Abstract
Inflammatory bowel disease (IBD) is a chronic inflammatory disease of the gastrointestinal tract of uncertain origin, which includes ulcerative colitis (UC) and Crohn’s disease (CD). The composition of gut microbiota may change in IBD affected individuals, but whether dysbiosis is the cause or the consequence of inflammatory processes in the intestinal tissue is still unclear. Here, the composition of the microbiota and the metabolites in stool of 183 subjects (82 UC, 50 CD, and 51 healthy controls) were determined. The metabolites content and the microbiological profiles were significantly different between IBD and healthy subjects. In the IBD group, Firmicutes, Proteobacteria, Verrucomicrobia, and Fusobacteria were significantly increased, whereas Bacteroidetes and Cyanobacteria were decreased. At genus level Escherichia, Faecalibacterium, Streptococcus, Sutterella and Veillonella were increased, whereas Bacteroides, Flavobacterium, and Oscillospira decreased. Various metabolites including biogenic amines, amino acids, lipids, were significantly increased in IBD, while others, such as two B group vitamins, were decreased in IBD compared to healthy subjects. This study underlines the potential role of an inter-omics approach in understanding the metabolic pathways involved in IBD. The combined evaluation of metabolites and fecal microbiome can be useful to discriminate between healthy subjects and patients with IBD.
Introduction
Inflammatory bowel disease (IBD) represents a group of chronic disorders that affect one or more parts of the intestine. Crohn’s disease (CD) and ulcerative colitis (UC) are considered the two major clinically defined forms1. IBD affects 1.5 million Americans, 2.2 million European, and several hundred thousand elsewhere in the world2. The highest incidences of CD and UC have been reported in northern Europe, the United Kingdom, and North America with a greater incidence of UC than CD3. However, the incidence of IBD is increasing in emerging populations such as Asian, probably due to changes in environmental factors. The peak incidence of IBD is between the second to fourth decades of life4. Incidence in established populations is similar in men and women, but is influenced by race and ethnicity.
The etiology of IBD is multifactorial and only partially known, resulting from genetic, immunological and environmental factors. The most accepted hypothesis for the pathogenesis of IBD is the development of an over aggressive adaptive immune response mediated by T cells to a subset of commensal enteric bacteria in genetically susceptible hosts5. Moreover, environmental factors, such as diet, life style and smoking play crucial roles in the development of disease6. In particular, the relationship between the metabolome and microbiota, such the association between butyrate and Ruminococcus and Butyricicoccus bacteria, has been shown to be of fundamental importance in elucidating IBD pathogenesis and in establishing targeted therapeutic strategies7,8. Twin-pair studies have shown differences in fecal metabolites and microbiome between patients with Crohn disease and their healthy siblings9,10. Moreover, the detection of metagenomics differences yielded the identification of disease-related biomarkers, which presumably derive from disease-associated alterations in the microbial flora11.
In this study, we have used a combined metabolomics and metagenomics approach to study the differences between CD and UC, and understand how these two pathologies arise. Fecal samples of IBD patients, with either CD and UC, and healthy subjects were analyzed by the means of metabolomics and metagenomics. Nuclear magnetic resonance spectroscopy (1H-NMR), gas chromatography mass spectrometry (GC-MS) and liquid chromatography in combination with quadrupole time-of-flight mass spectrometry (LC-QTOF-MS) were used to construct the metabolic profile of fecal samples, and 16 S rRNA gene sequencing data were produced from each biospecimen by the Illumina Hi-Seq platform to reveal the gut microflora composition. Finally, we investigated the correlation between measured metabolites and identified bacteria to better define the gut-microbiome metabolome during the development of IBD.
Results
Microbiota analysis
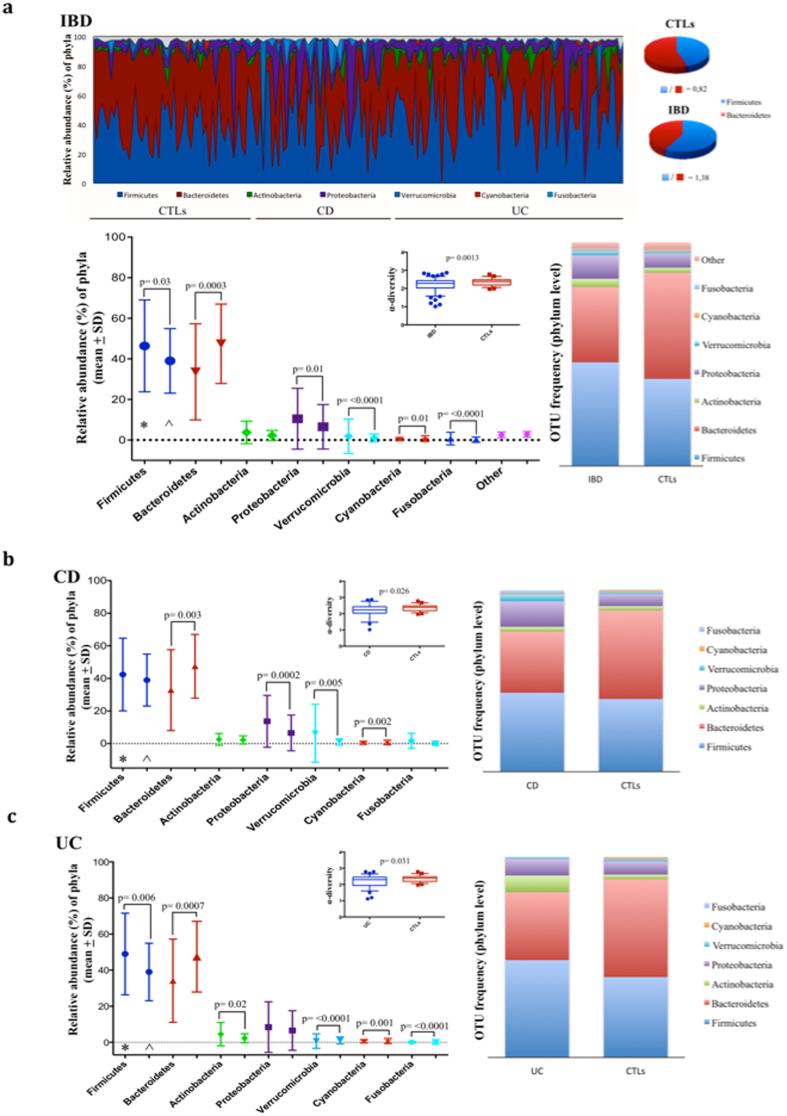
A total of 18,899,323 sequencing reads were obtained from the 183 fecal samples. The numbers of OTUs varied from 4,901 to 672,146. The mean differences in the α-Shannon values between the CTLs, the IBD, and the CD and UC groups were always statistically different (Fig. 1). The majority of the OTU sequences were assigned to seven main phyla: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia, Cyanobacteria and Fusobacteria. In Fig. 1 the relative abundance and frequency of the OTUs at the phylum level are shown for those OTUs that have at least 0.1% abundance. In the IBD group compared to the CTLs group, Firmicutes (46.39% vs 38.99%), Proteobacteria (10.49% vs 6.54%), Verrucomicrobia (1.90% vs 1.02%), and Fusobacteria (0.72% vs 0.19%) were significantly increased, whereas Bacteroidetes (33.63% vs 47.45%) and Cyanobacteria (0.73% vs 0.51%) were decreased. The Firmicutes:Bacteroidetes ratio was 0.82 in the CTLs group, but 1.38 in IBD patients (Fig. 1a).
Figure 1.
Microbiome taxonomic composition in IBD, CD, UC patients and control subjects (CTLs). Relative abundance at OTU frequency at phylum level, Firmicutes/Bacteroidetes ratio, and α-diversity are shown. The data are filtered by a frequency higher than 0.1% (a). Relative abundance of phyla and OTU frequency are shown in CD (*) (b) and UC (*) (c) patients compared to controls (CTLs, ^). Significant differences with p < 0.05 are shown. *patients; ^controls.
In CD patients only Bacteriodetes (32.83% vs 47.45%), Proteobacteria (13.67% vs 6.54%), Verrucomicrobia (3.76% vs 1.02%) and Cyanobacteria (0.38% vs. 0.73%) showed the same evidence of variation between the groups, and in UC patients only Proteobacteria did not attain the same significant difference. Additionally, in this group, Actinobacteria phylum (8.42% vs 2.26%) was significantly increased compared to CTLs (Fig. 1b,c).
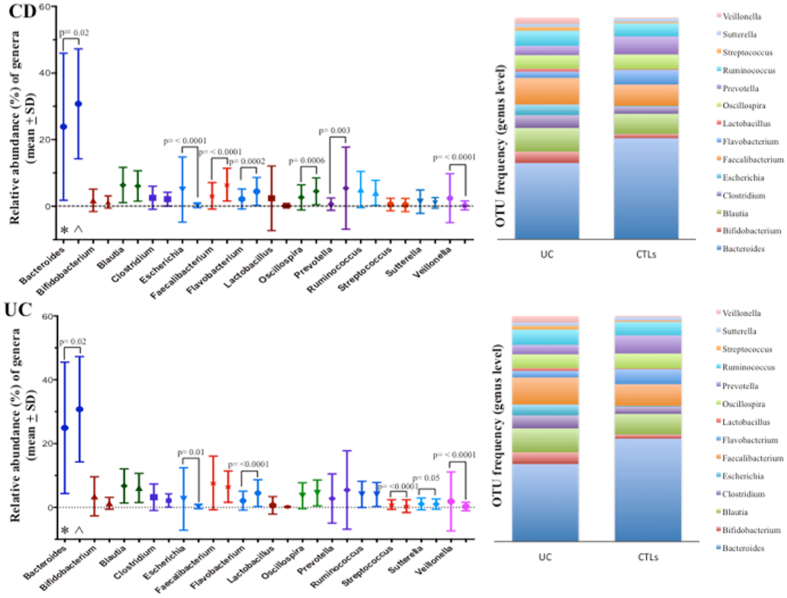
The relative abundances and frequency of the OTUs at order (Supplementary 1, Figs S1,2) and genus level (Supplementary 1, Fig. S3) are shown. At genus level, Escherichia, Faecalibacterium, Streptococcus, Sutterella and Veillonella all significantly increased in the IBD group, whereas Bacteroides, Flavobacterium, and Oscillospira geni all decreased. In CD patients, only Escherichia and Veillonella significantly increased, and, among the genera that decreased, also Prevotella decreased significantly. Interestingly, in this group the Faecalibacterium genus decreased compared to the control group, as opposed to what was observed in IBD as a whole and in the UC group. In UC patients, increase of the Faecalibacterium and decrease of the Oscillospira genera, respectively, were not statistically significant (Fig. 2).
Figure 2.
Microbiome taxonomic composition at genus level in CD, UC and controls subjects. Relative abundance of genera and OTU frequency are shown in CD (*) and UC (*) patients compared to controls (CTLs, ^). Significant differences with p < 0.05 are shown.
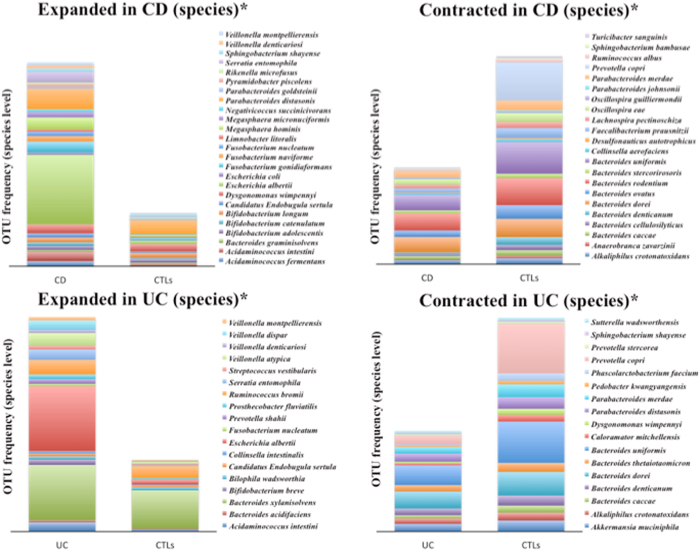
Finally, in Fig. 3 the distribution of the OTUs at the species level is shown for those that appeared increased and decreased in the CD and UC groups, respectively. In the CD group, 25 species increased and 22 species decreased, respectively, whereas in the UC group, 18 species increased and 17 species appeared reduced, respectively. Amongst Proteobacteria, Escherichia alberti resulted much more abundant than in controls (4.61% vs 0.24% in CD; 3.28% vs 0.24% in UC), and, among Bactroidetes, Prevotella copri was the main species reduced (0.02% vs 3.91% in CD; 0.68% vs 3.91% in UC) in both patient groups.
Figure 3.
Relative abundance of species in CD e UC patients compared to controls subjects. OTU frequency of species higher than 0.01% are indicated as expanded or contracted in patients groups and controls subjects. *Indicates that only levels of significance with p < 0.05 for the species indicated are shown.
Neither the diet nor the activity of disease did affect the microbiome composition. Regarding the localization of the disease, only the relative abundance of Fusobacteria correlated with the colon vs. the ileum localization of the Crohn’s disease. Finally, a significant correlation between abundance of Firmicutes and Verrucomicrobia and medications was observed in CD group, and between Actinobacteria and therapy in the UC group (see Supplementary 1, Table S1a,b,c).
Metabolomics analysis
GC-MS analysis
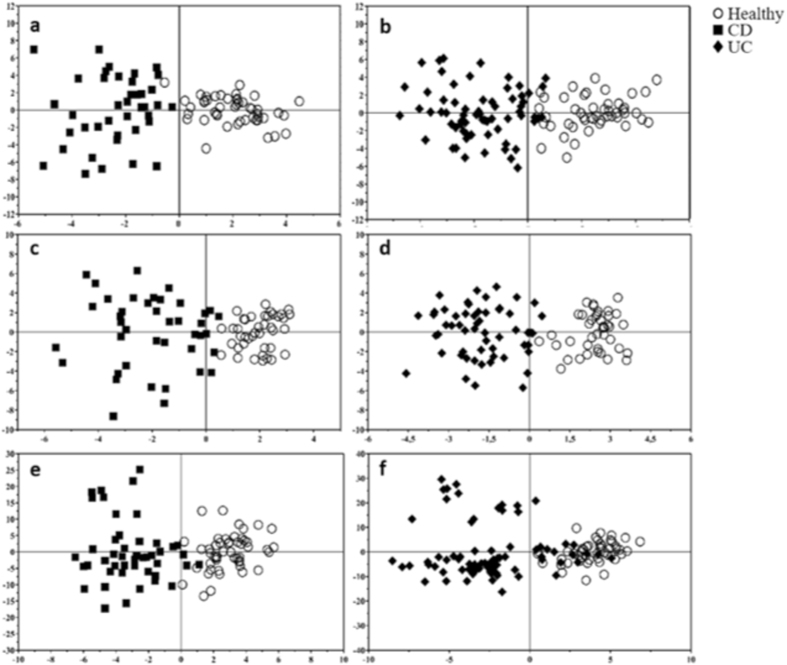
The OPLS-DA score plot demonstrates clear separation between both CD patients and healthy controls and UC patients and healthy controls (Fig. 4a,b). The quality of the models was evaluated using the corresponding Partial Least Square Discriminant Analysis (PLS-DA) models using a 7-fold cross-validation and permutation test over 400 times (Supplementary 1, Table S2). Moreover, to investigate if we could distinguish between the two pathological conditions, an OPLS-DA model was created. This comparison did not show good results, indicating an intrinsic similarity in metabolic profiles (Supplementary 1, Table S1). Discriminant metabolites were highlighted by the means of an S-plot and a Mann-Whitney test was carried out to find significant differences between the classes.
Figure 4.
OPLS-DA score plots. In the first column CD vs healthy comparisons are shown while the second column of plots contains UC vs healthy comparisons. Plots were obtained with GC-MS (a,b), 1H-NMR (c,d) and LC-MS/MS QTOF analysis (e,f).
Fourteen metabolites were detected to be responsible for the separation of the CD patients from the healthy control group (Fig. 5). Of these, eight were significantly increased in CD patients (alanine, beta-alanine, phenylacetic acid, 4-hydroxyphenylacetic acid, glyceric acid, phenylethylamine, putrescine and cadaverine) and six metabolites significantly decreased in CD patients (nicotinic acid, pantothenic acid, 3-methyladipic acid, 5β-coprostanol, 3-hydroxybutyric acid and hydrocinnamic acid).
Figure 5.
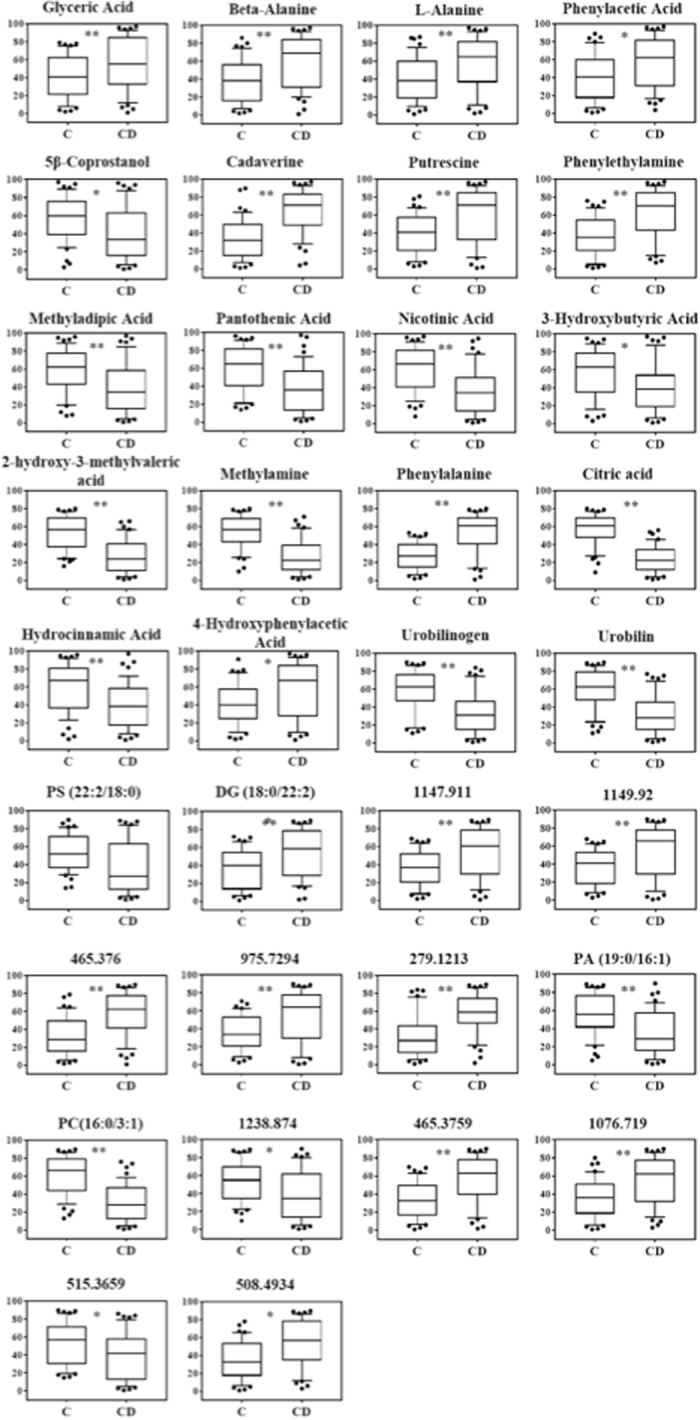
Statistically significant metabolites in CD vs healthy comparison. Discriminant metabolites obtained with the MVA, underwent to a Mann-Whitney test to determine which metabolites were statistically significantly variated. The resulted metabolites obtained are shown. Relative concentrations are represented in the y axis. * And **Indicates levels of significance with p < 0.05 and <0.01, respectively.
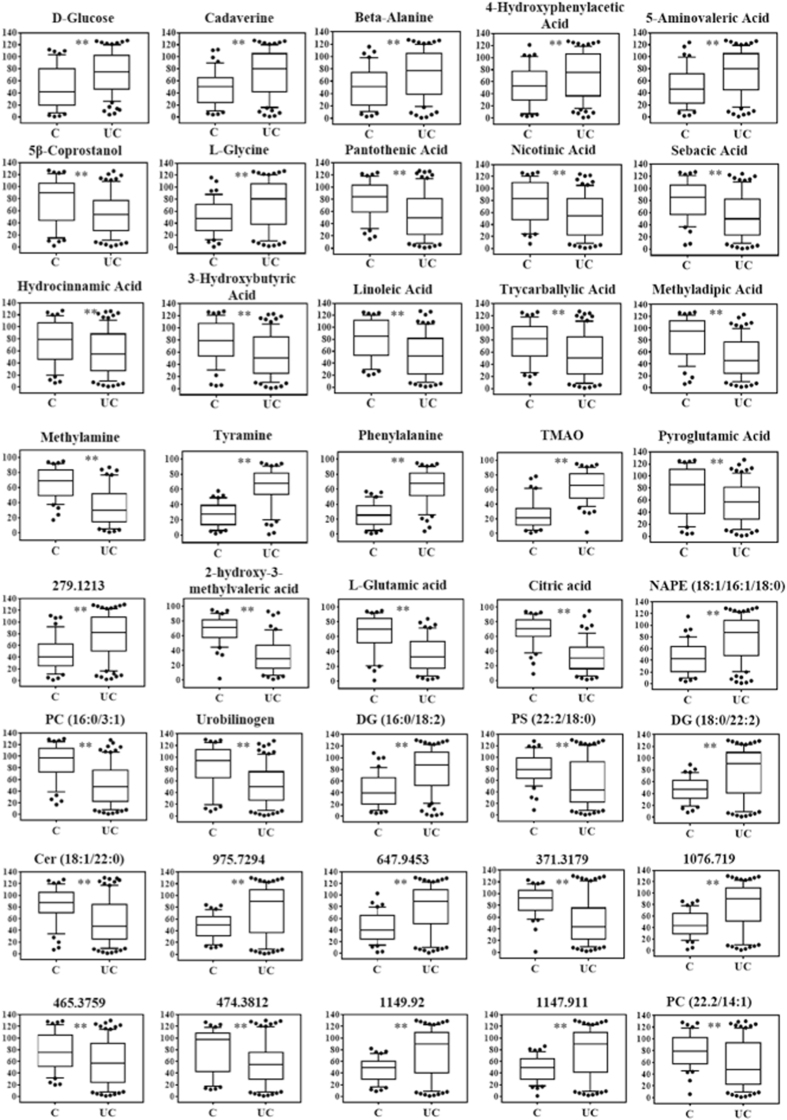
Regarding the comparison between UC patients and healthy controls, sixteen metabolites were able to discriminate between the two groups (Fig. 6), nine of which were in common with the CD patients-healthy controls comparison. Six metabolites significantly, 4-hydroxyphenylacetic acid, glucose, cadaverine and 5-aminovaleric acid), while 10 metabolites significantly decreased in UC patients (nicotinic acid, pantothenic acid, 3-methyladipic acid, pyroglutamic acid, 5β-coprostanol, 3-hydroxybutyric acid, hydrocinnamic acid, linoleic acid, sebacic acid and tricarballylic acid).
Figure 6.
Statistically significant metabolites in UC vs healthy comparison. Discriminant metabolites obtained with the MVA, underwent to a Mann-Whitney test to determine which metabolites were statistically significantly variated. The resulted metabolites obtained are shown. Relative concentrations are represented in the y axis. * And **Indicates levels of significance with p < 0.05 and <0.01, respectively.
1H-NMR analysis
Spectral resonances were assigned to individual metabolites on the basis of data published in the literature12,13 and using the 500 MHz library from Chenomx NMR suite 7.1 (Chenomx Inc., Edmonton, Alberta, Canada). OPLS-DA score plots demonstrated good separation between the healthy control group and CD and UC patients, respectively (Fig. 4c,d). However, just as with GC-MS data OPLS-DA was not able to distinguish between the two pathological conditions (UC and CD), indicating an intrinsic similarity in metabolic profiles (Supplementary 1, Table S2). In order to understand the actual trend of the metabolites, their relative concentrations were determined using the Chenomx NMR suite 7.1. The CD patients had a lower content of 2-hydroxy-3-methylvaleric acid, citric acid, and methylamine, but a higher content of cadaverine, putrescine and phenylalanine than the healthy controls (Fig. 5). The UC patients had significantly lower content of 2-hydroxy-3-methylvaleric acid, glutamic acid, citric acid, methylamine, but a higher content of cadaverine, trimethylamine-N-oxide (TMAO), tyramine and phenylalanine than the healthy controls (Fig. 6).
LC-QTOF-MS analysis
Peaks were identified and attributed to endogenous metabolites that included steroids, ceramides (Cer), phosphatidylserines (PS), phosphatidylcholine (PC), phosphatidylethanolamine (PE), diacylglycerols (DG), triacylglycerol (TG), n-acylphosphatidylethanolamines (NAPE) and other lipids.
OPLS-DA analysis displayed a clear separation between healthy subjects and both pathological classes (CD and UC) (Fig. 4e). As well as GC-MS and 1H-NMR analysis, the comparison between the two pathological classes did not show good separation (Supplementary 1, Table S2).
Seventeen metabolites were interpreted to be different between CD patients and healthy controls (Fig. 5). Using MS/MS fragmentation data, we were able to annotate six of them. DG (18:0/22:2) significantly increased in CD patients, while urobilin, PC (16:0/3:1), urobilinogen, PA (19:0/16:1) and PS (22:2/18:0) were found to be decreased.
Comparing UC patients with healthy controls, sixteen metabolites differentiated between the two classes. Nine of these were in common with the CD patients-healthy controls comparison. In particular, DG (16:0/18:2), DG (18:0/22:2) and NAPE (18:1/16:1/18:0) were identified to be significantly increased in UC patients feces, while PC (16:0/3:1), urobilinogen, PC (22.2/14:1) and Cer (18:1/22:0) were decreased in UC patients (Fig. 6).
Overall, neither the diet, nor the therapy, or the localization of the disease have had significant effect on the metabolome composition. In fact, the PLS-DA analysis for both UC and CD groups provided poor statistically significant results (see Supplementary 1, Table S3).
Pathways analysis
Metabolic pathways were built using the MetaboAnalyst version 3.014; only metabolites with significantly different (p < 0.05) concentrations between healthy patients and patients with CD and UC were used. The analysis showed how CD and UC significant metabolites were involved in different pathways as phenylalanine, glutathione and beta-alanine metabolism and pantothenate and CoA biosynthesis. The first three pathways are part of the amino acids metabolism while the last is involved in the metabolism of cofactor and vitamins. Moreover, the analysis demonstrated as UC metabolites were also implicated in methane metabolism and lysine degradation (Fig. 7a,b). The first is again comprised in the amino acids metabolism while the latter is classified as a part of the energy metabolism.
Figure 7.
Relevant metabolic pathways involved in CD (a) and in UC (b).
Correlation between metabolome and microbiome
The observation was restricted to the microbial genera whose relative abundance resulted statistically different among the different groups of subjects and to metabolites that were characterized by the highest discriminative power among the patients and healthy controls (Supplementary 1, Fig. S4).
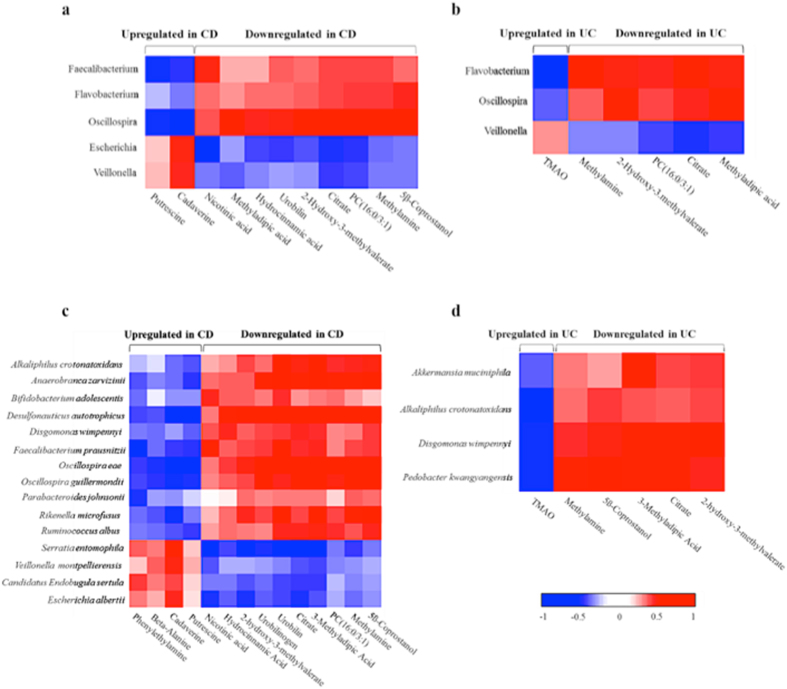
The Spearman correlation analysis revealed a strong association between five bacterial genera with 10 discriminant metabolites in CD patients (Fig. 8a). The most correlated genus was Oscillospira, particularly with hydrocinnamic acid, 3-methyladipic acid, 5β-coprostanol, citric acid, methylamine, 2-hydroxy-3-methylvaleric acid, PC (16:0/3:1) and urobilin (all positive correlations), while the genus negatively correlated with cadaverine and putrescine. In addition, the Faecalibacterium genus negatively correlated with the two biogenic amines, cadaverine and putrescine, while only nicotinic acid positively correlated. The Escherichia genus negatively correlated with nicotinic acid, citric acid and PC (16:0/3:1) and positively correlated with cadaverine. Lastly, a positive correlation was detected between the Flavobacterium genus and 5β-coprostanol and between the Veillonella genus and cadaverine.
Figure 8.
Inter-omic Spearman rank correlation between metabolites and bacterial genera and species. Spearman correlation between statistically different metabolites and bacterial genera was calculated both for CD (a) and UC (b). Spearman correlation between statistically different metabolites and bacterial species was calculated both for CD (c) and UC (d). Correlations with an r coefficient >0.5 are shown.
The Spearman correlation coefficient was less strong for UC than for CD patients (Fig. 8b). Only three bacterial genera correlated with six metabolites. Particularly, a strong positive correlation was observed between Flavobacterium genus and 3-methyladipic acid, 2-hydroxy-3-methylvaleric acid, citric acid, methylamine and PC (16:0/3:1), while this genus negatively correlated with trimethylamine-N-oxide (TMAO). UC patients also presented correlations between Oscillospira genus and 3-methyladipic acid, 2-hydroxy-3-methylvaleric acid and citric acid, and between Veillonella genus and citric acid.
Fifteen bacterial species showed a good correlation with 14 discriminant metabolites in Crohn disease patients (Fig. 8c, and Supplementary 1, Fig. S5). Seven species, Faecalibacterium prausnitzii, Oscillospira eae, Oscillospira guillermondii, Anaerobranca zavarzinii, Veillonella montpellierensis, Ruminococcus albus and Alkaliphilus crotonatoxidans belonging to the Firmicutes phylum, four, Desulphonauticus Autotrophicus, Serratia entomophila, Escherichia albertii and Candidatus Endobugula sertula to the Proteobacteria phylum, three, Dysgonomonas wimpennyi, Rikenella microfusus and Parabacteroides johnsonii to the Bacteroidetes phylum and finally only one, Bifidobacterium adolescentis to the Actinobacteria phylum. In particular, a strong positive correlation was seen between Oscillospira eae and 5β-coprostanol, 3-methyladipic acid, citric acid, methylamine, 2-hydroxy-3-methylvaleric acid, PC (16:0/3:1) and urobilin; between Oscillospira guillermondii and 5β-coprostanol, methylamine and PC (16:0/3:1); and finally between Desulphonauticus autotrophicus and 5β-coprostanol, 3-methyladipic acid, citric acid, methylamine and PC (16:0/3:1). On the other hand, a strong negative correlation between Faecalibacterium prausnitzii and phenylethylamine and between Desulphonauticus autotrophicus and putrescine and cadaverine was documented.
Evaluation of the metabolome-species relationship in UC patients showed good correlations between Pedobacter kwangyangensisvs and Dysgonomonas wimpennyi and 3-methyladipic acid, 5ß-Coprostanol, 2-hydroxy-3-methylvaleric acid, citric acid and methylamine, while only 3-methyladipic acid positively correlated with Akkermansia muciniphila species. A negative correlation was detected between TMAO and Pedobacter kwangyangensisvs, Dysgonomonas wimpennyi and Alkaliphilus crotonatoxidans species (Fig. 8d).
Discussion
It has been established that the gut microbiota is dysregulated in IBD, which leads to the modification of the bacterial metabolic activity15. In the clinical management of IBD, there are significant, inherent challenges in elucidating whether dysbiosis contributes to disease pathogenesis or whether, by contrast, it could be a secondary change associated with the inflammatory process, which may be driven by host genetics and environmental factors like diet.
In this study, we have applied a dual-omics approach to improve our current understanding of IBD, and in particular the host-microbial interactions, and the functional aspects of the gut microbiota.
Whereas it has been reported previously in IBD that there is a decrease in Firmicutes and Actinobacteria abundance, along with high levels of Proteobacteria compared to healthy subjects16, we did not find this in our patient cohort apart from the increase in Proteobacteria. Our cohort demonstrated, instead, increased levels of Firmicutes in IBD (with statistical significance only in the UC group) and Actinobacteria (with statistical significance in UC patients). Since in our analysis the relative abundance of Firmicutes and Actinobacteria also correlated with the IBD therapy, this aspect should be further investigated.
The bacterial complexity was significantly lower in IBD, CD and UC patients compared to control subjects, as previously reported. Taken together, these data may reflect at least in part the results obtained from the metabolomics analysis, which revealed characteristic clustering profiles among healthy subjects and patient categories (IBD, CD, and UC). Two biogenic amines, cadaverine and putrescine were found significantly increased in CD patients compared to healthy subjects. Polyamines are formed mainly by the decarboxylation of certain amino acids or by transamination of aldehydes or ketones, and are produced both by the host and by intestinal flora bacteria. While their specific role is still largely unknown, polyamines are involved in numerous physiological processes such as the preservation of membrane integrity and nucleic acids metabolism. Furthermore, they have a crucial role in the regulation of gene expression and translation17. Several studies demonstrated how polyamines are necessary for the division of epithelial cells18–21. In fact, the normal growth of the intestinal mucosa depends on the polyamines availability within the cell division crypts, and such substrates could be synthesized endogenously or absorbed at the lumen level. Elevated levels of polyamines seem to have a toxic effect and are associated with several diseases. It is believed that at the basis of this toxicity there is the oxidative stress caused by polyamines catabolism22. In our study, the Spearman correlation analysis demonstrated that the increase of the two polyamines negatively correlated with the amount of two bacterial genera belonging to the Firmicutes phylum, Faecalibacterium and Oscillospira. Moreover, cadaverine levels were also directly associated with the abundance of another Firmicutes genus, Veillonella, and the Proteobacterium Escherichia. Interestingly, Oscillospira and Faecalibacterium genera seems to have a protective action against inflammation23.
The increase of glyceric acid in the stool of patients with CD could be due to a release of triacylglycerols associated with the colon mucosa24. The high levels of amino acids, such as alanine, beta-alanine and phenylalanine in the feces of CD patients might, in part, result from malabsorption due to inflammation in these patients, or reflect an increase of the producing bacteria.
Two group B vitamins, nicotinic acid and pantothenic acid, significantly decreased in feces of CD patients25. Some studies report that pantothenic acid may have a protective effect against oxidative stress in mammalian tissues26. It seems that nicotinic acid might also have a protective effect on the mucosa of the colon, reducing inflammation27. The decreased levels of nicotinic acid in patients with CD may be due to a smaller presence of nicotinic acid producing bacteria. Decreased levels of this vitamin directly correlated with the reduced amount of the Faecalibacterium bacteria, particularly, with the decreased abundance of Faecalibacterium prausnitzii. It has already been demonstrated that Faecalibacterium has a protective role against inflammation of the colon mucosa28. Faecalibacterium prausnitzii abundance also inversely correlated with phenylethylamine. This amino acid is biosynthesized from the amino acid L-phenylalanine by enzymatic decarboxylation via the enzyme aromatic L-amino acid decarboxylase29. In addition to its presence in mammals, phenethylamine is found in many other organisms and foods, such as chocolate, especially after microbial fermentation. The concentration of phenylethylamine increased in feces of CD patients, suggesting that the depletion of Faecalibacterium prausnitzii could lead to an increase of some amine, which in turn could play a role in the pathogenesis of an inflammatory process30.
The amount of methylamine decreased in the aqueous fecal water extracts of patients with CD. This compound is derived from intestinal degradation of food components such as choline and carnitine by microbiota. The depletion of this microbiota-related metabolite correlated with the decrease of Oscillospira amount in feces, confirming the perturbation of the microbial homeostasis in patients with CD31.
In our study, 5β-coprostanol decreased in feces of CD patients. 5β-coprostanol derives from the catabolism of cholesterol by gut microbiota32. Again, the reduced amount of this metabolite correlated with the Oscillospira and Flavobacterium decrease in feces of CD patients. The low abundance of Oscillospira genus bacteria correlated also to the reduced levels of hydrocinnamic acid, 3-methyladipic acid, citric acid and 2-hydroxy-3-methylvaleric acid, confirming again the importance of this bacterium in the gut metabolism and wellness.
As several studies demonstrated, diacylglycerols, such as DG (18:0/22:2), are involved in the activation of the protein kinase C (PKC) in many cell types33,34. The upregulation of DG (18:0/22:2) in CD could suggest a modification of the protein kinase C (PKC) pathway. PKC plays a crucial role in many aspects of the gastrointestinal tract homeostasis, taking part in many physiological and pathological processes such as development, inflammation and tumorigenesis35.
Several others lipids, such as urobilin, urobilinogen, PC (16:0/3:1), PA (19:0/16:1) and PS (22:2/18:0), were found to be decreased in the same patients. Urobilinogen and urobilin are open tetrapyrroles deriving from bilirubin catabolism by gut microorganisms and are excreted with urines and feces36. The urobilin level reduction might be due to a significantly lower conversion rate of urobilin in CD patients as compared to the controls and this could indicate an altered entero-metabolic function in these patients37. The downregulation of phosphocholines, such as PC (16:0/3:1), could reflect a defect of PC synthesis or secretion. It has been known that several pathologies, like IBD or cancer, are correlated to the phospholipids pathways38. Cytosolic phospholipase A2 is activated by low concentration of Ca2+, and during this activation the lipase is transferred from the cytosol to the cell membrane to initialise the arachidonic acid cascade producing many inflammatory mediators39.
In addition to the previously described metabolites, fecal extracts derived from UC patients showed an increase in tyramine, TMAO, glycine, glucose, 5-aminovaleric acid, DG (16:0/18:2) and NAPE (18:1/16:1/18:0), and a decrease in glutamic acid, pyroglutamic acid, linoleic acid, sebacic acid and trycarballylic acid, PC (22.2/14:1), and Cer (18:1/22:0). Furthermore, in UC patients there was a decrease in 3-methyladipic acid, 2-hydroxy-3-methylvaleric acid, citric acid, methylamine and PC (16:0/3:1). The reduced levels of all these metabolites strongly correlated with the lower abundance of the Flavobacterium genus, belonging to the Bacteroidetes phylum. This genus also negatively correlated with TMAO levels. TMAO is generated by anaerobic bacteria through the digestion of dietary phosphatidylcholine and carnitine in a microbial-mammalian co-metabolic pathway. One previous study has found that plasma TMAO levels were significantly lower in UC population, suggesting that TMAO may be clinically useful in monitoring UC patients40.
3-Methyladipic acid, 2-hydroxy-3-methylvaleric acid and citric acid levels also positively correlated to Oscillospira abundance, suggesting a putative anti-inflammatory effect of Oscillospira in UC.
As previously described in the results, glucose increased in feces of UC patients. Glucose serves as an energy source for normal intestinal mucosa and its utilization is reduced during malnutrition and starvation, a common symptom observed in UC41. High levels of glucose thus indicate the inability of the colonic mucosal cells to utilise it for energy requirements.
The increased levels of NAPE (18:1/16:1/18:0) in UC patients suggest a modification of endocannabinoid system that has been demonstrated to be involved in the regulation of numerous gastrointestinal functions including gut homeostasis, modulating gastrointestinal motility, visceral sensation, and inflammation. Moreover, these metabolites have been recently implicated in IBD pathogenesis42,43. NAPEs are synthesized in the small intestine and after their synthesis are converted to the active NAEs. NAPEs are also converted into phosphatidic acid and anandamide by N-acylphosphatidylethanolamine-hydrolysing phospholipase D enzyme. Anandamide and 2-arachidonoylglycerol are endogenous bioactive lipids that bind and activate the cannabinoid receptors44. Decreased levels of anandamide and its synthesizing enzyme were found in the intestinal biopsies of UC patients45.
Our study combined the quantitative analysis of metabolites and microbiome in feces samples patients with CD and UC, and showed that this approach can be used to discriminate between healthy and diseased subjects. The results suggest that the detection of gut microbiota biomarkers in association with the comparative analysis of metabolites related to microbial metabolism or microbial-host co-metabolism could help to better understand the pathogenesis of IBD, and lead to the development of strategies for the early disease prediction and promote the development of novel targeted therapies.
Methods
Patients
The study was conducted in accordance with the principles of good clinical practice. The institutional ethics committee (Azienda Ospedaliero-Universitaria di Cagliari, Italy) approved the study, and written informed consent was obtained from each participant. All eligible UC (n = 82) and CD (n = 50) patients had their diagnosis confirmed by endoscopic, histological and radiographic data. Disease activity was verified by well-established criteria, including endoscopic grading according to the CDEIS and Rutgeerts scores for CD patients, and the Mayo score for UC patients46 (see Supplementary 1, Tables S5, S6). The healthy volunteers (n = 51) were recruited locally (Table 1). The exclusion criteria were as follows: (a) age >80 or <20 years; (b) use of antibiotics or probiotics in the last three months and (c) pregnancy. Each participant was given a sample collection kit with instructions. Hence, one fecal sample from each subject was collected and subsequently delivered to the laboratory within 3 hours. Samples were stored at −80 °C until use.
Table 1.
Subjects used for analysis.
| Groups | Healthy | Crohn Disease | Ulcerative Colitis | |
|---|---|---|---|---|
| Age | 40.7 ± 13 | 48.8 ± 13 | 47.3 ± 12 | |
| Sex | Male | 31 | 24 | 44 |
| Female | 20 | 26 | 38 | |
| Diet | Mediterranean | 49 | 50 | 82 |
| High-protein | 2 | — | — | |
| Smoke | Yes | 18 | 12 | 8 |
| Ex-smokers (>2 years) | 5 | 24 | 20 | |
| Alcohol | Yes | 28 | 15 | 10 |
| Coffee | Yes | 44 | 26 | 51 |
| Therapy | Mesalazine/Salazopyrin/ | — | 8 | 54 |
| Steroids | ||||
| Azatioprine | — | 9 | 15 | |
| Infliximab/Adalimumab | — | 27 | 9 | |
| No IBD therapy | 51 | 6 | 4 | |
| Lesion localization | Ileum-colon | — | 19 | — |
| Colon | — | 3 | — | |
| Ileum | — | 24 | — | |
| Ileum-caecum | — | 3 | — | |
| Rectum sigmoid | — | 1 | 21 | |
| Rectum | — | — | 16 | |
| Descending colon | — | — | 6 | |
| Rectum-sigmoid-descending colon | — | — | 14 | |
| Rectum-sigmoid-descending- transverse colon | — | — | 5 | |
| Pancolitis | — | — | 20 | |
| Total | 51 | 50 | 82 | |
Gut microbiota analysis
DNA extraction and quantification
DNA extraction from thawed fecal samples was performed using the QIAamp DNA stool MiniKit according to the instructions of the manufacturer (Qiagen), with minor modifications (see Supplementary 1).
Real-time quantitative PCR
Quantitative PCR (qPCR) was performed using degenerate primers encompassing the V3 and V4 hypervariable regions of the bacterial 16 S rRNA gene (see Supplementary 1).
Library preparation and sequencing
Barcoded amplicon libraries for the analysis on the Illumina MiSeq platform were generated using degenerate primers targeting the V3 and V4 hypervariable region of the bacterial 16 S rRNA gene and Nextera XT index kit (Illumina) (see Supplementary 1).
Sequence processing and data analysis
Analysis of the data generated on the Miseq System was carried out using the BaseSpace 16 S Metagenomics App (Illumina), whereas operational taxonomic unit (OTU) mapping to the Greengenes database (V.13.8) were performed using the Quantitative Insights Into Microbial Ecology (QIIME) platform (V.1.8.0). Sequences containing ambiguous or low-quality bases were filtered out using QIIME filter53. Remaining sequences were assigned to each sample according to the unique barcodes. Alpha diversity was estimated using Shannon index metric.
Metabolomics analysis
Stool samples extraction and preparation
Frozen feces (300 mg) were extracted with a solution of methanol\water (80:20) and the extract was divided in 3 aliquots for GC-MS1, H-NMR and LC-QTOF-MS analysis (see Supplementary 1).
Data processing
Chromatograms deriving from GC-MS and LC-QTOF-MS were processed to obtain a matrix of features present across all samples (see Supplementary 1).
Multivariate statistical analysis
Multivariate statistical data analysis was performed using SIMCA (version 14.0, Umetrics, Umea, Sweden). Principal components analysis (PCA) was used to identify the presence of outliers and to verify the influence of coffee, alcohol and smoke on metabolites composition of fecal samples. PCA is an unsupervised analysis that allows estimating and visualizing the distribution of the samples. Partial Least Square-Discriminant Analysis (PLS-DA) was performed to verify the influence of disease activity, disease localization and medications on metabolites composition. Orthogonal Partial Least Square-Discriminant Analysis (OPLS-DA) was performed in order to classify samples and elucidate metabolites able to differentiate the classes.
Univariate statistical analysis
GraphPad Prism software (version 7.01, GraphPad Software, Inc., CA, USA) was used to perform the univariate statistical analysis of the data and Spearman correlations between the microbiome and the metabolome. To verify the significance of metabolites obtained using multivariate statistical analysis and to find differences in the microbiome, the Kruskall-Wallis Mann and the Whitney test were performed.
Availability of data and material
All Illumina sequence data and all microbiome-metabolome correlation data from this study are available from the corresponding author on reasonable request. The datasets (classification, number of reads, percentages, Spearman correlations) supporting the conclusions of this work are included within the article as additional files (Supplementary information 2 and 3).
Ethical approval and consent to participate
The subjects gave informed consent, and the study protocol was approved by the Institution Ethics Committee (AOU Cagliari, Italy. Prot. NP/2014/3504; Prot. PG/2014/11480).
Electronic supplementary material
Acknowledgements
This work was supported by R.A.S. LR7/2007. Grant Code CRP 18488 (L.A.).
Author Contributions
A.M., L.A., P.U., and P.C. conceived the study, directed the project and designed the experiments. M.A.L. and I.I. obtained the samples and clinical details. T.C., V.P., S.O., and A.L.Z. performed microbiome experiments and data analysis. S.B. performed preliminary experiments for bacterial DNA quantitation. M.L.S., C.P., A.Mu., and S.L. performed metabolomics experiments and data analysis. M.L.S. and A.M. wrote the first draft of the manuscript, and L.A. and J.L.G. contributed to the final version. M.L.S., C.P., A.Mu., V.P., T.C., S.L., I.I., M.A.L., S.O., A.L.Z., J.L.G., P.U., P.C., L.A., and A.M. critically reviewed the data and the manuscript. All authors read and approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Luigi Atzori and Aldo Manzin contributed equally to this work.
A correction to this article is available online at https://doi.org/10.1038/s41598-018-23330-5.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10034-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/19/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
- 1.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nat. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Gunesh S, et al. The incidence of Crohn’s disease in Cardiff over the last 75 years: an update for 1996–2005. Alim Pharm Ther. 2008;27(3):211–219. doi: 10.1111/j.1365-2036.2007.03576.x. [DOI] [PubMed] [Google Scholar]
- 3.Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterol. 2012;142(1):46–54. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein CN, Rawsthorne P, Cheang M, Blanchard JF. A population-based case control study of potential risk factors for IBD. Am J Gastroenterol. 2006;101(5):993–1002. doi: 10.1111/j.1572-0241.2006.00381.x. [DOI] [PubMed] [Google Scholar]
- 5.Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369(9573):1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 6.Koloski NA, Bret L, Radford-Smith G. Hygiene hypothesis in inflammatory bowel disease: A critical review of the literature. World J Gastroenterol. 2008;14(2):165–173. doi: 10.3748/wjg.14.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nat. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 9.Jansson J, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory disease phenotype. Gastroenterol. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 11.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44(5):354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 12.Schicho R, et al. Quantitative metabolomic profiling of serum, plasma, and urine by 1H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J. Proteome Res. 2012;11(6):3344–3357. doi: 10.1021/pr300139q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wishart DS, et al. HMDB: the human metabolome database, Nucl. Acids Res. 2007;35(1):521–526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–27. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nat. 2013;504:451–45. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marteau P. Bacterial Flora in Inflammatory Bowel Disease. Dig Dis. 2009;27(1):99–103. doi: 10.1159/000268128. [DOI] [PubMed] [Google Scholar]
- 17.Timmons, J., Chang, E. T., Wang, J. Y. & Rao, J. N. Polyamines and Gut Mucosal Homeostasis. J Gastrointest Dig Syst. 2(Suppl 7), 001 (2012). [PMC free article] [PubMed]
- 18.Liu L, et al. Activation of TGF-β-Smad signaling pathway following polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2003;285(5):G1056–1067. doi: 10.1152/ajpgi.00151.2003. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, et al. Polyamine-modulated expression of c-myc plays a critical role in stimulation of normal intestinal epithelial cell proliferation. Am J Physiol Cell Physiol. 2005;288(1):C89–99. doi: 10.1152/ajpcell.00326.2004. [DOI] [PubMed] [Google Scholar]
- 20.Johnson LR. Regulation of gastrointestinal mucosal growth. Physiol Rev. 1988;68(2):456–502. doi: 10.1152/physrev.1988.68.2.456. [DOI] [PubMed] [Google Scholar]
- 21.Murphy GM. Polyamines in the human gut. Eur J Gastroenterol Hepatol. 2001;13:1011–114. doi: 10.1097/00042737-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 23.Konikoff T, Gophna U. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016;24(7):523–524. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 24.Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol. 2011;60:817–827. doi: 10.1099/jmm.0.028126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayo, B. & van Sinderen, D. Bifidobacteria: Genomics and Molecular Aspects. (Caister Acad. Press 2010).
- 26.Wojtczak L, Slyshenkov VS. Protection by pantothenic acid against apoptosis and cell damage by oxygen free radicals-the role of glutathione. Biofactors. 2003;17(1–4):61–73. doi: 10.1002/biof.5520170107. [DOI] [PubMed] [Google Scholar]
- 27.Singh N, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinken A, et al. Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J Bacteriol. 2014;196(18):3289–3302. doi: 10.1128/JB.01780-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berry MD. Mammalian central nervous system trace amines. Pharmacologic amphetamines, physiologic neuromodulators. J Neurochem. 2004;90(2):257–271. doi: 10.1111/j.1471-4159.2004.02501.x. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen OH, Rask-Madsen J. Mediators of Inflammation in Chronic Inflammatory Bowel Disease. Scand J Gastroenterol Suppl. 1996;216:149–159. doi: 10.3109/00365529609094569. [DOI] [PubMed] [Google Scholar]
- 31.Marchesi JR, et al. Rapid and Noninvasive Metabonomic Characterization of Inflammatory Bowel Disease. J Proteome Res. 2007;6(2):546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 32.Gerard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2014;3(1):14–24. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morotomi M, Guillem JG, LoGerfo P, Weinstein IB. Production of diacylglycerol, an activator of protein kinase C, by human intestinal microflora. Cancer Res. 1990;50(12):3595–3599. [PubMed] [Google Scholar]
- 34.Nishizuka Y. Studies and perspectives of protein kinase C. Science. 1986;233:305–313. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- 35.Yang L, Yan Y. Protein kinases are potential targets to treat inflammatory bowel disease. World J Gastrointest. Pharmacol Ther. 2014;5(4):209–217. doi: 10.4292/wjgpt.v5.i4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotal P, Fevery J. Quantitation of urobilinogen in feces, urine, bile and serum by direct spectrophotometry of zinc complex. Clin Chim Acta. 1991;202(1–2):1–9. doi: 10.1016/0009-8981(91)90250-G. [DOI] [PubMed] [Google Scholar]
- 37.Midtvedt T, et al. Increase of faecal tryptic activity relates to changes in the intestinal microbiome: analysis of Crohn’s disease with a multidisciplinary platform. PLoS One. 2013;8(6):e66074. doi: 10.1371/journal.pone.0066074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies JM, et al. Stool phospholipid signature is altered by diet and tumors. PloS one. 2014;9(12):e114352. doi: 10.1371/journal.pone.0114352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morita H, Nakanishi K, Dohi T, Yasugi E, Oshima M. Phospholipid turnover in the inflamed intestinal mucosa: arachidonic acid-rich phosphatidyl/plasmenyl-ethanolamine in the mucosa in inflammatory bowel disease. J Gastroenterol. 1999;34(1):46–53. doi: 10.1007/s005350050215. [DOI] [PubMed] [Google Scholar]
- 40.Wilson A, et al. Trimethylamine-N-oxide: A Novel Biomarker for the Identification of Inflammatory Bowel Disease. Dig Dis Sci. 2015;60(12):3620–3630. doi: 10.1007/s10620-015-3797-3. [DOI] [PubMed] [Google Scholar]
- 41.Balasubramanian K, et al. Metabolism of the colonic mucosa in patients with inflammatory bowel diseases: an in vitro proton magnetic resonance spectroscopy study. Magn Reson Imaging. 2009;27(1):79–86. doi: 10.1016/j.mri.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Alhouayek M, Muccioli GG. The endocannabinoid system in inflammatory bowel diseases: from pathophysiology to therapeutic opportunity. Trends Mol Med. 2012;18(10):615–625. doi: 10.1016/j.molmed.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Storr MA, Yüce B, Andrews CN, Sharkey KA. The role of the endocannabinoid system in the pathophysiology and treatment of irritable bowel syndrome. Neurogastroenterol Motil. 2008;20(8):857–868. doi: 10.1111/j.1365-2982.2008.01175.x. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Invest. 2014;124(8):3391–3406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Sabatino A, et al. The endogenous cannabinoid system in the gut of patients with inflammatory bowel disease. Mucosal Immunol. 2011;4(5):574–583. doi: 10.1038/mi.2011.18. [DOI] [PubMed] [Google Scholar]
- 46.Nikolaus S, Schreiber S. Diagnostics of inflammatory bowel disease. Gastroenterol. 2007;133(5):1670–1689. doi: 10.1053/j.gastro.2007.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.