Abstract
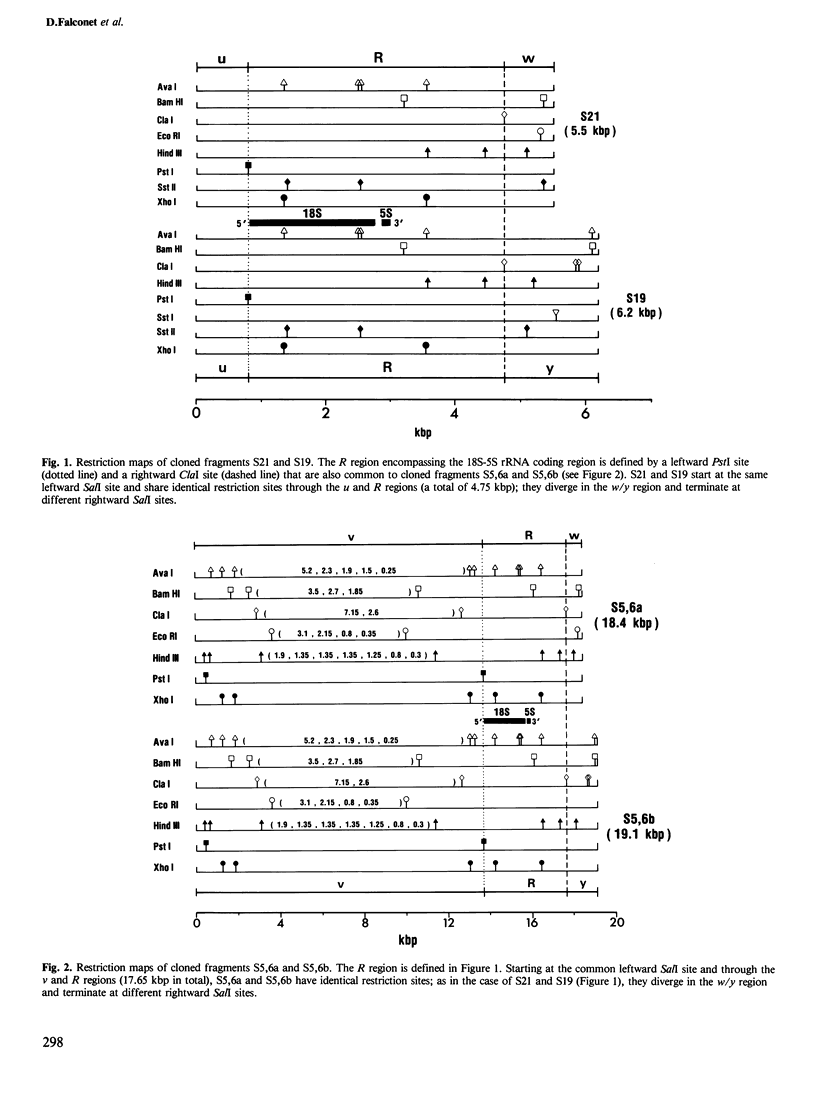
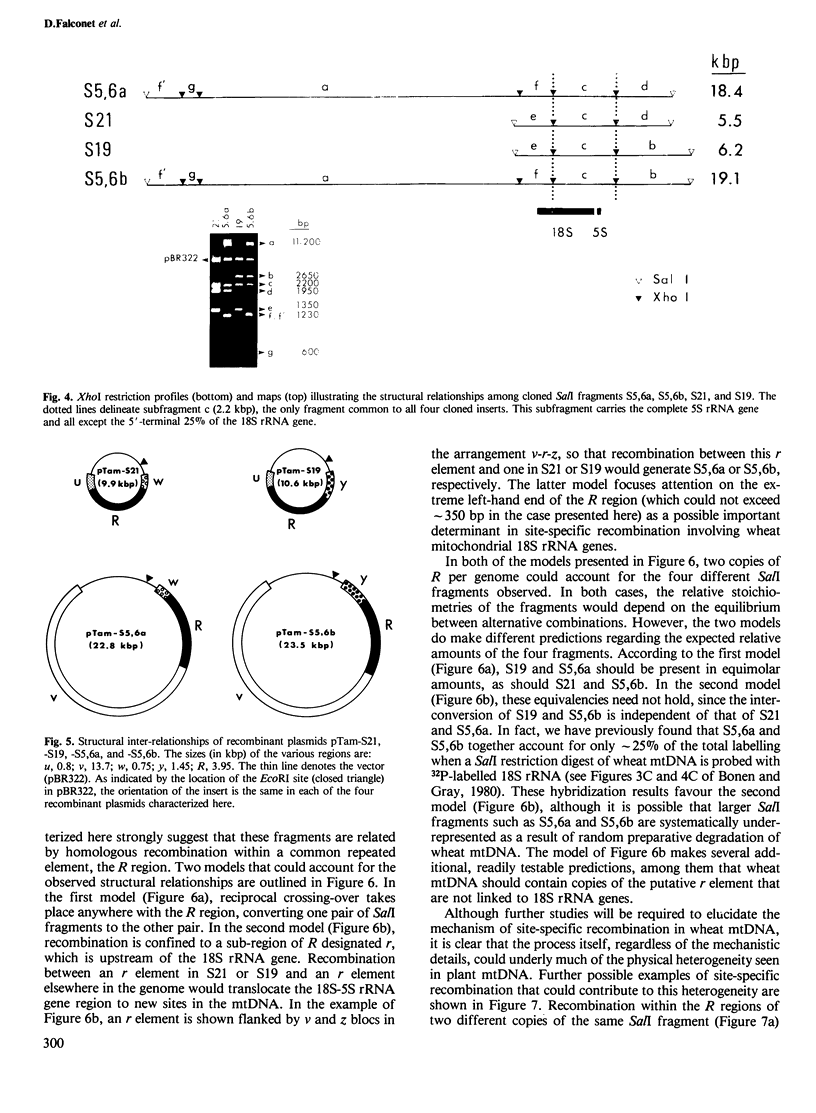
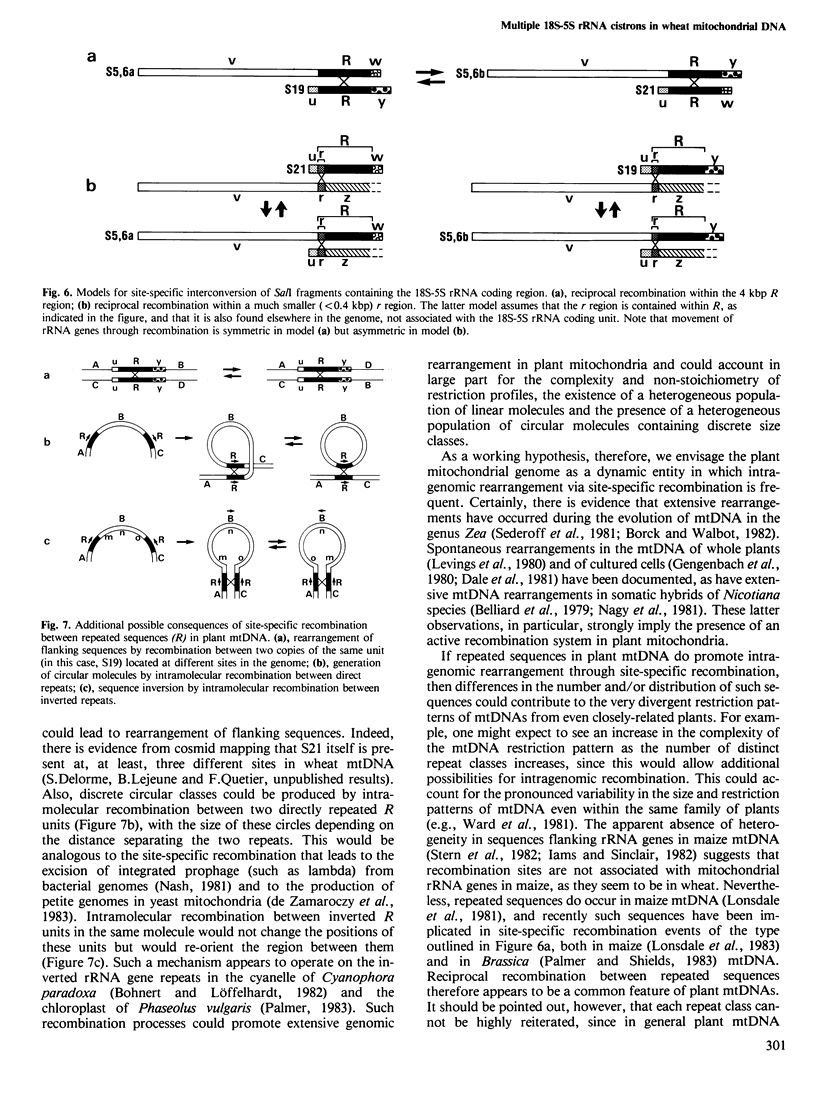
Closely linked genes for 18S and 5S rRNAs have been located on four different cloned SalI restriction fragments of wheat (Triticum aestivum L.) mitochondrial DNA. Restriction analysis has revealed that in each of the cloned fragments, the 18S and 5S rRNA genes are contained within the same basic structural unit, R, which is at least 4 kbp long. This unit is flanked by sequences designated u (0.8 kbp), v (13.7 kbp), w (0.7 kbp), and y (1.4 kbp), in the orientations v-R-w, v-R-y, u-R-w, and u-R-y in the four different SalI fragments. We conclude that 18S + 5S rRNA genes are located at several distinct sites in the wheat mitochondrial genome, and suggest that reciprocal intra- and/or intermolecular recombination between such repeated sequences could promote extensive genomic rearrangement and thus contribute to the physical heterogeneity that is a hallmark of most plant mitochondrial DNAs.
Keywords: plant mitochondrial DNA, repeated sequences, ribosomal RNA genes, site-specific recombination, wheat
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L., Gray M. W. Organization and expression of the mitochondrial genome of plants I. The genes for wheat mitochondrial ribosomal and transfer RNA: evidence for an unusual arrangement. Nucleic Acids Res. 1980 Jan 25;8(2):319–335. doi: 10.1093/nar/8.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck K. S., Walbot V. Comparison of the Restriction Endonuclease Digestion Patterns of Mitochondrial DNA from Normal and Male Sterile Cytoplasms of ZEA MAYS L. Genetics. 1982 Sep;102(1):109–128. doi: 10.1093/genetics/102.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennicke A. Mitochondrial DNA from Oenothera berteriana: PURIFICATION AND PROPERTIES. Plant Physiol. 1980 Jun;65(6):1207–1210. doi: 10.1104/pp.65.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. M., Duesing J. H., Keene D. Supercoiled mitochondrial DNAs from plant tissue culture cells. Nucleic Acids Res. 1981 Sep 25;9(18):4583–4593. doi: 10.1093/nar/9.18.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. M. Sequence homology among different size classes of plant mtDNAs. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4453–4457. doi: 10.1073/pnas.78.7.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontarnau A., Hernández-Yago J. Characterization of mitochondrial DNA in citrus. Plant Physiol. 1982 Dec;70(6):1678–1682. doi: 10.1104/pp.70.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. Mitochondrial genome diversity and the evolution of mitochondrial DNA. Can J Biochem. 1982 Mar;60(3):157–171. doi: 10.1139/o82-022. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iams K. P., Sinclair J. H. Mapping the mitochondrial DNA of Zea mays: Ribosomal gene localization. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5926–5929. doi: 10.1073/pnas.79.19.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Physicochemical characterization of mitochondrial DNA from pea leaves. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1830–1834. doi: 10.1073/pnas.69.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings C. S., 3rd, Kim B. D., Pring D. R., Conde M. F., Mans R. J., Laughnan J. R., Gabay-Laughnan S. J. Cytoplasmic Reversion of cms-S in Maize: Association with a Transpositional Event. Science. 1980 Aug 29;209(4460):1021–1023. doi: 10.1126/science.209.4460.1021. [DOI] [PubMed] [Google Scholar]
- Levings C. S., 3rd, Pring D. R. Restriction endonuclease analysis of mitochondrial DNA from normal and Texas cytoplasmic male-sterile maize. Science. 1976 Jul 9;193(4248):158–160. doi: 10.1126/science.193.4248.158. [DOI] [PubMed] [Google Scholar]
- Lonsdale D. M., Thompson R. D., Hodge T. P. The integrated forms of the S1 and S2 DNA elements of maize male sterile mitochondrial DNA are flanked by a large repeated sequence. Nucleic Acids Res. 1981 Aug 11;9(15):3657–3669. doi: 10.1093/nar/9.15.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda: the mechanism of conservation site specific recombination. Annu Rev Genet. 1981;15:143–167. doi: 10.1146/annurev.ge.15.120181.001043. [DOI] [PubMed] [Google Scholar]
- Sederoff R. R., Levings C. S., Timothy D. H., Hu W. W. Evolution of DNA sequence organization in mitochondrial genomes of Zea. Proc Natl Acad Sci U S A. 1981 Oct;78(10):5953–5957. doi: 10.1073/pnas.78.10.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Dyer T. A., Lonsdale D. M. Organization of the mitochondrial ribosomal RNA genes of maize. Nucleic Acids Res. 1982 Jun 11;10(11):3333–3340. doi: 10.1093/nar/10.11.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synenki R. M., Levings C. S., Shah D. M. Physicochemical characterization of mitochondrial DNA from soybean. Plant Physiol. 1978 Mar;61(3):460–464. doi: 10.1104/pp.61.3.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedel F., Quetier F. Physico-chemical characterization of mitochondrial DNA from potato tubers. Biochim Biophys Acta. 1974 Apr 10;340(4):374–387. doi: 10.1016/0005-2787(74)90059-8. [DOI] [PubMed] [Google Scholar]
- Wallace D. C. Structure and evolution of organelle genomes. Microbiol Rev. 1982 Jun;46(2):208–240. doi: 10.1128/mr.46.2.208-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. L., Anderson R. S., Bendich A. J. The mitochondrial genome is large and variable in a family of plants (cucurbitaceae). Cell. 1981 Sep;25(3):793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]
- Wells R., Birnstiel M. Kinetic complexity of chloroplastal deoxyribonucleic acid and mitochondrial deoxyribonucleic acid from higher plants. Biochem J. 1969 May;112(5):777–786. doi: 10.1042/bj1120777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F. Y., Wildman S. G. Simple procedure for isolation of satellite DNA's from tobacco leaves in high yield and demonstration of minicircles. Biochim Biophys Acta. 1972 Jan 18;259(1):5–12. doi: 10.1016/0005-2787(72)90468-6. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Faugeron-Fonty G., Bernardi G. Excision sequences in the mitochondrial genome of yeast. Gene. 1983 Mar;21(3):193–202. doi: 10.1016/0378-1119(83)90002-1. [DOI] [PubMed] [Google Scholar]