Abstract
DUF221 domain-containing genes (DDP genes) play important roles in developmental biology, hormone signalling transduction, and responses to abiotic stress. Therefore to understand their structural and evolutionary relationship, we did a genome-wide analysis of this important gene family in rice. Further, through comparative genomics, DDP genes from Oryza sativa subsp. (indica), nine different wild species of rice and Arabidopsis were also identified. We also found an expansion of the DDP gene families in rice and Arabidopsis which is due to the segmental duplication events in some of the gene family members. In general, a highly purifying selection was found acting on all the deduced paralogous and orthologous DDP gene pairs. The data from microarray and subsequent qRT-PCR analysis revealed that although several OsDDPs were differentially regulated under salinity stress, yet OsDDP6 was upregulated at all the developmental stages in salt tolerant rice genotype, FL478. Interestingly, OsDDP6 was found to be involved in proline metabolism pathway as indicated by protein network analysis. The diverse gene structures, varied transmembrane topologies and the differential expression patterns implied the functional diversity in DDP genes. Therefore, the comprehensive evolutionary analysis of DDP genes from different Oryza species and Arabidopsis performed in this study will provide the basis for further functional validation studies vis-à-vis DDP genes of rice and other plant species.
Introduction
Excess soil salinity is an important abiotic stress that adversely affects growth and development of plants. Rice is one of the most important cereals of the world. In general, rice plant is regarded as sensitive to salinity stress. High salinity stress causes severe impact on millions of hectares of agricultural land worldwide [1, 2]. Accumulation of salts in the irrigated soil is one of the prime factors that lowers water availability to root cells of rice plants due to their reduced osmotic potential, hence diminish growth, development and yield of rice [3,4]. Like other plants, the maintenance of osmotic and ionic homeostasis under salinity stress is a key challenge in rice too, which ultimately defines the salinity tolerance level of a particular rice genotype [5]. Therefore, there is an earnest need to develop salinity stress tolerant rice genotypes to overcome problems of salinity-related yield loss [6,7]. Plants manifest salinity stress responses in several domains such as morphological, physiological, biochemical and ultra-structural changes of cell to various molecular events [8–13]. Hence, the study of expression profiles, physio-chemical and structural properties of specific gene families as well as their expressions under a particular stress condition is the pre-requisite to uncover their specific functional roles [14]. Taking the advantage of sequenced genome, several gene families of rice under different abiotic stresses have been studied. Few examples are sodium/calcium exchanger [15], F-Box [16], SAP (Stress Associated Protein) [17], NAC (NAM, ATAF1/2 and CUC2) transcription factor [18], MADS-box (MCM1-agamous–deficient serum response factor) [19], GF14 [20], DREB (dehydration responsive element binding) [21], SnRK2 [22], TIFY [23], calcium-dependent protein kinase [24], WRKY [25], calcineurin B-like [26], MAPK (mitogen activated protein kinase) [27] and plasma-membrane intrinsic protein (PIP) gene families [28]. However, the unravelling of precise functions of many stress-responsive genes from different plants including Arabidopsis and rice remain one of the foremost challenges [29], and such gene products have been categorized as proteins containing hypothetical domains of unknown functions (DUFs). These domains are greatly conserved across genomes [30,31] indicating their biological importance. For example, DUF221 domain containing proteins are exclusively present in eukaryotic genomes [32]. DUF221 family (pfam accession: 02714) belongs to anoctamin superfamily (pfam accession: cl21726), which also contains another family, anoctamin/calcium-activated chloride channels (pfam 04547). DUF221 is homologous to domains present in calcium-activated chloride channels anoctamin/TMEM16 [33]. Although anoctamins are calcium-activated, all of them are not ion channels, some members are phospholipid scrambles which translocate the phospholipids in a lipid bilayer [32]. The DUF221 domain containing proteins (DDPs) are found mostly as transmembrane protein families in combination with other functional domains, as implied by the probable role of DUF221 in membrane integration of the host protein [34]. The first indication for the existence of DDPs as gene families in plants came from the model plant Arabidopsis [35]. This unique domain has been found in many stress-responsive genes of many plants. For example, this domain has been found to be present in a dehydration-responsive gene, ERD4, of Brassica juncea [34]; drought associated genes (e.g. LOC_Os12g39320) of rice [36]; osmotic shock-responsive influx cation channel gene, AtCSC1 in Arabidopsis, where this domain helps in the permeability to Ca2+ ions under hyperosmotic shock [37]. Recently, Yuan et al. [35] have characterized a hyper-osmolality-gated calcium-permeable channel, OSCA1 (homolog of AtCSC1) from Arabidopsis which increases intracellular Ca2+ levels under hyperosmotic stress. Since this protein and other members from the same family possess DUF221 domain, it indicates that this domain may also play a direct or indirect roles in osmosensing. This signature domain has also been detected in a vacuolar membrane protein, PenV of Penicillium chrysogenum which is associated with beta-lactum biosynthesis [38]. Moreover, this domain has been discovered in the virulence genes of fungus, Colletotrichum higginsianum [39]. However, little is known about the functions of DUF221 domain containing genes in plants. Except, Li et al. [40], who have done a preliminary analysis of DUF221 gene family (named as OsOSCA family in their study) in rice, the status of this gene family is largely unexplored in rice too. Therefore, having seen an obvious role of genes possessing DUF221 domain in especially hyperosmotic stress, we did an in-depth study of DUF211 gene family in different Oryza species. Additionally, we also studied its transcriptional regulation in relation to salinity stress in rice with the aim of understanding its possible roles in the tolerant glycophytic rice genotype (FL478).
Materials and methods
Identification, nomenclature and characterization of DDP genes
Arabidopsis DDP amino acid sequences were downloaded from UniPort (http://www.uniprot.org/). Rice DDP amino acid sequences were obtained from RGAP databases (http://rice.plantbiology.msu.edu/). The local Hidden Markov Model-based searches (HMMER: http://hmmer.janelia.org/) was built from all the known DDP proteins. Apart from Hidden Markov Models analysis, we also used ‘DUF221 domain’ as keyword searches in Gramene [41] and Phytozome [42] for the identification of DDP gene family members in the genomes of different plant species. All the retrieved sequences were scanned and curated using Pfam (http://Pfam.sanger.ac.uk/), InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) and NCBI Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) for the authentication of the presence of DUF221 domain.
The number of ESTs (Expression sequence tags) for OsDDP genes were determined using data of Rice Genome Annotation Project [43], whereas their functional annotations were verified by AmiGO 2 [44]. To provide a simple nomenclature, members of the studied gene family from rice were named from 1 to10 as per their appearance in chronological order (from top to bottom on the respective chromosomes) on the chromosomes. The genes were denominated as OsDDP1 to OsDDP10 (Oryza sativa DUF221 Domain containing Protein). To denote the splice variants, the Arabic numbers were added after “.” sign. Similar nomenclature was adopted for DDP gene family of all other plant species studied herein (S1 Table).
Several parameters such as length, isoelectric point (pI) and post-translational modifications and grand average of hydropathicity (GRAVY) values [45] of all the DDP protein sequences of rice, other Oryza species (O. barthi, O. brachyantha, O. glaberrima, O. glumaepatula, O. longistaminata, O. meridionalis, O. nivara, O. punctate, O. rufipogon and O. sativa (indica) and Arabidopsis were determined using Expasy server (www.expasy.org).
Chromosomal organization, segmental duplication, detection of introns and exons, alternative splicing
Genomic distribution of OsDDPs and AtDDPs was depicted using MapChart 2.30 with default parameters [46] and the orientations of these genes were identified from Gramene [41] and TAIR 10 [47] respectively. Segmental duplications of DDP gene family of rice and Arabidopsis for the detection of homologous genomic regions were determined from their corresponding duplication data in Plant Genome Duplication Database [48]. Alternative splicing of each gene was determined from plant ensemble server.
Number of introns and exons in the individual DDP gene were determined by aligning the corresponding genomic and coding sequences. Number of introns in alternative splice forms was determined from the Gramene web server [41]. Synteny blocks were visualized using VISTA (VISualization Tool for Alignments) [49].
Identification of transmembrane topology and domain organization of DDP proteins from rice and Arabidopsis
The putative transmembrane domains and signal peptides for the longest ORF (open reading frame) of each of the DDP proteins were identified by PROTTER version 1.0 [50]. Besides, DUF221 domain and other conserved domains were detected by Pfam (http://Pfam.sanger.ac.uk/), InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) and NCBI Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Motif analysis and phylogenetic relation among DDP proteins
All the OsDDP protein sequences were scanned using MEME Suite 4.10.1/MAST software [51] for the identification of conserved motifs, with the same parameters as used by Mondal et al. [52]. Functional annotations of these motifs were performed using HHpred (http://toolkit.tuebingen.mpg.de/hhpred), and their sequence logos were generated with WebLogo [53]. The phylogenetic relationship among the various DDP proteins was analysed using ClustalW [54] and the dendrogram was constructed using MEGA 6 [55] by neighbour-joining method [56], with all default parameters except a bootstrap of 1000 replicates. The orthologous genes are enlisted in S2 Table.
Estimation of Ka/Ks ratio
Segmentally duplicated genes from rice and Arabidopsis were used to estimate the extent of selection pressure on them using codeml program based PAL2NAL (http://www.bork.embl.de/pal2nal/). The Ka/Ks ratios of orthologous genes of three OsDDPs from different plant species as well as orthologues of all OsDDPs from the wild species of rice were determined. Likewise, Ka/Ks ratios of the orthologues of three AtDDPs were estimated from different plant species. Three genes each of rice and Arabidopsis were selected on the basis of the presence of their orthologues in all the studied plant species. For each orthologous gene pairs, Ka/Ks ratio was estimated, the average ratio values were plotted. The age of the duplicated genes was calculated as done by Jami et al. [57].
Gene expression analysis with microarray data
Microarray-based expression profiles of OsDDPs and AtDDPs were retrieved from publicly available Affymetrix microarray data (51 K and 22 K Affymetrix gene chips respectively, experimental Ids: Os-00001 for rice and AT-00120 for Arabidopsis). Expression patterns were studied in 29 and 37 different tissues as well as at 9 and 10 developmental stages of rice and Arabidopsis respectively, by retrieving the log2-transformed affymetrix data on respective arrays using Genevestigator database tool [58]. Likewise, the expression patterns of DDPs in salinity tolerant rice cultivar FL478 as well as Arabidopsis under salinity stress were studied in roots and shoots by retrieving the corresponding log2-transformed fold change values. Heat maps were generated by means of the MeV software package [59] with average linkage hierarchical clustering using Euclidean distance metric for tissues and developmental stages, and Pearson correlation for stress. The expression datasets retrieved from the Genevestigator and subsequently used for generating the heat maps are provided in S3 Table and S4 Table for rice and Arabidopsis, respectively.
Identification of miRNAs targeting DDPs of rice and its wild species
To predict miRNAs that may target DDPs, the individual cDNA sequences of DDP genes of rice were used as input in psRNATarget [60] against all the rice mature miRNAs that were reported in miRbase [61]. The predicted miRNAs are enlisted in S5 Table.
Plant material, stress treatment and detection of transcriptional regulation by qRT-PCR (quantitative real time polymerase chain reaction)
The FL478, a salinity tolerant rice cultivar [3], was used for the gene expression study through qRT-PCR. Seeds were surface sterilized with sodium hypochlorite (3%) and were germinated on the germination papers. Then plants were raised up to the reproductive stage under greenhouse conditions in pots. The salt stress (EC of ~10 ds/m) was applied to the pot-grown rice plants for 24 h at different developmental stages namely: tillering, booting, flag leaf, panicle initiation and milk stages (young grain with white liquid). Simultaneously, control samples were obtained at each developmental stage of the rice plant treated with distilled water for 24 h. The tissues after harvesting from the stressed as well as control rice plants were immediately frozen and kept at -80°C until further use.
RNA isolation, cDNA synthesis and qRT-PCR analysis were performed according to the protocol described by Ganie et al. [3]. For qRT-PCR study, 3 biological replicates per sample were used with 3 technical replicates for each biological replicate.
It is worth mentioning here that we used the diluted cDNA (1:100), reverse transcribed from total RNA with oligodT primer using Superscript II (Invitrogen), as a template for quantifying the OsDDPs. For the quantification of expression of miRNAs targeting OsDDPs, cDNA synthesised from total RNA (as used for OsDDPs) instead of cDNA synthesised from miRNA was used. The primers were designed manually with actin as endogenous control. Primers used for qRT-PCR study are given in S6 Table. Statistical analyses were conducted using the SAS 9.4 software (SAS Institute Inc., NC, U.S.A).
Cis-element analysis
Almost 2 kb of upstream sequence from the translation start site of all the 10 OsDPPs and DDPs from different wild rice species were analyzed by PLACE database [62] for abiotic stress-responsive cis-elements. Further to determine the interactions with the other proteins, the corresponding protein sequence of differentially regulated OsDDP proteins were analysed using STRING software version 9.1 [63] to reveal their functional interactions with other proteins.
Results
Identification and structural analysis of DDP gene family in rice, its wild species and Arabidopsis
Based on the presence of DUF221 domain, a genome-wide investigation led to identification of 10 members of DDP gene family each in O. sativa (japonica), O. barthi, O. brachyantha, O. glaberrima, O. longistaminata, O. meridionalis, O. nivara, O. rufipogon and O. sativa (indica); whereas only nine members were found in O. glumaepatula and O. punctata. All the DDP proteins in rice were found to possess a single DUF221 domain which was also reported by Li et al. [40]. In addition, majority of the identified proteins showed DUF4463 and RSN1_TM domains upstream of DUF221 domain. It was also found that rice DDPs displayed similar organization of DUF221 and other two domains when compared with their orthologues in different wild rice species as well as Arabidopsis. Further, all the DDP proteins except OsDDP8 in rice showed positive GRAVY scores indicating their transmembrane nature (S1 Table). Functional annotation further confirmed that DDP proteins were membrane bound proteins.
Chromosomal distribution and duplication of rice and Arabidopsis DDP genes
The distribution of DDP genes across rice and Arabidopsis genomes was relatively uneven in the sense that not all the chromosomes contained the loci coding for DDPs (S1A and S1B Fig). For example, chromosome 2, 4, 6, 8 and 9 of rice as well as 2 and 5 of Arabidopsis did not have any DDP genes. Ten and 15 loci in rice and Arabidopsis, identified in our study, encoded for 18 OsDDP and 28 AtDDP proteins respectively, indicating that these genes have undergone alternative splicing. For example, six DDP genes in rice and seven DDP genes in Arabidopsis had alternative splicing (S1 Table). The DDP genes from wild species of rice, except O. brachyantha, O. glaberrima, O. longistaminata, O. punctata and O. sativa (indica), also had alternative splicing as indicated by the disproportion in the number of loci and the protein products (S1 Table). The number of DDP genes was found to be almost identical among rice and its wild species. Compared to rice, Arabidopsis was found to contain 15 DDP genes, which could be attributed to the higher number of gene duplication events in latter. In contrast to the number (11) as reported by Li et al. [40], the number of DDP genes in rice in our study was found to be only 10. This is because we rejected LOC_Os03g04450 (OsOSCA4.1) of Li et al. [40] as it failed to show any DUF221 domain by NCBI Conserved Domain Database. Further only one pair of rice OsDDPs (20%) was found to have block duplication, while as, no gene pairs were found to be segmentally duplicated (S1A Fig). The block duplicates of OsDDP3 and OsDDP10 were found on chromosome 3 and 12 respectively. In case of Arabidopsis, six pairs of genes formed by nine genes (60%) were found to be duplicated as block, however like rice, no segmental duplicates could be found. The 10 block duplicates localized on chromosome 1, 3 and 4 with maximum number (4) of duplicated block on chromosome 1 and 4 (S1B Fig). In addition, it was found that almost all the DDP gene members were located in the syntenic regions across the different Oryza species (S2 Fig).
Phylogenetic analysis among the paralogous and orthologous DDPs
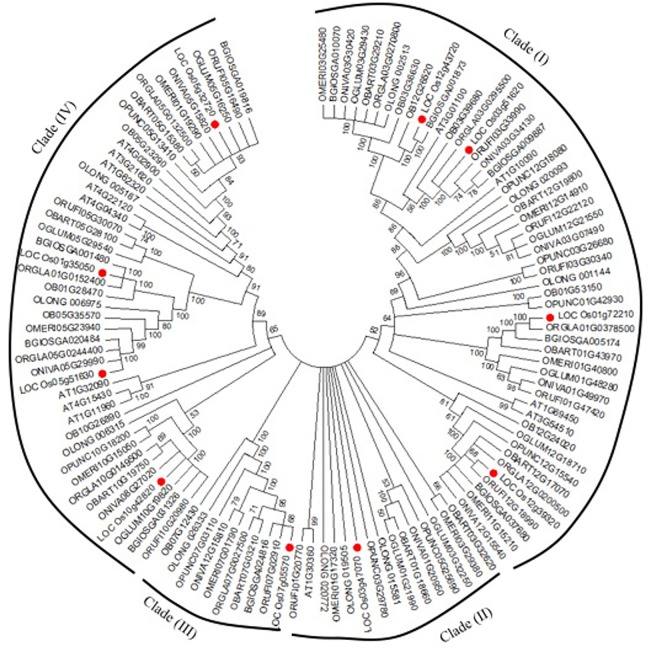
With the purpose of evaluating the evolutionary relationship among the DDP proteins of rice, its different wild species and Arabidopsis, a phylogenetic analysis was performed. Firstly, a phylogenetic analysis was performed among the 10 rice DDPs (Fig 1). The pair-wise amino acid similarities among OsDDPs varied from 27 to 81% (S7 Table). It can be seen from the Fig1 that OsDDP1 and OsDDP6 proteins, having the maximum similarity (81%) between them, occurred as the closet pair in Group A of phylogenetic tree, whereas OsDDP1 and OsDDP8 were the farthest in the group due to low similarity (32%). It was also observed that the segmentally duplicated pair, OsDDP3-OsDDP10 occupied the same sub-group. OsDDP proteins with the similar membrane topology or the similar pattern of sequence motifs were found to be present in the same group of the phylogenetic tree. Next, the phylogenetic relationship among the DDP proteins of rice, its wild species and Arabidopsis was evaluated. For this, multiple sequence alignment of all DDP proteins was performed to construct a dendrogram that segregated them into four major clades (Fig 2). Based on the similar membrane topology and intron-exon structure, the DDPs from the orthologous species were found to occupy the same clades of dendrogram. DDPs of all the species were distributed in all the 4 clades except Arabidopsis and O. glumaepatula which were not present in the clade III, the smallest among the 4 clades. The clade I was found to contain maximum number of 51 DDP members while as, clade III was represented by the minimum of 10 DDPs. No clade or sub-clade was found to be specific to any particular species. It was observed that OsDDPs from O. sativa (japonica) were more closely related to those of O. sativa (indica), O. glaberrima, O. rufipogon and O. barthi, whereas O. longistaminata and O. punctata were found to be less closely related to O. sativa (japonica) sp.
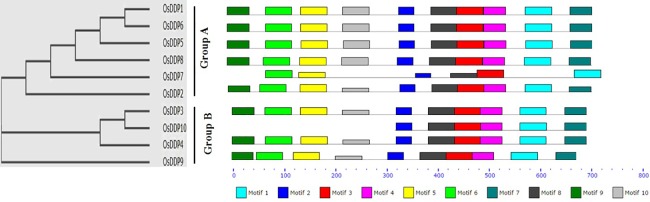
Fig 1. Phylogenetic tree of OsDDP proteins along with their different motifs.
Ten different motifs were found in MEME analysis. The proteins, except OsDDP7 in Group A and OsDDP10 in Group B, with the similar motif organization are found in the same groups of tree. Segmental duplicated pair (OsDDP3-OsDDP10) can be seen in the same sub-group. The details of the 10 motifs can be seen in S10 Table.
Fig 2. Dendrogram of DDP proteins from rice, different wild rice species, O. sativa (indica) and Arabidopsis.
DDP protein sequences were aligned using ClustalW and the dendrogram was generated using MEGA6 software by the Neighbor-joining method with 1000 bootstrap replicates. DDP proteins are categorized into 4 different clades on the basis of sequence homology as described in the text.
Estimating the age and selection pressure for duplicated gene pairs
In order to estimate the approximate evolutionary age of segmentally duplicated paralogous of DDP gene pairs in Arabidopsis and rice, we employed the number of synonymous substitutions per synonymous site (Ks). The nucleotide sequences of the duplicated gene pair, OsDDP3-OsDDP10, in rice have a Ks value of 0.8116, indicating that this gene pair might have duplicated~62.4 MYA (million years ago). Likewise, the duplicated gene pairs AtDDP2-AtDDP14, AtDDP6-AtDDP14, AtDDP9-AtDDP13, AtDDP12-AtDDP6 and AtDDP8-AtDDP7 in Arabidopsis showed Ks values of 1.9492, 3.0995, 0.8802, 2.1362 and 3.1159 respectively, signifying that these block duplications might have occurred approximately 65, 103, 29, 71 and 104 MYA.
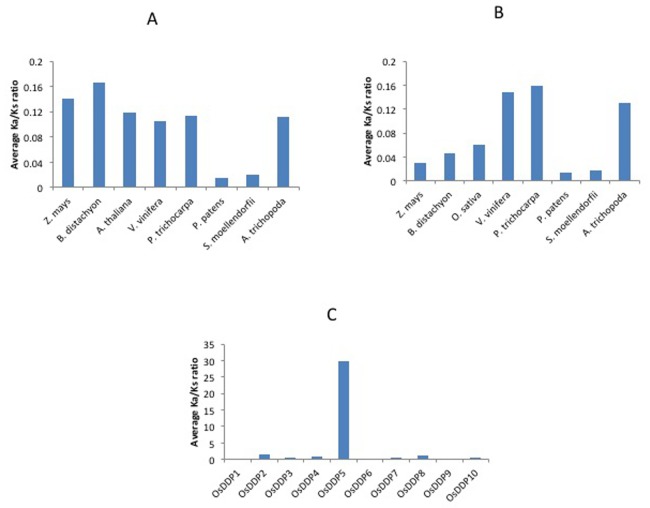
When Ka/Ks for a gene pair is equal to 1, it is said to be going through neutral evolution, while gene pairs undergoing positive or negative purifying selection have Ka/Ks >1 or <1, respectively [64]. All the rice and Arabidopsis segmentally duplicated paralogous DDP gene pairs as well as their orthologous gene pairs in different plant species exhibited Ka/Ks <1 (Fig 3 and S8 Table). However, five OsDDPs (OsDDP2, OsDDP4, OsDDP5, OsDDP7 and OsDDP8) exhibited a Ka/Ks >1 with some of their wild rice orthologues, whereas the rest five OsDDPs (OsDDP1, OsDDP3, OsDDP6, OsDDP9 and OsDDP10) showed Ka/Ks <1 for all their corresponding wild rice orthologues (S8 Table).
Fig 3. Analysis of evolutionary pressure on DDP genes.
Averaged Ka/Ks ratio between orthologous DDP genes of rice (A) Arabidopsis (B) and different selected plant species. Averaged Ka/Ks ratio between orthologous DDP genes of rice, its different wild species and O. sativa (indica) rice (C). The Ka/Ks ratio of each pair is given in S8 Table.
Sequence conservation among the DDP proteins
DUF221 domain is significantly conserved across the different eukaryotic genomes [30]. To find out its conservation, DDP protein sequences from rice were aligned. It was found that DUF221 domain region was the highly conserved region across the OsDDP sequences (S3 Fig). Apart from DUF221 domain, many other localized regions were also found to be conserved in DDPs across the different species (S4 Fig). To confirm the identity of these locally conserved regions, MEME search helped us to identify 10 different motifs which are fairly conserved among rice DDP protein sequences (Fig 1, S9 and S10 Tables). To affirm if the motifs obtained from the MEME analysis are similar to any of the known protein motifs, we did HHpred analysis. It was observed that these novel motifs did not show any significant similarity with the known motifs (S10 Table).
Cis-motifs in the DDP gene promoters
An extensive analysis of 2kb upstream promoter region (from the translation initiation codon) of DDP genes from rice, its nine different wild species and O. sativa (indica) facilitated the identification of conserved and over-represented consensus cis-motifs involved in abiotic stresses especially salt and osmotic/dehydration stresses. Twenty different salt and osmotic stress-responsive cis-elements were clearly found in the promoters of DDPs (S11 Table). Among them, MYCCONSENSUSAT is the most enriched cis-element in all the studied promoter sequences, followed by ACGTATERD1 and MYBCORE. These three cis-elements along with other three elements (GT1GMSCAM4, MYB1AT and MYB2CONSENSUSAT) were present in the promoter regions of all the DDP sequences. Interestingly, cis-elements ABREATRD22 was found to be absent from the promoters of all O. nivara and O. longistaminata DPP genes. None of the promoters of DDP gene possessed all the 20 identified motifs. The average number of the identified cis-elements was found to be almost similar in all the DDPs of rice and its wild species (S5 Fig).
miRNAs-DDP gene family networking
In an attempt to find out the miRNAs targeting the DDP family members of rice and its wild species, psRNATarget predicted only a few members of DDP gene family from all the Oryza species that were targeted by conserved miRNAs (S5 Table). For an example, in rice, OsDDP6 and OsDDP10 were targeted by miRNAs belonging to three different families. Five members of osa-miR818 family (osa-miR818a, b, c, d and e) as well as osa-miR1436 were predicted to target OsDDP6 by cleavage and inhibition of translation respectively, whereas osa-miR6248 was predicted to inhibit the translation of OsDDP10. Similarly, in case of wild rice species, the number of DDP family members targeted by miRNAs ranged from minimum of two each in O. barthi and O. glumaepatula to maximum of five in O. meridionalis, whereas the number of miRNAs targeting the different DDP gene family members ranged from minimum of three (targeting four different DDP genes in O. longistaminata) to maximum of ten (targeting five different DDP genes in O. meridionalis). Interestingly, miR1436 and different members of miR818 family were found to target the DDP genes from most of the Oryza species studied except O. longistaminata, O. punctata and O. brachyantha. Further, it was found that a particular DDP gene (e.g. OsDDP6) was targeted by multiple miRNAs.
Transmembrane topology and intron-exon structure of DDPs from rice, wild species of rice and Arabidopsis
DDP genes encode integral membrane proteins with multiple predicted transmembrane (TM) helices to act as transport channels. Topological structure prediction using Protter software and hydrophobicity analyses showed the presence of TM helices in all DDPs (S6 Fig). The number of TM helices varied from a minimum of 7 (OsDDP2) to maximum of 11 (OsDDP10) in rice, and from 9 (AtDDP1, AtDDP2, AtDDP4, AtDDP5, AtDDP6, AtDDP7, AtDDP9, AtDDP11, AtDDP12, AtDDP13 and AtDDP15) to 11 (AtDDP3, AtDDP8 and AtDDP10) in Arabidopsis. Similarly for wild species of rice, it was varied from 7 (OloDDP5, OloDDP7) to 11 (ObaDDP5, OglaDDP2, OglDDP5, OmeDDP2, OsiDDP1, OsiDDP2, OsiDDP5). TM helices in DDPs were found to be located in the amino acid sequence between 6–27, 87–111, 145–165, 357–381, 401–429, 449–468, 549–580, 601–621 and 627–647. Besides the TM helices, another modular architecture of DDPs was the ‘loop region’ which was found to be located between the amino acid residues of 166 to 356 amino acid residues. It was observed from the TM topology that all the DDPs had two distinct clusters of TM helices joined by a large hydrophilic intracellular loop. However, as an exception, the two clusters of TM helices in ObarDDP1 protein of O. barthi were joined by an extracellular loop. In general, lesser number of TM helices was located in the N-terminal cluster than in the C-terminal cluster. DUF221 domain represents the C-terminal cluster containing in general six predicted TM helices.
Structural analysis of all DDP genes implied that the number of introns varied from minimum of 4 in rice (OsDDP3) to maximum of 16 in Arabidopsis (AtDDP5) (S7 Fig). In Arabidopsis, AtDDP15 was the only member found to be intron-less. The average number of introns in DDP gene family were found to be conserved (approximately nine introns) among all the studied species with a maximum average of 11.2 introns in O. meridionalis and a minimum of nine introns in case of O. glaberrima. Additionally, variations in lengths of exons, introns and UTRs (untranslated region) were observed. None of the DDP genes of rice were found to lack either 5´ or 3´ UTRs, whereas in Arabidopsis AtDDP5 and AtDDP7 were found to be lacking 5´ UTR, and another pair (AtDDP6 and AtDDP11) lacking both 5´ and 3´ UTRs. However, the absence of either one or both the UTRs was found more frequently among the 10 species of Oryza with O. glaberrima, O. longistaminata and O. sativa subsp. indica lacking both the UTRs. The segmentally duplicated DDP genes of both rice and Arabidopsis were found to have a different organisation as well as dissimilar number and length of introns and exons. Orthologous genes with almost similar number of introns and exons were found to be present in the same clade of dendrogram. However, the organisation of introns and exons was not coherent in all of them.
Expression profiles of OsDDPs and AtDDPs
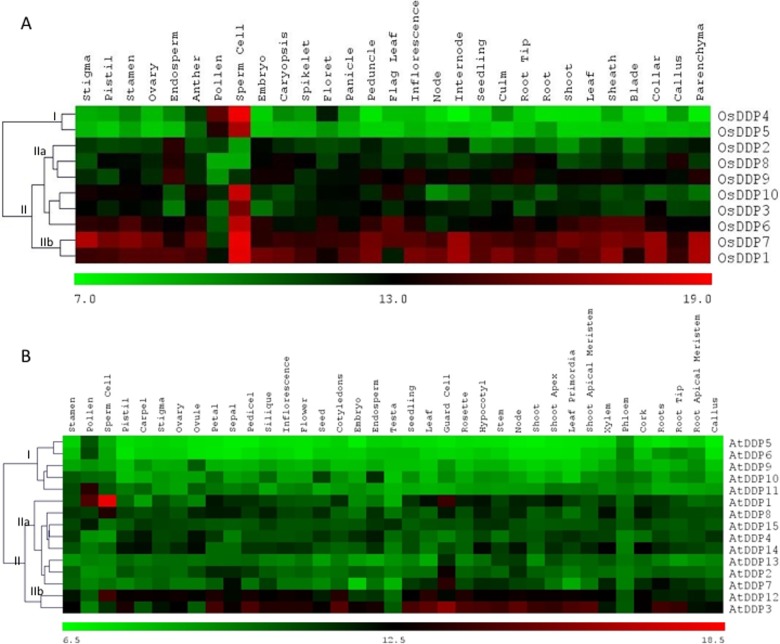
Microarray-based spatio-temporal expression profiles of OsDDPs and AtDDPs in various tissues and organs, at different developmental stages as well as under salinity stress were analysed using publicly available microarray data of Genevestigator. In both rice as well as Arabidopsis, the microarray expression profiling of DDP gene family showed that the members of this family had varied expression patterns. For an example, in rice, OsDDP4 and OsDDP5 (in cluster I) exhibited the lowest level of expression in all the studied tissues (except sperm cells, in which all the OsDDPs showed high expression except OsDDP2 and OsDDP8) (Fig 4A). However, in contrast, the expression of OsDDP1 and OsDDP7 (in cluster-IIb) was the highest in all the tissues. The expression of all other genes in cluster-IIa was almost low among all the tissue. Some of the OsDDPs were found to exhibit a moderately high to high expression level specific to certain tissues. They were OsDDP3, OsDDP10, OsDDP5 and OsDDP4 (also showed moderately high expression in pollen tissue) which showed high sperm cell-specific expression; OsDDP2, OsDDP8 (specific to callus and root tip) and OsDDP9 showed moderately high expression specific to endosperm and some vegetative tissues.
Fig 4. Microarray-based expression pattern of DDP genes in different tissues/organs.
Expression profile of DDP genes generated from microarray data indifferent tissues/organs of (A) rice and (B) Arabidopsis. The heatmaps represent hierarchical clustering (using Euclidean distance metric) of average log signal values of DDP genes and were generated using MeV software package. The color bar below the heat maps represent relative expression values with green color representing the lowest expression while as, red the highest expression level. Cluster-grouping represented by Roman numbers followed by letters as discussed in the text.
In Arabidopsis, however, the expression of DDPs in different tissues was not as diverse as was observed in rice. In general, a very low level of expression was observed for all the AtDDPs among all the tissues except AtDDP3 and AtDDP12 (in cluster-IIb of Fig 4B) which showed moderately high level of expression in most of the tissues. The expression of AtDDPs in cluster-I was found to be uniformly lowest. It was also found that while most of the AtDDPs exhibited a near uniformly low level of expression, higher tissue-specific expression of some AtDDPs was also observed. For instance, AtDDP1 was found to be specifically expressed in guard cells and in some male reproductive tissues such as pollen and sperm cell. Similarly, the expression of AtDDP7 was also found to be guard cell specific.
Besides, the microarray-based expression of rice and Arabidopsis DDPs were also studied at different stages of development. Like their expression in different rice tissues, OsDDP4 and OsDDP5 (in cluster-I of Fig 5A) exhibited the lowest level of expression across the different stages of development too. Likewise, the expression of OsDDP1, OsDDP7 and OsDDP6 were also the highest and near constant across the 9 developmental stages. These 3 genes were highly expressed OsDDP9 in cluster-IIb. All the other genes in cluster IIa showed almost uniform moderately low level of expression.
Fig 5. Microarray-based expression pattern of DDP genes during development.
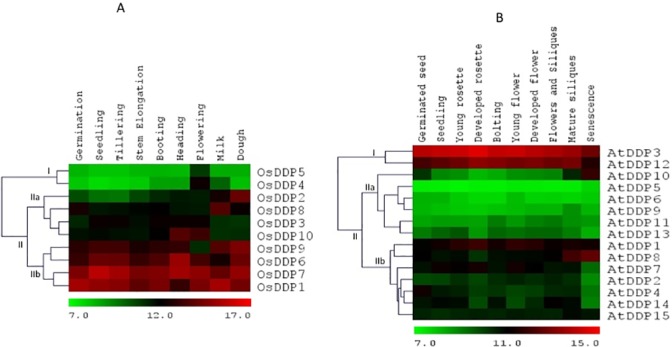
Heat maps showing expression patterns of DDP genes from rice (A) and Arabidopsis (B) at different developmental stages as indicated. Heat maps represent hierarchical clustering (using Euclidean distance metric) of average log signal values of DDP genes and were generated using MeV software package. The color bar below the heat maps represent relative expression values with green color representing the lowest expression while as, red the highest expression level. Cluster-grouping represented by Roman numbers followed by letters as discussed in the text.
In Arabidopsis, the expression of AtDDP3 and AtDDP12 (in cluster-Ia of Fig 5B) was the uniformly higher across the 10 different developmental stages than other genes. On the other hand, except the moderately high expression of AtDDP10 only at senescence stage, all the genes in cluster-IIa showed very low level of expression across the different developmental conditions with AtDDP5 showed the lowest level of expression. Expression of AtDDP1, AtDDP7 and AtDDP8 (in cluster-IIb) was found to be specific only to some developmental stages. All other genes in this cluster showed moderately low level of expression.
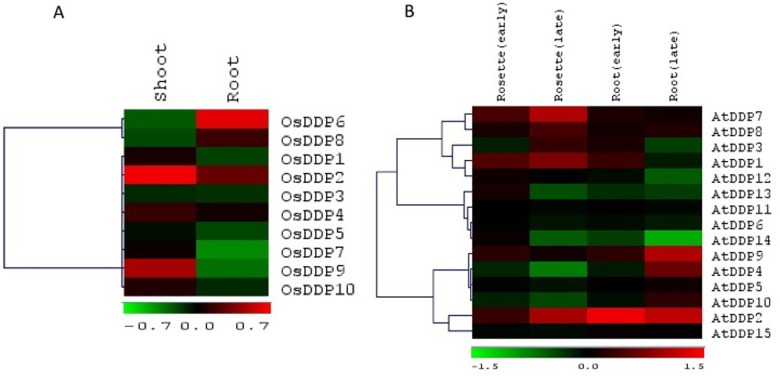
In order to identify the members that are responsive to salinity stress, we analysed the expression of OsDDPs and AtDDPs under salinity stress. In case of rice, we exploited the microarray data for the roots and shoots of salt tolerant cultivar FL478 under salt stress. As shown in Fig 6A, OsDDP2 and OsDDP9 were highly up-regulated in shoots, while as, their expressions in roots were moderately up-regulated and down-regulated respectively. However, OsDDP4 was up-regulated and down-regulated in shoots and roots respectively. In the roots of FL478, OsDDP6 was highly up-regulated, whereas its expression in shoots was very low. Similarly, the expression of OsDDP8 was moderately up-regulated in roots but down-regulated in shoots. All other OsDDPs displayed low to moderately low level of expression both in roots and shoots under salinity stress. In general, all OsDDPs except OsDDP6 and OsDDP8 showed higher level of expression in shoots than roots of FL478.
Fig 6. Microarray-based expression pattern of DDP genes under salinity stress.
Expression analyses of DDP genes of rice (A) and Arabidopsis (B) under salt stress in shoots and roots as indicated above the heat map. Weighted average linkage method and Pearson correlation distance metric were used for hierarchical clustering of DDP genes. The color bar below the heat maps represent relative expression values with green color representing the lowest expression while as, red the highest expression level.
In Arabidopsis, microarray-based expression data of AtDDPs in root and shoot (rosette) was retrieved from AtGenExpress, Genevestigator. As shown in Fig 6B, the expression of AtDDP1, AtDDP2, AtDDP7 and AtDDP8 in rosette leaf of Arabidopsis was up-regulated from the early phase to late phase of salt stress, whereas the expression of the rest AtDDPs was down-regulated, except AtDDP7which exhibited unaltered expression in roots. In the roots, however, the expression of AtDDP4, AtDDP9 and AtDDP10 was seen to be up regulated from the early to late periods of the stress, whereas AtDDP1¸ AtDDP2, AtDDP12 and AtDDP14 were found to be down-regulated from the early to late phase of the stress. Salt stress was not however found to alter the expression of AtDDP12 in rosette. All other AtDDPs were found to be expressed at a low level under salinity in the roots. Moreover, the expression of AtDDP2, under salinity, was found to be the highest, whereas the expression of AtDDP14 was the lowest, among all the AtDDPs in both rosette and shoot. Additionally, the expression of four genes such as AtDDP5, AtDDP6, AtDDP11 and AtDDP15 was found to be significantly invariable in both rosette and roots under salinity stress.
qRT-PCR-based expression analysis of OsDDPs and of miRNAs targeting OsDDPs
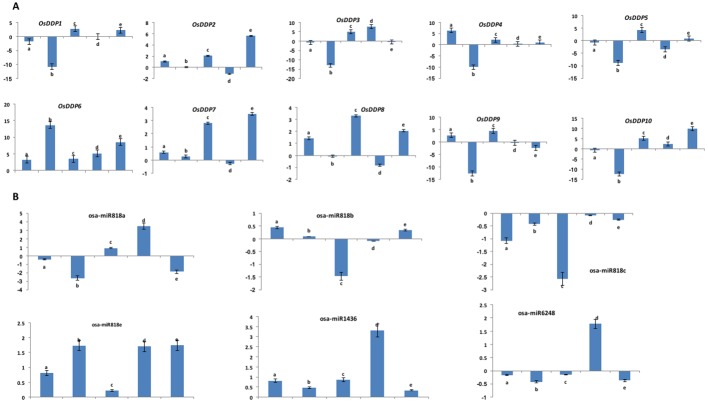
To gain insights into the better understanding about how OsDDP genes changed their expression in response to salinity stress as well as to validate their altered gene expression as shown by microarray-based expression patterns in FL478, we analyzed the expression profiles of the 10 OsDDPs as well as six miRNAs by qRT-PCR at different developmental stages of FL478. Our results indicated that all the 10 genes were up-regulated at one or more developmental stages of FL478 under stress conditions (Fig 7A). Among them, OsDDP6 was found to be up-regulated across all the developmental stages. On the contrary, microarray data indicated OsDDP2 to be the most responsive to salinity. All the genes, except OsDDP3 and OsDDP9 were up-regulated at flag leaf and milk developmental stages, whereas all the genes barring OsDDP6 and OsDDP7 were down-regulated at the booting stage. At the vegetative phase (tillering), only OsDDP1, OsDDP3, OsDDP5 and OsDDP10 were found to be down-regulated. It was also observed that OsDDP4 displayed the highest level of expression at tillering stage, OsDDP6 at booting and milk stages, OsDDP3 at panicle initiation and flag leaf stages, whereas OsDDP10 showed the highest level of expression at milk stage. Moreover, as shown in S1 Table, EST analysis showed that the number of ESTs for the OsDDP gene members varied from 5 (OsDDP5) to 73 (OsDDP8).
Fig 7. qRT-PCR based expression profiles.
Mean fold change (log2 scale) in expression of all DDP gene members (A) and miRNAs predicted for DDP genes (B) in salt tolerant FL478 rice genotype at different developmental stages under salinity stress. Values on Y-axis represent the relative transcript abundance (log2 scale). qRT-PCR analysis of both DDP genes and miRNAs was done with cDNA template generated from the harvested tissues of salt stressed (200 mM NaCl) and unstressed (control) plants as mentioned in materials and methods. As osa-miR818d could not be amplified, it was not hence included in the qRT-PCR analysis. Only osa-miR818c showed the opposite expression as that of its target (OsDDP6). Error bars represent the standard deviation (±), n = 3. Different letters indicate significant differences at P ≤ 0.05. Different alphabets indicate different developmental stages (a = tillering, b = booting, c = flag leaf, d = panicle initiation, and e = grain with white milk stages).
The expression analysis of different miRNAs targeting DDP genes showed that none of the studied miRNAs, except osa-miR818c, showed a completely reverse expression pattern to that of their targets across all the developmental stages under salinity stress (Fig 7B). Instead, they showed reverse abundance in their transcript levels only at particular stages of development. For an example, osa-miR818e and osa-miR1436 showed up-regulation across all the developmental stages studied which was not coherent with the expression level of their target gene OsDDP6. Among the miRNAs targeting OsDDP6, only osa-miR818c showed reverse regulation at all the stages. Similarly, osa-miR6248 showed the reverse level of expression as that of its target OsDDP10, but only at flag-leaf and milk stages.
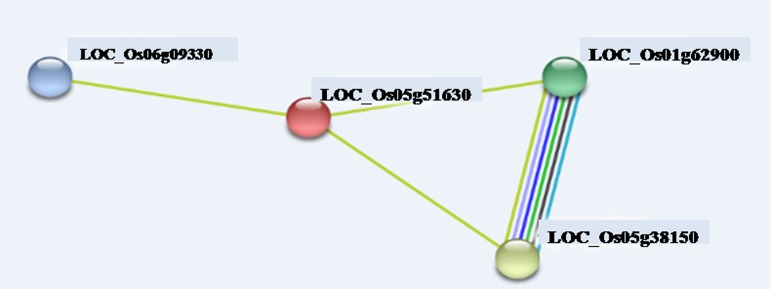
Having seen that OsDDP6 was up-regulated at all the developmental stages, we were then interested to know its interaction with the other genes. Therefore, to identify the protein network that consists of proteins interacting with the differentially expressed gene i.e. OsDDP6 [LOC_Os05g51630], direct and indirect interactions between these proteins were derived using the STRING search tool. The network connecting up-regulated proteins is shown in Fig 8, and indicates that there were four nodes (representing proteins) and four edges (representing interactions). The interacted proteins were ubiquitin-conjugating enzyme [LOC_Os06g09330], amino acid kinase [LOC_Os01g62900] and P5CS (plays a key role in proline biosynthesis, leading to osmoregulation in plants). This interaction clearly indicated that OsDDP6 is involved in salinity tolerance. It was also found that there were two KEGG pathways in the network namely ‘arginine and proline metabolism’ and other one was amino acid metabolism.
Fig 8. Protein interaction network of LOC_Os05g51630.
In the network generated by STRINGV9.1, each node represents a protein and each edge represent an interaction, coloured by evidence type (see STRING website for colour legend). The original graphic output was modified to name the LOC number of the gene.
Expression of genes lying on duplicated segments of rice and Arabidopsis genomes
The rigorous search in the Plant Genome Duplication Database [48] revealed that 2 and 9 genes were present on duplicated segments of chromosomes in rice and Arabidopsis respectively (S1 Fig). In order to understand the functional diversification of duplicated DDP genes, the microarray-based expression profiles for 1 and 6 pairs of duplicated genes in rice and Arabidopsis respectively, were compared under salinity stress, in different tissues as well as across several developmental stages (S8 Fig). Based on their microarray based expression profiles, the two segmentally duplicated paralogous genes (OsDDP3 and OsDDP10) in rice showed almost identical expression pattern in all the tissues and across the developmental conditions. However, they exhibited expression divergence under the stress conditions due to the specifically moderate expression of OsDDP10 in shoots. In Arabidopsis, the duplicated gene pairs showed high divergence in expression as compared to rice. The paralogous pairs AtDDP2-AtDDP14 and AtDDP9-AtDDP13 showed divergence only with respect to stress. A high divergence in expression domain of AtDDP12-AtDDP6 gene pair was observed, as the expression of AtDDP12 was specific to different vegetative and reproductive tissues as well as had a slightly different expression pattern than AtDDP6 under stress conditions. Comparison of expression patterns of AtDDP8-AtDDP7 pair revealed that this pair had undergone divergence in expression due to the highly specific expression of only AtDDP8 at late reproductive and senescence stages. Likewise, AtDDP13-AtDDP11 also differentially expressed under different vegetative and reproductive tissues. Expression profile of AtDDP6-AtDDP14 duplicated pair exhibited tissue and early stress specific expression.
Discussion
Plants acquire the tolerance to environmental stresses by orchestrating the changes of various events including gene expression [65]. However, the exact functions of some gene products, categorized as proteins containing hypothetical domains of unknown functions (DUFs), are not studied well. One such domain is DUF221. The highly conserved DDPs are osmoregulatory cation channels permeable to calcium [35, 37], and therefore may be associated with the abiotic stress responses such as salinity.
Since most of the orthologous DDP gene family members were located on the same order of chromosome among the different Oryza species (S2 Table), it was intriguing to know if these orthologous genes were present as syntenic blocks in chromosomal regions. We found through VISTA alignments that almost all the orthologous genes were present in syntenic blocks. For example, in case of OsDDP3 (S2 Fig), almost 1.5 kb intergenic region on 5′-end and 3.5 kb intergenic region on 3′-end were found to be highly conserved across the Oryza species, indicating this region to be a syntenic block. However, considerable differences were found for O. punctata and O. brachyantha genomes which could be due to their different genome types, BB- and FF-genomes respectively. The similar length of conserved intergenic region surrounding MIR319a locus in orthologous species of Arabidopsis has already been reported [66]. Further, to assure the genetic colinearity in the analyzed syntenic region, we found that a gene (Os03g0673600) next to the 5′-end of the OsDDP3 was consistently present in the similar order of location across the Oryza species with BB- and FF-genomes, confirming our assumption to form the synthenic region. The similarity among these genes found next to the 5′-ends of the OsDDP3 orthologues ranged from 70–95%.
The higher number with different lengths of DDP transcripts per gene loci (S7 Fig, S1 Table) identified in different Oryza species in this study indicates the presence of splice variants that may be associated with functional diversity of DDPs. The duplicated gene pairs of rice and Arabidopsis (except AtDDP7 and AtDDP8) were found to have different number of splice forms which may add to the divergence in their expression. The alternatively spliced DDP transcripts could be regulated in tissue-, developmental-, or stress-specific manner and hence allow the organisms to fine-tune the gene expression more efficiently [67].
DUF221 domain has been reported to be highly conserved in sequence across the plant genomes [30], which is also confirmed in our study (S3 Fig). The other locally conserved regions (motifs) other than DUF221 domain (S9 Table) identified through the multiple sequence alignments of DDPs did not show a considerable similarity with any of the known protein motifs. However, they may act as scaffolds for the DDPs and be associated with the calcium transport by some means, thereby adding to the functional divergence of DDPs.
All DDPs are transmembrane calcium permeable channels. Their transmembrane nature was confirmed by higher GRAVY scores, which is a characteristic physiochemical property of transmembrane proteins [45]. The membrane topologies and motif patterns of OsDDP channel proteins were in agreement with their clustering in the phylogenetic tree (Fig 1). Proteins within a single group (Group A or B) of phylogenetic tree shared the similar pattern of different sequence motifs and membrane topologies except OsDDP7 and OsDDP10 in Group A and B respectively. In general, cytoplasmic loops of Group A differed from that of Group B, except OsDDP2, possess 4 folds as compared to 3 folds (except OsDDP3) in Group B (S6 Fig). The clustering of OsDDPs was found to be corroborated with the clustering of OsOSCAs as obtained by Li et al. [40]. Similar type of results for NCX membrane-protein family have also been obtained very recently [15]. As reported by of Hou et al. [37], DUF221 domain was found to represent the C-terminal cluster of TM helices by us too.
To infer the further structural changes in the DDP genes during the course of evolution, the intron-exon structures of all DDP genes were analysed. Although the average number of introns in DDP gene family was found to be conserved among all the studied species, a high variation in structural organisation and size of introns as well as exons was found (S7 Fig), implying that these gene structures might have undergone shuffling during the course of evolution [68]. Similar diversification in structural organisation of auxin transporter gene families has been documented in maize recently [69]. Further, the expression divergence of segmental duplicated DDP genes of both rice and Arabidopsis was found to be complemented by the diversified structural organisation of their introns and exons.
The phylogenetic analysis of DDP orthologues from rice, its wild species and Arabidopsis showed that almost all the orthologous DDPs, which shared the similar structural organisations, clustered together with a convincing bootstrap value (1000) (Fig 2). The dendrogram also revealed that almost all the segmental duplicates of Arabidopsis were clustered together into single clade (clade IV). This outcome further substantiates the gene duplication in Arabidopsis during evolution, which might eventually allow the protein functional diversity by adaptive evolution [70]. However, the duplicated pair (OsDDP3-OsDDP10) of rice was found scattered in clade I and clade II, which might possibly be due to the more sequence homology between OsDDP10 of this pair and its orthologous sequences which all were found to be in the same clade. Based on their related structural organization, several DDP proteins from rice and Arabidopsis were found to be in the same subgroups implying a common ancestral origin. Similar results with phylogeny are also documented elsewhere [71, 72]. Moreover, despite the fair degree of homology among the O. sativa (japonica), different wild rice species as well as O. sativa (indica), it was inferred from the dendrogram that japonica sub-species was more closely related to indica sub-species, O. glaberrima, O. rufipogon and O. barthi. These species were found phylogenetically closer in some other earlier studies [40,73,74]. The phylogenetic results obtained here are in agreement with the fact that even closely related Oryza species harbour remarkable changes in the genome [74].
Duplications inplant genomes play a central role in species diversification, and hence generating the platform needed for adaptive evolution [75]. The difference in the number of DDP genes in the studied plant species prompted us to address if this difference could be attributed to the gene duplication. The number of DDP genes was found to be almost identical in rice (with a single segmental duplication in DDP gene family) and its wild species (S1 Table). However, Arabidopsis genome, although almost three times smaller than that of rice, was found to contain 5 DDP genes more than rice genome, which could be ascribed to the higher number of clad-specific genomic segmental duplication events indicot Arabidopsis [76,77]. The similar expansion of different gene families in Arabidopsis as compared to rice is well documented in literature [19,71,78]. Similarly, segmental duplications have been reported in various other gene families of rice [15,19,72] and Arabidopsis [15,29,79]. It is notable to cite that members of DDP gene family in segmental duplicated regions of rice and Arabidopsis shared 50–78% identity at the amino acid sequence level (S7 Table). Similar level of similarity has also been observed among the duplicated genes of other gene families [19,57].
The gene pairs present on the segmentally duplicated regions of chromosome can acquire functional diversity in terms of neo-functionalization or pseudo-functionalization [80,81]. To address the diversity in the function of duplicated DDP gene pairs of rice and Arabidopsis, their expression patterns in different tissues as well as developmental stages and under salinity stress were compared (S8 Fig). The expression divergence of duplicated gene pair in rice during stress indicates that the duplication is significant for the stress response. The divergence in expression of duplicated genes in some other gene families of rice has also been reported [19, 72]. In Arabidopsis, the exhibition of expression divergence by all the duplicated paralogues indicates that they have undergone neo-functionalization or sub-functionalization. Neo-functionalization in duplicated Arabidopsis genes has also been reported [82]. The divergence in expression of segmentally duplicated orthologous DDP pairs in rice and Arabidopsisis is evident from the fact that all these duplicated gene pairs tend to be derived from the duplication events that date back from the appearance of the crucifers to divergence between monocots and dicots (29–104 MYA). Hence, our data corroborates with the fact that the degree of expression divergence of duplicates is proportional to their divergence times [83].
The approximate age of segmentally duplicated DDP paralogues of rice and Arabidopsis was estimated from their corresponding Ks values [84]. The approximate age (64.2 MY) of duplicated gene pair (OsDDP3-OsDDP10) of rice implicates that this duplication dates back to the divergence of poaceae from the common ancestor ~55–70 MYA [85]. The approximate ages of 29, 65 and 71 MY of gene pairs AtDDP9-AtDDP13, AtDDP2-AtDDP14 and AtDDP12-AtDDP6 indicated that duplication of these pairs could have occurred before the appearance of crucifers~24–40 MYA [86] (S8 Table).
From the Ka/Ks analysis, it was observed that all the duplicated paralogous gene pairs of rice and Arabidopsis undergo purifying (negative) selection, as all of them showed Ka/Ks <1 (S8 Table). The orthologous gene pairs from both the species were also found to be under purifying selection. However, twenty orthologous DDP gene pairs of rice and some of its wild species exhibited a Ka/Ks >1, indicating a strong positive selection for these pairs. The reason for this high ratio could not be known, however, it might be due to the changes that cultivated rice had accumulated during the course of domestication and divergence from its wild species [87]. The purifying selection on this gene family indicates its crucial role in the plant biology, and therefore unfits in the form of alterations in this gene family would be eliminated by default [88].
To get the insights into the spatio-temporal expression patterns of the DDP genes in rice and its different wild species under abiotic stress conditions, detection of regulatory abiotic stress-responsive cis-elements in their promoter regions are pre-requisite. Therefore, we restricted ourselves to find only stress related cis-elements in the DDP genes of rice and its different orthologous of different Oryza species. The presence of 20 salt and osmotic stress-responsive cis-elements in the promoters of DDPs (S11 Table) indicated the indispensable role of this gene family in response to such environmental constraints. The high enrichment of dehydration-responsive cis-elements such as, MYCCONSENSUSAT [89], ACGTATERD1 [90] and MYBCORE [91] in all the analyzed DDPs supports their role in salt and osmotic stress. The average number of the identified cis-elements was found to be almost similar in all the DDPs of rice and its wild species which might also indicate at their purifying selection as signified by the low Ka/Ks values of their orthologues. Moreover, the promoter regions of segmentally duplicated gene pair (OsDDP3-OsDDP10) in rice showed a different pattern (both in terms of type and number of motifs) of the identified cis-motifs (S11 Table), which might be a key factor in determining their expression divergence. The identified cis-elements might alter the expression of DDP genes in association with their putative transcription factors (TFs) which could ultimately culminate in the stress tolerance through the modulation of intracellular Ca2+ levels [92]. The identified motifs can be further analysed through in-depth molecular studies for understanding the transcriptional regulation of DDP genes by the putative TFs that bind such motifs.
Next, we attempted to identify if there was any miRNA-target module among the DDPs. Only 2 members (OsDDP6 and OsDDP10) were predicted to be targeted by miRNAs belonging to 3 different families (S5 Table). OsDDP6 was likely targeted by the members of osa-miR818 family (except osa-miR818f) and by osa-miR1436; whereas osa-miR6248 was predicted to target OsDDP10. Members of osa-miR818 are important for the regulating post embryonic development [93]. The miR818 of barley and osa-miR1436, targeting OsDDP6, are reported to be possibly regulated by abiotic stress such as drought and salt [94,95]. Likewise, osa-miR6248 has been reported to be differentially regulated under arsenate stress [96]. Hence, these results implicate that OsDDP6 and OsDDP10 might play an important role in abiotic stress responses. Additionally, the divergence in expression of OsDDP10 from its segmentally duplicated partner OsDDP3 might also be in part due to its regulation by osa-miR1436. Further, we also predicted few more DDP ortholouges in wild species of rice that were targeted by miRNAs (S5 Table). Interestingly, most of the DDP members from the rice and different wild rice species, targeted by the same miRNA, were found to be orthologues of each other (S2 Table). This indicated that different orthologues had conserved their regulation by same miRNA even during the course of domestication of rice. Targeting of DDP genes from the eight out of 11studied Oryza species only by miR1436 and different members of miR818 family suggested the functional redundancy of these miRNAs in regulating the Ca2+ levels across different species under different developmental and stress conditions. The finding that a particular DDP gene was targeted by multiple miRNAs is supported by the fact that a single gene can be regulated by multiple miRNAs [97,98] and therefore suggests that these miRNAs act in a functionally redundant manner [99].
Analyzing the important characteristic of gene expression pattern across the broad spectrum of different tissues, developmental stages and stress conditions would provide a vital insight into the physiological and developmental functions of DDPs. Hence, we exploited the microarray datasets for knowing the possible functions of DDPs from rice and Arabidopsis. It was found that while many DDPs maintain their transcripts at either constantly high or low levels across different tissues and developmental stages, several others expressed specifically in different tissues and at particular stages of development. This type of expression pattern has also been obtained for other gene families of rice and Arabidopsis [19,79]. In rice, the high level of expression of almost all the OsDDPs, especially OsDDP3, OsDDP10, OsDDP5 and OsDDP4, in sperm cells indicated their role in reproduction and hence in the early embryogenesis (Fig 4A). Uniformly, high expression of OsDDP1, OsDDP7 and OsDDP6 in all studied tissues implicates that these 3 genes might be involved in fundamental physiological functions. The highly specific expression of OsDDP4 in pollen tissue highlights its role in male reproductive development. The endosperm-specific moderately high expression of OsDDP2, OsDDP8 and OsDDP9 indicates towards their involvement in the nourishment and the development of embryo. The expression of DDPs in different tissues of Arabidopsis, however, was not as diverse and as high as observed in rice (Fig 4B). The similar difference in the expression pattern of histone chaperone gene families of rice and Arabidopsis was also observed [100]. The moderately high expression of AtDDP3 and AtDDP12 across almost all the studied tissues specified their commitment to some basal cellular functions. In addition, the guard cell-specific expression of AtDDP1 and AtDDP7 pointed out their possible role in stomata dynamics. Besides, the very high level of expression of AtDDP1 in sperm cell and pollen is an indicative of its concern with the male reproductive development.
The microarray-based expression patterns of OsDDPs and AtDDPs were also studied at different stages of development (Fig 5A and 5B). In rice, the highly expressed OsDDP1, OsDDP7 and OsDDP6 during developmental stages corroborated with their constantly high expression across the different tissues, and hence pointing towards their role in the development of rice plant. Moreover, the higher level of expression of OsDDP2, OsDDP8 and OsDDP10 specifically during the reproductive stages is an indication of their probable role in grain yield. Although the expression of OsDDP3 was uniform and moderately high across the developmental stages, the specific expression of OsDDP10 during the heading stage might partly be accredited to the segmental duplication and subsequent divergence in this gene pair. Similarly, in case of Arabidopsis, the highest expression of AtDDP3 and AtDDP12 across the development of the plant was in agreement with their similar degree of expression across the tissues. The high level of expression of AtDDP8 and AtDDP10 at the senescence stage indicates that they can be regulated by ethylene and abscisic acid, the two principal hormones involved in senescence [101, 102]. Owing to their possible roles in senescence, these two genes may also play a role in augmenting the grain yield [103] and in protecting the plants from stress conditions [104]. In addition, the specific expression of AtDDP1and AtDDP7during the vegetative and early reproductive stages indicated their possible involvement in transition from vegetative to reproductive phase of development.
Analysis of microarray-based expression profiles of DDP gene family members in rice and Arabidopsis led to the identification of some individual members which might play crucial roles for the tolerance of salinity stress (Fig 6A and 6B). In rice, microarray-based expression profiles were analyzed in the root and shoot of FL478 under salinity stress condition. The up-regulation of OsDDP6 and OsDDP2 in roots under salinity indicates that the two genes might act in concert as the sensors of overwhelming salinity at the site of contact with the surrounding environment.
We also validated the expression profiles of rice DDP genes by qRT-PCR at different developmental stages of FL478 (a salinity stress tolerant genotype) under salinity stress. It was found that the salinity tolerance of FL478 at different developmental stages might be partly contributed by one or more members of OsDDP family highly expressing at those stages (Fig 7A). The high level of expression of these calcium channel encoding genes is reminiscent of the fact that cytosolic Ca2+ concentration is crucial for regulating the response to stress conditions [92,105,106]. The upregulation of OsDDP2, OsDDP6, OsDDP4, OsDDP9 and OsDDP10 under different stages of development in this study corroborated with their microarray-based expression. However, in general OsDDP6 was found to be exhibiting the higher level of expression under salinity stress than OsDDP2 (showing the high overall expression in both root and shoot in microarray-based data) which might be due to the fine-tuning in the regulation of its expression by osa-miR1436 as well as different members of osa-miR818. Hence, OsDDP6 can prove as an important candidate in salt tolerance breeding of susceptible rice genotypes. Like their microarray-based expression pattern, the expression of segmentally duplicated gene pair such as OsDDP3-OsDDP10 was also found to be diverged across different developmental stages. Further, the upregulation of most of the OsDDPs at the flag leaf (particularly OsDDP3) and milk stage (especially OsDDP6 and OsDDP10) indicates their crucial role in regulating the grain filling and grain size, and hence in the yield under salinity stress. Moreover, the up-regulation of only OsDDP6 and OsDDP7 at booting stage indicates that these 2 members partly take care of tolerating the salt stress at this stage. The highest expression of OsDDP3 at the panicle initiation/emergence indicates its role at this relatively more salinity sensitive stage, and hence might contribute to the more grain numbers under salinity. The contributory duty of salinity tolerance at vegetative stage has been bestowed especially to OsDDP4.
The expression of OsDDP2, OsDDP6, OsDDP7, OsDDP9 and OsDDP10 at vegetative stage was found to be supported by the results of Li et al. [40], who studied the expression of OsDDPs (as OsOSCAs in their study) only at four-leaf stage of rice plants (the early tillering stage) under abiotic stress conditions. Though all the mentioned genes were up-regulated under salinity stress in their study too, the level of expression and the down-regulation of OsDDP1 in our study might be due to the different rice genotypes used. Further, the results from EST analysis of OsDDPs (S1 Table) were almost in accordance with our expression profiling results, as the gene members which had low number of ESTs mostly exhibited low level of expression than those with higher number of ESTs. However, some discrepancies in this relationship might be ascribed to the different rice genotype in our study. In spite of the differential regulation of the DPP genes under salinity stress found in this study, more comprehensive molecular and biochemical characterization of putative DDP gene members needs to be done so as to establish their role in salinity and other stress conditions.
Moreover, the expression profiles of all miRNAs, baring osa-miR818c, showed the similar trends as shown by their targets OsDDP6 and OsDDP10 under salinity stress conditions (Fig 7B). Although osa-miR818e and osa-miR1436 were predicted to target OsDDP6 (up-regulated under salinity at all the developmental stages), they were found probably not to be regulating its transcript levels as indicated by their up-regulation during salinity. In contrast, the down-regulation of osa-miR818c across all the developmental stages under salinity stress indicated that OsDDP6 might be the target of osa-miR818c. The down-regulation of osa-miR818a and osa-miR818b at different developmental conditions may possibly augment the role of osa-miR818c in negatively regulating the transcript levels of OsDDP6. Such negative regulation was also found in case of osa-miR393a and its target gene TIR1 under salinity stress (3). In addition, the down-regulation of osa-miR6248 at flag-leaf and milk stages implied that this miRNA might be regulating its target OsDDP10 at only these two specific stages, whereas its regulation at other developmental stages might be attributed to some other aspects such as promoter methylation [3]. The miRNAs found herein that showed altered expression than their target genes under salinity stress, however, must be analysed by functional genomics studies for verifying the regulation of OsDDP6 and OsDDP10 by them.
It is noteworthy to mention here that we used cDNA reverse transcribed from total RNA with oligo dT primer (using SuperScript II) as a template for quantifying both the OsDDPs and the respective miRNAs. The quantification of miRNAs using cDNA prepared from total RNA is reported for the first time which was possible as the studied miRNAs were located either in exons or UTRs, which gave us a chance to exploit the usefulness of this technique. This is quite legitimate because miRNA transcripts possess a polyA tail [107, 108] that can be primed with oligo dT in SuperScript II. Additionally, miRNA genes are also transcribed by RNA Pol II like any other protein coding genes [109]. Therefore, this approach of quantifying miRNA transcripts may prove economical and more efficient than what is normally done using costly commercial kits (using mature miRNA sequence as one primer and a universal primer as another). The efficiency of this approach can be understood by that any probe (most commonly mature miRNA), for example in RNA blot, for quantifying the transcript levels of a particular member of a family (that has identical mature sequences in all or most of its members) would or would not give the information specifically about the miRNA member that was actually aimed at. Therefore, to avoid such bottlenecks, the allele specific primers can be designed from the distinct and specific regions of corresponding member after alignment of all the members (precursor sequences) of a miRNA family that have identical mature sequences. Mature sequence can be used as one primer (forward/reverse) and the other primer of the pair can be designed from a region specifically. However, it is also must to mention that this approach has a drawback of not quantifying the miRNAs that are intronic.
In conclusion, this study presents an in-depth account of the DDP gene family in rice as well its comprehensive comparative evaluation in different wild species of rice and Arabidopsis. The comparative genomics and phylogenetic analysis revealed the fair degree of conservation of this family in the form of high purifying selection. This study also encourages further in-depth molecular investigations targeting some of the differentially regulated OsDDP members (OsDDP2 and OsDDP6) and the miRNAs (osa-miR818c) for enhancing the performance of rice and other plant species under high salinity leading to the subsequent agricultural development.
Supporting information
(A) Chromosomal distribution and segmental duplication of rice DDP genes. (B) Genomic distribution and expansion of DDP genes on Arabidopsis chromosomes. Bars on chromosomes denote the genes while as, red spots on the chromosomes are centromeres. Gene segmental duplications are shown with dotted lines between the duplicated gene pairs.
(TIF)
Conserved regions are indicated in pink (90% identity over a 100-bp window). The relative position of the Os-DDP3 and synteny block region is also indicated.
(TIFF)
Multiple sequence alignment of DUF221 domain from DDP proteins of rice. The bar line indicates the DUF221 domain. The gene ids can be seen in S2 Table.
(TIF)
Multiple sequence alignment of DDP proteins of selected species is generated using CLUSTAL X and visualized in Jalview. The gene ids of the different species can be seen in S2 Table.
(TIF)
Bar diagram showing that the average number of cis-elements in the 2 kb promoter regions of DDP genes of rice and its wild species as well as O. sativa (indica) is almost uniform.
(TIF)
Predicted topological structures of all DDP family members of rice, wild rice and Arabidopsis. Topological structures were predicted using Protter v1.0. For splice variants, topological architecture for only the largest transcript has been provided. Finger-like projections represent loops joining two clusters of TMs. The right side of each structure represents the extra-cellular region while as, left side the intra-cellular region.
(JPG)
Typical structure of different DDP genes. Lengths are drawn according to the size of genes. Black boxes indicate the coding exons, lines joining them indicate the introns which were not drawn to scale. The white color boxes represent the non-coding exons, whereas the white boxes at the ends represent the UTRs. It can be seen that the average number of introns in DDP gene families of different species was found to be conserved (approximately 9 introns). However, frequent length changes in exons, introns and UTRs can be seen.
(TIFF)
The line graphs represent the microarray-based expression profiles for one and six pairs of duplicated genes in rice (A) and Arabidopsis (B) respectively, under salinity, in different tissues as well as across several developmental stages. The expression profile of every duplicated gene pair is shown in three different line graphs to represent tissues, developmental stages and salinity from left to right.
(TIFF)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The Ka/Ks ratios for segmental duplicates of rice and Arabidopsis are also given.
(XLSX)
The sequence logos were generated using WebLogo.
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
The authors are grateful to Dr K. V. Bhat, Head, Division of Genomic Resources and Director, NBPGR, New Delhi for providing the facility. SAG is grateful to Department of Biotechnology, Govt. of India, New Delhi for providing the fellowship.
Data Availability
All data are given as Supporting Information files along with this paper.
Funding Statement
The experiment was conducted under in-house funding. There was no support from external funding.
References
- 1.Rengasamy P. Soil processes affecting crop production in salt-affected soils. Funct Plant Biol. 2010; 37:613–620. [Google Scholar]
- 2.Ganie SA, Karmakar J, Roychowdhury R, Mondal TK, Dey N. Assessment of genetic diversity in salt-tolerant rice and its wild relatives for ten SSR loci and one allele mining primer of salT gene located on 1st chromosome. Plant Syst Evol. 2014; 300:1741–1747. [Google Scholar]
- 3.Ganie SA, Dey N, Mondal TK. Promoter methylation regulates the abundance of osa-miR393a in contrasting rice genotypes under salinity stress. Funct Integr Genom. 2015; 16:1–11. [DOI] [PubMed] [Google Scholar]
- 4.Molla KA, Debnath AB, Ganie SA, Mondal TK. Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice (Oryza sativa L.). BMC Plant Biol. 2015; 15:122 doi: 10.1186/s12870-015-0498-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soni P, Kumar G, Soda N, Singla-Pareek SL, Pareek A. Salt overly sensitive pathway members are influenced by diurnal rhythm in rice. Plant Signal Behav. 2013; 8:e24738 doi: 10.4161/psb.24738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pani DR, Sarangi SK, Misra RC, Pradhan SK, Subudhi SK, Mondal TK. Performance of rice germplasm (Oryza sativa L.) under coastal saline conditions. J Indian Soc Coastal Agric Res. 2013; 31:11–20. [Google Scholar]
- 7.Ganie SA, Borgohain MJ, Kritika K, Talukdar A, Pani DR, Mondal TK. Assessment of genetic diversity of Saltol QTL among the rice (Oryza sativa L.) genotypes. Physiol Mol Biol Plants. 2016; 22:107–114. doi: 10.1007/s12298-016-0342-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dat J, Vandenabeele S, Vranova E, Van Montagu M, Inzé D, Van Breusegem F. Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci. 2000; 57:779–795. doi: 10.1007/s000180050041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajendran K, Tester M, Roy SJ. Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 2009; 32:237–249. doi: 10.1111/j.1365-3040.2008.01916.x [DOI] [PubMed] [Google Scholar]
- 10.Liu S, Zheng L, Xue Y, Zhang Q, Wang L, Shou H. Over-expression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice. J Plant Biol. 2010; 53:444–452. [Google Scholar]
- 11.Ruan SL, Ma HS, Wang SH, Fu YP, Xin Y, Liu WZ, et al. Proteomic identification of OsCYP2, a rice cyclophilin that confers salt tolerance in rice (Oryza sativa L.) seedlings when overexpressed. BMC Plant Biol. 2011; 11:34 doi: 10.1186/1471-2229-11-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar G, Kushwaha HR, Panjabi-Sabharwal V, Kumari S, Joshi R, Karan R, et al. Clustered metallothionein genes are co-regulated in rice and ectopic expression of OsMT1e-P confers multiple abiotic stress tolerance in tobacco via ROS scavenging. BMC Plant Biol. 2012; 12:107 doi: 10.1186/1471-2229-12-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Wang J, Liu H, Zou D, Zhao H. Influence of natural saline-alkali stress on chlorophyll content and chloroplast ultrastructure of two contrasting rice (Oryza sativa L. japonica) cultivars. Aust J Crop Sci. 2013; 7:289–292. [Google Scholar]
- 14.Sharoni AM, Nuruzzaman M, Satoh K, Moumeni A, Attia K, Venuprasad R, et al. Comparative transcriptome analysis of AP2/EREBP gene family under normal and hormone treatments, and under two drought stresses in NILs setup by Aday Selection and IR64. Mol Genet Genomics. 2012; 287:1–19. doi: 10.1007/s00438-011-0659-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh AK, Kumar R, Tripathi AK, Gupta BK, Pareek A, Singla-Pareek SL. Genome-wide investigation and expression analysis of Sodium/Calcium exchanger gene family in rice and Arabidopsis. Rice. 2015; 8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007; 143:1467–1483. doi: 10.1104/pp.106.091900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vij S, Tyagi AK. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger (s) in rice and their phylogenetic relationship with Arabidopsis. Mol Genet Genomics. 2006; 276:565–575. doi: 10.1007/s00438-006-0165-1 [DOI] [PubMed] [Google Scholar]
- 18.Fang Y, You J, Xie K, Xie W, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics. 2008; 280:547–563. doi: 10.1007/s00438-008-0386-6 [DOI] [PubMed] [Google Scholar]
- 19.Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007; 8:242 doi: 10.1186/1471-2164-8-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F, Li Q, Sun L, He Z. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 2006; 13:53–63. doi: 10.1093/dnares/dsl001 [DOI] [PubMed] [Google Scholar]
- 21.Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, et al. OsDREB genes in rice, Oryza sativa L. encode transcription activators that function in drought‐, high‐salt- and cold‐responsive gene expression. The Plant J. 2003; 33:751–763. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 2004; 16:1163–1177. doi: 10.1105/tpc.019943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye H, Du H, Tang N, Li X, Xiong L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol. 2009; 71:291–305. doi: 10.1007/s11103-009-9524-8 [DOI] [PubMed] [Google Scholar]
- 24.Ray S, Agarwal P, Arora R, Kapoor S, Tyagi AK. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol Genet Genomics. 2007; 278:493–505. doi: 10.1007/s00438-007-0267-4 [DOI] [PubMed] [Google Scholar]
- 25.Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008; 49:865–879. doi: 10.1093/pcp/pcn061 [DOI] [PubMed] [Google Scholar]
- 26.Gu Z, Ma B, Jiang Y, Chen Z, Su X, Zhang H. Expression analysis of the calcineurin B-like gene family in rice (Oryza sativa L.) under environmental stresses. Gene. 2008; 415:1–12. doi: 10.1016/j.gene.2008.02.011 [DOI] [PubMed] [Google Scholar]
- 27.Rohila JS, Yang Y. Rice mitogen‐activated protein kinase gene family and its role in biotic and abiotic stress response. J Integr Plant Biol. 2007; 49:751–759. [Google Scholar]
- 28.Guo L, Wang ZY, Lin H, Cui WE, Chen J, Liu M, Gu H. Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 2006; 16:277–286. doi: 10.1038/sj.cr.7310035 [DOI] [PubMed] [Google Scholar]
- 29.Kushwaha HR, Singh AK., Sopory SK, Singla-Pareek SL, Pareek A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics. 2009; 10:200 doi: 10.1186/1471-2164-10-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Häuser R, Pech M, Kijek J, Yamamoto H, Titz BR, Naeve F, et al. Hughes Diarmaid, ed. "RsfA (YbeB) Proteins are conserved ribosomal silencing factors. PLoS Genet. 2012; 8:e1002815 doi: 10.1371/journal.pgen.1002815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodacre NF, Gerloff DL, Uetz P. Protein domains of unknown function are essential in bacteria. MBio. 2014; 5:e00744–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunner JD, Lim NK, Schenck S, Duerst A, Dutzler R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 2014; 516:207–212. doi: 10.1038/nature13984 [DOI] [PubMed] [Google Scholar]
- 33.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008; 134:1019–1029. doi: 10.1016/j.cell.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rai A, Suprasanna P, D'Souza SF, Kumar V. Membrane topology and predicted RNA-binding function of the 'early responsive to dehydration (ERD4)' plant protein. PLoS ONE. 2012; 7:e32658 doi: 10.1371/journal.pone.0032658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan F, Yang H, Xue Y, Kong D, Ye R, Li C, et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature. 2014; 514:367–371. doi: 10.1038/nature13593 [DOI] [PubMed] [Google Scholar]
- 36.Shaik R, Ramakrishna W. Bioinformatic analysis of epigenetic and microRNA mediated regulation of drought responsive genes in rice. PLoS ONE. 2012; 7:e49331 doi: 10.1371/journal.pone.0049331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou C, Tian W, Kleist T, He K, Garcia V, Bai F, et al. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 2014; 24:632–635. doi: 10.1038/cr.2014.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernández-Aguado M, Teijeira F, Martín JF, Ullán RV. A vacuolar membrane protein affects drastically the biosynthesis of the ACV tripeptide and the beta-lactam pathway of Penicillium chrysogenum. Appl Microbiol Biotechnol. 2013; 97:795–808. doi: 10.1007/s00253-012-4256-0 [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Zhao D, Zheng L, Hsiang T, Wei Y, Fu Y, et al. Identification of virulence genes in the crucifer anthracnose fungus Colletotrichum higginsianum by insertional mutagenesis. Microb Pathog. 2013; 64:6–17. doi: 10.1016/j.micpath.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Yuan F, Wen Z, Li Y, Wang F, Zhu T, et al. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol. 2015; 15:261 doi: 10.1186/s12870-015-0653-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaiswal P, Ni J, Yap I. Gramene: a bird’s eye view of cereal genomes. Nucleic Acids Res. 2006; 34:D717–D723. doi: 10.1093/nar/gkj154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40: D1178–D1186. doi: 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013; 6:4 doi: 10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO Hub, Web Presence Working Group. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009; 25:288–289. doi: 10.1093/bioinformatics/btn615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982; 157:105–132. [DOI] [PubMed] [Google Scholar]
- 46.Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered. 2002; 93:77–78. [DOI] [PubMed] [Google Scholar]
- 47.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012; 40: D1202–D1210. doi: 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013; 41:D1152–1158. doi: 10.1093/nar/gks1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics Nucleic Acids Res. 2004; 32:W273–W279. doi: 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014; 30: 884–886. doi: 10.1093/bioinformatics/btt607 [DOI] [PubMed] [Google Scholar]
- 51.Bailey T, Williams N, Misleh C, Li W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006; 34:W369–W373. doi: 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mondal TK, Ganie SA, Rana MK, Sharma TR. Genome-wide analysis of zinc transporter genes of maize (Zea mays). Plant Mol Biol Rep. 2014; 32:605–616. [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30:2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 57.Jami SK, Clark GB, Ayele BT, Ashe P, Kirti PB. Genome-wide comparative analysis of annexin superfamily in plants. PLoS ONE. 2012; 7: e47801 doi: 10.1371/journal.pone.0047801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008; 2008:420747 doi: 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998; 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011; 39:W155–W159. doi: 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. doi: 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999; 27:297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011; 39:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner A. Selection and gene duplication: a view from the genome. Genome Biol. 2002; 3:1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mondal TK, Ganie SA. Identification and characterization of salt responsive miRNA-SSR markers in rice (Oryza sativa). Gene. 2014; 535:204–209. doi: 10.1016/j.gene.2013.11.033 [DOI] [PubMed] [Google Scholar]
- 66.Warthmann N, Das S, Lanz C, Weigel D. Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol Biol Evol. 2008; 25:892–902. doi: 10.1093/molbev/msn029 [DOI] [PubMed] [Google Scholar]
- 67.Reddy AS, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013; 25:3657–3683. doi: 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kolkman JA, Stemmer WP. Directed evolution of proteins by exon shuffling. Nat Biotechnol. 2001; 19:423–428. doi: 10.1038/88084 [DOI] [PubMed] [Google Scholar]
- 69.Yue R, Tie S, Sun T, Zhang L, Yang Y, Qi J, et al. Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE. 2015; 10:e0118751 doi: 10.1371/journal.pone.0118751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J, Clegg MT, Jiang T. Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica Genomes. Plant Physiol. 2004; 135: 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics. 2012; 13:544 doi: 10.1186/1471-2164-13-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma M, Singh A, Shankar A, Pandey A, Baranwal V, Kapoor S, et al. Comprehensive expression analysis of rice armadillo gene family during abiotic stress and development. DNA Res. 2014; 21: 267–283. doi: 10.1093/dnares/dst056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yi QM, Deng WG, Xia ZP, Pang HH. Polymorphism and genetic relatedness among wild and cultivated rice species determined by AP‐PCR analysis. Hereditas. 1995; 122:135–141. [DOI] [PubMed] [Google Scholar]
- 74.Chen J, Huang Q, Gao D, Wang J, Lang Y, Liu T, Chen C. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat Commun. 2013; 4:1595 doi: 10.1038/ncomms2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009; 183: 557–564. doi: 10.1111/j.1469-8137.2009.02923.x [DOI] [PubMed] [Google Scholar]
- 76.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004; 101: 9903–9908. doi: 10.1073/pnas.0307901101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu J, Wang J, Lin W, Li S, Li H, Zhou J, et al. The Genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005; 3:e38 doi: 10.1371/journal.pbio.0030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol. 2003; 3:17 doi: 10.1186/1471-2148-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mustafiz A, Singh AK, Pareek A, Sopory SK, Singla-Pareek SL. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct Integr Genomics. 2011; 11:293–305. doi: 10.1007/s10142-010-0203-2 [DOI] [PubMed] [Google Scholar]
- 80.Lynch M Conery JS. The evolutionary fate and consequences of duplicate genes. Sci. 2000; 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- 81.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002; 3:827–37. doi: 10.1038/nrg928 [DOI] [PubMed] [Google Scholar]
- 82.Jamsheer KM, Mannully CT, Gopan N, Laxmi A. Comprehensive evolutionary and expression analysis of FCS-Like zinc finger gene family yields insights into their origin, expansion and divergence. PLoS ONE. 2015; 10:e0134328 doi: 10.1371/journal.pone.0134328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gu Z, Nicolae D, Lu HH, Li WH. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 2002; 18:609–613. [DOI] [PubMed] [Google Scholar]
- 84.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA. 2009; 106:5737–5742. doi: 10.1073/pnas.0900906106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Shi X, Hao B, Ge S, Luo J. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol. 2005; 165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x [DOI] [PubMed] [Google Scholar]
- 86.Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004; 16:1667–1678. doi: 10.1105/tpc.021345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sweeney M, McCouch S. The complex history of the domestication of rice. Ann Bot. 2007; 100: 951–957. doi: 10.1093/aob/mcm128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koonin EV. The logic of chance, the: the nature and origin of biological evolution Upper Saddle River: 2011; FT Press. [Google Scholar]
- 89.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signalling. Plant Cell. 2003; 15:63–78. doi: 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K,Yamaguchi‐Shinozaki K. Two different novel cis‐acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark‐induced senescence. Plant J. 2003; 33:259–270. [DOI] [PubMed] [Google Scholar]
- 91.Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell (1993) 5:1529–1539. doi: 10.1105/tpc.5.11.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reddy AS, Ali GS, Celesnik H, Day IS. Coping with stresses: roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell. 2011; 23:2010–2032. doi: 10.1105/tpc.111.084988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luo YC, Zhou H, Li Y, Chen JY, Yang JH, Chen YQ, et al. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 2006; 580:5111–5116. doi: 10.1016/j.febslet.2006.08.046 [DOI] [PubMed] [Google Scholar]
- 94.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008; 8:25 doi: 10.1186/1471-2229-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deng P, Wang L, Cui L, Feng K, Liu F, Du X, et al. Global identification of microRNAs and their targets in barley under salinity stress. PloS One. 2015; 10:e0137990 doi: 10.1371/journal.pone.0137990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Q. Novel miRNAs in the control of arsenite levels in rice. Funct Integr Genomics. 2012; 12:649–658. doi: 10.1007/s10142-012-0282-3 [DOI] [PubMed] [Google Scholar]
- 97.Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3' untranslated region. Oncogene. 2010; 29:2302–2308. doi: 10.1038/onc.2010.34 [DOI] [PubMed] [Google Scholar]
- 98.Hashimoto Y, Akiyama Y, Yuasa Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS ONE. 2013; 8:e62589 doi: 10.1371/journal.pone.0062589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007; 134:1051–1060. doi: 10.1242/dev.02817 [DOI] [PubMed] [Google Scholar]
- 100.Tripathi AK, Singh K, Pareek A, Singla-Pareek SL. Histone chaperones in Arabidopsis and rice: genome-wide identification, phylogeny, architecture and transcriptional regulation. BMC Plant Biol. 2015; 15:42 doi: 10.1186/s12870-015-0414-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aneja M, Gianfagna T, Ng E. The roles of abscisic acid and ethylene in the abscission and senescence of cocoa flowers. Plant Growth Regul. 1999; 27:149–155. [Google Scholar]
- 102.Trivellini A, Ferrante A, Vernieri P, Serra G. Effects of abscisic acid on ethylene biosynthesis and perception in Hibiscus rosa-sinensis L. flower development. J Exp Bot. 2011; 62: 5437–5452. doi: 10.1093/jxb/err218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buchanan-Wollaston V. Senescence processes in plants. Ann Bot. 2008; 101:197–197. [Google Scholar]
- 104.Munné-Bosch S, Alegre L. Die and let live: leaf senescence contributes to plant survival under drought stress. Funct Plant Biol. 2004; 31:203–216. [DOI] [PubMed] [Google Scholar]
- 105.McAinsh MR, Pittman JK. Shaping the calcium signature. New Phytol. 2009; 181:275–294. doi: 10.1111/j.1469-8137.2008.02682.x [DOI] [PubMed] [Google Scholar]
- 106.DeFalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ sensors in plant signalling. Biochem J. 2010; 425:27–40. [DOI] [PubMed] [Google Scholar]
- 107.Szarzynska B, Sobkowiak L, Pant BD, Balazadeh S, Scheible WD, Mueller-Roeber B, et al. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 2009; 37:3083–3093. doi: 10.1093/nar/gkp189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ganie SA, Mondal TK. Genome-wide development of novel miRNA-based microsatellite markers of rice (Oryza sativa) for genotyping applications. Mol Breed. 2015; 35:51–60. [Google Scholar]
- 109.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004; 23:4051–4060. doi: 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Chromosomal distribution and segmental duplication of rice DDP genes. (B) Genomic distribution and expansion of DDP genes on Arabidopsis chromosomes. Bars on chromosomes denote the genes while as, red spots on the chromosomes are centromeres. Gene segmental duplications are shown with dotted lines between the duplicated gene pairs.
(TIF)
Conserved regions are indicated in pink (90% identity over a 100-bp window). The relative position of the Os-DDP3 and synteny block region is also indicated.
(TIFF)
Multiple sequence alignment of DUF221 domain from DDP proteins of rice. The bar line indicates the DUF221 domain. The gene ids can be seen in S2 Table.
(TIF)
Multiple sequence alignment of DDP proteins of selected species is generated using CLUSTAL X and visualized in Jalview. The gene ids of the different species can be seen in S2 Table.
(TIF)
Bar diagram showing that the average number of cis-elements in the 2 kb promoter regions of DDP genes of rice and its wild species as well as O. sativa (indica) is almost uniform.
(TIF)
Predicted topological structures of all DDP family members of rice, wild rice and Arabidopsis. Topological structures were predicted using Protter v1.0. For splice variants, topological architecture for only the largest transcript has been provided. Finger-like projections represent loops joining two clusters of TMs. The right side of each structure represents the extra-cellular region while as, left side the intra-cellular region.
(JPG)
Typical structure of different DDP genes. Lengths are drawn according to the size of genes. Black boxes indicate the coding exons, lines joining them indicate the introns which were not drawn to scale. The white color boxes represent the non-coding exons, whereas the white boxes at the ends represent the UTRs. It can be seen that the average number of introns in DDP gene families of different species was found to be conserved (approximately 9 introns). However, frequent length changes in exons, introns and UTRs can be seen.
(TIFF)
The line graphs represent the microarray-based expression profiles for one and six pairs of duplicated genes in rice (A) and Arabidopsis (B) respectively, under salinity, in different tissues as well as across several developmental stages. The expression profile of every duplicated gene pair is shown in three different line graphs to represent tissues, developmental stages and salinity from left to right.
(TIFF)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The Ka/Ks ratios for segmental duplicates of rice and Arabidopsis are also given.
(XLSX)
The sequence logos were generated using WebLogo.
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
All data are given as Supporting Information files along with this paper.