Abstract
Programmed cell death (PCD) is a prerequisite for successful development and it limits the spread of biotrophic pathogens in a rapid hypersensitive response at the site of infection. KDEL-tailed cysteine endopeptidases (KDEL CysEP) are a subgroup of papain-type cysteine endopeptidases expressed in tissues undergoing PCD. In Arabidopsis, three KDEL CysEPs (AtCEP1, AtCEP2, and AtCEP3) are expressed. We have previously shown that AtCEP1 is a factor of basal resistance to powdery mildew caused by the biotrophic ascomycete Erysiphe cruciferarum, and is expressed in spatiotemporal association with the late fungal development on Arabidopsis leaves. The endoplasmic reticulum-localized proenzyme of AtCEP1 was further visualized at the haustorial complex encased with callose. The AtCPR5 gene (CONSTITUTIVE EXPRESSION OF PR GENES 5) is a regulator of expression of pathogenesis related genes. Loss of AtCPR5 leads to spontaneous expression of chlorotic lesions which was associated with enhanced expression of AtCEP1. We used the atcpr5-2 mutant plants and the atcep1 atcpr5-2 double mutants harboring a non-functional reporter (PCEP1::pre-pro-3xHA-EGFP-KDEL) for visualization of AtCEP1 promoter activity. We found the specific up-regulation of AtCEP1 in direct neighborhood of spreading leaf lesions thus likely representing cells undergoing PCD. Furthermore, we found a strong resistance of atcpr5 mutant plants against infection with E. cruciferarum. Loss of AtCEP1 had no obvious influence on the strong resistance of atcpr5-2 mutant plants against infection with E. cruciferarum. However, the area of necrotic leaf lesions associated with E. cruciferarum colonies was significantly larger in atcpr5-2 as compared to atcep1 atcpr5-2 double mutant plants. The presence of AtCEP1 thus contributes to AtCPR5-controlled PCD at the sites of powdery mildew infection.
Introduction
Programmed cell death (PCD) is a genetically determined, highly regulated process in all multicellular organisms and a prerequisite for successful development. PCD eliminates tissues and cells serving temporary functions during development such as tapetum cells in anthers and suspensor cells connecting the embryo to the mother plant or nucellus cells of a mature ovule [1–4]. Plants furthermore limit the spread of fungal or bacterial pathogens under execution of PCD at the site of infection in a mechanism called the hypersensitive response (HR) [5].
Diverse classes of proteases are involved in PCD, including cysteine proteases, serine proteases, aspartic proteases and metalloproteases [6,7]. A unique group of papain-type cysteine endopeptidases (CysEPs) is specific for plant PCD and characterized by a C-terminal KDEL endoplasmic reticulum (ER) retention signal (KDEL CysEPs) with RcCysEP from castor bean (Ricinus communis) as the founding member [8–10]. KDEL CysEPs are not present in mammals or fungi, but are ubiquitous in plants [11]. KDEL CysEPs are synthesized as pre-pro-enzymes and are co-translationally transferred into the ER, where the pre-sequence signal peptide is removed. KDEL CysEPs can be stored as enzymatically inactive pro-enzymes in ER-derived compartments [10,12–14]; upon acidification, the KDEL CysEPs are released and the pro-sequence together with the C-terminal KDEL endoplasmtic reticulum retention signal are removed for activation of the enzyme [10,15] (as described in detail previously [13]). The mature, enzymatically active KDEL CysEPs exhibit unusual broad substrate specificity. KDEL CysEPs are unique in being able to digest not only cytoplasmic components in tissues that collapse during final stages of PCD; they furthermore are able to digest extensins that form the basic scaffold for cell wall formation (as described in detail previously [16]). The broad substrate specificity is due to the active site cleft of the KDEL CysEPs that accepts a wide variety of amino acids including proline and glycosylated hydroxyproline of the hydroxyproline rich glycoproteins of the cell wall [17]. The respective amino acids, which are decisive for this generally more open appearance of the active site cleft, together with the amino acids defining the catalytic pocket are highly conserved among all known KDEL CysEPs [11].
In Arabidopsis, three KDEL CysEPs—CEP1 (At5g50260), CEP2 (At3g48340), and CEP3 (At3g48350)—have been identified that are expressed in tissues undergoing developmental PCD (as described in detail previously [13,16]). Furthermore, CEP1 was found to be expressed in late response to biotic stress stimuli in the leaf (as described in detail previously [18]). Two CEP1 T-DNA insertion lines (SAIL_158_B06 and SALK_01306, both carrying the T-DNA insertion within the 3rd exon) showed enhanced susceptibility to powdery mildew caused by the biotrophic ascomycete Erysiphe cruciferarum. A translational fusion protein of CEP1 with a three-fold hemaglutinin-tag and the green fluorescent protein under control of the endogenous CEP1 promoter (PCEP1::pre-pro-3xHA-EGFP-AtCEP1-KDEL) rescued the pathogenesis phenotype demonstrating the function of CEP1 in restriction of powdery mildew disease. atcep1 knockout plants transformed with the non-functional reporter including EGFP without the mature CEP1 subunit (PCEP1::pre-pro-3xHA-EGFP-KDEL) retained susceptibility to E. cruciferarum. The spatiotemporal CEP1-reporter expression during fungal infection together with microscopic inspection of the interaction phenotype suggested a function of CEP1 in controlling late stages of the compatible interaction including late epidermal cell death (as described in detail previously [18]).
Defense responses in Arabidopsis are regulated by multiple signal transduction pathways in which salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) function as key signaling molecules. Mutants such as cpr5 constitutively activate these defense pathways [19]. The CPR5 gene (CONSTITUTIVE EXPRESSION OF PR GENES 5, At5g64930) is a regulator of expression of pathogenesis related genes and participates in signal transduction pathways involved in plant defense and development such as leaf senescence or flowering. It codes for nuclear envelope membrane protein [20]. Loss of CPR5 leads to spontaneous expression of chlorotic lesions and reduced trichome development [21,22]. The cpr5 plants were found to be constitutively resistant to virulent pathogens such as the bacterial pathogen Pseudomonas syringae and the oomycete Hyaloperonospora arabidopsidis [19,21]. We found in public expression data that CEP1 (At5g50260, Affymetrix ATH1 probe set ID 248545_at; GEO accession GSE5745; this is unpublished work) is constitutively up-regulated in cpr5 mutants [www.genevestigator.com; 23]. We used the cpr5-2 mutant allele that has a point mutation in the fourth exon leading to a premature stop codon (Trp477stop) [24, 25] in order to analyze a possible contribution of the CEP1 upregulation to chlorotic leaf lesions in cpr5. We measured an increase in CEP1 expression in cpr5-2 mutant, which coincided with the appearance of leaf lesions. The expression of CEP1 was particularly evidenced in leaf cells that surround the chlorotic lesions and presumably underwent cell death. Furthermore, we found a strong resistance of cpr5-2 against infection with E. cruciferarum and studied the pathogenesis and cell death phenotypes in cep1 cpr5-2 double mutants as compared to the single mutants. This suggests a contribution of CEP1 to CPR5-controlled cell death.
Materials and methods
Arabidopsis mutant and reporter plants
We used homozygous knockout mutants for cep1 (SAIL_158_B06; T-DNA insertion within the third exon) [18]. For imaging the functional proenzyme of CEP1 by confocal laser scanning microscopy (CLSM), we used the functional reporter PCEP1::pre-pro-3xHA-EGFP-AtCEP1-KDEL that rescued the cep1 knockout phenotype when expressed in cep1 [18].
A homozygous cpr5-2 mutant allele (NASC stock code N3770) with a point mutation in the fourth exon leading to a stop codon (Tpr477stop) [25] was obtained and confirmed by sequencing: a 653 bp fragment comprising half of the fourth exon including the stop codon and six bp of the 3’UTR was amplified by PCR using the primers cpr5-2 fw GGCTCCTCGTAAGTGTCTTCAGC and cpr5-2 rv GGTCTGACTATGCTTGAGACGAG. The resulting PCR product was sequenced using the primers cpr5-2 seq fw CGATCATCAGGTACGAAGC and cpr5-2 seq rv GTCACGTTTATAGGACCG. Homozygous cep1 cpr5-2 double mutant plants were obtained by crossing. In order to monitor CEP1 promotor activity in cells of the cpr5-2 mutant background, we made homozygous double mutants by crossing cpr5 mutants and cep1 knockout mutants harboring the non-functional reporter PCEP1::pre-pro-3xHA-EGFP-KDEL that exhibits a translational fusion protein of the necessary CEP1 targeting sequences together with a three-fold hemaglutinin-tag and the green fluorescent protein EGFP but lacking the mature CEP1 subunit under control of the endogenous CEP1 promoter [18].
Leaf infection with powders mildew and symptoms rating
Inoculation and assessment of disease progression was carried out as described [18].
Necrotic leaf area measurement using fluorescence microscopy
Callose depositions in Arabidopsis cells were visualized by methyl blue (Sigma Aldrich Chemie GmbH, München, Germany) staining. For this purpose, leaves of cpr5-2 single mutant plants and of cep1 cpr5-2 double mutant plants, respectively, were harvested 5 days post inoculation (dpi) with E. cruciferarum and were discoloured in ethanol-acetic acid glacial (EtOH-HAc; 6:1). The discoloured leaves were rinsed with distilled water and transferred into 67 mM K2HPO4 buffer for 10 minutes followed by incubation for 2 h in the staining solution (0.05% methyl blue in 67 mM K2HPO4) in the dark and by direct analysis using fluorescence microscopy (Olympus BX61TRF, Japan).
Only areas of necrotic leaf lesions associated with powdery mildew colonies were measured using their fluorescence intensity. Areas as measured by fluorescence intensity under excitation by 340 nm wave length thus represented necrotic cells, but also other callose-enriched structures such as encasements of established haustoria. Sixty five fluorescing areas detected on 19 leaves on cpr5-2 single mutant plants and on cep1 cpr5-2 double mutant plants, respectively, were photographed and the fluorescing areas were quantified using ImageJ graphical analysis software version 1.48v [26]. To this end, the colour threshold was set to hide noise signals of unspecific adsorbed staining. Regions of specific fluorescence signals were selected and their areas were measured. Subsequently the data were corrected for the callose depositions belonging to powdery mildew-induced papillae and encasements of established haustoria. For this purpose, the average areas of ten powdery mildew colonies that were not associated to necrotic leaf lesions, on five leaves of cpr5-2 single mutant plants and of cep1 cpr5-2 double mutant plants, respectively, were measured using their fluorescence intensity. These values were subtracted from the fluorescent areas of necrotic leaf lesions associated with powdery mildew colonies on cpr5-2 single mutant plants and on cep1 cpr5-2 double mutant plants, respectively, thus obtaining the area of a chlorotic leaf lesion associated with a powdery mildew colony.
qRT-PCR
Primers used for qRT-PCR are: ACT8 qRT fw: TGAGACCTTTAATTCTCCAGCTATG; ACT8 qRT rv: CCAGAGTCCAACACAATACCG; CEP1 qRT fw: TCAGCCTGTTTCTGTTGCTATT; CEP1 qRT rv: CATCTCCCGGTAAACACTCC; CEP2 qRT fw: GCTGTTGCAAACCAACCTG; CEP2 qRT rv: TTCCACAAGATCCCGTAAACA; CEP3 qRTfw: GCTCACCAGCCTGTCTCTGT; CEP3 qRT rv: CGCATTCTCCGATAAACACA. qRT-PCR was carried out as described [18].
Confocal laser scanning microscopy
Staining of callose and imaging of the non-functional (PCEP1::pre-pro-3xHA-EGFP-KDEL) or functional (PCEP1::pre-pro-3xHA-EGFP-AtCEP1-KDEL) reporter plants was carried out as described [18].
Results
The proenzyme of CEP1 is associated with the haustorial complex encased in callose
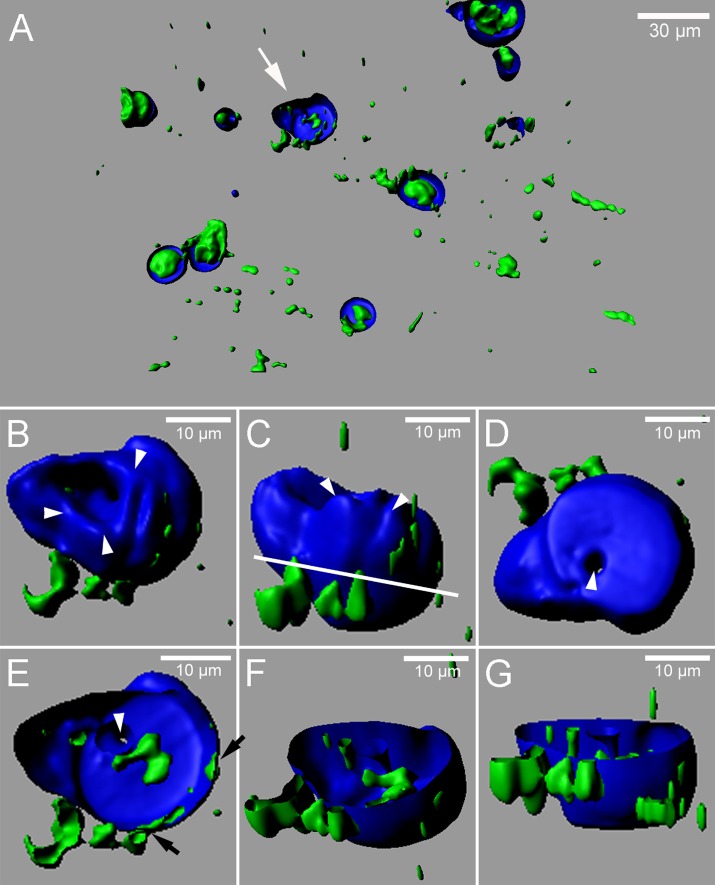
We previously described that cells attacked by Erysiphe cruciferarum express the proenzyme of CEP1 from the functional reporter PCEP1::pre-pro-3xHA-EGFP-AtCEP1-KDEL in the endoplasmic reticulum (ER) and accumulate it around haustoria. This led to the speculation that CEP1 could limit haustorium functions, which could explain enhanced growth of E. cruciferarum on cep1 mutant plants [18]. To get a more detailed impression of whether CEP1 could indeed closely associate with haustoria, we combined live cell imaging of PCEP1::pre-pro-3xHA-EGFP-AtCEP1-KDEL reporter plants with imaging of callose after staining with the aniline dye methyl blue at 11 dpi. Three dimensional reconstructions of fluorescence signals recorded by CLSM suggested that the proenzyme of CEP1 is closely associated with callose-encased haustoria and can be even found within encasements of haustorial complexes (Fig 1A). Closer inspection of fluorescence signals suggested that the CEP1-containing ER is present on both sides of the callose encasement, the cytoplasmic side and the haustorial side (Fig 1B–1G). The ER seemed to either span the encasement or the ER that was associated with the haustorium was enclosed during development of the encasement. In some haustorial encasements, GFP signals were strong but we were unable to further resolve the position of the GFP signal within or around the haustorial complex.
Fig 1. Three-dimensional reconstruction of fluorescence signals from the functional proenzyme of CEP1 (green) and callose depositions (blue) in epidermal cells of Arabidopsis invaded by haustoria of E.cruciferarum.
(A) Overview about several interaction sites with callose-encased haustorial complexes in the epidermis of Arabidopsis leaves at 11 dpi. CEP1 proenzyme is visible in epidermal cells and in and around encased haustoria that are shown in an optical section and reconstructed in 3 dimensions for lower z-sections. (B-G) Accumulation of the CEP1 proenzyme at the side and within a haustorium that is encased with callose. The same haustorium is also highlighted with an arrow in A. (B) View from the cell surface. At this site, little CEP1-loaded ER is visible at the periclinal cell cortex (arrowheads) (C) View from the side of the haustorium. Arrowheads indicate the rim of the encasement closest to the periclinal cell cortex (D) Bottom up view. Arrowhead indicates incomplete closure of the callose encasement (E-G) Callose-encased haustorium cut at the line indicated in panel C viewed from different angels. Arrowhead indicates incomplete closure of the callose encasement. Black arrows indicate sites where CEP1 signals are visible at both the cytosolic and the haustorial side of the encasement. Original pictures were recorded by CLSM and reconstructed into a 3-demensional picture by the IMARIS software (Bitplane).
CEP1 is specifically upregulated in cpr5-2 mutants in leaf cells undergoing cell death that surround the chlorotic lesions
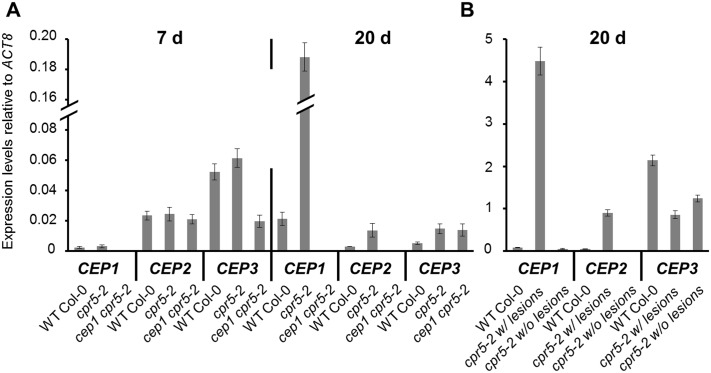
In publicly accessible global gene expression analysis of the Arabidopsis atcpr5 mutant (GEO accession GSE5745) displayed in Genevestigator [23], we found that CEP1 transcripts are significantly more abundant in two-week-old cpr5-2 mutants than in wild type. CPR5 is considered as a negative control element of plant programmed cell death and effector-triggered immunity through cell cycle control [27]. We used the cpr5-2 mutant allele that has a point mutation in the fourth exon leading to a stop codon (Trp477stop) [24,25]. Quantitative RT-PCR confirmed up-regulation of CEP1 in the cpr5-2 mutant allele correlating to the appearance of leaf lesions. CEP1 was not expressed to higher values in leaves of seven day old cpr5-2 seedlings before lesions were visible (Fig 2A left). An about ten-fold up-regulation of CEP1 was found in leaf rosettes of 20 day old cpr5-2 plants and correlated in time with the occurrence of spontaneous leaf lesions (Fig 2A right). On the other hand, CEP2 and CEP3 exhibited no strong expression in 20 day old rosette leaves in connection with the appearance of leaf lesion formation (Fig 2A). When we separated leaf rosettes of 20 day old cpr5-2 mutants into leaves with and without visible necrotic lesions, only necrotic leaves showed about 50-fold up-regulation of CEP1 transcripts (Fig 2B). Again, CEP2 and CEP3 did not show comparably high expression in leaves with necrotic lesions as compared to CEP1.
Fig 2. CEP1 is specifically upregulated in cpr5 mutants in leaf cells undergoing cell death that surround the chlorotic lesions.
Relative gene expression of CEP1, CEP2 and CEP3 was measured in 7 and 20 days old seedlings of wild type Col-0 as well as cpr5-2 single and cep1 cpr5-2 double mutant plants. Four leaves with visible lesions from four different plants of each genotype, respectively, were collected in one tube for isolation of RNA (A). Eight—twelve leaf pieces with or without lesions from six different plants of each genotype, respectively, were collected in one tube for isolation of RNA (B). qRT-PCR was performed with CEP1, CEP2 and CEP3 gene-specific primers. Expression levels were relatively set to the reference gene ACT8. Error bars represent standard deviations due to three technical replicates. The results were similarly reproduced in a second independent experiment (biological replicate). Furthermore, the data for the cep1 cpr5-2 double mutant plants were similarly reproduced with a second independent line. cpr5-2 w/lesions, leaf pieces with lesions; cpr5-2 w/o lesions, leaf pieces without lesions.
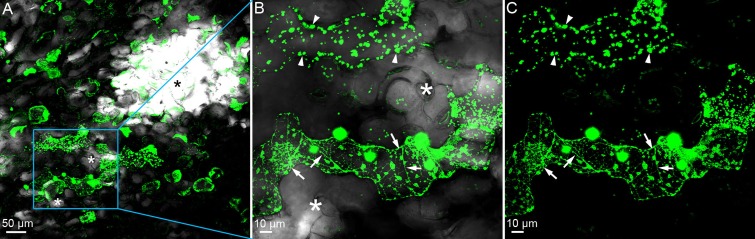
We made homozygous double mutants by crossing cep1 knockout mutants harboring the non-functional reporter PCEP1::pre-pro-3xHA-EGFP-KDEL and cpr5-2 mutants in order to visualize leaf cells that would express CEP1 (Fig 3) and to study the impact of cep1 knockout mutation on the cpr5-mediated resistance or cell death phenotype (see below). Strikingly, promoter activity of CEP1 was exclusively confined to cells in direct neighborhood of chlorotic lesions (Fig 3A, asterix). Leaf cells in areas more distant to the chlorotic lesion exhibited only autofluorescence but no specific GFP signal as identified by wavelength scanning with the confocal laser scanning microscope. The pattern of pre-pro-3xHA-EGFP-KDEL fluorescence showed partially a reticular pattern typical for the ER and partially a punctate pattern of disintegrated ER in cells that likely underwent PCD during spreading of lesions (Fig 3B and 3C).
Fig 3. CEP1 is specifically expressed in leaf cells undergoing cell death that surround the chlorotic lesions.
A. The white area (black asterisk *) indicates a chlorotic lesion in the terminal stage; the blue box indicates the representative tissue, where the enlarged image in B and C can be found. A and B. The white asterisks (*) indicate cells in spreading chlorotic lesion presumably undergoing programmed cell death. B. Merge of bright field image and EGFP. B and C. The pattern of fluorescence shows partially a reticular pattern typical for the endoplasmic reticulum (arrows) and partially a punctate pattern suggesting endoplasmatic reticulum disintegration in cells that likely underwent PCD (arrow heads). A, B and C. EGFP indicates the expression of the non-functional reporter construct PCEP1::pre-pro-3xHA-EGFP-KDEL.
cpr5-2 mutants are powdery mildew resistant
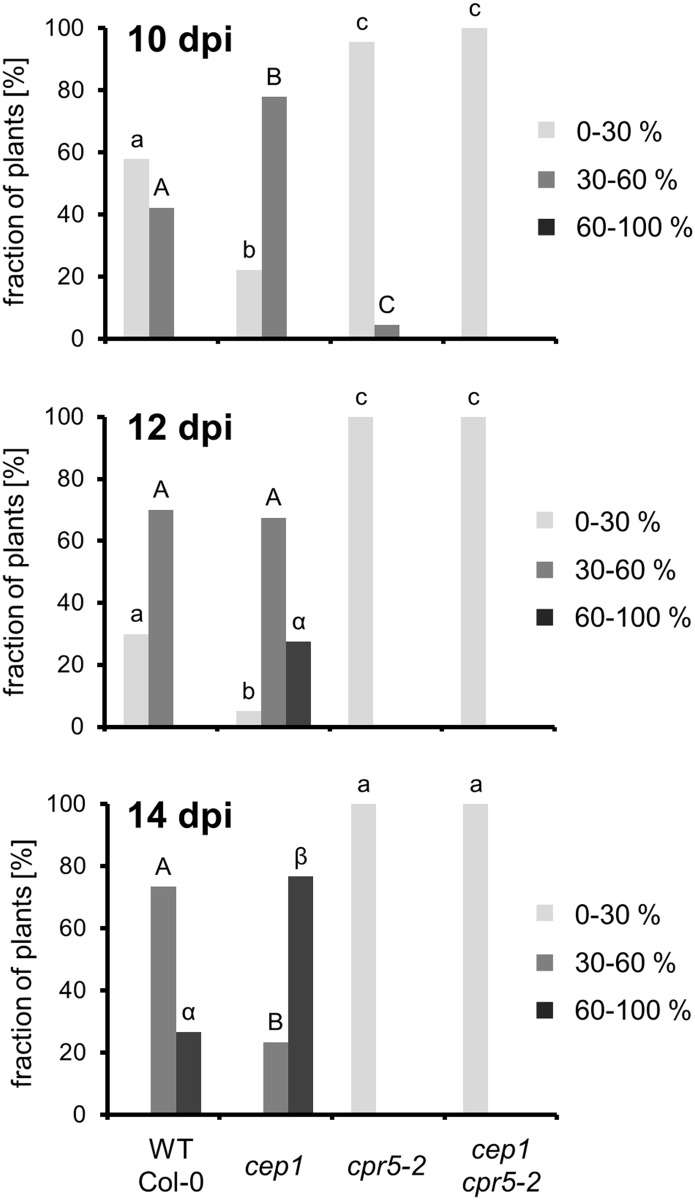
We observed a strong resistance of cpr5-2 mutant plants against infection with E. cruciferarum (Figs 4 and 5). Arabidopsis susceptibility to E. cruciferarum was scored by visual examination of the whole plant 10, 12 and 14 day after inoculation (Fig 4). The wild type Col-0 control plants developed the usual symptoms within 10 and 14 day after inoculation by exhibiting in time an increasing leaf area covered by powdery mildew symptoms with approximately 75% infected leaves in the category of 30–60% symptomatic leaf area and 25% in the category of >60% symptomatic leaf area14 day after inoculation. The cep1 knockout plants again exhibited more severe symptoms as compared to wild type with approximately 25% infected leaves in the category of 30–60% symptomatic leaf area and 75% in the category of >60% symptomatic leaf area 14 days after inoculation. In marked contrast to these susceptible genotypes, the cpr5-2 mutant plants showed exclusively leaves with powdery mildew symptoms in the category of <30% symptomatic leaf area throughout the investigation time (Figs 4 and 5).
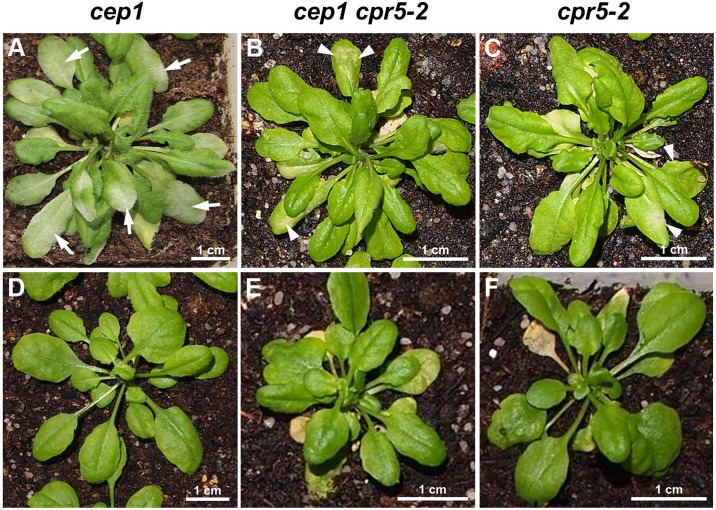
Fig 4. cpr5-2 mutants are powdery mildew resistant.
Disease symptoms of wild type, cep1 and cpr5-2 single mutants and cep1 cpr5-2 double mutant plants upon leaf infection with E. cruciferarum were scored after visual inspection of the whole plant 10, 12 and 14 days post inoculation (dpi). Distribution of infected leaves in the three categories was carried out as described [18]. Infected leafs were distributed in the three categories <30%, 30–60%, and >60% diseased leaf area [18]. Columns of the same colour marked with different letters indicate statistically different groups (p < 0.05), and columns marked with identical letters indicate groups not statistically different (p > 0.05) according to the ANOVA- and Duncan test, and represent the frequency of plants distributed in the three categories of susceptibility. Data represent the respective means of eight experiments from independent inoculation events of the mutants with the corresponding parent background control. Each experiment comprised five plants per line.
Fig 5. Powdery mildew disease phenotypes caused by the biotrophic ascomycete Erysiphe cruciferarum.
(A) cep1 knockout mutants developed the usual symptoms upon infection with powdery mildew (arrows). (B and C) In contrast, the cep1 cpr5-2 double mutant plants and the cpr5 single mutant plants are resistant to E. cruciferarum; necrotic lesions are indicated by white arrow heads; please note the scaling bar: cpr5 and cep1 cpr5-2 mutants are dwarf. Plants in (A-C) are shown 11 days post low density inoculation. (D-F) Non-infected plants at the time of inoculation.
CEP1 contributes to powdery mildew induced leaf lesions on cpr5-2
We analyzed the homozygous cep1 cpr5-2 double mutant obtained by crossing the cep1 knockout mutants harboring the non-functional reporter PCEP1::pre-pro-3xHA-EGFP-KDEL and the cpr5-2 mutants, in order to study the impact of cep1 knockout mutation on the cpr5-mediated resistance or cell death phenotype as compared to the single mutants. Interestingly, loss of CEP1 had no obvious influence on the strong resistance of cpr5-2 mutant plants against infection with E. cruciferarum as analyzed by macroscopic symptoms rating (Figs 4 and 5). All leaves of the cep1 cpr5-2 double mutant fell into the category of <30% symptomatic leaf area throughout the investigation time. The cpr5-2 mutant phenotype appeared thus epistatic over the cep1 mutant phenotype (Figs 4 and 5).
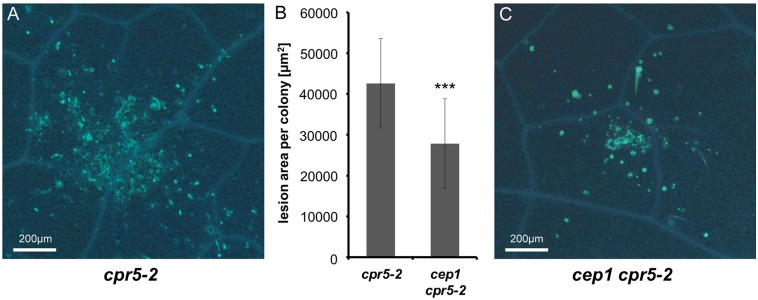
Our results suggested a strong activity of the CEP1 promoter in spatiotemporal association with lesions developing in cpr5-2 mutant plants (Fig 3). We therefore undertook a microscopic analysis of cpr5-2 single mutant plants in comparison to cep1 cpr5-2 double mutant plants at 5 days post inoculation with E. cruciferarum, when the powdery mildew growth still allows for the observation of single fungal colonies often associated with necrotic leaf lesions. Callose depositions were visualized by methyl blue staining in Arabidopsis cells at papillae, encasements of established haustoria as well as whole cells that die in the course of leaf lesion formation or epidermal hypersensitive response (Fig 6). The area of each necrotic leaf lesion associated with an E. cruciferarum colony was measured by fluorescence intensity. These average necrotic leaf areas were significantly larger in cpr5-2 single mutant plants (Fig 6A and 6B) as compared to cep1 cpr5-2 double mutant plants (Fig 6B and 6C). The presence of CEP1 thus contributed to the powdery mildew induced PCD and supported necrotic lesion formation.
Fig 6. CEP1 contributes to powdery mildew induced leaf lesions on cpr5-2.
(A) Representative leaf lesion on cpr5-2 single mutant plants. (B) Comparison of the necrotic leaf lesion area per colony on cpr5-2 single and on cep1 cpr5-2 double mutant plants. Columns show the area of a necrotic leaf lesion per E. cruciferarum colony (five spores/mm2) five days post inoculation (dpi) measured by fluorescence intensity. The areas showing fluorescence above a certain threshold intensity were measured. Data present the mean of 65 lesions on 19 leaves on cpr5 mutant plants and on cep1 cpr5-2 mutant plants, respectively. Error bars are standard deviations. Differences between cpr5 single and cep1 cpr5-2 double mutants are highly significant after two sided Student’s t-test (p < 0.001, ***). (C) Representative leaf lesion on cep1 cpr5-2 double mutant plants.
Discussion
CEP1 is associated with encased haustorial complexes
Callose encasements of haustorial complexes is considered as a basal defense reaction of Arabidopsis against powdery mildew fungi, which cannot fully suppress this response [28]. Similarly, we interpreted spatiotemporal association of CEP1 expression with late pathogenesis of E. cruciferarum as a contribution to basal resistance that limits fungal colonization success even in compatible interaction [18]. Plant ER has been observed before in close spatial proximity of developing and mature haustorial complexes [28–32]. Here, we visualized the presumable proenzyme ER-resident CEP1 around and within haustorial encasements at 11 days post inoculation. We further speculate that CEP1 could be released from a leaky ER or be actively secreted during late development or senescence of haustoria and come into contact with the haustorium. Direct activity of the protease on the haustorial complex or function in late epidermal cell death (as described in detail previously [18]) possibly explains the function of CEP1 in restricting late fungal development.
CEP1 is de-regulated in association with CPR5-controlled PCD
Publicly available gene expression data drew our attention to CPR5 as a possible regulator of CEP1 expression. Our targeted expression analyses of CEP1 in young asymptomatic and older leaves showing spontaneous leaf lesion development supported that CEP1 is overexpressed during de-regulated PCD in cep5 mutants. With the findings that CPR5 controls cell cycle and PCD during development and effector triggered immunity to leaf pathogens of Arabidopsis [24,27] and the observation that CEP1 contributes to pathogen-induced epidermal cell death, it appeared possible that CEP1 supports an CPR5-controlled cell death program. Indeed, similar to CEP1, CPR5 controlled E. cruciferarum-induced leaf cell death albeit in the opposite direction. This was associated with near-complete powdery mildew resistance of cpr5-2 mutants, which appears, based on previously published and our data, generally resistant to many hemibiotrophic and biotrophic pathogens [21,27,33,34].
CEP1 contributes to CPR5-controlled PCD at sites of powdery mildew infection
Double loss of function mutants of CPR5 and CEP1 retained powdery mildew resistance and spontaneous lesion phenotype observed in the cpr5-2 single mutant. Hence, the cpr5-2 mutant phenotype was largely epistatic over that of the cep1 phenotype in terms of symptom formation of powdery mildew. However, microscopic inspections of the size of lesions that were associated with powdery mildew microcolony formation showed that CEP1 indeed contributed to lesion formation in cpr5-2 mutants challenged by E. cruciferarum. Because these lesions have never been observed to a comparable extent in Col-0 before when challenged by E. cruciferarum and extended into the mesophyll tissue of atcpr5 mutants, we conclude that CPR5 negatively controls a powdery mildew triggered form of PCD. This type of PCD is simultaneously supported by CEP1, such that the cep1 cpr5-2 double mutant showed limited spreading of pathogen triggered cell death before leaf lesions became visible by naked eye. Together this supports that CEP1 acts in pathogen-triggered and CPR5-controlled leaf cell death although it does not represent a central switch in that type of PCD because visible lesions still appeared to an extent not readily distinguishable from that of the atcpr5 single mutant. This study was carried out in an accession of Arabidopsis susceptible to powdery mildew. Since CPR5 acts in effector triggered immunity [24,27], it would be interesting to see if CEP1 also contributes to the hypersensitive response that is mediated by endogenous or ectopically expressed powdery mildew resistance genes [35,36] in Arabidopsis. Spatial association of CEP1 expression with powdery mildew infection sites or CPR5 controlled cell death reactions might be interesting in context of other genes that function in spatial association with cell death. For instance, the salicylic acid receptors NPR3 and NPR4 support or restrict cell death via the abundance of NPR1 in a manner that depends on salicylic acid gradients in spatial distance to the actual site of lesion formation [37]. It might be thus interesting to study CEP functions in further mutants that show alteration of spontaneous or pathogen-triggered cell death.
Conclusion
Papain-type CysEPs have diverse functions in plant defense to pathogens [38]. Our data further suggest that the papain-type KDEL CysEP CEP1 fulfills its function in plant defense during late development of E. cruciferarum in close spatial association with the fungal haustorium and haustorial callose encasements. Additionally, CPR5 controls resistance to powdery mildew and PCD in response to infection by E. cruciferarum. In cpr5 mutants, CEP1 is overexpressed in spatiotemporal association with spontaneous cell death. CEP1 contributes to CPR5-controlled PCD triggered by E. cruciferarum but is not required to express high level powdery mildew resistance of cpr5 mutants.
Acknowledgments
We are grateful to Dr. Erika Isono (Technische Universität München, Center of Life and Food Sciences Weihenstephan) for the qRT-PCR primer design.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by grants from the Deutsche Forschungsgemeinschaft [SFB924 to CG (A07) and RH (B08)] and by the Technische Universität München within the funding programme Open Access Publishing. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Senatore A, Trobacher CP, Greenwood JS (2009) Ricinosomes predict programmed cell death leading to anther dehiscence in tomato. Plant Physiol 149: 775–790. doi: 10.1104/pp.108.132720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z et al. (2014) The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26: 2939–2961. doi: 10.1105/tpc.114.127282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López-Fernández MP, Maldonado S (2015) Programmed cell death in seeds of angiosperms. J Integr Plant Biol 57: 996–1002. doi: 10.1111/jipb.12367 [DOI] [PubMed] [Google Scholar]
- 4.Zhou L-Z, Höwing T, Müller B, Hammes UZ, Getl C, Dresselhaus T (2016) Expression analysis of KDEL CysEPs programmed cell death markers during reproduction in Arabidopsis. Plant Reprod 29: 265–272. doi: 10.1007/s00497-016-0288-4 [DOI] [PubMed] [Google Scholar]
- 5.Dickman MB, Fluhr R (2013) Centrality of host cell death in plant-microbe interactions. Annu. Rev Phytopathol 51: 543–570. doi: 10.1146/annurev-phyto-081211-173027 [DOI] [PubMed] [Google Scholar]
- 6.Beers EP, Jones AM, Dickerman AW (2004) The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65: 43–58. doi: 10.1016/j.phytochem.2003.09.005 [DOI] [PubMed] [Google Scholar]
- 7.Schaller A (2004) A cut above the rest: the regulatory function of plant proteases. Planta 220: 183–197. doi: 10.1007/s00425-004-1407-2 [DOI] [PubMed] [Google Scholar]
- 8.Schmid M, Simpson D, Kalousek F, Gietl C (1998) A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta 206: 466–475. doi: 10.1007/s004250050423 [DOI] [PubMed] [Google Scholar]
- 9.Schmid M, Simpson D, Gietl C (1999) Programmed cell death in castor bean endosperm is associated with the accumulation and release of a cysteine endopeptidases from ricinosomes. Proc Natl Acad Sci USA. 96: 14159–14164. doi: 10.1073/pnas.96.24.14159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmid M, Simpson DJ, Sarioglu H, Lottspeich F, Gietl C (2001) The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc Natl Acad Sci USA 98: 5353–5358. doi: 10.1073/pnas.061038298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hierl G, Vothknecht U, Gietl C (2012) Programmed cell death in Ricinus and Arabidopsis: the function of KDEL cysteine peptidases in development. Physiol Plant 145: 103–113. doi: 10.1111/j.1399-3054.2012.01580.x [DOI] [PubMed] [Google Scholar]
- 12.Toyooka K, Okamoto T, Minamikawa T (2000) Masstransport of a KDEL- tailed cysteine protease (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicleis involved in protein mobilization in germinating seeds. J Cell Biol 148: 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hierl G, Höwing T, Isono E, Lottspeich F, Gietl C (2014) Ex vivo processing for maturation of Arabidopsis KDEL-tailed cysteine endopeptidase2 (AtCEP2) pro-enzyme and its storage in endoplasmic reticulum derived organelles. Plant Mol Biol 84: 605–620. doi: 10.1007/s11103-013-0157-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano RT, Yamada K, Bednarek P, Nishimura M, Hara-Nishimura I (2014) ER bodies in plants of the Brassicales order: biogenesis and association with innate immunity. Front Plant Sci 5: 73 doi: 10.3389/fpls.2014.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M et al. (2014) Curr. Biol. 24: 931–40. [DOI] [PubMed] [Google Scholar]
- 16.Helm M, Schmid M, Hierl G, Terneus K, Tan M, Lottspeich F et al. (2008) KDEL-tailed cysteine endopeptidases involved in programmed cell death, inter-calation of new cells and dismantling of extension scaffolds. Am J Bot 95: 1049–1062. doi: 10.3732/ajb.2007404 [DOI] [PubMed] [Google Scholar]
- 17.Than ME, Helm M, Simpson DJ, Lottspeich F, Huber R, Gietl C (2004) The2.0-Å crystal structure of the KDEL-tailed cysteine endopeptidase from germinating endosperm of Ricinus communis confirms its function in the final stage of programmed cell death. J Mol Biol 336: 1103–1116. [DOI] [PubMed] [Google Scholar]
- 18.Höwing T, Huesmann C, Hoefle C, Nagel M-K, Isono E, Hückelhoven R et al. (2014) Endoplasmic reticulum KDEL-tailed cysteine endopeptidases 1of Arabidopsis (AtCEP1) is involved in pathogen defense. Front Plant Sci 5: 58 doi: 10.3389/fpls.2014.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr–induced resistance in Arabidopsis. Plant Cell 12: 2175–2190. doi: 10.1105/tpc.12.11.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirik V, Bouyer D, Schobinger U, Bechtold N, Herzog M, Bonneville JM et al. (2001) CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biol 11: 1891–1895. [DOI] [PubMed] [Google Scholar]
- 21.Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1- independent resistance. Plant Cell 9: 1573–1584. doi: 10.1105/tpc.9.9.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brininstool G, Kasili R, Simmons LA, Kirik V, Hulskamp M, Larkin JC (2008) Constitutive Expressor Of Pathogenesis-related Genes5 affects cell wall biogenesis and trichome development. BMC Plant Biol 8: 58 doi: 10.1186/1471-2229-8-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray data base and analysis toolbox. Plant Physiol 136: 2621–2632. doi: 10.1104/pp.104.046367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boch J, Verbsky ML, Robertson TL, Larkin JC. Kunkel BN (1998) Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol Plant Microbe Interact 11: 1196–1206. doi: 10.1094/MPMI.1998.11.12.1196 [Google Scholar]
- 25.Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasband WS, ImageJ US National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2014.
- 27.Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R et al. (2014) A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe 16: 787–794. doi: 10.1016/j.chom.2014.10.005 Cell, 6: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Micali CO, Neumann U, Grunewald D, Panstruga R, O'Connell R (2011) Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol 13: 210–226. doi: 10.1111/j.1462-5822.2010.01530.x [DOI] [PubMed] [Google Scholar]
- 29.Leckie CP, Callow JA, Green JR (1995) Reorganization of the endoplasmic reticulum in pea leaf epidermal cells infected by the powdery mildew fungus Erysiphe pisi. New Phytol 131: 211–221. doi: 10.1111/j.1469-8137.1995.tb05722.x [Google Scholar]
- 30.Koh S, Andre A, Edwards H, Ehrhardt D, Somerville S (2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 44: 516–529. doi: 10.1111/j.1365-313X.2005.02545.x [DOI] [PubMed] [Google Scholar]
- 31.Eichmann R, Dechert C, Kogel K-H, Hückelhoven R (2006) Transient over-expression of barley BAX Inhibitor-1 weakens oxidative defence and MLA12-mediated resistance to Blumeria graminis f.sp hordei. Molecular Plant Pathol 7: 543–552. doi: 10.1111/j.1364-3703.2006.00359.x [DOI] [PubMed] [Google Scholar]
- 32.Eichmann R, Hückelhoven R (2008) Accommodation of powdery mildew fungi in intact plant cells. J Plant Physiol 165: 5–18. doi: 10.1016/j.jplph.2007.05.004 [DOI] [PubMed] [Google Scholar]
- 33.Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE (2001) Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J 26: 409–420. [DOI] [PubMed] [Google Scholar]
- 34.Orjuela J, Deless EF, Kolade O, Cheron S, Ghesquiere A, Albar L (2013) A recessive resistance to rice yellow mottle virus is associated with a rice homolog of the CPR5 gene, a regulator of active defense mechanisms. Mol Plant Microbe Interact 26: 1455–1463. doi: 10.1094/MPMI-05-13-0127-R [DOI] [PubMed] [Google Scholar]
- 35.Maekawa T, Kracher B, Vernaldi S, Ver Loren van Themaat E, Schulze-Lefert P (2012) Conservation of NLR-triggered immunity across plant lineages. Proc Natl Acad Sci USA 109: 20119–20123. doi: 10.1073/pnas.1218059109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M et al. (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120. doi: 10.1126/science.291.5501.118 [DOI] [PubMed] [Google Scholar]
- 37.Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–233. doi: 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misas-Villamil JC, van der Hoorn RA, Doehlemann G (2016) Papain-like cysteine proteases as hubs in plant immunity. New Phytol 212: 902–907. doi: 10.1111/nph.14117 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.