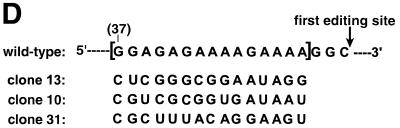Abstract
Several mitochondrial mRNAs of the kinetoplastid protozoa do not encode a functional open reading frame until they have been edited through the addition or deletion of U nucleotides at specific sites. Genetic information specifying the location and extent of editing is present on guide RNAs (gRNAs). The sequence adjacent to most mRNA editing sites has a high purine content which previously has been proposed to facilitate the editing reaction through base-pairing to a poly(U) tail at the 3′ end of the gRNA. We demonstrate here that gRNA binding alone is insufficient to create an editing site and that the mRNA sequence near an editing site is an additional determinant affecting the efficiency of the reaction.
INTRODUCTION
Part of the genetic information specifying the location of a U-insertion or -deletion within mRNAs of the kinetoplastid protozoa has been demonstrated to be carried on guide RNAs (gRNAs). A gRNA binds to its cognate mRNA immediately 3′ of an editing site and also contains a sequence, complementary to a block of edited mRNA, which functions as a guide for the insertion and deletion of the appropriate number of U nucleotides (1–3). Editing of the mRNA is believed to be initiated by an endonucleolytic cleavage at the first unpaired nucleotide 5′ of the gRNA–mRNA duplex. U nucleotides are added or removed from the 3′ end of the 5′ cleavage fragment, and a ligase activity rejoins the two mRNA fragments, completing one editing cycle. gRNAs isolated from the mitochondria also contain non-genomically encoded U nucleotides at the 3′ end (4). It has previously been suggested that the binding of the U-tail of the gRNA to the pre-edited sequence increases the stability of the gRNA–mRNA duplex and positions the 5′ endonucleolytic mRNA cleavage fragment (Fig. 1) for the subsequent U addition/deletion and ligation reactions (3,4).
Figure 1.
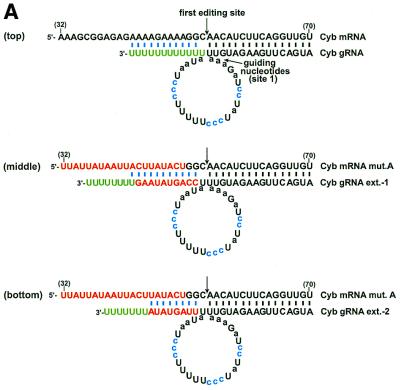
gRNA-binding is not sufficient for editing. (A) Base-pairing of a cytochrome b (Cyb) gRNA and mRNA transcript. A poly(U) tail that is added to the 3′ end of the gRNA by a TUTase activity within the editing extract (green) has the potential to interact (blue) with the purine-rich pre-edited sequence. Mutations (mut.) to the mRNA that would disrupt this interaction and two compensatory extensions (ext.) to the 3′ end of the gRNA are shown in red. The A→C substitutions within the gRNA that stimulate the in vitro reaction are highlighted in blue and the guiding nucleotides are in lower case. The first editing site and the corresponding guiding nucleotides are indicated. (B) The primer extension reaction used to detect in vitro U-insertions within the first editing site. A primer annealed immediately 3′ of the first cytochrome b editing site is extended by a number of radiolabeled dA nucleotides corresponding to the number of U nucleotides inserted during editing. Addition of ddG terminates the reaction. (C) Cytochrome b mRNA transcripts containing the wild-type and mutated pre-edited sequence were assayed for gRNA-directed U-insertions using no added gRNA (–), and gRNAs with and without (wt) a compensatory 3′ extension. The marker for correct editing results from the extension of a template containing three insertions at the first editing site (+3). Additional bands are artifacts of the assay (ar) as indicated by their presence in the non-extract treated lanes. (D) The wild-type cytochrome b editing domain and the sequence of three RNAs cloned from the random RNA pool. The sequence that was made random is shown in square brackets. (E) Assay of the above RNAs for gRNA-directed U-insertions within the first editing site.
The sequence surrounding most mRNA editing sites is very purine-rich, consistent with the possibility that pairing of the poly(U) tail of the gRNA is important to the editing reaction (1). We demonstrate here, however, that mutations to the purine-rich pre-edited sequence of a Leishmania tarentolae mRNA are not compensated for by mutations to the gRNA that should have restored pairing. We further demonstrate that although a purine sequence is important to the editing reaction, it has a role that is independent of base-pairing with the gRNA. The results suggest that additional genetic information specifying efficient editing is contained within the pre-edited region of the mRNA.
MATERIALS AND METHODS
Editing assays
Mitochondrial editing extract was prepared from L.tarentolae (UC strain) as previously described (5,6). The editing assays to detect gRNA-directed U-insertions within the ND7 and cytochrome b mRNA transcripts have also been previously described (6,7). T7 RNA polymerase was used to directly transcribe the gRNA and mRNA transcripts from oligodeoxynucleotides listed below (8). The template for the transcription of the cytochrome b mRNA transcript containing the wild-type pre-edited sequence was amplified from oligodeoxynucleotide 714333 using primers 851787 and 17704. The templates encoding the cytochrome b mRNA with the mutated and random pre-edited sequence were similarly amplified from oligodeoxynucleotides 961573 and 100901, respectively. The extension of oligodeoxynucleotide 1017134, which has three T nucleotides at the first ND7 editing site, was used to generate the +3 marker for the primer extension assay, and oligodeoxynucleotide 666615 was used to generate the marker for the assay for editing of the cytochrome b transcripts.
Oligodeoxynucleotides
Templates for the transcription of gRNAs and mRNA.
wt ND7 mRNA (wild-type pre-edited sequence),
5′-GTCCGAGAGCGAAGAAACATAATGTTCTTTTTATATTTATCGTGTAGTCGGCTAAATAAAATTTAACCTATAGTGAGTCGTATTA-3′;
ND7 mRNA mut. A,
5′-GTCCGAGAGCGAAGAAACATAATGTTCGGAAGATAAGAATCGTGTAGTCGGCTAAATAAAATTTAACCTATAGTGAGTCGTATTA-3′;
ND7 mRNA mut. B,
5′-GTCCGAGAGCGAAGAAACATAATGTTCGCAAGATAAGAA-TCGTGTAGTCGGCTAAATAAAATTTAACCTATAGTGAGTCGTATTA-3′;
ND7 gRNA (no 3′ extension),
5′-TAATCATACTCCATATTTATTAATTATCTAACATTATGATTCTCCCTATAGTGAGTCGTATTA-3′;
ND7 gRNA ext.-1,
5′-TGCATAATCATACTCCATATTTATTAATTATCTAACATTATGATTCTCCCTATAGTGAGTCGTATTA-3′;
cytochrome b gRNA (no 3′ extension),
5′-TATTAGGGAAAAAGGGATAGGATCTTTAACATCTTCAAGTCATATGTGCCTATAGTGAGTCGTATTA-3′;
cytochrome b gRNA ext.-1,
5′-CTTATACTGGATATTAGGGAAAAAGGGATAGGATCTTTAACATCTTCAAGTCATATGTGCCTATAGTGAGTCGTATTA-3′;
cytochrome b gRNA ext.-2,
5′-TATACTAAATATTAGGGAAAAAGGGATAGGATCTTTAACATCTTCAAGTCATATGTGCCTATAGTGAGTCGTATTA-3′.
Primers and templates for PCR.
851787,
5′-TAATACGACTCACTATAGGGATAAATTTAATTTAAATTTTAAATAATTATA-3′;
714333,
5′-GGGATAAATTTAATTTAAATTTTAAATAATTATAAAAGCGGAGAGAAAAGAAAAGGCAACATCTTCAGGTTGTTTATTACGAGTATATGGTGTAGG-3′;
961573,
5′-GGGATAAATTTAATTTAAATTTTAAATAATTATATTATTATAATTACTTATACTGGCAACATCTTCAGGTTGTTTATTACGAGTATATGGTGTAGG-3′;
100901,
5′-GGGATAAATTTAATTTAAATTTTAAATAATTATANNNNNNNNNNNNNNNGGCAACATCTTCAGGTTGTTTATTACG-3′;
17704,
5′-CCTACACCATATACTCGTAAT-3′.
Assay markers. 1017134,
5′-AAATATAAAAAGTTTAACATTATGTTTCTTCGCTCTCGGAC-3′;
666615,
5′-GAAAAGAAAAGGCTTTAACATCTTCAGGTTGTTTA-TTACGAGTATATGGTGTAG-3′.
RESULTS
Base-pairing of the mRNA with the gRNA is not sufficient for in vitro editing
In vitro assays have previously been described for gRNA-directed editing of L.tarentolae cytochrome b and ND7 mRNA transcripts (6,7). The binding of the cognate gRNAs, encoding the insertion of three U nucleotides into the first editing site of these transcripts, are indicated (Figs 1 and 2). Although gRNAs isolated from the mitochondria have a non-genomically encoded 3′ poly(U) tail, the gRNAs used for the in vitro editing reactions were not synthesized with a 3′ poly(U) tail because it does not significantly affect the reaction efficiency (7). However, since a terminal uridylyltransferase activity (TUTase) within the editing extract adds U nucleotides to the 3′ end of the gRNAs, the possibility of a base-pairing interaction with the purine-rich pre-edited sequence could not be excluded.
Figure 2.
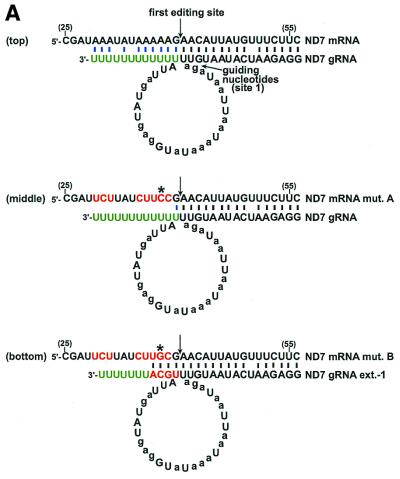
A single nucleotide substitution restores editing. (A) Base-pairing of ND7 gRNA and mRNA transcripts. The asterisk indicates the location of a single nucleotide substitution that can restore in vitro editing. The remaining features are as described for Figure 1. (B) The primer extension assay was performed to detect U-insertions within the first ND7 editing site. The extension was terminated by the addition of ddC.
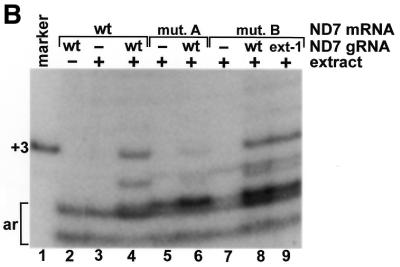
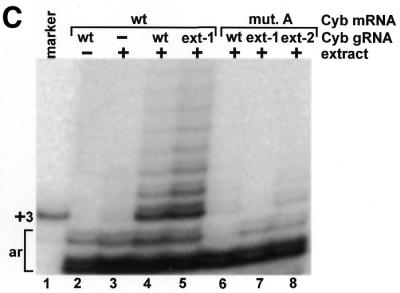
A primer extension reaction was used to detect the gRNA-directed in vitro U-insertions within the first editing site of a cytochrome b mRNA transcript (Fig. 1B). A DNA primer complementary to sequence immediately downstream of the first editing site was annealed to a DNA template amplified from the extract-treated mRNA transcripts (6). Extensions were performed with a DNA polymerase in the presence of [α-32P]dATP and the reactions were terminated by the incorporation of a dideoxynucleotide immediately after the first editing site. A synthetic cytochrome b template containing three thymidines within the first editing site was used in the primer extension reaction as a marker for the incorporation of the correct number of gRNA-directed U insertions (Fig. 1C, lane 1). In the absence of extract treatment (Fig. 1C, lane 2), a primer extension product consistent with correct editing of the cytochrome b mRNA transcript was not detected; only artifacts of the primer extension reaction are visible (Fig. 1C, labeled ar). The intensity of the lower artifact band is directly proportional to the amount of template put into the reactions and can be used to normalize the editing signal (6). As previously described, in vitro editing of the cytochrome b transcript was stimulated by mutation of an A–U sequence on the gRNA that appeared to inhibit the reaction (6) (Fig. 1A, blue nucleotides). In the presence of the editing extract and a gRNA containing this mutation, a predominant primer extension band indicative of correct editing was visible (Fig. 1C, lane 4). There was also a ladder of extension products that was presumably derived from sub-populations of transcripts having different numbers of inserted U nucleotides, and an explanation for this misediting has previously been proposed (6).
Mutation of the purine-rich sequence flanking the third to 15th cytochrome b mRNA editing sites (Fig. 1A, nucleotides 32–51), which would inhibit base-pairing of the U-tail of the gRNA, inhibited 98 ± 2% (n = 3) of the gRNA-directed U-insertions within the first editing site (Fig. 1C, lane 6). However, editing was not rescued by a 3′ extension on the gRNA that should have restored base-pairing to the pre-edited cytochrome b mRNA transcript (Fig. 1A, middle, and C, lane 7). Incubation of the radiolabeled 3′ extended gRNA with the editing extract indicated that it had approximately the same extent of U-additions to the 3′ end as the non-extended gRNA (data not shown). The 3′ extended gRNA also directed editing of the mRNA transcript containing the wild-type purine sequence as well as the non-extended gRNA (Fig. 1C, lanes 4 and 5). This indicates that the failure of the 3′ extended gRNA to rescue editing of the mutated mRNA was not a result of the artificial extension causing it to misfold. Incubation of the radiolabeled mRNA transcripts with the editing extract also indicated that the decreased editing of the mutated sequence did not result from a decreased stability (data not shown).
Since the average length of the U-tail on the gRNAs isolated from the mitochondria is ∼15 nt (4), it is possible that the 3′ extended gRNA failed to rescue in vitro editing because the increased stability of the artificial duplex was inhibitory. This was previously demonstrated for the U-deletion type editing catalyzed by a mitochondrial extract prepared from Trypanosoma brucei (9). However, a second gRNA, with a shorter 3′ extension that could interact with the mutated pre-edited sequence with binding energy approximating that of a 13 nt poly(U) tail binding to the wild-type pre-edited sequence, also failed to significantly rescue editing (Fig. 1A, bottom, and C, lane 8). In addition, a second set of substitution mutations to the purine-rich sequence of the cytochrome b mRNA transcript was also inhibitory to the editing reaction and could not be rescued with a gRNA containing a compensatory 3′ extension (data not shown).
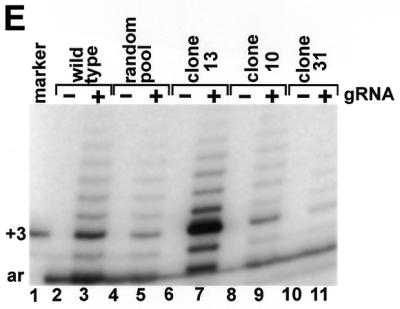
The sequence requirement within the pre-edited region of the cytochrome b mRNA was further explored. Deletion of the first 5 nt from the 5′ end of the pre-edited region of the cytochrome b mRNA transcript (Fig. 1A, nucleotides 32–36) had no effect on the reaction (data not shown). The adjacent 15 purines of the pre-edited region (Fig. 1D, nucleotides 37–51) were replaced with an equal representation of each of the four nucleotides, and this population was edited almost as efficiently as the wild-type sequence (Fig. 1E, lanes 2–5). This indicates that the absolute sequence requirement would be limited to only a few of the randomized positions.
Several RNAs were cloned from the random RNA pool and used as substrates in the editing reaction (Fig. 1E). Clone 13 was a 9-fold (±3, n = 3) better substrate for the U-insertion reaction than the wild-type cytochrome b sequence in spite of the fact that it has fewer purines and would be predicted to have less affinity for a 3′ poly(U) tail on the gRNA (10). In addition, clones 10 and 31 both have eight purines and both are predicted to bind similarly to the 3′ end of the gRNA, yet clone 10 was a 4-fold (±1, n = 3) better substrate for the editing reaction. This further emphasizes the fact that the sequence 5′ of the editing site influences the reaction and the effect cannot be solely explained by a base-pairing requirement with the 3′ end of the gRNA.
ND7 is the only other L.tarentolae transcript that has been demonstrated to be edited in vitro (7). To determine whether a purine sequence is important for the editing of mRNA transcripts other than cytochrome b, the purines within the ND7 editing domain were also mutated, and U-insertions occurring within the first editing site were assayed with a primer-extension reaction. As previously described (7), the presence of both extract and a gRNA containing three first site guiding nucleotides led to the incorporation of three U nucleotides (Fig. 2B, lane 4). A smaller population of RNAs containing an incorrect number of insertions was also formed in the editing reaction.
Substitution of eight purines upstream of the first editing site of the ND7 transcript with C or U, which would inhibit base-pairing of the U-tail of the gRNA (Fig. 2A, middle), inhibited 80% of the editing reaction (±2%, n = 3) (Fig. 2B, lane 6). Substitution of a single G within the mutated pre-edited region restored editing to that of the wild-type RNA (Fig. 2B, lane 8). However, the binding of this sequence to the U-tail of the gRNA would still be expected to be unfavorable (Fig. 2A, middle), and it is therefore unlikely that the increased editing is related to binding of the gRNA. Addition of a compensatory 3′ extension to the gRNA that should have restored base-pairing had no further stimulatory effect (Fig. 2A, bottom, and B, lane 9). Therefore, inhibition of the in vitro editing reaction caused by mutations to the purines flanking the editing sites of two different mRNAs cannot be explained solely on the basis of a requirement to interact with the 3′ end of the gRNA.
The gRNA-independent U-insertion reaction also has a sequence specificity
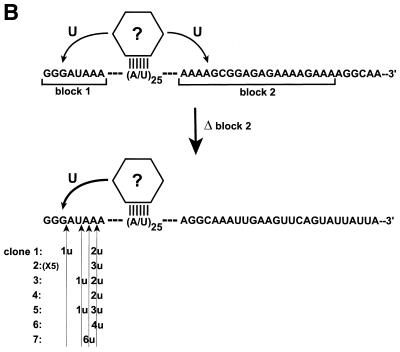
In addition to the gRNA-directed reaction, a cytochrome b mRNA transcript has been demonstrated to have U nucleotides inserted by the fractionated editing extract independent of gRNA (5,11). This reaction is most consistent with a model in which a component of the editing machinery interacts with an A–U sequence element present within the 5′ untranslated sequence of the cytochrome b mRNA (6) (Fig. 3). There is also indirect evidence indicating that the same interaction occurs in vivo (6). The majority (85%) of the gRNA-independent insertions within the cytochrome b transcript are approximately evenly distributed between the sequence immediately 5′ and 3′ of the A–U element (5); these sequences are within a purine-rich 8 nt block at the 5′ end of the mRNA (Fig. 3B, block 1) and within the purine-rich pre-edited sequence (Fig. 3B, block 2).
Figure 3.
The gRNA-independent U-insertion reaction also has a sequence specificity. (A) Sequence of the cytochrome b mRNA transcript used for the gRNA-independent reaction. The critical 34 nt A–U sequence is underlined, the pre-edited region is boxed and the mutations to the endogenous gRNA binding site are indicated in lower case. (B) The gRNA-independent reaction is most consistent with a model in which components of the editing complex (designated by ?) interact with an A–U sequence element (6). The majority of the gRNA-independent U-insertions occur on either side of the A–U sequence (5). The locations of these insertions are bracketed and labeled blocks 1 and 2, respectively. Deletion of the sequence in block 2 results in the gRNA-independent insertions being restricted entirely to the 5′ end of the A–U sequence. The location and number of the gRNA-independent U-insertions within the 11 cloned RNAs containing insertions are indicated. Four clones having U-insertions identical to clone 2 were identified (X5).
Since the gRNA-directed and gRNA-independent reactions appear to exploit an overlapping set of enzymes (6), the gRNA-independent reaction provides an additional means of testing the hypothesis that the editing machinery has a mRNA sequence preference independent of gRNA binding. Deletion of the purine-rich cytochrome b mRNA pre-edited sequence (Fig. 3B, block 2) resulted in a sequence more abundant in pyrimidines being placed immediately 3′ of the A–U element. This mutation was previously demonstrated not to inhibit the gRNA-independent reaction (5), but the location of the U-insertions occurring within this RNA was not determined.
The gRNA-independent U-insertions occur at a low level necessitating that the extract-treated RNAs with U-insertions be enriched prior to cloning and sequencing. A previously described procedure based on 4-thiouridine incorporation and passage through an organomercury matrix was used to enrich for extract-treated RNAs containing U-insertions (5). In the absence of the purine-rich pre-edited sequence, U-insertions were not detected 3′ of the A–U elements in any of the 11 clones with insertions (Fig. 3, bottom). The U-insertions were confined to the purine-rich sequence 5′ of the A–U element. This is in contrast to RNAs containing the wild-type pre-edited sequence (5), and suggests that a component of the U-insertion machinery has a mRNA sequence preference independent of gRNA binding.
DISCUSSION
Phylogenetic analysis indicates that there is a strong bias for purines within the sequence immediately flanking insertion editing sites of T.brucei mRNAs (12) and several explanations for this bias have been suggested. Part of the bias may have resulted from an evolutionary constraint to maintain accurate editing (12). In order for a gRNA to pair with pyrimidines flanking the mRNA editing sites, there would be a requirement for complementary purines within the gRNA, and these additional purines could potentially create editing ambiguities by acting as guiding nucleotides for U-insertions. Alternatively, it had previously been proposed that base-pairing of the 3′ poly(U) tail of the gRNA to the pre-edited region stabilizes the gRNA–mRNA duplex (4). In support of this proposal, tethering of the 3′ end of the gRNA to the mRNA sequence upstream of the editing sites can stimulate the deletion editing catalyzed by a T.brucei mitochondrial extract (9). Likewise, increasing the tethering of the 3′ end of an ND7 gRNA to the mRNA stimulated the U-insertion editing catalyzed by a L.tarentolae extract (13). However, the interpretation of this later study is complicated because the guiding nucleotides of the ND7 gRNA were also mutated, and mutation of the guiding nucleotides of a L.tarentolae cytochrome b gRNA has been demonstrated to stimulate insertion editing independent of increased tethering (6).
The results presented here do not necessarily exclude earlier explanations for the sequence preference within the mRNA pre-edited sequence. Rather, the results suggest that there is an additional sequence constraint within the pre-edited mRNA region. There are currently only two L.tarentolae mRNAs that are edited in vitro (6,7), and we have demonstrated for both mRNAs that mutation of the pre-edited sequence can significantly affect editing in a manner that does not correlate with the ability to base pair with the 3′ end of the gRNA. Since several independent mutations were assayed, it is unlikely that the inhibitory mRNA mutations could all result from unintended detrimental interactions with the gRNA. Likewise, the mutation in clone 13 stimulated editing by almost an order of magnitude relative to the wild-type cytochrome b mRNA sequence (Fig. 1E) and this is difficult to reconcile with a gRNA binding effect. The existence of an additional sequence constraint is further supported by the U-insertion reaction that occurs independent of gRNA also having a sequence preference (Fig. 3).
The sequence constraint for U-insertion editing within the pre-edited region is small. This is indicated both by the randomized cytochrome b pre-edited sequence being edited within a factor of two of the wild-type level (Fig. 1E) and by a single purine substitution to a mutated ND7 mRNA being able to rescue editing (Fig. 2B). While this manuscript was in preparation, it was reported that U-deletion editing is stimulated if the mRNA sequence immediately upstream of the editing sites is not paired with the gRNA, but a sequence preference was not reported (9). However, the RNA sequence and structural requirements for U-insertion and deletion editing are known to differ (14), and the sequence preference described here may be another difference. This is supported by the phylogenetic analysis indicating that there is a stronger sequence bias surrounding sites of U-insertion in T.brucei than there is at sites of U-deletion (12).
The additional sequence constraint within the pre-edited region could be critical to define the location of a proper site of editing and to limit inappropriate editing. The current view of editing site recognition is that the first unpaired nucleotide following the duplex formed by the binding of the gRNA to the mRNA defines an editing site (1–3). Since gRNAs can act in cis (13) and intramolecular duplexes would be expected to be common in mRNAs, additional specificity elements would be required to limit inappropriate editing. The purines within the editing domain may partially fulfill this function. In addition, as the sequence of the pre-edited mRNA can dramatically affect editing efficiency, this sequence may be one means through which some transcripts could have been evolved to become preferentially edited or regulated.
In T.brucei, a transcript encoding part of the ATPase 6 pre-mRNA is edited in vitro. However, an ND7 transcript is not edited by this system unless the pre-edited sequence of the ND7 transcript is substituted with the ATPase 6 editing domain (3). It was suggested that base-pairing of the U-tail of the gRNA would be optimal with the ATPase 6 pre-edited sequence as it is more purine-rich than the corresponding ND7 sequence. Although this is a possible explanation for the chimeric RNA being edited, the alternative suggested here also needs to be considered.
Acknowledgments
ACKNOWLEDGEMENTS
Supported by NIH grant R29-AI41138. L.M.O. was supported by American Heart Association post-doctoral fellowship MN/9920509Z.
References
- 1.Blum B., Bakalara,N. and Simpson,L. (1990) A model for RNA editing in kinetoplastid mitochondria: ‘guide’ RNA molecules transcribed from maxicircle DNA provide the edited information. Cell, 60, 189–198. [DOI] [PubMed] [Google Scholar]
- 2.Seiwert S.D., Heidmann,S. and Stuart,K. (1996) Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell, 84, 831–841. [DOI] [PubMed] [Google Scholar]
- 3.Kable M.L., Seiwert,S.D., Heidmann,S. and Stuart,K. (1996) Activation of guide RNA-directed editing of a cytochrome b mRNA. Science, 273, 1189–1195. [DOI] [PubMed] [Google Scholar]
- 4.Blum B. and Simpson,L. (1990) Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the pre-edited region. Cell, 62, 391–397. [DOI] [PubMed] [Google Scholar]
- 5.Brown L.M., Burbach,B.J., McKenzie,B.A. and Connell,G.J. (1999) A cis-acting A–U sequence element induces kinetoplastid U-insertions. J. Biol. Chem., 274, 6295–6304. [DOI] [PubMed] [Google Scholar]
- 6.Oppegard L.M., Kabb,A.L. and Connell,G.J. (2000) Activation of guide RNA-directed editing of a cytochrome b mRNA. J. Biol. Chem., 275, 33911–33919. [DOI] [PubMed] [Google Scholar]
- 7.Byrne E.M., Connell,G.J. and Simpson,L. (1996) Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J., 15, 6758–6765. [PMC free article] [PubMed] [Google Scholar]
- 8.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Reyes J., Zhelonkina,A., Rusche,L. and Sollner-Webb,B. (2001) Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol., 21, 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freier S.M., Kierzek,R., Jaeger,J.A., Sugimoto,N., Caruthers,M.H., Neilson,T. and Turner,D.H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connell G.J., Byrne,E.M. and Simpson,L. (1997) Guide RNA-independent and guide RNA-dependent uridine insertion into cytochrome b mRNA in a mitochondrial lysate from Leishmania tarentolae. Role of RNA secondary structure. J. Biol. Chem., 272, 4212–4218. [DOI] [PubMed] [Google Scholar]
- 12.Burgess M.L. K. and Stuart,K. (2000) Sequence bias in edited kinetoplastid RNAs. RNA, 6, 1492–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapushoc S.T. and Simpson,L. (1999) RNA, in vitro uridine insertion RNA editing mediated by cis-acting guide RNAs. RNA, 5, 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz-Reyes J., Rusche,L.N., Piller,K.J. and Sollner-Webb,B. (1998) T.brucei RNA editing: adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol. Cell, 1, 401–409. [DOI] [PubMed] [Google Scholar]