassezia and can be caused by a variety of species including M. globosa, M. sympodialis, and M. furfur (Prohic et al., 2016). Malassezia has also been implicated in the pathogenesis of a variety of other immune mediated skin disorders such as seborrheic dermatitis, atopic dermatitis and psoriasis. These diseases are not consistently associated with higher burden of Malassezia on the lesional skin of affected individuals, but may be due in part to inappropriate immune reaction to Malassezia. Patients with atopic dermatitis have increased circulating IgE to Malassezia compared to healthy controls (Scalabrin et al., 1999). In vitro experiments have also shown that Malassezia activates C-lectin receptors including Mincle and Dectin-2, and interestingly, mast cells from patients with atopic dermatitis have decreased Dectin-1 expression compared to healthy controls (Ishikawa et al., 2013; Ribbing et al., 2011; Yamasaki et al., 2009).
Chronic skin wounds contain a heterogeneous mix of fungi that are likely distinct from healthy skin flora and may influence healing. In a study of 100 diabetic foot ulcers, Cladosporium and Candida were the most abundant fungi identified and Malassezia was rarely detected (Kalan et al., 2016). The authors demonstrated that two highly abundant fungi obtained from wounds (Candida albicans and Trichosporon asahii) may form a biofilm when co-cultured in vitro with selected bacteria, although whether this biofilm formation occurs in vivo or if it alters wound healing is not known.
PROSPECTS FOR THERAPEUTIC MANIPULATION OF FUNGAL COMMUNITIES
If fungi are part of the healthy microbiota and are altered in disease, perhaps contributing to pathologies, it is attractive to imagine that therapeutic manipulation of the fungal microbiota could be a useful approach to treatment or prevention of disease. In fact, in the early 1900’s French microbiologist Henri Boulard observed people in Southeast Asia using a tea made from skins of tropical lychee and mangosteen fruits to alleviate symptoms of cholera. He isolated a yeast from this material and named it after himself, Saccharomyces boulardii, although it is now generally considered a subspecies of S. cerevisiae (Saccharomyces cerevisiae var. boulardii). Genomic sequencing reveals loss of multiple genes and elements relative to more common strains of S. cerevisiae (Khatri et al., 2017). S. boulardii is widely available and used today as a probiotic, and data are reasonably good that it has beneficial effects in combating certain intestinal bacterial pathogens including enterohemorrhagic Escherichia coli, Clostridium difficile, Vibrio cholera, and Helicobacter pylori infections (Kelesidis and Pothoulakis, 2012). Diverse mechanisms of protection have been reported, although rarely directly tested. For example, a secreted phosphatase is reported to dephosphorylate E. coli endotoxin, but whether this is important for the fungus’ therapeutic efficacy against E. coli is unknown (Buts et al., 2006). Similarly, a secreted protease inactivates Clostridium difficile toxins A and B in vitro and in vivo, but whether this protease is required for the protective effects of S. boulardii against C. difficile has not yet been fully established (Castagliuolo et al., 1999). S. boulardii may have broad immunomodulatory functions that influence resistance to pathogens; the fungus appears to boost overall intestinal IgA production, although how is still unclear (Qamar et al., 2001).
β-glucan purified from S. cerevisiae cell walls is promoted as a nutritional supplement helpful for weight loss, cancer treatment and prevention, boosting immune responses against infection, lowering cholesterol, and radioprotection (Saber et al., 2017; Volman et al., 2008). However, little is established mechanistically to support such claims at this time. Injectable soluble β-glucans are being investigated as adjunctive immune-potentiating agents together with antibodies or chemotherapy in cancer therapy (Thomas et al., 2017a). This appears to work primarily through complement mobilization and activation/recruitment of neutrophils to tumor (Chan et al., 2016). Oral delivery of particulate glucans has mimicked some of the benefits of injectable soluble β-glucans in animal models, and at least one study has observed that orally-delivered particulate glucans are taken up by intestinal phagocytes, trafficked to bone marrow and lymphoid organs, and ultimately are released as soluble β-glucan fragments in circulation which can activated neutrophils to kill tumor cells (Figure 1) (Hong et al., 2004). Whether endogenous commensal fungi, at “normal” levels or during fungal blooms, are similarly converted into immune-activating circulating β-glucan fragments is not clear. Clinically, β-glucan blood assays are generally used to detect invasive fungal disease (White et al., 2017), and fungal colonization of the intestines (without invasive disease) may contribute to the variable baseline levels of circulating β-glucans detected.
Figure 1. Alterations in commensal fungi may affect the host in diverse ways.
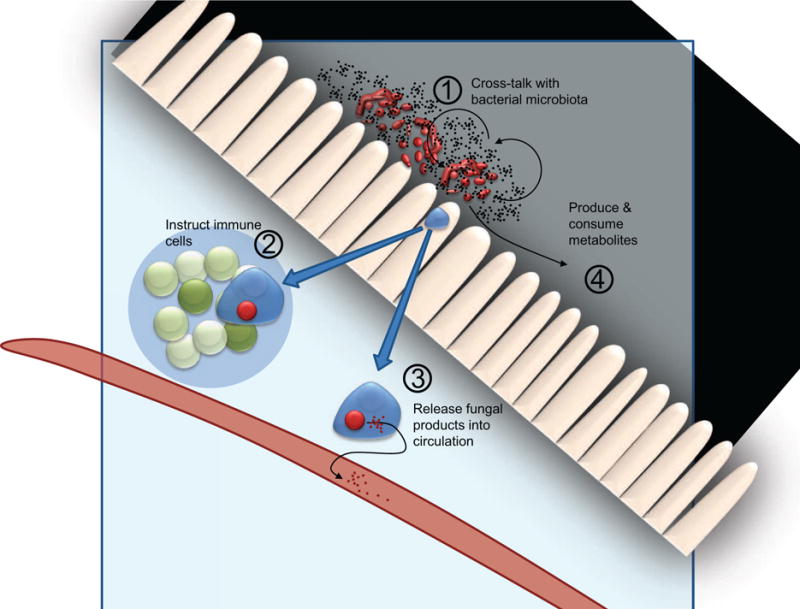
Changing intestinal fungi either by adding (probiotic) or subtracting (anti-fungal drugs) organisms can alter the makeup of the bacterial microbiota (1). Fungi detected by the intestinal immune system can lead to inflammatory or tolerant immune responses and can direct immune cell trafficking (2). Phagocytes digesting fungi can release fungal-derived molecules into circulation that may have immunoregulatory effects (3). Finally, intestinal fungi release and consume metabolites that can lead to activation or suppression of immune responses.
Oral antifungal medications are another potential tool to therapeutically alter gut fungal communities. However, treatment of mice with an antifungal medication such as fluconazole or amphotericin does not result in elimination or near-elimination of commensal gut fungi. Rather, the relative distribution of fungal species is altered as some fungi decrease in abundance while others (presumably resistant to the antifungal medication) actually increase in relative and even absolute abundance. We have observed that disruption of fungal communities by antifungal medications enhances the severity of DSS colitis and allergic airway disease in mice (Wheeler et al., 2016).
Fungal communities affect, and are affected by bacterial communities (Figure 1). In studies profiling fungal and bacterial microbiota, significant positive and negative associations between specific types of fungi and bacteria are commonly noted. For example, in a recent analysis of bacterial and fungal organisms in fecal samples from patients with Crohn’s disease, a statistically significant association was observed between Candida tropicalis, Serratia marcescens, and Escherichia coli (Hoarau et al., 2016). Interestingly, the investigators noted that these three organisms can be coaxed into forming a mixed biofilm which they hypothesize could facilitate their co-colonization of the intestines. It has also been reported in an animal model that S. boulardii interacts with and alters commensal bacterial microbiota (Yu et al., 2017) (Figure 1). Similarly, a recent study demonstrated that oral administration of the dietary yeast Candida kefyr (also known as Candida pseudotropicalis or Kluyveromyces marxianus) was protective in mouse models of colitis and experimental autoimmune encephalomyelitis (Takata et al., 2015). This was accompanied by alterations in the bacterial microbiota, and transfer of the microbiota into new animals could confer protection that was associated with an increase in mesenteric lymph node regulatory T cells and reduction in lamina propria Th17 cells.
Oral antibiotic treatment is often associated with fungal expansion in the gastrointestinal tract, indicating that in the steady-state, bacterial communities keep fungi (especially Candida spp.) in check. In this context, animal models suggest that overgrowth of Candida can alter the recovery of the bacterial microbiota after antibiotic treatment is stopped (Erb Downward et al., 2013; Mason et al., 2012). Similarly, treating animals with oral anti-fungal drugs has been reported to alter the bacterial microbiota (Wheeler et al., 2016). The mechanisms by which bacteria and fungi regulate each other in commensal communities are undoubtedly diverse and may offer novel targets for manipulating the microbiome.
Like any other organism, fungi consume and release metabolites and are thus expected to influence the metabolome of a microbial community (Figure 1). A recent study looking at intestinal metabolites produced when germ-free mice were colonized with S. cerevisiae noted an increase in purine metabolism leading to increased uric acid production that the investigators could experimentally link to the ability of S. cerevisiae to exacerbate colitis in their animals (Chiaro et al., 2017). A Rhodotorula did not have this affect in the animals, suggesting that this is not an effect that is common to all fungi. However, whether other fungi influence purine metabolism to affect disease and whether this effect is significant in a steady-state mixed microbial community remains to be established. Tantalizingly, the investigators noted a positive correlation between ASCA (linked to IBD in patients, as discussed above) and uric acid levels in the serum of healthy adults, suggesting a link between altered immune interactions with fungi and uric acid production.
CONCLUSIONS
Fungal members of microbial communities on mucosal surfaces are part of the normal ecology of our bodies. We have evolved to tolerate these passengers and likely to make use of them in diverse ways that we are only beginning to understand. Future studies designed to further our understanding of how fungi interact with the microbiome and our immune systems may lead to novel therapeutic approaches to fighting infection, treating cancer, and managing health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Like bacteria, fungi are part of the microbiomes found on body surfaces, and interest is growing in how these organisms contribute to health and disease. Limon et al. review recent developments in identifying members of the fungal microbiota as well as understanding how our immune systems interact with commensal fungi.
References
- Bliss JM, Basavegowda KP, Watson WJ, Sheikh AU, Ryan RM. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr Infect Dis J. 2008;27:231–235. doi: 10.1097/INF.0b013e31815bb69d. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Mills DA. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl Environ Microbiol. 2013;79:2519–2526. doi: 10.1128/AEM.03870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti L, De Caro F, Boccia G, Cavallo P, Capunzo M. Surveillance of nosocomial infections: a preliminary study on yeast carriage on hands of healthcare workers. J Prev Med Hyg. 2008;49:63–68. [PubMed] [Google Scholar]
- Burge HA. An update on pollen and fungal spore aerobiology. J Allergy Clin Immunol. 2002;110:544–552. doi: 10.1067/mai.2002.128674. [DOI] [PubMed] [Google Scholar]
- Buts JP, Dekeyser N, Stilmant C, Delem E, Smets F, Sokal E. Saccharomyces boulardii produces in rat small intestine a novel protein phosphatase that inhibits Escherichia coli endotoxin by dephosphorylation. Pediatr Res. 2006;60:24–29. doi: 10.1203/01.pdr.0000220322.31940.29. [DOI] [PubMed] [Google Scholar]
- Cabello H, Torres A, Celis R, El-Ebiary M, Puig de la Bellacasa J, Xaubet A, Gonzalez J, Agusti C, Soler N. Bacterial colonization of distal airways in healthy subjects and chronic lung disease: a bronchoscopic study. Eur Respir J. 1997;10:1137–1144. doi: 10.1183/09031936.97.10051137. [DOI] [PubMed] [Google Scholar]
- Cassone A, Cauda R. Candida and candidiasis in HIV-infected patients: where commensalism, opportunistic behavior and frank pathogenicity lose their borders. AIDS. 2012;26:1457–1472. doi: 10.1097/QAD.0b013e3283536ba8. [DOI] [PubMed] [Google Scholar]
- Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun. 1999;67:302–307. doi: 10.1128/iai.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AS, Jonas AB, Qiu X, Ottoson NR, Walsh RM, Gorden KB, Harrison B, Maimonis PJ, Leonardo SM, Ertelt KE, et al. Imprime PGG-Mediated Anti-Cancer Immune Activation Requires Immune Complex Formation. PLoS One. 2016;11:e0165909. doi: 10.1371/journal.pone.0165909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, Bushman FD, Collman RG. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaro TR, Soto R, Zac Stephens W, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Lucht L, Tipton L, Rogers MB, Fitch A, Kessinger C, Camp D, Kingsley L, Leo N, Greenblatt RM, et al. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med. 2015;191:932–942. doi: 10.1164/rccm.201409-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PLoS One. 2012;7:e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S, Hemarajata P, Versalovic J. The human gut microbiome and body metabolism: implications for obesity and diabetes. Clin Chem. 2013;59:617–628. doi: 10.1373/clinchem.2012.187617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, Curtis JL. Bacterial Topography of the Healthy Human Lower Respiratory Tract. MBio. 2017;8 doi: 10.1128/mBio.02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollive S, Chen YY, Grunberg S, Bittinger K, Hoffmann C, Vandivier L, Cuff C, Lewis JD, Wu GD, Bushman FD. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One. 2013;8:e71806. doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drell T, Lillsaar T, Tummeleht L, Simm J, Aaspollu A, Vain E, Saarma I, Salumets A, Donders GG, Metsis M. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One. 2013;8:e54379. doi: 10.1371/journal.pone.0054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 2014;9:e90899. doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, Huffnagle GB. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep. 2013;3:2191. doi: 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, Koh AY. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 2015;21:808–814. doi: 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, NIHISCCS Program et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev. 2012;25:106–141. doi: 10.1128/CMR.00021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrow JS, Webster J. Quetelet’s index (W/H2) as a measure of fatness. Int J Obes. 1985;9:147–153. [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halwachs B, Madhusudhan N, Krause R, Nilsson RH, Moissl-Eichinger C, Hogenauer C, Thallinger GG, Gorkiewicz G. Critical Issues in Mycobiota Analysis. Front Microbiol. 2017;8:180. doi: 10.3389/fmicb.2017.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. MBio. 2016;7 doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Yan J, Baran JT, Allendorf DJ, Hansen RD, Ostroff GR, Xing PX, Cheung NK, Ross GD. Mechanism by which orally administered beta-1,3-glucans enhance the tumoricidal activity of antitumor monoclonal antibodies in murine tumor models. J Immunol. 2004;173:797–806. doi: 10.4049/jimmunol.173.2.797. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Underhill DM. Striking a balance: fungal commensalism versus pathogenesis. Curr Opin Microbiol. 2013;16:366–373. doi: 10.1016/j.mib.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinali G, Arthur I, Normand AC, Giraldo A, et al. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database–the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med Mycol. 2015;53:313–337. doi: 10.1093/mmy/myv008. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Itoh F, Yoshida S, Saijo S, Matsuzawa T, Gonoi T, Saito T, Okawa Y, Shibata N, Miyamoto T, Yamasaki S. Identification of distinct ligands for the C-type lectin receptors Mincle and Dectin-2 in the pathogenic fungus Malassezia. Cell Host Microbe. 2013;13:477–488. doi: 10.1016/j.chom.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WL, Program NCS, Segre JA, Kong HH. Diverse Human Skin Fungal Communities in Children Converge in Adulthood. J Invest Dermatol. 2016;136:2356–2363. doi: 10.1016/j.jid.2016.05.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joossens S, Reinisch W, Vermeire S, Sendid B, Poulain D, Peeters M, Geboes K, Bossuyt X, Vandewalle P, Oberhuber G, et al. The value of serologic markers in indeterminate colitis: a prospective follow-up study. Gastroenterology. 2002;122:1242–1247. doi: 10.1053/gast.2002.32980. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalan L, Loesche M, Hodkinson BP, Heilmann K, Ruthel G, Gardner SE, Grice EA. Redefining the Chronic-Wound Microbiome: Fungal Communities Are Prevalent, Dynamic, and Associated with Delayed Healing. MBio. 2016;7 doi: 10.1128/mBio.01058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelesidis T, Pothoulakis C. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therap Adv Gastroenterol. 2012;5:111–125. doi: 10.1177/1756283X11428502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri I, Tomar R, Ganesan K, Prasad GS, Subramanian S. Complete genome sequence and comparative genomics of the probiotic yeast Saccharomyces boulardii. Sci Rep. 2017;7:371. doi: 10.1038/s41598-017-00414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Nunez G, Shibuya A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2) Cell Host Microbe. 2014;15:95–102. doi: 10.1016/j.chom.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RS, Harris CA, Small CB, Moll B, Lesser M, Friedland GH. Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. N Engl J Med. 1984;311:354–358. doi: 10.1056/NEJM198408093110602. [DOI] [PubMed] [Google Scholar]
- Kohler JR, Casadevall A, Perfect J. The spectrum of fungi that infects humans. Cold Spring Harb Perspect Med. 2014;5:a019273. doi: 10.1101/cshperspect.a019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Kramer R, Sauer-Heilborn A, Welte T, Guzman CA, Abraham WR, Hofle MG. Cohort Study of Airway Mycobiome in Adult Cystic Fibrosis Patients: Differences in Community Structure between Fungi and Bacteria Reveal Predominance of Transient Fungal Elements. J Clin Microbiol. 2015;53:2900–2907. doi: 10.1128/JCM.01094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Bihan M, Yooseph S, Methe BA. Analyses of the microbial diversity across the human microbiome. PLoS One. 2012;7:e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori G, Lamas B, Richard ML, Brandi G, da Costa G, Hoffmann TW, Di Simone MP, Calabrese C, Poggioli G, Langella P, et al. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn’s Disease Patients. J Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, Aagaard KM. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun. 2014;5:3889. doi: 10.1038/ncomms4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar Rodriguez M, Perez D, Javier Chaves F, Esteve E, Marin-Garcia P, Xifra G, Vendrell J, Jove M, Pamplona R, Ricart W, et al. Obesity changes the human gut mycobiome. Sci Rep. 2015;5:14600. doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun. 2012;80:3371–3380. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motooka D, Fujimoto K, Tanaka R, Yaguchi T, Gotoh K, Maeda Y, Furuta Y, Kurakawa T, Goto N, Yasunaga T, et al. Fungal ITS1 Deep-Sequencing Strategies to Reconstruct the Composition of a 26-Species Community and Evaluation of the Gut Mycobiota of Healthy Japanese Individuals. Front Microbiol. 2017;8:238. doi: 10.3389/fmicb.2017.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog. 2014;10:e1003996. doi: 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson KH, Koljalg U. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One. 2006;1:e59. doi: 10.1371/journal.pone.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73:30–38. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S. Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol. 2016;55:494–504. doi: 10.1111/ijd.13116. [DOI] [PubMed] [Google Scholar]
- Qamar A, Aboudola S, Warny M, Michetti P, Pothoulakis C, LaMont JT, Kelly CP. Saccharomyces boulardii stimulates intestinal immunoglobulin A immune response to Clostridium difficile toxin A in mice. Infect Immun. 2001;69:2762–2765. doi: 10.1128/IAI.69.4.2762-2765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbing C, Engblom C, Lappalainen J, Lindstedt K, Kovanen PT, Karlsson MA, Lundeberg L, Johansson C, Nilsson G, Lunderius-Andersson C, Scheynius A. Mast cells generated from patients with atopic eczema have enhanced levels of granule mediators and an impaired Dectin-1 expression. Allergy. 2011;66:110–119. doi: 10.1111/j.1398-9995.2010.02437.x. [DOI] [PubMed] [Google Scholar]
- Saber A, Alipour B, Faghfoori Z, Yari Khosroushahi A. Cellular and molecular effects of yeast probiotics on cancer. Crit Rev Microbiol. 2017;43:96–115. doi: 10.1080/1040841X.2016.1179622. [DOI] [PubMed] [Google Scholar]
- Scalabrin DM, Bavbek S, Perzanowski MS, Wilson BB, Platts-Mills TA, Wheatley LM. Use of specific IgE in assessing the relevance of fungal and dust mite allergens to atopic dermatitis: a comparison with asthmatic and nonasthmatic control subjects. J Allergy Clin Immunol. 1999;104:1273–1279. doi: 10.1016/s0091-6749(99)70024-2. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding, C. Fungal Barcoding Consortium Author, L. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, Hill CF, Zalar P, de Hoog GS, Crous PW. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud Mycol. 2007;58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. Fungal microbiota dysbiosis in IBD. Gut. 2016 doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz DA, Meyerholz DK, Welsh MJ. Origins of cystic fibrosis lung disease. N Engl J Med. 2015;372:351–362. doi: 10.1056/NEJMra1300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudfeld CR, Dasenbrook EC, Merz WG, Carroll KC, Boyle MP. Prevalence and risk factors for recovery of filamentous fungi in individuals with cystic fibrosis. J Cyst Fibros. 2010;9:110–116. doi: 10.1016/j.jcf.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Tomita T, Okuno T, Kinoshita M, Koda T, Honorat JA, Takei M, Hagihara K, Sugimoto T, Mochizuki H, et al. Dietary Yeasts Reduce Inflammation in Central Nerve System via Microflora. Ann Clin Transl Neurol. 2015;2:56–66. doi: 10.1002/acn3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Iliev ID, Brown J, Underhill DM, Funari VA. Mycobiome: Approaches to analysis of intestinal fungi. J Immunol Methods. 2015;421:112–121. doi: 10.1016/j.jim.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Sadjadian P, Kollmeier J, Lowe J, Mattson P, Trout JR, Gargano M, Patchen ML, Walsh R, Beliveau M, et al. A randomized, open-label, multicenter, phase II study evaluating the efficacy and safety of BTH1677 (1,3-1,6 beta glucan; Imprime PGG) in combination with cetuximab and chemotherapy in patients with advanced non-small cell lung cancer. Invest New Drugs. 2017a;35:345–358. doi: 10.1007/s10637-017-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, Sela DA, Muller AJ, Mullin JM, Albert K, et al. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res. 2017b;77:1783–1812. doi: 10.1158/0008-5472.CAN-16-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Valenza G, Tappe D, Turnwald D, Frosch M, Konig C, Hebestreit H, Abele-Horn M. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros. 2008;7:123–127. doi: 10.1016/j.jcf.2007.06.006. [DOI] [PubMed] [Google Scholar]
- van Woerden HC, Gregory C, Brown R, Marchesi JR, Hoogendoorn B, Matthews IP. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect Dis. 2013;13:69. doi: 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velegraki A, Cafarchia C, Gaitanis G, Iatta R, Boekhout T. Malassezia infections in humans and animals: pathophysiology, detection, and treatment. PLoS Pathog. 2015;11:e1004523. doi: 10.1371/journal.ppat.1004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesty A, Biswas K, Taylor MW, Gear K, Douglas RG. Evaluating the Impact of DNA Extraction Method on the Representation of Human Oral Bacterial and Fungal Communities. PLoS One. 2017;12:e0169877. doi: 10.1371/journal.pone.0169877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volman JJ, Ramakers JD, Plat J. Dietary modulation of immune function by beta-glucans. Physiol Behav. 2008;94:276–284. doi: 10.1016/j.physbeh.2007.11.045. [DOI] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe. 2016;19:865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PL, Price JS, Posso RB, Barnes RA. An evaluation of the performance of the Dynamiker(R) Fungus (1–3)-beta-D-Glucan Assay to assist in the diagnosis of invasive aspergillosis, invasive candidiasis and Pneumocystis pneumonia. Med Mycol. 2017 doi: 10.1093/mmy/myx004. [DOI] [PubMed] [Google Scholar]
- Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, Filkins LM, O’Toole GA, Moulton LA, Ashare A, et al. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome. 2014;2:40. doi: 10.1186/2049-2618-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Matsumoto M, Takeuchi O, Matsuzawa T, Ishikawa E, Sakuma M, Tateno H, Uno J, Hirabayashi J, Mikami Y, et al. C-type lectin Mincle is an activating receptor for pathogenic fungus, Malassezia. Proc Natl Acad Sci U S A. 2009;106:1897–1902. doi: 10.1073/pnas.0805177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AM, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, Bluemel S, Hartmann P, Xu J, Koyama Y, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017 doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Zhao XK, Cheng ML, Yang GZ, Wang B, Liu HJ, Hu YX, Zhu LL, Zhang S, Xiao ZW, et al. Saccharomyces boulardii Administration Changes Gut Microbiota and Attenuates D-Galactosamine-Induced Liver Injury. Sci Rep. 2017;7:1359. doi: 10.1038/s41598-017-01271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zheng W, Zhao G, Zhang H, Wang X, Guo Y, Qin C, Shi Y. Peripheral Lymphoid Volume Expansion and Maintenance Are Controlled by Gut Microbiota via RALDH+ Dendritic Cells. Immunity. 2016;44:330–342. doi: 10.1016/j.immuni.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


